Abstract
The stable states of differentiated cells are now known to be controlled by dynamic mechanisms that can easily be perturbed. An adult cell can therefore be reprogrammed, altering its pattern of gene expression, and hence its fate, to that typical of another cell type. This has been shown by three distinct experimental approaches to nuclear reprogramming: nuclear transfer, cell fusion and transcription-factor transduction. Using these approaches, nuclei from ‘terminally differentiated’ somatic cells can be induced to express genes that are typical of embryonic stem cells, which can differentiate to form all of the cell types in the body. This remarkable discovery of cellular plasticity has important medical applications.
In the early embryo of vertebrates, totipotent cells have the potential to differentiate and give rise to cells that function in specific tissues, ultimately forming an entire organism, including the extra-embryonic tissues, such as the placenta. This process of cell specification is controlled by the interplay of endogenous and exogenous factors (see page 713). At the blastocyst stage of the early embryo, the cells of the inner cell mass (from which embryonic stem (ES) cell lines are derived1,2) are pluripotent: they are able to form each of the three germ layers — the endoderm, ectoderm and mesoderm. Eventually, cells that are committed to each of these germ layers specialize to give rise to the tissues of the adult body, such as the brain, intestine or cardiac muscle. These differentiated adult cells generally do not switch fates; for example, hepatocytes do not spontaneously become cardiomyocytes.
Several classic studies, however, suggested that ‘committed’ cells of the embryo are ‘plastic’, because the fate of these cells can change when they are explanted and exposed to a different microenvironment. In one of these studies, cells from the imaginal discs of Drosophila melanogaster pupae were serially transplanted into the abdomen of an adult fly, and ‘transdetermination’ was observed: cells that were originally destined to form genital structures gave rise to leg or head structures and, eventually, on subsequent transplantations, to wings3,4. Although such switches in cell fate occurred at a low frequency, these experiments by Hadorn3 and Gehring4 provided evidence that explanted cells are surprisingly plastic. In another elegant study5, cells were transplanted from quails to chickens: these cells were sufficiently similar to be able to participate in normal development on transplantation but were histologically distinct, enabling them to be tracked. Using this property, Le Lievre and Le Douarin5 showed that explanted neural crest cells adopt new fates (bone, cartilage and connective tissue) that are dictated by their new cellular neighbourhood and not their original location in the avian embryo. One caveat of these findings is that the fate of single cells could not be followed. But, as early as the mid-1960s, such embryonic cell transplantation experiments suggested that the generally stable state of a specialized cell was plastic and could be altered in response to the extracellular environment.
It was long thought that when a cell differentiates, it loses chromosomes or permanently inactivates genes that it no longer needs. Why would a specialized cell maintain the potential to reactivate genes typical of another cell type? This would seem to be a risky mechanism, given the possibility that genes could be inappropriately activated. Yet three approaches to nuclear reprogramming — nuclear transfer, cell fusion and transcription-factor transduction (described in detail below) — have shown conclusively in a defined specialized cell type (that is, in a cell that has been carefully determined to be differentiated) that cell fate can be reversed, returning the cell to an embryonic state (Fig. 1). These three experimental models therefore provide evidence that, with few exceptions (such as homologous recombination in lymphocytes), highly specialized somatic cells retain all of the genetic information that is needed for them to revert to ES cells and that the genes of the somatic cells have not been permanently inactivated. In addition, the three approaches show that, although the differentiated state of a cell is generally stable, cellular ‘memory’ is dynamically controlled and subject to changes induced by perturbations in the stoichiometry of the transcriptional regulators present in the cell at any given time.
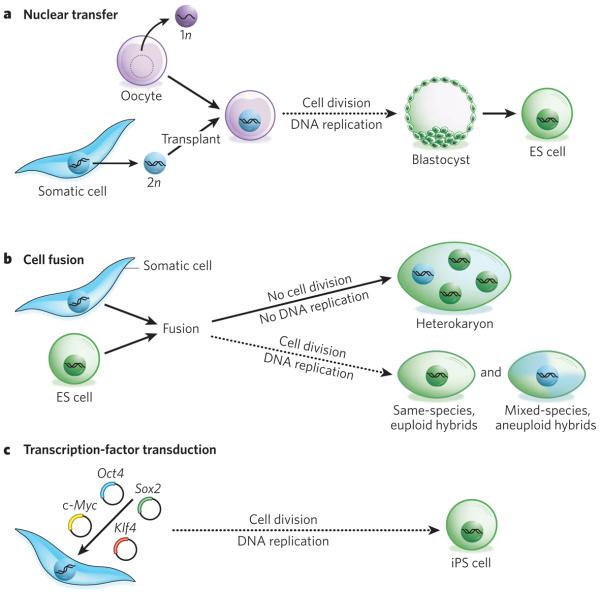
Figure 1. Three approaches to nuclear reprogramming to pluripotency.
a, Nuclear transfer. In this approach, the nucleus of a somatic cell (which is diploid, 2n) is transplanted into an enucleated oocyte. In the environment of the oocyte, the somatic cell nucleus is reprogrammed so that the cells derived from it are pluripotent. From this oocyte, a blastocyst is generated, from which embryonic stem (ES)-cell lines are derived in tissue culture. If development is allowed to proceed to completion, an entire cloned organism is generated. b, Cell fusion. In this approach, two distinct cell types are combined to form a single entity. The resultant fused cells can be heterokaryons or hybrids. If the fused cells proliferate, they will become hybrids, and on division, the nuclei fuse to become 4n (that is, twice the number of chromosomes in a somatic cell) or greater. If the cells are derived from the same species, their karyotype will remain euploid (that is, they will have balanced sets of chromosomes); however, if they are from different species, they will be aneuploid, as chromosomes will be lost and rearranged. Heterokaryons, by contrast, are short-lived and do not divide. As a result, they are multinucleate: the nuclei from the original cells remain intact and distinct, and the influence of one genotype on another can be studied in a stable system in which no chromosomes are lost. If the heterokaryons are of mixed species, the gene products of the two cell types can be distinguished. By altering the nuclear ratio in the fusion, and hence the stoichiometry of the regulators provided by each type of cell, the heterokaryon is reprogrammed towards the desired cell type (Fig. 3). Culture medium also has a role and needs to have a composition favoured by the desired cell type. Dashed arrows indicate slower processes (involving multiple rounds of cell division) than solid arrows (no division). c, Transcription factor transduction. This approach can be used to form induced pluripotent stem (iPS) cells, which have similar properties to ES cells and can be generated from almost any cell type in the body through the introduction of four genes (Oct4, Sox2, Klf4 and c-Myc) by using retroviruses. The pluripotent state is heritably maintained, and vast numbers of cells can be generated, making this approach advantageous for clinical applications. 1n, haploid.
Recent studies show that pluripotent stem cells with properties similar to ES cells (called iPS cells) can be induced readily from differentiated somatic cells. This finding has led to great excitement regarding the potential of these cells for improving the understanding and treatment of disease and has highlighted the need for a better mechanistic understanding of the reprogramming process. Insight is needed into which regulators are required for iPS cells to be reliably and efficiently generated and induced to differentiate towards the specialized cell fate of interest. To achieve this goal, all three approaches to nuclear reprogramming need to be enlisted.
This Review provides a historical perspective on the key findings (Fig. 2) that led to the discovery of cellular plasticity, discussing studies using each of the three approaches to nuclear reprogramming in turn. It also indicates the questions that must be answered before nuclear reprogramming can fulfil its potential in medical applications.
