Abstract
Objective
To evaluate whether HIV-1 clade C (HIV-C) envelope variations that arise during disease progression in rhesus macaque model reflect changes that occur naturally in human infection.
Design
An infant macaque was infected with SHIV-1157i, an R5 tropic clade C SHIV (SHIV-C) which expresses a primary HIV-C envelope derived from an infected human infant, and monitored over a five-year period. Genetic variation of the V1-V5 envelope region, which is the main target for humoral immune responses, derived from the infected macaque and infant was examined.
Methods
V1-V5 envelope region were cloned and sequenced from longitudinal PBMC samples collected from the infected macaque and infant. Phylogenetic analysis (phylogenetic tree, diversity, divergence, ratio of non-synonymous (dN) and synonymous substitution (dS) and dN distribution) were performed. Plasma RNA viral load, CD4+ T-cell count, changes in the length of V1-V5 region, putative N-linked glycosylation sites (PNGSs) number and distribution were also measured.
Results
Phylogenetic analysis revealed that changes in the macaque closely reflected those of infant during disease progression. Similar distribution patterns of dN and hot spots were observed between the macaque and infant. Analysis of PNGSs revealed several common variations between the virus populations in the two host species. These variations correlate with decline of CD4 count in the macaque and might be linked with disease progression.
Conclusion
SHIV-C infection of macaque is a relevant animal model for studying variation of primary HIV-C envelope during disease progression and could be used to analyze the selection pressures that are associated with those changes.
Keywords: SHIV, HIV-1, evolution, rhesus macaque, disease progression
Introduction
The V1-V5 region of HIV-1 envelope glycoprotein is the focus of numerous studies due to its critical role in viral pathogenesis and immune evasion [1–7]. Because of its hyper-variable nature, majority of the potential vaccines targeting the envelope have thus far failed to elicit sterilizing immunity against heterologous viral challenge in animal studies and human clinical trials [8–14]. Thus, it is important to better understand the changes of V1-V5 region that occur during the course of disease and the factors that contribute to these changes.
Non-human primate models that utilize SHIV [15–20] had been used to study clade B and laboratory-adapted strain envelope variants during the course of disease [21–23]. It has been suggested that there is a close similarity in the envelope evolution during SHIV infection of non-human primates and HIV-1 infection of humans; similar envelope mutations were observed over time in a laboratory worker accidentally infected with HIV-1 IIIB and a macaque experimentally infected with a SHIV expressing the identical envelope [22, 24]. However, HIV-1 IIIB is an X4 laboratory-adapted clade B virus whose envelope evolution may not be representative of that of R5 tropic primary isolates and only a small number of sequences from both the macaque and human were analyzed. Several studies have also indicated that the env of different HIV-1 clades can evolve differently under selective pressure [1, 25, 26]. To adequately determine if SHIV infection of macaques is a viable model for assessing envelope evolution, SHIV constructs using env from recently transmitted and biologically relevant, primary HIV-1 isolates must be tested. The evolutionary biology of the R5 SHIV-C in this current study, SHIV-1157i, represents such a virus.
We have been following a cohort of mostly HIV-C infected mother/infant pairs in Lusaka, Zambia. Among them, infant 1157i, a slow-progressor, was followed prospectively and viruses derived longitudinally from the infant have been extensively characterized [27]. SHIV-1157i was generated with an R5 tropic env derived from infant 1157i at 6 months old. An infant macaque was infected with SHIV-1157i and monitored through disease progression until it died from AIDS after ~5 years [28, 29]. Over this period, we tracked the changes of V1-V5 region from the time of inoculation to euthanasia of the infected macaque. Together with data previously gathered for infant 1157i, we were able to compare the genetic variation of the V1-V5 region of this particular primary clade C isolate during disease progression in both species [27].
Since our study utilized a clade C primary R5 tropic env instead of an X4 laboratory-adapted clade B strain, and we analyzed over 300 viral envelope sequences, our investigation represents a more in depth comparison of viral evolution during disease progression. This study has provided a unique opportunity to identify potential envelope mutations in the infected macaque which may associate with and predict future disease progression in the infected infant.
Methods
Construction of SHIV-1157i
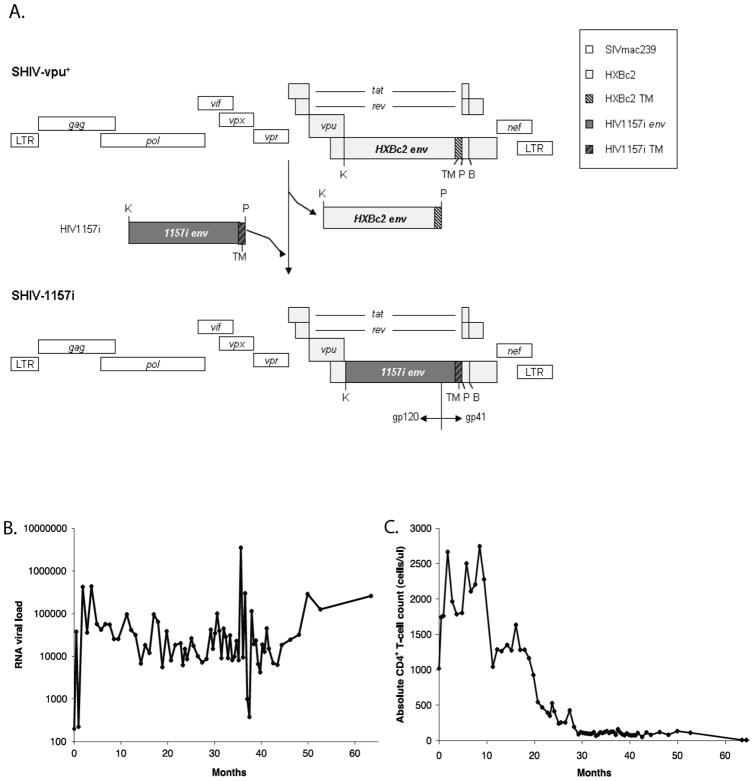
SHIV-1157i contains env of a primary HIV-C isolate from a 6 month old Zambian infant 1157i. PuvI (P) was introduced into the 3′ half of SHIV-vpu+ proviral DNA. The 2.0-kb KpnI (K)-PvuI fragment of HIV1157i was amplified to replace the corresponding region of SHIV-vpu+ env. The modified 3′ half was ligated with the 5′ half of SHIV-vpu+ proviral DNA to form full-length SHIV-1157i (Fig. 1A) [28, 29].