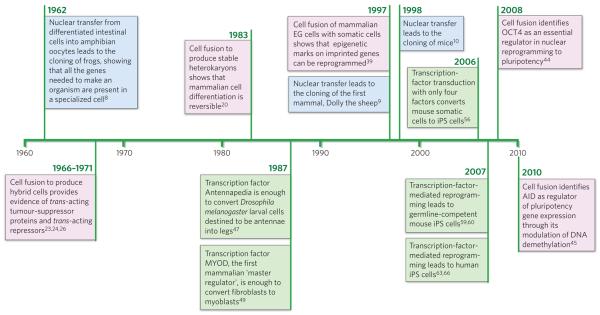
Figure 2. Timeline of discoveries in nuclear reprogramming.
Three approaches to nuclear reprogramming are described: nuclear transfer (blue), cell fusion (pink) and transcription-factor transduction (green). These complementary approaches have provided synergistic insights for almost 50 years and continue to inform the understanding of nuclear reprogramming and influence medical advances. EG cell, embryonic germ cell.
Nuclear transfer
When a nucleus from a differentiated somatic cell, such as an intestinal cell, is transplanted into an enucleated oocyte, nuclear reprogramming is initiated, leading to the generation of an entire individual, which is a genetically identical clone of the original somatic cell. Such nuclear-transfer experiments, also known as cloning (Fig. 1), have shown definitively that all of the genes required to create an entire organism are present in the nucleus of the specialized cell and can be activated on exposure to reprogramming factors present in the oocyte. In other words, cell specialization involves a change in gene expression, not in gene content, and the process of differentiation can be fully reversed. In this section, we present a historical overview of the findings from nuclear-transfer studies and the insight into cellular differentiation and gene regulation that researchers have gained from this approach.
In amphibians
Elegant cloning studies in frogs provided the first conclusive evidence that genes are not lost or permanently silenced during cell specialization. Briggs and King were the first to show6, in 1952, that the transfer of nuclei from early blastocysts into enucleated oocytes resulted in clones, in this case of swimming tadpoles. But they had difficulty reproducing this finding with cells from more specialized tissues, which often gave rise to abnormal tissues, and they interpreted this limitation as evidence for a loss of plasticity as cells differentiate. Gurdon undertook the same experiment7,8 with a different frog species and succeeded in transferring nuclei from highly specialized tadpole intestinal cells into ultraviolet-light-irradiated oocytes, obtaining not only tadpoles but also normal adult frogs. The choice of frog species was crucial but so was the interpretation of the results. Although the incidence was low, Gurdon interpreted his findings as evidence that the process of cell specialization did not require irreversible nuclear changes. A decade after Briggs and King's findings were published, Gurdon's reports7,8 that differentiation might be reversible attracted immediate attention and excitement but were also controversial. First, the low frequency (~1%) with which a normal adult was obtained after nuclear transfer from adult somatic cells into enucleated oocytes (compared with 30% for nuclei from embryonic cells or after serial transfer; that is, two sequential nuclear-transfer experiments) suggested that the clones could have arisen from a subpopulation of contaminating cells, possibly residual intestinal stem cells. Second, the experiments could not be replicated in other species. These obstacles were overcome — but not for more than three decades.
In mammals
Why did the cloning of mammals not succeed until 1997? It was certainly not for lack of trying. The first successfully cloned mammal was Dolly the sheep, made by fusing a mammary cell with an enucleated oocyte (by using a fusogenic electrical pulse)9. Wilmut and colleagues were the first to clone a mammal9, owing, in part, to their strategy. They used unfertilized oocytes as recipients, together with donor cells that had been induced to exit the cell cycle by serum deprivation in culture, which they postulated would change the chromatin structure in a manner conducive to nuclear reprogramming. A year later, Wakayama and colleagues’ technical prowess and persistence, in conjunction with the use of an enucleation pipette designed to deliver piezoelectric pulses, led to the cloning of mice10. This instrument enabled the nucleus to be removed from the mouse oocyte, which is more delicate than that of the sheep, and replaced with the nucleus of a somatic cell. This process of somatic-cell nuclear transfer (SCNT) was soon replicated in many laboratories. Importantly, Jaenisch and colleagues finally put to rest the argument that the ‘reprogramming’ resulted from the presence of contaminating stem cells or progenitor cells11,12. They obtained definitive evidence that the fate of nuclei from highly specialized, purified olfactory neurons, or from B cells in which the immunoglobulin locus had been rearranged, could be reversed to produce a mouse clone11,12.
SCNT is possible not only with oocytes but also with fertilized eggs (or zygotes), showing that the reprogramming factors are still present at this stage of development13. Notably, intact nuclei are not removed in either method of nuclear transfer. Instead, the complex of the condensed chromosomes and the spindle is carefully extracted at cell-cycle phases at which the nuclear envelope has broken down (during meiosis in oocytes or metaphase in zygotes undergoing mitosis) so that reprogramming factors that are normally localized to the nucleus are dispersed in the cytosol.
In addition to sheep and mice, a wide range of species have now been successfully cloned using SCNT, ranging from domesticated animals such as dogs and goats, and their hybrids such as mules, to wild animals such as African wildcats and wolves14. Furthermore, nuclei from frozen tissues have been transplanted successfully into enucleated oocytes a decade after tissue freezing. Such experiments were carried out with the aim of eventually using cryopreserved cells for therapeutic purposes or of cloning extinct animals15. A low efficiency of nuclear cloning (1–2%) is still typical of mice, which are the most widely used experimental animal model14. It is notable that ES cells (nuclear-transfer-derived ES cells (ntES cells)) can be generated with much higher efficiencies (~20%) from blastocysts formed by SCNT16. And, although primates have not yet been cloned, ntES cells have been successfully generated from non-human primates17.
Mechanistic insights
Cloned mice with apparently normal gross anatomy can have numerous abnormalities, which raises concerns about the fidelity of SCNT for generating cloned organisms or cells without phenotypic defects. Common abnormalities include aberrant gene expression in embryos, telomere elongation, obesity in adults, impaired immune systems and, often, increased cancer susceptibility and premature death14. To overcome these problems, various technical modifications have been tested in mice, including attempts at chemically activating oocytes to make them more responsive, changing the timing of enucleation, inhibiting cytokinesis and using cell fusion instead of nuclear injection, but these alterations have led to only modest (that is, still 1–3% at best) increases in the frequency of cloned animals14.
The efficiency of generating cloned mice increases substantially (from 1% to ~20%) when the cell source for nuclei is ntES cells rather than somatic cells, an equivalent efficiency to using normal ES cells to generate a cloned mouse16. Indeed, the approach of using ntES cells enabled researchers to clone mice from T cells and B cells11. The ntES cells are lines of ES cells derived from a cloned animal and therefore have undergone many rounds of cell division in culture. These findings suggest that the process of nuclear reprogramming is substantially enhanced by passage through an ES-cell state. Notably, cloning efficiency also differs depending on the cell type, the developmental stage, and the mouse strain of the nuclear donor.
The developmental defects in cloned animals are presumed to result, in part, from problems with the fidelity of genomic reprogramming18, owing to a failure to erase ‘epigenetic memory’ completely, a term used here to define heritable effects on gene expression that are not due to differences in DNA sequence. Epigenetic factors that contribute to the maintenance of gene expression patterns include regulators of DNA methylation, histone modifications and replacements, and ATP-dependent chromatin remodellers (see pages 721 and 728). Most such modifiers of chromatin structure do not recognize specific DNA sequences. Specific factors guide these modifiers to their targets, and feedback loops maintain the required balance of such targeting factors. Both modifiers and targeting factors therefore have crucial roles.