Fig. 1.
(A). Pvu I (P) was introduced into the 3′ half of SHIV-vpu+ (28) proviral DNA. The 2.0 Kb Kpn I - Pvu I fragment of HIV-1 1157i (spanning most of gp120 as well as the entire gp41 extracellular domain and the transmembrane region [TM]) was amplified to replace the corresponding region in the SHIV-vpu+ envelope. The modified 3′ half of SHIV-vpu+ was ligated with the 5′ half of SHIV-vpu+ proviral DNA to form the full length SHIV-1157i [28, 29]. (B) Plasma viral RNA load and (C) absolute CD4+ T-cell counts.
Animals and animal care
An infant Indian rhesus macaque (Macaca mulatta), RPn-8, was inoculated intravenously with 6 ml cell-free supernatant from 293T cells transfected with the infectious molecular clone, SHIV-1157i, and followed prospectively with complete blood counts, T-cell subset analyses, viral RNA load determinations, and clinical exams [28]. This animal was kept according to National Institutes of Health guidelines on the care and use of laboratory animals at the Yerkes National Primate Research Center (YNPRC). The facility is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. All experiments were approved by the Animal Care and Use Committees of the YNPRC and the Dana-Farber Cancer Institute.
Plasma viral RNA load
RNA was isolated from plasma using QiaAmp Viral Mini Kit (Qiagen), and vRNA loads were measured by quantitative reverse transcriptase PCR for SIV gag sequences [30]. The detection limit was 50 viral RNA copies/ml of plasma [28].
PCR, cloning and sequencing
Sample collection, cloning and sequencing of infant 1157i V1-V5 env region have been described previously [27, 31]. Genomic DNA of infected macaque PBMC was extracted from 6, 20, 37, 50 and 64 months post-inoculation samples using Genomic DNA Purification Kit (Gentra Systems). Nested PCR was used to amplify a 1.1 kb fragment spanning the V1-V5 env region. First round PCR was performed with the primers ENF1 (5′ GATGCATGAGGATATAATCAGTTTATGGGA 3′) and ENR1 (5′ ATTGATGCTGCGCCCATAGTGCT 3′). Second round PCR was performed with the primers 1157i-DraIII (5′ TTGACTCCACTCTGTGTCACTTTAAAG 3′) and 1157i-AvrII-AS (5′ TGCTATTCCTAGGGGCTTGATTTCTAC 3′). To minimize PCR bias, first round PCR products were generated in duplicates and combined to be used as templates for the second round PCR. Amplified fragments were cloned into pSP72 NL4-3 A-S-Av after digestion with Dra III and Avr II and sequenced with dideoxy terminators (ABI BigDye Kit).
Sequence analyses
Sequence alignment was carried out on the translated amino-acid sequence in ClustalW [32], as implemented in BioEdit 7.0.9.0. Neighbor-joining phylogenetic analyses were done in MEGA using the Kimura 2-parameter distance to explore genealogical relationships among infant 1157i and macaque RPn-8 V1-V5 sequences, and support for the nodes was evaluated with bootstrap.
Variations in genetic diversity, genetic divergence, the number and location of putative N-linked glycosylation sites (PNGSs), and the length of the V1-V5 fragment were analyzed. Viral genetic diversity was estimated as the average nucleotide difference between sequences within a contemporaneous set, and genetic divergence was calculated as the average genetic distance to the earliest viral population collected for the human or macaque, respectively. The number and location of PNGSs was estimated using N-GlycoSite from the Los Alamos National Laboratory.
The instantaneous rates of nonsynonymous (dN) and synonymous substitutions (dS) were compared to evaluate the role of natural selection. Estimates of dN, dS, and dN/dS for each time point were obtained in Datamonkey using the Fixed Effects Likelihood procedure [33].
Results
HIV-1157i and SHIV-1157i infection courses
The HIV-C V1-V5 region used to construct SHIV-1157i was derived from a cloned viral isolate of infant 1157i obtained at 6 months old. This child was infected in-utero, delivered naturally with normal birth weight and breast-fed till 20 months old. Both the child and his mother were anti-retroviral drug naïve. Infant 1157i is designated as a slow-progressor since he remains clinically asymptomatic throughout the study period, with a CD4+ T-cell count of 811 cells/ul at ~8 years old. Analyses of viral isolates from this infant have been published previously [27], and the viral sequences are included here for comparison.