The potency of epigenetic regulation is exemplified by the finding that queen and worker honeybees are clones. Despite their identical DNA, queens and workers have different behaviours, morphologies and reproductive capacities. They differ because some larvae, the future queens, ingest royal jelly. The composition of royal jelly is unknown, but its effects can largely be mimicked by decreasing the levels of a single repressive epigenetic regulator, the DNA methyltransferase DNMT3 (ref. 19). The frequency of abnormalities in cloned animals that have been generated by nuclear transfer suggests that nuclear reprogramming is incomplete and that a better understanding of the mechanisms of gene regulation, particularly those of epigenetic memory, is required. The combined impact of diverse epigenetic changes and how they relate to gene expression changes and cellular memory is a subject of intense investigation and is of interest for the basic understanding of cellular plasticity and for the potential of using ES cells from SCNT-derived embryos in clinical applications.
Cell fusion
Cell fusion involves fusing two or more cell types to form a single entity. This allows the impact of one genome on another to be studied, and in this way the existence of trans-acting repressors and tumour-suppressor proteins was uncovered in the late 1960s (Figs 1 and 2). Approximately a decade later, cell-fusion studies provided the first evidence that the differentiated state of mammalian somatic cells is not fixed and irreversible but, instead, is dictated by the balance of regulators and requires continuous regulation (refs 20-22). Such studies could not be taken further until recent molecular technologies were developed, at which point cell-fusion experiments showed that the pluripotent state can dominate the differentiated state under certain conditions, leading to previously silent genes becoming activated. This approach is now being revived as a potent way of elucidating the regulatory mechanisms, such as DNA demethylation, that are required for nuclear reprogramming. In this section, we discuss the conversion of one type of differentiated somatic cell to another type and then the conversion of differentiated somatic cells to pluripotent cells.
Induction of genes typical of a particular somatic cell
Cell fusion can generate hybrids or heterokaryons (Fig. 1b). Hybrids proliferate, causing the nuclei of the original cells to fuse, whereas heterokaryons do not proliferate and therefore contain multiple distinct nuclei. Nearly four decades ago, fusion experiments showed that when a differentiated cell such as a mouse fibroblast was fused with a hamster melanocyte or a rat hepatocyte, melanin and tyrosine amino-transferase, respectively, ceased to be synthesized23-25. These pioneering studies provided novel evidence that gene expression is regulated not only by cis-acting DNA elements but also by trans-acting repressors. A few years later, cell hybrids also provided compelling evidence that there are trans-acting tumour-suppressor proteins, because in some cases in which non-cancerous cells and tumour cells were fused, the ‘normal’ state dominated the transformed state, preventing tumour formation. This suppression of malignancy did not result from oncogene loss, because on prolonged proliferation the transformed phenotype re-emerged26. In a handful of studies, genes that had been silent in one cell type were activated27,28; however, to select for hybrids, they must be dividing, which leads to nuclear fusion, loss and rearrangement of chromosomes, and aneuploidy if the hybrids are of mixed species. As a result, it was unclear from experiments using these proliferative cell hybrids whether the observed gene activation was caused by the loss of a gene encoding a repressor or the action of an activator.
In 1983, the first definitive evidence that previously silent genes could be activated in mammalian cells was obtained by H.M.B. and colleagues, by producing heterokaryons20, which are short-lived, non-dividing, multinucleate fusion products of two distinct cell types. If the cell types are from different species, their gene products can be distinguished, and nuclear reprogramming can be assessed. This demonstration of nuclear reprogramming was at first met with incredulity, because the prevailing dogma held that the differentiated state of mammalian cells was fixed and irreversible. By using heterokaryons, the problems of chromosome loss and rearrangement that were typical of hybrids were overcome, because in the absence of proliferation the nuclei of the two cell types remained distinct and intact.
Early heterokaryon studies had shown nuclear swelling and DNA and RNA synthesis, but silent genes had not been activated29, presumably as a result of the cell type chosen. These studies involved chicken erythrocytes, which have a nucleus but are among the most specialized and difficult cells to reprogram. Activation of previously silent genes was first detected in heterokaryons of muscle cells and amniotic cells20. In this study, to increase gene dosage and avoid cell division, mouse muscle cells were selected as the fusion partner, as they are naturally multinucleate and post-mitotic. Human amniotic cells were chosen as the other fusion partner, because their embryonic origin indicated that these cells might be more plastic than other cell types. In the resultant heterokaryons, directed differentiation was observed, and several human muscle proteins were expressed, indicating that muscle genes had been activated in non-muscle cells20,21.
Subsequently, heterokaryons formed from mouse muscle syncytia and diverse cell types, including human fibroblasts (which arise from the mesoderm), hepatocytes (from the endoderm) and keratinocytes (from the ectoderm), demonstrated that previously silent muscle genes could be activated in cells representative of all three embryonic lineages20,21. The relative ratio of the nuclei, or the gene dosage, contributed by the two cell types dictated the direction of reprogramming21,30. DNA replication was not required, and DNA methylation status was crucial to the outcome in heterokaryon studies31-33. The frequency and kinetics of reprogramming also differed among cell types21. That previously silent genes could be activated in muscle-cell-containing heterokaryons was rapidly corroborated by others34, and this was also found to be the case for other cell types. For example, erythroid-specific and hepatocyte-specific genes were activated in fibroblast-derived nuclei present in non-dividing, mixed-species heterokaryons35,36. Together, these heterokaryon experiments showed that the expression of previously silent genes typical of diverse differentiated mammalian cell types could be induced in other differentiated cell types, even in vivo37,38. Furthermore, they showed that the differentiated state was not fixed and irreversible but, instead, was continuously regulated by the balance of regulators present at any given time21,22, providing strong evidence for nuclear plasticity.
Induction of pluripotency genes in somatic cells
With the advent of improved molecular tools, there has been renewed interest in using cell fusion, as an approach to studying pluripotency and the regulatory mechanisms that are involved. To avoid the problems of aneuploidy, all hybrid studies cited below fused cells of the same species. Tada, Surani and colleagues were the first to show nuclear reprogramming of somatic cells in proliferative hybrids. They fused female embryonic germ cells, which are pluripotent stem cells derived from primordial germ cells, with thymocytes from adult mice39. They then investigated which sequences of DNA had been demethylated, by using DNA methylation-sensitive restriction enzymes, and whether certain imprinted and non-imprinted genes from the somatic genome had been activated. Furthermore, they showed that their fused tetraploid cells were pluripotent: the cells could contribute to the three germ layers in chimaeric embryos.
Tada and colleagues then showed that somatic cells can acquire a pluripotent state after being fused with ES cells40. They fused ES cells from male mice with thymocytes from female mice that contained a GFP reporter transgene driven by the promoter of mouse Oct4 (also known as Pou5f1), and they found that genes on the inactive X chromosome and the Oct4–GFP reporter construct of thymocytes were re-activated. With ES cells as a fusion partner, in contrast to germ cells (described above), imprinted genes in fused tetraploid cells were not demethylated. Subsequently, the same research group generated intersubspecies hybrid cells with Mus musculus domesticus ES cells and M. musculus molossinus thymocytes41. There are frequent DNA sequence polymorphisms between the genomes of these two subspecies, which allowed the researchers to monitor the origin of the RNA and DNA in hybrid clones. Using this elegant approach, they showed that the reprogrammed somatic genome in hybrids with ES cells becomes hyperacetylated at histones H3 and H4, whereas the lysine residue at position 4 of H3 becomes globally hyper-dimethylated and hyper-trimethylated, an indication that the epigenome was converted to a pluripotent state.
Cowan and colleagues extended this work by showing that nuclear reprogramming of human somatic cells can be achieved by fusing them with human ES cells in tetraploid hybrids in a 1:1 ratio42. Smith and colleagues subsequently demonstrated that, in mice, overexpression of Nanog, which encodes a pluripotency transcription factor, substantially enhanced fusion-based nuclear reprogramming43. Although, in general, such nuclear-reprogramming analyses were limited (with some notable exceptions41) because only a few gene products could be distinguished, these experiments clearly showed that, after proliferation and selection in culture, regulators of pluripotency can override regulators of cell differentiation.
Heterokaryons of mixed species allow more comprehensive studies of nuclear reprogramming than same-species cell fusions, as all gene expression changes can be monitored on the basis of species-specific differences. In addition, in mixed-species heterokaryons, chromosome loss, rearrangement and aneuploidy, which confound the interpretation of results obtained with mixed-species, proliferative hybrids, do not occur. However, it seemed counter-intuitive to use heterokaryons for studying the induction of pluripotency in somatic cells, because ES cells divide rapidly and extensively. Therefore, it seemed probable that the induction of growth arrest that follows cell fusion in heterokaryons, which are non-dividing, would lead to a loss of pluripotency and to differentiation. In contrast to expectations, results from two laboratories using mixed-species heterokaryons have shown that pluripotency genes such as Oct4 and Nanog are activated and their promoters demethylated within 1 day of fusion when mouse ES cells are fused with human B cells or with human fibroblasts44,45.
Heterokaryons are ideally suited to analyses of the earliest molecular events that underlie reprogramming. The key to such gene expression analyses is the ability to purify the small proportion of heterokaryons in the population (~1%) immediately after fusion, which can be achieved by flow cytometry. By contrast, in the experiments that have been carried out so far, hybrids have proliferated extensively and been drug selected before being analysed, so it has not been possible to assess the earliest changes in gene expression. The efficiency of inducing pluripotency depends on the cell types used to form the heterokaryons (in the example above, 16% for B cells and 70% for fibroblasts)44,45, and differences in the timing of gene activation and DNA demethylation are observed. Presumably, these differences arise because the ratio of the nuclei from the original cells and the balance of transcriptional regulators that are present dictate the extent to which cells can be reprogrammed towards pluripotency (as seen previously for gene activation in somatic cells using heterokaryons20-22,32).
The rapid rate of reprogramming detected in heterokaryons as opposed to hybrids makes them useful for elucidating the molecular mechanisms that are required for initiating reprogramming to a pluripotent state, by using loss-of-function and gain-of-function approaches (Fig. 3). For example, when mouse ES cells that have lost Oct4 expression are fused to form heterokaryons with human B cells, the B cells are not reprogrammed, showing that OCT4 is required for reprogramming towards a pluripotent state44. In another example, using heterokaryons, H.M.B. and colleagues recently elucidated a novel role for AID, an enzyme known to deaminate cytosine residues. They uncovered an active mechanism that is essential for DNA demethylation and for the induction of nuclear reprogramming of fibroblasts towards pluripotency45 (see ref. 46 for a review). An enzyme that removes methyl groups from cytosine bases has yet to be discovered. These new data suggest that, instead, a DNA repair mechanism may operate in mammals, in which a methylated base or nucleotide is exchanged for an unmethylated one. These studies exemplify the potential of cell fusion to provide mechanistic insights into the roadblocks, such as DNA demethylation, to the successful reprogramming of somatic cells by SCNT or transcription-factor transduction.
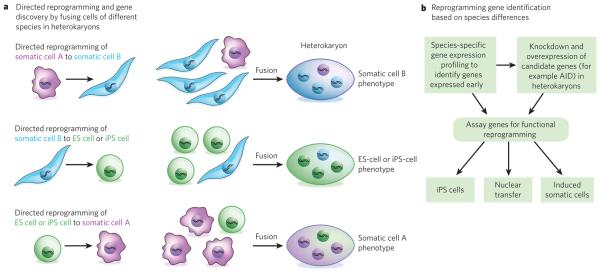
Figure 3. Investigating the genes involved in nuclear reprogramming by using mixed-species heterokaryons.
Cell fusion leads to nuclear reprogramming towards a specific phenotype, which is dictated by the nuclear ratio of the fused cell types in heterokaryons, which do not divide. When, for example, cells from humans and mice are fused in a skewed ratio (such as 1:3) (a), the human cells will generally be reprogrammed towards the mouse cell phenotype (three examples are shown). To uncover which genes are involved in this process at the onset of reprogramming (b), genome-wide species-specific gene expression profiling can be carried out on the three types of heterokaryon shown. In this way, the transcripts of human genes that are induced soon after fusion can be identified, and the effects of knocking down these candidate genes (loss of function) or overexpressing them (gain of function) these candidate genes can also be tested45. The function of these genes can then be validated by assays that assess whether they are required for nuclear transfer or for generating iPS cells or induced somatic cells. For example, assays can test whether expression of the genes identified in the heterokaryons with an ES-cell or iPS-cell phenotype (a, centre) enhances, or is required for, the generation of iPS cells or for reprogramming by nuclear transfer. The genes identified in the heterokaryons with a somatic cell phenotype (a, top and bottom) can be tested to uncover whether they enhance, or are required for, the conversion of iPS cells or ES cells into a particular somatic cell type or the conversion of one type of somatic cell into another type. Such experiments will increase the understanding of the molecular regulators of nuclear reprogramming and therefore improve the safety and efficacy of cells produced for therapeutic purposes.
Transcription-factor transduction
The overexpression of a single transcription factor in somatic cells was unexpectedly found to activate cohorts of genes that were typical of other somatic cell types, first in D. melanogaster47,48 and subsequently in mammals49, leading to remarkable changes in cell fate. More surprisingly, it was discovered that pluripotency can be regained by numerous differentiated somatic cell types through overexpressing just four transcription-factor-encoding genes. These cells, known as iPS cells, are the strongest example so far of the plasticity of cells in response to a disruption in the stoichiometry of their transcriptional regulators. Human iPS cells can be used to generate cells for tissue repair or replacement while avoiding the ethical and immunological issues that are inherent in the use of ES cells (Fig. 4). Furthermore, because these cells can be derived from a patient's own cells, they give researchers the ability to model human diseases and screen drug candidates in vitro in an unprecedented manner. In this section, we provide a historical perspective on the findings from transcription-factor transduction experiments, which first involved single factors and then groups of factors. In the past four years, it has been shown that increasing the expression of four proteins can reprogram a differentiated somatic cell to become a pluripotent cell, potentially revolutionizing medicine and highlighting the importance of nuclear reprogramming.
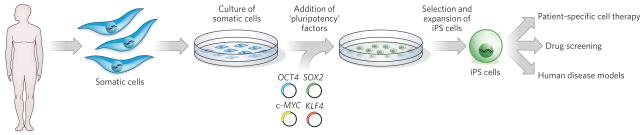
Figure 4. Applications of iPS cells.
To generate iPS cells, fibroblasts (or another type of adult somatic cell) are transduced with retroviruses encoding four pluripotency factors (SOX2, KLF4, c-MYC and OCT4)56,63. Fully reprogrammed iPS cells have similar properties to ES cells. They are competent to form teratomas on injection into mice and are capable of generating progeny. A patient's cells can be used to derive iPS cells, which can then be induced to undergo differentiation into various types of somatic cell, all with the same genetic information as the patient. For example, dopaminergic neurons could be generated from the cells of a patient with Parkinson's disease and then transplanted to replace those neurons that have been lost. These differentiated cells can also be used in disease models for studying the molecular basis of a broad range of human diseases that are otherwise difficult to study (for instance, those that affect brain cells) and for screening the efficacy and safety of drug candidates for treating these diseases.