SHIV-1157i was constructed as described (Fig. 1A) [29] and inoculated intravenously into the infant macaque RPn-8. Systemic infection was achieved after inoculation with a plasma viral RNA load of ~3.7 x 104 copies/ml detected at 2 weeks post inoculation (pi) and remained at ~1.0 x 104 copies/ml or higher for most of the study period (Fig. 1B). CD4+ T-cells steadily declined over time, reaching <200 cells/ul at ~28 months pi (Fig. 1C) and remained low till euthanasia at 64 months pi due to AIDS, as characterized by opportunistic infections [28]. PBMC were collected longitudinally throughout the study and samples from 6, 20, 37, 50 and 64 months pi were selected for this study. The V1-V5 region was cloned and sequenced. A total of 161 V1-V5 env clones were obtained from all time points, ~32 clones per time point.
Phylogenetic relationship among V1-V5 sequences from macaque and human
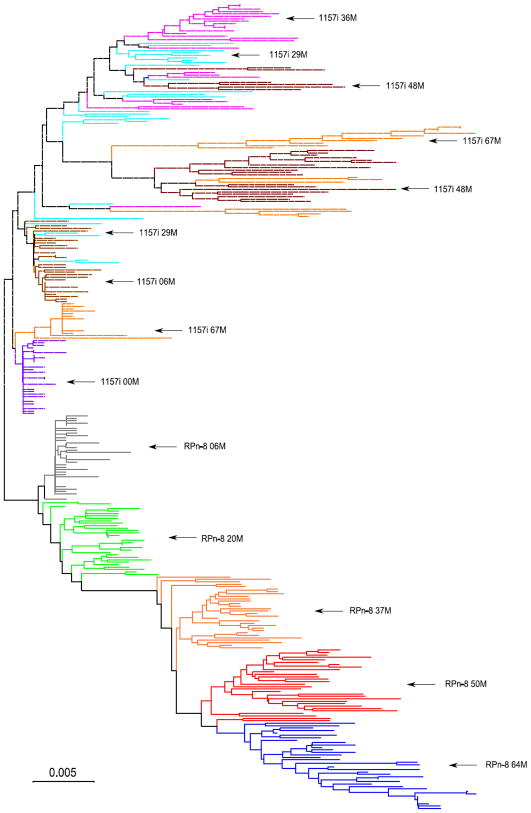
The neighbor-joining phylogeny clustered all envelope sequences derived from the infected macaque in a monophyletic clade (Fig. 2), consistent with the fact that they all derived from a single infectious molecular clone, SHIV-1157i. Both the macaque and infant portions of the tree resemble the idealized trees expected for viruses evolving under continuous immune pressure [34]. Sequences collected at later time points concentrated on the longer branches of the tree. The distribution of sequences from later time points indicates that the phylogenetic structure of the tree is more complicated in the infant than in the macaque. Infant sequences from the 67-month time point are scattered throughout the tree, with some sequences clustered with earlier lineages. In contrast, all macaque sequences from the last time point were found in a monophyletic clade.
Fig. 2.
Consensus tree from a Neighbor-Joining bootstrap analysis showing phylogenetic relationships among viral samples derived from the longitudinal follow-up of the macaque (RPn-8, solid lines) and infant (1157i, dashed lines) in this study. Labels indicate the source and time of sample collection. For example, RPn-8 20M corresponds to viral sequences coming from the macaque, and collected 20 months post-inoculation. Cut-off value for the condensed tree was set at 75%.
Longitudinal variation in diversity and divergence
Constraints within the viral env may hamper certain amino acid changes, regardless of any unique selective pressure by the individual host species, because of the cost to replicative fitness of the virus. As such, env changes we observed in infant 1157i during viral evolution may be similarly reflected in the macaque. To determine whether longitudinal variation of the V1-V5 region in isolates from RPn-8 and 1157i have similarities, we compared changes in the diversity and divergence of all viral populations sampled from each host. Viral genetic diversity measures the level of polymorphism found within a viral population at the nucleotide level, whereas divergence measures the differentiation of a population relative to the original strain. In the case of macaque, divergence was calculated relative to the sequence of the single clone inoculated experimentally, whereas in the infant, divergence was calculated as the average genetic distance between all sequences within the viral population at a given time point and all sequences from the viral population at birth.
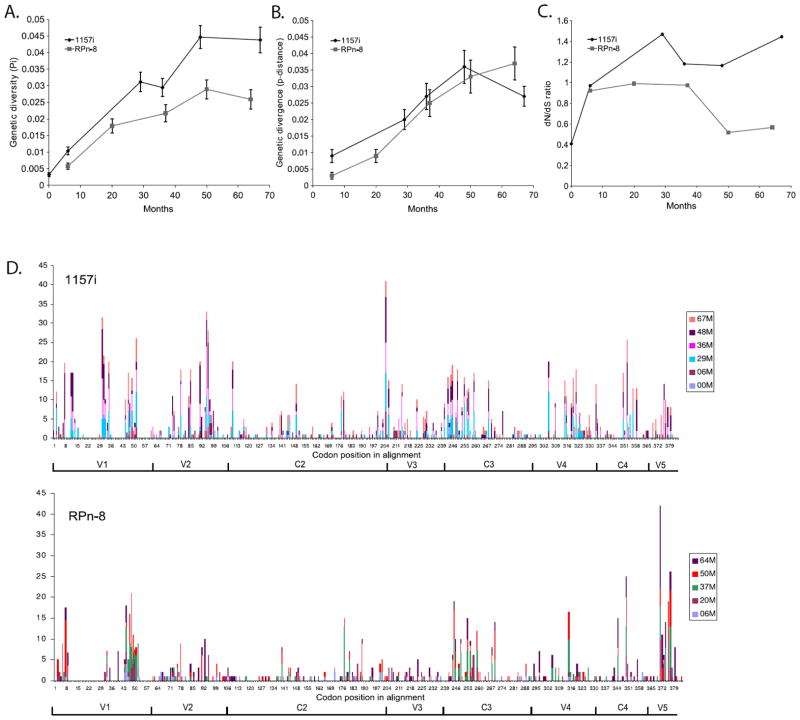
Our results indicate that diversity and divergence followed similar trajectories in both the macaque and infant at early time points but differ at later time points (Fig. 3A & B). In the case of infant, diversity increased quasi-monotonically over time, reaching an average pair-wise distance of 4.3% at 67 months after birth. Diversity in the macaque increased in parallel with the infant for the first 20 months. Thereafter, diversity gradually leveled off, reached its peak of an average genetic distance of 2.8% in the population collected 50 months pi, and decreased slightly at the time of euthanasia (Fig. 3A).