Conversion of one somatic-cell fate to another
The fate of a cell can be altered by forced expression of single tissue-specific transcription factors. Gehring and colleagues were the first to show this47 in 1987: in D. melanogaster larvae, ectopic overexpression of a homeotic gene, Antennapedia, under the control of a heat-shock gene promoter led to a change in body plan, with an additional set of legs being formed instead of antennae. Even more striking was the finding by Gehring48 almost a decade later that ectopic expression of eyeless (known as Pax6 in mice), a master controller of a cascade of 2,500 genes, led to the development of functional eyes on the legs, wings and antennae of D. melanogaster. In mice, the first tissue-specific master regulatory transcription factor was identified by Weintraub and colleagues49 in 1987. They found that it was possible to induce a phenotypic conversion to the myogenic lineage by expressing a single muscle helix–loop–helix protein MYOD49. The cloning of Myod followed astute observations by Taylor and Jones50 in 1979, who noticed that the filamentous structures in fibroblast cultures treated with 5-azacytidine, a demethylating anticancer agent, were not fungal contamination but, instead, syncytial myotubes. In addition, in 2004, the mouse C/EBP family of transcription factors was shown by Graf and colleagues to have a key role in the conversion of one blood cell type to another (from lymphocytes to macrophages)51, and when the gene encoding the transcription factor PAX5 was removed from B cells, these cells reverted to less specialized progenitors52. For a comprehensive review of such trans-differentiation induced by transcription factors, see ref. 53. It should be noted that, in mammals, altering the expression of single transcription factors generally results in the phenotype of somatic cells changing only to that of closely related cell types, so the effects of transcription factors are highly context dependent54,55.
Conversion of somatic cells to pluripotent cells
Although the elegant fusion experiments by Tada, Surani and colleagues39 clearly showed that ES cells and embryonic germ cells contain factors that can induce reprogramming and pluripotency in somatic cells, attempts by many investigators to identify master regulators of the ES-cell state have failed. As a result, the prevailing view until about four years ago was that nuclear reprogramming to a pluripotent state is a highly complex process that might entail the cooperation of up to 100 factors. Consequently, when S.Y. and colleagues showed56,57 that a combination of only four transcription factors could generate ES-cell-like pluripotent cells from mouse fibroblasts, the results elicited excitement, as well as scepticism. They used retroviral vectors to introduce into mouse embryonic and adult fibroblasts a mini-complementary-DNA library of 24 genes expressed by ES cells, and these genes were then tested for their collective ability to induce pluripotency. Pluripotency was assayed by examining for activation of a reporter gene construct containing the promoter of Fbx15 (also known as Fbxo15), a gene previously identified as being specific to ES cells58. Clones in which the Fbx15 promoter was activated produced a reporter protein that rendered them resistant to the drug neomycin, and these drug-resistant clones had similar morphology, growth properties and gene expression characteristics to ES cells. More importantly, after injection into mice, they were capable of forming teratomas (tumours that include cells of all three germ layers), indicating their pluripotency. Rather than determining the contribution of each factor singly or in subgroups, factors were progressively eliminated from the pool one at a time. As a result, the authors identified four key factors that sufficed to induce pluripotency in fibroblasts — OCT4, SOX2, KLF4 and c-MYC — and they named these pluripotent cells iPS cells56.
Owing to the simplicity of the approach, and because iPS cells initially failed to produce adult chimaeric mice, many investigators were dubious about their validity as stem cells with ES-cell-like properties. However, within a year, two research groups independently showed that overexpression of the four factors generated cells capable of forming adult chimaeras and generating functional germ cells59,60. Subsequent studies showed that iPS cells could be generated without overexpressing c-MYC61,62. Human iPS cells were obtained within a year by inducing overexpression of the same combination of factors63-65 or different, but overlapping, cocktails of factors66. Strikingly, transgenes encoding the four factors need to be present only when iPS cells are being generated. When these cells have become established, the retroviral transgenes become silenced, and the endogenous genes encoding the four factors become activated. Hence, the self-renewal of iPS cells and maintenance of their pluripotency rely entirely on endogenous gene expression, of the genes encoding OCT4, SOX2, KLF4 and c-MYC, suggesting that iPS cells have undergone almost-complete reprogramming.
Technical advances in iPS-cell generation
The four factors that were initially identified can now be substituted with different factors or with certain small molecules. But the original finding — that a set of factors is required — holds true, and certain key factors such as OCT4 cannot be omitted. A pluripotent stem-cell state has now been induced in a plethora of differentiated cell types, inclu ding pancreatic β cells, keratinocytes, hepatocytes and stomach cells67.
The various cell types are, however, converted to a pluripotent state with varying (generally low) frequencies, indicating that (as is the case for nuclear transfer, heterokaryon and single-transcription factor experiments) reprogramming is context dependent and the cell type affects the capacity to become an iPS cell. Efforts have been directed towards improving the proportion of retroviral-vector-transduced cells that is converted. For example, exposure to a hypoxic environment or vitamin C has been shown to increase the frequency of iPS-cell generation68,69. In addition, disrupting the signalling pathways mediated by the tumour-suppressor protein p53 or the cell-cycle regulator INK4A removes cell-cycle control checkpoints, and iPS cells are then generated more rapidly70-74. Certain microRNAs have also been described to enhance the efficiency of reprogramming75. Furthermore, stem cells or progenitor cells seem to be more readily reprogrammed to become iPS cells than more specialized cells of a certain lineage, as has been shown for haematopoietic cells76. Despite these advances in our ability to generate iPS cells, all transduced cells do not become iPS cells. This is particularly clear from a study by Wernig, Jaenisch and colleagues77, who produced transgenic mice from iPS cells that were established using doxycycline-inducible vectors. When mouse fibroblasts were isolated from these doxycycline-regulated transgenic mice, the frequency of iPS cells generated from them was 100 fold greater than the frequency with which the original iPS cells were generated but was still unexpectedly low77. A similar finding was reported with a doxycycline-inducible, excisable piggyBac transposon system encoding all four of the initially identified factors78. On the basis of these findings, many researchers now think that reprogramming during iPS-cell generation entails stochastic (that is, random) events79,80.
Changing approach
The multiple-factor approach to reprogramming that is used to generate iPS cells has led to a shift from attempting to identify and use a single ‘master’ regulatory transcription factor to the use of multiple factors in the reprogramming of mammalian cell fate. Using the principles and experimental approach that S.Y. and colleagues determined for generating iPS cells, two research groups (led by Melton and by Wernig) have deciphered the individual contributions of a panoply of transcription factors, identifying three that can convert one cell type to another (factors that can convert, for example, exocrine cells into insulin-producing endocrine cells in vivo and fibroblasts into excitatory neurons in vitro)81,82. Many groups are now trying to deconvolve large numbers of transcription factors and identify sets that can be used to convert one type of somatic cell to another type or to a progenitor or stem cell that could be used to regenerate a tissue. In addition, analysing mixed-species heterokaryons might help to uncover previously unidentified or unexpected genes and mechanisms involved in reprogramming cells from stably differentiated states to a pluripotent state or in the reverse direction83-86 (Fig. 3). Gaining a better understanding of the reprogramming process and how epigenetic memory is established could reduce the chances of generating tumorigenic cells and therefore enhance the usefulness of iPS cells, providing safer sources for cell therapy in the future.