Fig. 3.
Changes in (A) genetic diversity and (B) divergence over time for infant 1157i and macaque RPn-8. Genetic diversity is calculated from the average number of nucleotide differences within a given time point. Genetic divergence is calculated from the average number of changes between each time point and the initial population. (C) non-synonymous and synonymous (dN/dS) ratio over time for infant 1157i and macaque RPn-8. (D) Estimated number of observed non-synonymous substitutions per codon within V1-V5 region in infant 1157i and macaque RPn-8. Results represented are cumulative of all time points for human or macaque. M represents either months of age in human or months post inoculation in macaque. Numbers of the horizontal axis correspond to amino acid position within the sequence alignment. All variable loops and constant regions within the alignment are labeled.
Sequence divergence in the infant increased in parallel with its diversity until it reached its maximum of 3.6% at 48 months. This was ensued by a noticeable decrease to 2.7% at 67 months. This decrease in divergence coincided with the re-emergence of viral lineages derived from earlier sequences shown by our phylogenetic results. Interestingly, divergence in the macaque increased monotonically throughout the study, and reached 3.7% by 64 months pi (Fig. 3B).
Synonymous and non-synonymous changes in macaque and human
To evaluate the role played by natural selection, we compared the ratio of dN/dS (Fig. 3C). Briefly, dN/dS ratios are expected to be ~0 under strong purifying selection, ~1 under neutrality, and >1 in cases where positive Darwinian selection is the major evolutionary force driving variation in the gene studied. In the infant, the dN/dS ratio was >1 in the majority of the populations sampled, ranging from 0.97 at 6 months to 1.44 at 67 months of age. In contrast, the dN/dS ratio in the macaque remained <1 till 50 months pi where it declined to 0.52. These data suggest that selective pressure was stronger in the infant than in the macaque. We then compared the distribution of these changes in the viral populations from the infant and macaque by plotting the number of observed dN per codon per time point (Fig. 3D). The comparison shows that despite the differences in the number of changes and selective pattern, amino acid replacements accumulated in similar regions in the infant and macaque. These regions include the variable loops as well as in the supposedly conserved C3 and C4 regions.
Longitudinal changes in the number of N-linked glycosylation sites in V1-V5
It has been suggested that lengthening of the V1-V5 region and an increase in PNGSs correlate with slower disease progression [3, 21]. To see if this held true in our study, we examined the change in number of PNGSs in RPn-8 and found that it declined over time from 26 PNGSs present in the inoculum to a mean of 21 PNGSs by 37 months pi (Table 1). This was followed by a gradual recovery to a mean of 25 PNGSs by 64 months pi. The V1-V5 length in RPn-8 followed a similar trend (Table 1). In contrast to the macaque, variation in PNGSs and V1-V5 length in the infant showed no directional pattern.
Table 1.
Possible N-glycosylation site numbers and length of V1-V5 region.
| Sample | n | V1 Length | V2 Length | C2 Length | V3 Length | C3 Length | V4 Length | C4 Length | V5 Length | Total V1-V5 length |
|---|---|---|---|---|---|---|---|---|---|---|
| 1157i 00M | 31 | 21 (21 - 21) | 42 (42 - 42) | 99 (99 - 99) | 35 (35 - 35) | 52 (52 - 52) | 28 (28 - 28) | 40 (40 - 40) | 15 (15 - 15) | 332 (332 - 332) |
| 1157i 06M | 29 | 23 (20 - 28) | 42 (40 - 42) | 99 (99 - 99) | 35 (35 - 35) | 52 (52 - 52) | 25 (20 - 28) | 40 (40 - 40) | 15 (15 - 15) | 331 (322 - 339) |
| 1157i 29M | 26 | 30 (22 - 38) | 41 (39 - 43) | 99 (99 - 99) | 35 (35 - 35) | 51 (50 - 52) | 26 (20 - 28) | 40 (40 - 40) | 15 (15 - 15) | 337 (329 - 349) |
| 1157i 36M | 26 | 31 (17 - 36) | 42 (40 - 42) | 99 (99 - 99) | 35 (35 - 35) | 51 (50 - 52) | 23 (20 - 28) | 40 (40 - 40) | 15 (15 - 15) | 336 (319 - 344) |
| 1157i 48M | 25 | 32 (28 - 38) | 41 (39 - 42) | 99 (99 - 99) | 35 (35 - 35) | 52 (50 - 52) | 27 (23 - 30) | 40 (40 - 40) | 14 (12 - 15) | 340 (335 - 347) |
| 1157i 67M | 32 | 26 (22 - 49) | 41 (40 - 42) | 99 (99 - 99) | 35 (35 - 35) | 52 (50 - 52) | 28 (27 - 30) | 40 (40 - 40) | 15 (13 - 15) | 335 (327 - 362) |
| SHIV1 157i | 1 | 22 | 42 | 99 | 35 | 52 | 28 | 40 | 15 | 333 |
| RPn-8 06M | 34 | 22 (22 - 22) | 42 (42 - 42) | 99 (99 - 99) | 35 (35 - 35) | 52 (52 - 52) | 28 (28 - 28) | 40 (40 - 40) | 15 (15 - 15) | 333 (333 - 333) |
| RPn-8 20M | 31 | 21 (21 - 22) | 42 (42 - 42) | 99 (99 - 99) | 35 (35 - 35) | 52 (52 - 52) | 28 (28 - 28) | 40 (40 - 40) | 15 (15 - 15) | 332 (332 - 333) |
| RPn-8 37M | 30 | 22 (21 - 22) | 39 (39 - 39) | 99 (99 - 99) | 35 (35 - 35) | 52 (52 - 52) | 26 (24 - 28) | 40 (40 - 40) | 15 (12 - 15) | 327 (325 - 330) |
| RPn-8 50M | 30 | 21 (15 - 27) | 39 (39 - 39) | 99 (99 - 99) | 35 (35 - 35) | 52 (52 - 52) | 28 (24 - 28) | 40 (40 - 40) | 13 (13 - 15) | 327 (320 - 334) |
| RPn-8 64M | 36 | 28 (27 - 29) | 39 (39 - 40) | 99 (99 - 99) | 35 (35 - 35) | 52 (52 - 52) | 27 (24 - 28) | 40 (40 - 40) | 15 (14 - 15) | 334 (331 - 337) |
| Sample | n | V1 PNGS | V2 PNGS | C2 PNGS | V3 PNGS | C3 PNGS | V4 PNGS | C4 PNGS | V5 PNGS | Total V1-V5 PNGS |
|---|---|---|---|---|---|---|---|---|---|---|
| 1157i 00M | 31 | 3 (3 - 3) | 3 (2 - 3) | 8 (7 - 8) | 1 (1 - 1) | 3 (2 - 3) | 4 (3 - 4) | 2 (1 - 2) | 2 (2 - 2) | 26 (25 - 26) |
| 1157i 06M | 29 | 3 (2 - 4) | 3 (1 - 3) | 7 (7 - 8) | 1 (0 - 2) | 3 (3 - 3) | 4 (3 - 4) | 2 (1 - 2) | 2 (1 - 2) | 25 (23 - 27) |
| 1157i 29M | 26 | 4 (3 - 6) | 2 (1 - 3) | 7 (6 - 8) | 1 (1 - 1) | 2 (2 - 3) | 4 (2 - 4) | 2 (1 - 2) | 2 (2 - 2) | 24 (21 - 27) |
| 1157i 36M | 26 | 4 (3 - 6) | 3 (2 - 3) | 6 (5 - 7) | 1 (1 - 1) | 2 (2 - 3) | 4 (3 - 4) | 2 (1 - 2) | 1 (1 - 2) | 23 (20 - 25) |
| 1157i 48M | 25 | 4 (3 - 5) | 2 (1 - 3) | 7 (6 - 8) | 1 (1 - 1) | 2 (1 - 3) | 4 (4 - 4) | 1 (0 - 2) | 1 (0 - 2) | 23 (20 - 25) |
| 1157i 67M | 32 | 4 (3 - 7) | 3 (2 - 3) | 7 (6 - 8) | 1 (1 - 1) | 3 (1 - 3) | 4 (3 - 4) | 2 (1 - 2) | 2 (1 - 2) | 25 (21 - 28) |
| SHIV1 157i | 1 | 3 | 3 | 8 | 1 | 3 | 4 | 2 | 2 | 26 |
| RPn-8 06M | 34 | 3 (2 - 3) | 3 (2 - 3) | 8 (7 - 8) | 1 (1 - 1) | 3 (3 - 3) | 4 (3 - 4) | 2 (1 - 2) | 2 (2 - 2) | 25 (24 - 26) |
| RPn-8 20M | 31 | 3 (2 - 3) | 3 (3 - 3) | 7 (6 - 8) | 1 (1 - 1) | 3 (2 - 3) | 4 (4 - 4) | 2 (1 - 2) | 1 (0 - 1) | 24 (23 - 25) |
| RPn-8 37M | 30 | 3 (2 - 3) | 2 (2 - 2) | 6 (5 - 7) | 1 (0 - 1) | 3 (3 - 3) | 4 (3 - 4) | 2 (2 - 2) | 1 (1 - 1) | 21 (19 - 23) |
| RPn-8 50M | 30 | 2 (2 - 3) | 2 (1 - 2) | 7 (6 - 7) | 1 (0 - 1) | 3 (1 - 3) | 4 (4 - 4) | 2 (2 - 2) | 1 (0 - 2) | 22 (20 - 24) |
| RPn-8 64M | 36 | 4 (3 - 4) | 2 (2 - 3) | 7 (6 - 7) | 1 (1 - 1) | 3 (2 - 3) | 4 (3 - 5) | 2 (2 - 2) | 2 (1 - 2) | 25 (23 - 27) |
Numer of samples (n). Numbers reported are to the nearest whole number. Average (Min-Max)
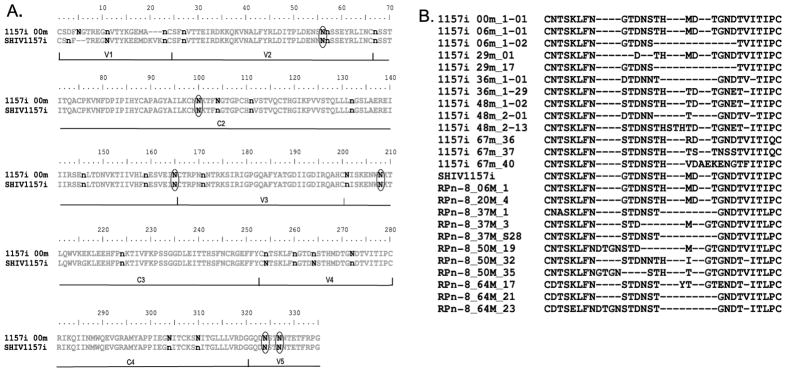
In both the macaque and infant, the majority of the PNGSs were distributed in identical regions. With 17 PNGSs in RPn-8 and 14 PNGSs in 1157i that were relatively conserved throughout the infection (Fig. 4A). These conserved PNGSs may have important roles in the structural integrity or receptor binding capability of envelope. However, differences in PNGS do exist between the macaque and infant. The macaque had only 9 PNGSs that were variable throughout the infection, compared with 12 PNGSs in the infant. Interestingly, 6 of these variable sites (N56, 100, 165, 208, 324 and 327) were identical between the macaque and infant.