Future challenges
The history of nuclear reprogramming is colourful and encompasses almost 50 years of research in which increasingly sophisticated tools have become available. Each of the three main approaches is distinct; however, each informs the others, and the findings are synergistic. In each case, if the balance of regulators is tipped to favour pluripotency massively — by immersing a small somatic cell nucleus in a mixture of oocyte regulators (nuclear transfer), an excess of ES-cell factors (cell fusion) or an extremely large amount of four pluripotency proteins (transcription-factor transduction) — then the epigenome is altered, and the expression of pluripotency genes is induced in an otherwise stably differentiated cell. It follows that, to maintain a cell's pluri-potent state, there must be mechanisms such as feedback loops and autoregulation of key positive and negative transcriptional regulators of pluripotency (such as OCT4) to attain critical threshold levels of these transcriptional regulators. Similar thresholds of differentiation-specific transcription factors (such as MYOD) maintain the phenotypic identity of somatic cells such as muscle cells. In this respect, differentiation and pluripotency are controlled by mechanisms similar to those for prokaryotes, which were described decades ago by Jacob and Monod and by Ptashne87-89. Despite this conceptual framework, much remains to be learned about the molecular basis of both transcriptional memory and epigenetic memory before reprogramming is fully understood and its potential can be harnessed. Systematic studies using all three approaches to elucidate the genes and mechanisms that control reprogramming will enhance the fidelity and efficiency of reprogramming to a pluripotent state and to desired differentiated states.
A comparison of the three approaches to nuclear reprogramming reveals certain common features. These include the demethylation of pluripotency gene promoters and the activation of Oct4, Nanog and a battery of other ES-cell-specific genes. In addition, cell ‘rejuvenation’ is evident from the lengthening of telomeres and the increased activity of telomerase90-92. For all three approaches, some somatic cell types are more readily reprogrammed than others. And, in female cells, genes on the inactive X chromosome are reactivated39,93-95, although this reversal of relatively stable methylated states is often incomplete.
The three approaches differ in technical feasibility, time required for reprogramming, efficiency of pluripotency gene induction at the single cell level, and cell yield (Fig. 5). These features can be differentially exploited to study the mechanisms underlying the conversion to pluripotency and to obtain enough cells for therapeutic applications. Nuclear transfer is characterized by rapid reprogramming18 and is ideally suited to elucidating the fundamental principles of early embryonic development and reproductive biology, as well as yielding a wealth of ES cells for therapeutic applications. By contrast, cell fusion is technically simple. When cell fusion is used to form mixed-species hetero karyons, which do not proliferate, pluripotency genes are activated quickly and can be activated with high frequency45. This approach is therefore particularly well suited to elucidating the molecular mechanisms that control the onset of nuclear reprogramming, but it does not yield clinically useful cells. Transcription-factor transduction does yield such cells. The ease with which iPS cells are produced in abundance in laboratories around the world, their potential for studying the mechanisms underlying human disease, and their usefulness for drug discovery and cell therapy is currently unparalleled67.
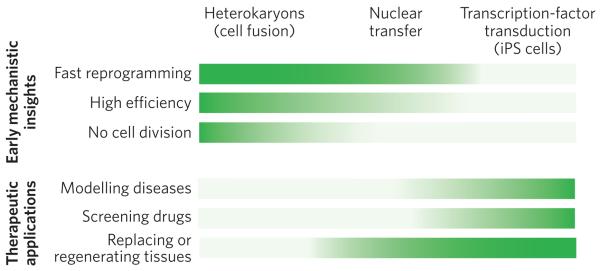
Figure 5. Comparison of the advantages of the three approaches to nuclear reprogramming.
The three approaches to reprogramming somatic cells differ in their technical difficulty, speed of reprogramming, efficiency of inducing pluripotency, and cell yield. Therefore, each approach is better suited for studies that provide early mechanistic insights (top) or for therapeutic applications (bottom). The greater the intensity of the colour, the more advantageous the technique. For gaining mechanistic insights (top) into the onset of reprogramming, heterokaryons are particularly advantageous, for three main reasons. First, they are quickly reprogrammed to express pluripotency genes (1 to 2 days ). This is also the case for nuclear transfer. By contrast, it takes weeks to generate iPS cells. Second, reprogramming by cell fusion is highly efficient. When mouse ES cells are fused with human fibroblasts, up to 70% of heterokaryons (enriched by fluorescence-activated cell sorting) activate the expression of pluripotency genes within 1 day. It is technically challenging (and therefore inefficient) to carry out nuclear transfer in mice, so it is difficult to use this approach for large-scale molecular analyses. Furthermore, the efficiency of generating iPS cells by transcription-factor transduction is low, about 0.01–0.1%. Third, cell division does not occur in heterokaryons. It also does not occur after nuclear transfer during the time when pluripotency genes are induced, allowing active mechanisms that induce pluripotency gene expression to be studied because this induction is independent of cell division and DNA replication; passive mechanisms may accompany cell division (for example dilution of DNA methyltransferases). By contrast, many rounds of cell division are required to generate iPS cells. For therapeutic applications (bottom), iPS cells are particularly advantageous, for three main reasons. First, diseases can readily be modelled using iPS cells derived from patients, overcoming the ethical issues and problems with immunological rejection that are inherent in obtaining human ES cells for studying disease. Skin fibroblasts can be readily obtained from the skin of an individual with a particular heritable disease, induced to become pluripotent in vitro and then induced to undergo differentiation to become the cell type of interest (for example a specific kind of cardiac cell). The pathways underlying a disease state (that is, gene expression and signalling) can thus be studied in cells that are not easily accessible in living humans. Second, drug screening can be carried out in vitro using these iPS-cell-based disease models to determine whether therapeutic drug candidates ameliorate or correct aberrant pathways. Third, for certain diseases, cell therapy might soon be used to regenerate or replace defective tissues, with the caveat that the tumorigenic potential, which is in part due to viral vector integration, must be overcome. Both nuclear transfer (leading to ES-cell production) and transcription-factor transduction (to produce iPS cells) have a high cell yield, which is important for cell therapy applications.
The three approaches may also differ in their underlying molecular mechanisms; for example, DNA demethylation is a crucial step in reprogramming by all three approaches. So far, no consensus DNA demethylase has been identified in mammals. A passive, stochastic DNA demethylation mechanism has been proposed to operate in the generation of iPS cells: in this mechanism, as the cells are propagated, the DNA is progressively demethylated, owing to a failure of maintenance methylation after DNA replication79. By contrast, an active DNA demethylation mechanism that is replication independent and mediated by AID has been shown in heterokaryons45. After nuclear transfer, similarly to heterokaryons, pluripotency genes are activated and their promoters demethylated in the absence of DNA replication or cell division18. Further studies are required to determine whether these are fundamental differences in nuclear-reprogramming mechanisms. Future studies using all three approaches will improve the current understanding of nuclear reprogramming and cell differentiation, which is essential to advance cell therapy, the modelling of human disease in vitro and the search for therapeutic agents96-98.
Acknowledgements
We apologize to those whose work is not cited because of space constraints. We are grateful to our colleagues J. Gurdon, T. Graf, J. Pomerantz and T. Tada, as well as to members of our laboratories for their insightful comments on the manuscript and especially to N. Bhutani and S. Corbel for figure design. We also thank our funding sources: S.Y. acknowledges support from the Japanese Ministry of Education, Culture, Sports, Science & Technology, the Japan Science and Technology Agency and the National Institute of Biomedical Innovation (Japan); and H.M.B. acknowledges support from the National Institutes of Health (grants AG009521, AG020961 and HL096113), MDA (4320), the Juvenile Diabetes Research Foundation (34-2008-623), LLS (6025-09), the California Institute for Regenerative Medicine (RT1-01001 and RB1-02192) and the Baxter International Foundation.