Fig. 4.
(A) V1-V5 consensus sequence of infant 1157i at 0 month of age and inoculation strain SHIV-1157i is shown. Putative N-linked glycosylation sites (PNGSs) within sequences from all time points were located as described in Methods. n represents relatively conserved PNGSs in macaque or human over time. N represents variable PNGSs in macaque or human over time. Common variable PNGSs between macaque and human are circled. Variable regions and constant regions are shown along the bottom of the sequences. (B) sequence alignment of V4 in infant 1157i and macaque RPn-8 at different time points. Sequences were aligned using Bioedit 7.0.9.0. Sequences represented here are examples from each time points. These are not consensus sequences and do not represent all observed variations within V4.
The temporal dynamics of the variations in PNGSs were similar for both the infant and macaque. For instance, sites N100, 208, 324 and 327 fluctuated in both 1157i and RPn-8 over time. For site N56, changes in the infant were observed starting from 6 months. In comparison with the macaque, the identical PNGS was ablated at 37 months but gradually reemerged by 64 months pi. Another site with similar changes between 1157i and RPn-8 is N165. Prevalence of the PNGS at N165 reached its lowest level by 36 months old in the infant before remerging. Likewise, this site was reduced in the macaque at 20 months but, contrary to the infant, this PNGS was completely ablated by 37 months pi and never reemerged from later time points in the macaque. We also noticed a pattern unique to the macaque, where the prevalence of several PNGSs, including N56, was reduced near 37 month pi but gradually re-emerged at later time points.
Discussion
In the present study, we analyzed changes in the V1-V5 envelope region in viral populations sampled longitudinally from an R5 SHIV-C infected macaque with those from a Zambian infant. Longitudinal variation in sequence diversity and divergence in HIV-1 infection in humans has been intensely studied, and we found that SHIV infection in this macaque followed the general pattern described for HIV-1 infection in humans [35]: an initial phase where both diversity and divergence increased linearly up to the 20 month sampling interval; an intermediate phase where diversity stabilized but divergence continued to increase up to the 50 month sampling interval; a late phase where divergence and diversity decreased or stabilized from the 50 month sample onwards. Our sampling intervals did not allow us to precisely determine the time transitions between the different phases.
In humans naturally infected with HIV-1, differential rates of disease progression have been linked with viral genetic diversity and dN/dS ratios. In particular, higher dN/dS estimates appear to correlate with slower rates of disease progression, as is the case for having a higher number of sites inferred to be evolving under positive Darwinian selection or having high adaptation rates [36, 37]. Estimates of genetic diversity and dN/dS were all higher in the infant, a slow progressor, which is in agreement with previous observations [38–40]. These data suggests that ablation of the macaque’s immune responses at later time points resulted in a drastic reduction of selective pressure on the viral populations. In comparison, the infant’s immune status appears to be healthy and exert a significant selective pressure on the viral populations, in fact, the child remains asymptomatic at age 10.
The distribution of dN along the V1-V5 region for the infant and macaque was located primarily within the variable loops. But our analysis also indicated an unexpected variable domain located in the α2-helix of C3 in the infant and macaque viruses. Alterations within this domain correlate with low CD4 counts in the macaque, suggesting that it may contain an immune epitope and could affect the biological properties of the virus [41–45]. This phenomenon could be clade specific, as the homologous region is reported to be more conserved in clade B [1, 25, 41, 46]. As reported by other groups, emergence of deletions within V4 was also observed in the infant and macaque in this study (Fig. 4B) [22, 24, 47].