Footnotes
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 2.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl Acad. Sci. USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hadorn E. Constancy, variation and type of determination and differentiation in cells from male genitalia rudiments of Drosophila melanogaster in permanent culture in vivo [in German with English abstract] Dev. Biol. 1966;13:424–509. doi: 10.1016/0012-1606(66)90058-3. [DOI] [PubMed] [Google Scholar]
- 4.Gehring W. Clonal analysis of determination dynamics in cultures of imaginal disks in Drosophila melanogaster. Dev. Biol. 1967;16:438–456. doi: 10.1016/0012-1606(67)90058-9. [DOI] [PubMed] [Google Scholar]
- 5.Le Lievre CS, Le Douarin NM. Mesenchymal derivatives of the neural crest: analysis of chimaeric quail and chick embryos. J. Embryol. Exp. Morphol. 1975;34:125–154. [PubMed] [Google Scholar]
- 6.Briggs R, King TJ. Transplantation of living nuclei from blastula cells into enucleated frogs' eggs. Proc. Natl Acad. Sci. USA. 1952;38:455–463. doi: 10.1073/pnas.38.5.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gurdon JB. The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J. Embryol. Exp. Morphol. 1962;10:622–640. [PubMed] [Google Scholar]
- 8.Gurdon JB. Adult frogs derived from the nuclei of single somatic cells. Dev. Biol. 1962;4:256–273. doi: 10.1016/0012-1606(62)90043-x. In this study, using nuclear transfer, differentiated intestinal cells in amphibians were shown to retain all of the genetic information to produce an entire frog. [DOI] [PubMed] [Google Scholar]
- 9.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. In this study, the first cloned mammal, Dolly the Sheep, was generated using nuclear transfer. [DOI] [PubMed] [Google Scholar]
- 10.Wakayama T, Perry AC, Zuccotti M, Johnson KR, Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394:369–374. doi: 10.1038/28615. In this study, the first cloned mice, the most widely used experimental animal, were generated using nuclear transfer. [DOI] [PubMed] [Google Scholar]
- 11.Hochedlinger K, Jaenisch R. Monoclonal mice generated by nuclear transfer from mature B and T donor cells. Nature. 2002;415:1035–1038. doi: 10.1038/nature718. [DOI] [PubMed] [Google Scholar]
- 12.Eggan K, et al. Mice cloned from olfactory sensory neurons. Nature. 2004;428:44–49. doi: 10.1038/nature02375. [DOI] [PubMed] [Google Scholar]
- 13.Egli D, Rosains J, Birkhoff G, Eggan K. Developmental reprogramming after chromosome transfer into mitotic mouse zygotes. Nature. 2007;447:679–685. doi: 10.1038/nature05879. [DOI] [PubMed] [Google Scholar]
- 14.Thuan NV, Kishigami S, Wakayama T. How to improve the success rate of mouse cloning technology. J. Reprod. Dev. 2010;56:20–30. doi: 10.1262/jrd.09-221a. [DOI] [PubMed] [Google Scholar]
- 15.Wakayama S, et al. Production of healthy cloned mice from bodies frozen at −20 degrees C for 16 years. Proc. Natl Acad. Sci. USA. 2008;105:17318–17322. doi: 10.1073/pnas.0806166105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang X, et al. Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nature Genet. 2007;39:295–302. doi: 10.1038/ng1973. [DOI] [PubMed] [Google Scholar]
- 17.Byrne JA, et al. Producing primate embryonic stem cells by somatic cell nuclear transfer. Nature. 2007;450:497–502. doi: 10.1038/nature06357. [DOI] [PubMed] [Google Scholar]
- 18.Simonsson S, Gurdon J. DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei. Nature Cell Biol. 2004;6:984–990. doi: 10.1038/ncb1176. [DOI] [PubMed] [Google Scholar]
- 19.Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319:1827–1830. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- 20.Blau HM, Chiu CP, Webster C. Cytoplasmic activation of human nuclear genes in stable heterocaryons. Cell. 1983;32:1171–1180. doi: 10.1016/0092-8674(83)90300-8. This paper shows that differentiated mammalian cells are plastic: their differentiated state can be reversed by fusing them to another cell to form a stable, non-dividing heterokaryon. [DOI] [PubMed] [Google Scholar]
- 21.Blau HM, et al. Plasticity of the differentiated state. Science. 1985;230:758–766. doi: 10.1126/science.2414846. [DOI] [PubMed] [Google Scholar]
- 22.Blau HM, Baltimore D. Differentiation requires continuous regulation. J. Cell Biol. 1991;112:781–783. doi: 10.1083/jcb.112.5.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davidson RL, Ephrussi B, Yamamoto K. Regulation of pigment synthesis in mammalian cells, as studied by somatic hybridization. Proc. Natl Acad. Sci. USA. 1966;56:1437–1440. doi: 10.1073/pnas.56.5.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiss MC, Chaplain M. Expression of differentiated functions in hepatoma cell hybrids: reappearance of tyrosine aminotransferase inducibility after the loss of chromosomes. Proc. Natl Acad. Sci. USA. 1971;68:3026–3030. doi: 10.1073/pnas.68.12.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ringertz NR, Savage RE. Cell Hybrids. Academic; 1977. [Google Scholar]
- 26.Harris H, Miller OJ, Klein G, Worst P, Tachibana T. Suppression of malignancy by cell fusion. Nature. 1969;223:363–368. doi: 10.1038/223363a0. [DOI] [PubMed] [Google Scholar]
- 27.Peterson JA, Weiss MC. Expression of differentiated functions in hepatoma cell hybrids: induction of mouse albumin production in rat hepatoma-mouse fibroblast hybrids. Proc. Natl Acad. Sci. USA. 1972;69:571–575. doi: 10.1073/pnas.69.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davidson RL. Regulation of melanin synthesis in mammalian cells: effect of gene dosage on the expression of differentiation. Proc. Natl Acad. Sci. USA. 1972;69:951–955. doi: 10.1073/pnas.69.4.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harris H, Watkins JF, Ford CE, Schoefl GI. Artificial heterokaryons of animal cells from different species. J. Cell Sci. 1966;1:1–30. doi: 10.1242/jcs.1.1.1. [DOI] [PubMed] [Google Scholar]
- 30.Pavlath GK, Blau HM. Expression of muscle genes in heterokaryons depends on gene dosage. J. Cell Biol. 1986;102:124–130. doi: 10.1083/jcb.102.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiu CP, Blau HM. Reprogramming cell differentiation in the absence of DNA synthesis. Cell. 1984;37:879–887. doi: 10.1016/0092-8674(84)90423-9. [DOI] [PubMed] [Google Scholar]
- 32.Miller SC, Pavlath GK, Blakely BT, Blau HM. Muscle cell components dictate hepatocyte gene expression and the distribution of the Golgi apparatus in heterokaryons. Genes Dev. 1988;2:330–340. doi: 10.1101/gad.2.3.330. [DOI] [PubMed] [Google Scholar]
- 33.Chiu CP, Blau HM. 5-Azacytidine permits gene activation in a previously noninducible cell type. Cell. 1985;40:417–424. doi: 10.1016/0092-8674(85)90155-2. [DOI] [PubMed] [Google Scholar]
- 34.Wright WE. Induction of muscle genes in neural cells. J. Cell Biol. 1984;98:427–435. doi: 10.1083/jcb.98.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baron MH, Maniatis T. Rapid reprogramming of globin gene expression in transient heterokaryons. Cell. 1986;46:591–602. doi: 10.1016/0092-8674(86)90885-8. [DOI] [PubMed] [Google Scholar]
- 36.Spear BT, Tilghman SM. Role of α-fetoprotein regulatory elements in transcriptional activation in transient heterokaryons. Mol. Cell Biol. 1990;10:5047–5054. doi: 10.1128/mcb.10.10.5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johansson CB, et al. Extensive fusion of haematopoietic cells with Purkinje neurons in response to chronic inflammation. Nature Cell Biol. 2008;10:575–583. doi: 10.1038/ncb1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weimann JM, Charlton CA, Brazelton TR, Hackman RC, Blau HM. Contribution of transplanted bone marrow cells to Purkinje neurons in human adult brains. Proc. Natl Acad. Sci. USA. 2003;100:2088–2093. doi: 10.1073/pnas.0337659100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tada M, Tada T, Lefebvre L, Barton SC, Surani MA. Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. EMBO J. 1997;16:6510–6520. doi: 10.1093/emboj/16.21.6510. This report shows that fusing embryonic germ cells with somatic cells results in the reprogramming of epigenetic marks on imprinted genes in the somatic cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tada M, Takahama Y, Abe K, Nakatsuji N, Tada T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr. Biol. 2001;11:1553–1558. doi: 10.1016/s0960-9822(01)00459-6. [DOI] [PubMed] [Google Scholar]