N-glycosylation plays a critical role in immune evasion, and increases in the number of PNGS have been linked with immune resistance [2, 7, 48, 49]. Although we found no clear pattern of change for the number of PNGSs in the infant, we did observed an interesting pattern of decreasing PNGSs after inoculation and a gradual recovery at later time points in the macaque. Reductions of PNGSs number in the macaque between 6 and 37 months pi coincide with its rapid decline in CD4 count and the onset of AIDS. Whether the eventual recovery of PNGSs at the last time point is due to immune escape or increase in replication fitness is unclear [50–53].
Besides sharing similar PNGS distribution patterns and hot spots, we observed several similar PNGS variations within viral populations of the infant and macaque. In particular, N56 that locates at the C-terminal of V2, and N165 that locates within C2. Alteration of PNGS at the C-terminal of V2 has been reported in animals infected by clade B SHIV [21, 22, 54]. A recent study also demonstrated that removal of PNGS at this position increases HIV-1 89.6 sensitivity to neutralizing antibodies [55], thus suggesting that N56 could be a common immunological epitope for both clade B and C viruses [56, 57]. It is surprising to find a variable PNGS, N165, within a constant region in the infant and macaque. Changes at this site coincide with the macaque’s persistently low CD4 counts and were shown by others to affect the virus sensitivity to monoclonal antibodies [29, 58]. Ablation of a similar site has also been reported in patients [7]. It is possible that N165 is an immune epitope and ablation of it might enhance the infectivity or kinetics of the escape viruses.
In order to eliminate the possibility that any of the clones were the result of in-vitro PCR recombination, phylogenetic analyses were performed to distinguish between PCR recombination and actual in-vivo recombination events. Briefly, with in-vivo recombination one would observe an accumulation of mutations after a recombination event on a given branch within an estimated phylogeny. However, with PCR recombination there would be no such additional substitutions on the recombinant branch and therefore the clones would be non-unique and identical to parental clones. Application of this test revealed a negligible number of clones were indeed generated by PCR recombination. In fact, only two and four clones, respectively, were identified for the macaque and infant that showed no additional substitutions along a branch in the estimated phylogeny (data not shown). Thus, PCR recombination occurred on a minor scale and did not adversely affect our analyses. Limiting dilution PCR (LDPCR) was not feasible in this study due to insufficient materials for most of the time points analyzed. Nevertheless, LDPCR was carried out on the last time point sampled of the macaque and revealed clones with similar phylogenetic distributions that were consistent with those obtained by the regular PCR approach. No inherent differences were observed between the two procedures.
Taken together, our data suggest that genetic evolution of an R5 SHIV generated from the envelope of a primary isolate reflects changes that occur in HIV-1 during disease progression in the infected infant. The infected macaque has a relatively faster disease progression in comparison with the infant, but a much slower disease progression compare with other SHIVs such as SHIV89.6P. This could be due to the differences in selection forces between the two species. Our data also imply that these changes occur in a compressed time frame since the infected macaque progressed to AIDS while the child has remained asymptomatic. Although there is no identical individual amino acid mutation comparable with previously reported clade B studies, mutations do seem to occur in similar regions. In addition, even though it is difficult to make a direct comparison of the time course between human and macaque in disease progression, mutations observed in our infected macaque at later time points could be indicators of future disease progression in the child. A further understanding of factors which caused these genetic changes could provide beneficial insights for future HIV-1 vaccine designs, and the significance of these changes on the biological function of the envelope is currently under investigation. The caveat for our study is the small sample size and the observation may be limited to this human and macaque pair. However, this is a prospective study and the first study to our knowledge demonstrating the evolutionary similarity of a primary clade C envelope between a naturally infected human with an experimentally infected macaque. This study also re-enforces the previous observation that HIV-1 envelope undergoes similar changes in human and monkey during disease progression [22].
Acknowledgments
This work was supported by PHS grants, CA75903, TW001429 and NCRR COBRE grant RR15635 to C.W.; P01AI48240 to R.M.R., C.W., and R.A.R.; R01 DE12937, R01 DE0160354, and R37AI34266 to R.M.R.; P.L. was supported by an FWO postdoctoral research fund.
Footnotes
Author’s contributions: F.Y.T performed experiments, analyzed data and wrote the manuscript. F.G.H, D.C.T., P.L. and H.Z. helped analyze data. R.A.R oversaw the animal aspects of the study. R.M.R. and C.W. oversaw all aspects of this project. All authors provided critical input on the manuscript.