- 41.Kimura H, Tada M, Nakatsuji N, Tada T. Histone code modifications on pluripotential nuclei of reprogrammed somatic cells. Mol. Cell. Biol. 2004;24:5710–5720. doi: 10.1128/MCB.24.13.5710-5720.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cowan CA, Atienza J, Melton DA, Eggan K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science. 2005;309:1369–1373. doi: 10.1126/science.1116447. [DOI] [PubMed] [Google Scholar]
- 43.Silva J, Chambers I, Pollard S, Smith A. Nanog promotes transfer of pluripotency after cell fusion. Nature. 2006;441:997–1001. doi: 10.1038/nature04914. [DOI] [PubMed] [Google Scholar]
- 44.Pereira CF, et al. Heterokaryon-based reprogramming of human B lymphocytes for pluripotency requires Oct4 but not Sox2. PLoS Genet. 2008;4:e1000170. doi: 10.1371/journal.pgen.1000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhutani N, et al. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. This report shows that the enzyme AID is essential for the demethylation of DNA and for the induction of pluripotency by forming heterokaryons. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Agarwal S, Daley GQ. AID for reprogramming. Cell Res. 2010;20:253–255. doi: 10.1038/cr.2010.30. [DOI] [PubMed] [Google Scholar]
- 47.Schneuwly S, Klemenz R, Gehring WJ. Redesigning the body plan of Drosophila by ectopic expression of the homoeotic gene Antennapedia. Nature. 1987;325:816–818. doi: 10.1038/325816a0. [DOI] [PubMed] [Google Scholar]
- 48.Gehring WJ. The master control gene for morphogenesis and evolution of the eye. Genes Cells. 1996;1:11–15. doi: 10.1046/j.1365-2443.1996.11011.x. [DOI] [PubMed] [Google Scholar]
- 49.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 50.Taylor SM, Jones PA. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell. 1979;17:771–779. doi: 10.1016/0092-8674(79)90317-9. [DOI] [PubMed] [Google Scholar]
- 51.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 52.Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 2007;449:473–477. doi: 10.1038/nature06159. [DOI] [PubMed] [Google Scholar]
- 53.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 54.Farah MH, et al. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- 55.Schafer BW, Blakely BT, Darlington GJ, Blau HM. Effect of cell history on response to helix-loop-helix family of myogenic regulators. Nature. 1990;344:454–458. doi: 10.1038/344454a0. [DOI] [PubMed] [Google Scholar]
- 56.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. This report shows that the introduction of four transcription factors into somatic mouse cells is sufficient to make these cells (now known as iPS cells) pluripotent. [DOI] [PubMed] [Google Scholar]
- 57.Yamanaka S. Strategies and new developments in the generation of patient-specific 57 stem cells. Cell Stem Cell. 2007;1:39–49. doi: 10.1016/j.stem.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 58.Tokuzawa Y, et al. Fbx15 is a novel target of Oct3/4 but is dispensable for embryonic stem cell self-renewal and mouse development. Mol. Cell. Biol. 2003;23:2699–2708. doi: 10.1128/MCB.23.8.2699-2708.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wernig M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 60.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 61.Nakagawa M, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nature Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 62.Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 63.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. This report shows that human somatic cells can be made pluripotent (converted to iPS cells) solely by introducing four transcription factors. [DOI] [PubMed] [Google Scholar]
- 64.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 65.Aasen T, et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nature Biotechnol. 2008;26:1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- 66.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 67.Yamanaka S. A fresh look at iPS cells. Cell. 2009;137:13–17. doi: 10.1016/j.cell.2009.03.034. [DOI] [PubMed] [Google Scholar]
- 68.Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell. 2009;5:237–241. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 69.Esteban MA, et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6:71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 70.Li H, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marion RM, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Utikal J, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kawamura T, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hong H, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nature Biotechnol. 2009;27:459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eminli S, et al. Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nature Genet. 2009;41:968–976. doi: 10.1038/ng.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wernig M, et al. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nature Biotechnol. 2008;26:916–924. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Woltjen K, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hanna J, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature. 2009;460:49–52. doi: 10.1038/nature08180. [DOI] [PubMed] [Google Scholar]
- 81.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to β-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vierbuchen T, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Palermo A, et al. Nuclear reprogramming in heterokaryons is rapid, extensive, and bidirectional. FASEB J. 2009;23:1431–1440. doi: 10.1096/fj.08-122903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang F, Pomerantz JH, Sen G, Palermo AT, Blau HM. Active tissue-specific DNA demethylation conferred by somatic cell nuclei in stable heterokaryons. Proc. Natl Acad. Sci. USA. 2007;104:4395–4400. doi: 10.1073/pnas.0700181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pomerantz JH, Mukherjee S, Palermo AT, Blau HM. Reprogramming to a muscle fate by fusion recapitulates differentiation. J. Cell Sci. 2009;122:1045–1053. doi: 10.1242/jcs.041376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Terranova R, Pereira CF, Du Roure C, Merkenschlager M, Fisher AG. Acquisition and extinction of gene expression programs are separable events in heterokaryon reprogramming. J. Cell Sci. 2006;119:2065–2072. doi: 10.1242/jcs.02945. [DOI] [PubMed] [Google Scholar]
- 87.Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 88.Ptashne M. A Genetic Switch: Gene Control and Phage Lambda. Blackwell Science; 1986. [Google Scholar]
- 89.Blau HM. Differentiation requires continuous active control. Annu. Rev.Biochem. 1992;61:1213–1230. doi: 10.1146/annurev.bi.61.070192.010025. [DOI] [PubMed] [Google Scholar]
- 90.Rideout WM, Eggan K, Jaenisch R. Nuclear cloning and epigenetic reprogramming of the genome. Science. 2001;293:1093–1098. doi: 10.1126/science.1063206. [DOI] [PubMed] [Google Scholar]
- 91.Wakayama T, et al. Cloning of mice to six generations. Nature. 2000;407:318–319. doi: 10.1038/35030301. [DOI] [PubMed] [Google Scholar]
- 92.Marion RM, et al. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009;4:141–154. doi: 10.1016/j.stem.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 93.Eggan K, et al. X-chromosome inactivation in cloned mouse embryos. Science. 2000;290:1578–1581. doi: 10.1126/science.290.5496.1578. [DOI] [PubMed] [Google Scholar]
- 94.Nolen LD, et al. X chromosome reactivation and regulation in cloned embryos. Dev. Biol. 2005;279:525–540. doi: 10.1016/j.ydbio.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 95.Maherali N, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 96.Dimos JT, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 97.Park IH, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Daley GQ. Stem cells: roadmap to the clinic. J. Clin. Invest. 2010;120:8–10. doi: 10.1172/JCI41801. [DOI] [PMC free article] [PubMed] [Google Scholar]