References
- 1.Gaschen B, Taylor J, Yusim K, Foley B, Gao F, Lang D, et al. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 2.Geyer H, Holschbach C, Hunsmann G, Schneider J. Carbohydrates of human immunodeficiency virus. Structures of oligosaccharides linked to the envelope glycoprotein 120. J Biol Chem. 1988;263:11760–11767. [PubMed] [Google Scholar]
- 3.Sagar M, Wu X, Lee S, Overbaugh J. Human immunodeficiency virus type 1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. J Virol. 2006;80:9586–9598. doi: 10.1128/JVI.00141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saunders CJ, McCaffrey RA, Zharkikh I, Kraft Z, Malenbaum SE, Burke B, et al. The V1, V2, and V3 regions of the human immunodeficiency virus type 1 envelope differentially affect the viral phenotype in an isolate-dependent manner. J Virol. 2005;79:9069–9080. doi: 10.1128/JVI.79.14.9069-9080.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stevceva L, Yoon V, Anastasiades D, Poznansky MC. Immune responses to HIV Gp120 that facilitate viral escape. Curr HIV Res. 2007;5:47–54. doi: 10.2174/157016207779316396. [DOI] [PubMed] [Google Scholar]
- 6.Thorner AR, Barouch DH. HIV-1 Vaccine Development: Progress and Prospects. Curr Infect Dis Rep. 2007;9:71–75. doi: 10.1007/s11908-007-0025-0. [DOI] [PubMed] [Google Scholar]
- 7.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 8.Flynn NM, Forthal DN, Harro CD, Judson FN, Mayer KH, Para MF. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J Infect Dis. 2005;191:654–665. doi: 10.1086/428404. [DOI] [PubMed] [Google Scholar]
- 9.Gilbert PB, Peterson ML, Follmann D, Hudgens MG, Francis DP, Gurwith M, et al. Correlation between immunologic responses to a recombinant glycoprotein 120 vaccine and incidence of HIV-1 infection in a phase 3 HIV-1 preventive vaccine trial. J Infect Dis. 2005;191:666–677. doi: 10.1086/428405. [DOI] [PubMed] [Google Scholar]
- 10.Greenier JL, Van Rompay KK, Montefiori D, Earl P, Moss B, Marthas ML. Simian immunodeficiency virus (SIV) envelope quasispecies transmission and evolution in infant rhesus macaques after oral challenge with uncloned SIVmac251: increased diversity is associated with neutralizing antibodies and improved survival in previously immunized animals. Virol J. 2005;2:11. doi: 10.1186/1743-422X-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mascola JR, Sambor A, Beaudry K, Santra S, Welcher B, Louder MK, et al. Neutralizing antibodies elicited by immunization of monkeys with DNA plasmids and recombinant adenoviral vectors expressing human immunodeficiency virus type 1 proteins. J Virol. 2005;79:771–779. doi: 10.1128/JVI.79.2.771-779.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pitisuttithum P, Gilbert P, Gurwith M, Heyward W, Martin M, van Griensven F, et al. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J Infect Dis. 2006;194:1661–1671. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 13.Xu R, Srivastava IK, Kuller L, Zarkikh I, Kraft Z, Fagrouch Z, et al. Immunization with HIV-1 SF162-derived Envelope gp140 proteins does not protect macaques from heterologous simian-human immunodeficiency virus SHIV89.6P infection. Virology. 2006;349:276–289. doi: 10.1016/j.virol.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 14.Rasmussen RA, Ong H, Song R, Chenine AL, Ayash-Rashkovsky M, Hu SL, et al. Efficacy of a multigenic protein vaccine containing multimeric HIV gp160 against heterologous SHIV clade C challenges. AIDS. 2007;21:1841–1848. doi: 10.1097/QAD.0b013e32828684ea. [DOI] [PubMed] [Google Scholar]
- 15.Hayami M, Igarashi T. SIV/HIV-1 chimeric viruses having HIV-1 env gene: a new animal model and a candidate for attenuated live vaccine. Leukemia. 1997;11 (Suppl 3):95–97. [PubMed] [Google Scholar]
- 16.Karlsson GB, Halloran M, Li J, Park IW, Gomila R, Reimann KA, et al. Characterization of molecularly cloned simian-human immunodeficiency viruses causing rapid CD4+ lymphocyte depletion in rhesus monkeys. J Virol. 1997;71:4218–4225. doi: 10.1128/jvi.71.6.4218-4225.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luciw PA, Pratt-Lowe E, Shaw KE, Levy JA, Cheng-Mayer C. Persistent infection of rhesus macaques with T-cell-line-tropic and macrophage-tropic clones of simian/human immunodeficiency viruses (SHIV) Proc Natl Acad Sci U S A. 1995;92:7490–7494. doi: 10.1073/pnas.92.16.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reimann KA, Li JT, Voss G, Lekutis C, Tenner-Racz K, Racz P, et al. An env gene derived from a primary human immunodeficiency virus type 1 isolate confers high in vivo replicative capacity to a chimeric simian/human immunodeficiency virus in rhesus monkeys. J Virol. 1996;70:3198–3206. doi: 10.1128/jvi.70.5.3198-3206.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shibata R, Maldarelli F, Siemon C, Matano T, Parta M, Miller G, et al. Infection and pathogenicity of chimeric simian-human immunodeficiency viruses in macaques: determinants of high virus loads and CD4 cell killing. J Infect Dis. 1997;176:362–373. doi: 10.1086/514053. [DOI] [PubMed] [Google Scholar]
- 20.Vlasak J, Ruprecht RM. AIDS vaccine development and challenge viruses: getting real. AIDS. 2006;20:2135–2140. doi: 10.1097/QAD.0b013e328010beb5. [DOI] [PubMed] [Google Scholar]
- 21.Blay WM, Gnanakaran S, Foley B, Doria-Rose NA, Korber BT, Haigwood NL. Consistent patterns of change during the divergence of human immunodeficiency virus type 1 envelope from that of the inoculated virus in simian/human immunodeficiency virus-infected macaques. J Virol. 2006;80:999–1014. doi: 10.1128/JVI.80.2.999-1014.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hofmann-Lehmann R, Vlasak J, Chenine AL, Li PL, Baba TW, Montefiori DC, et al. Molecular evolution of human immunodeficiency virus env in humans and monkeys: similar patterns occur during natural disease progression or rapid virus passage. J Virol. 2002;76:5278–5284. doi: 10.1128/JVI.76.10.5278-5284.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kraft Z, Derby NR, McCaffrey RA, Niec R, Blay WM, Haigwood NL, et al. Macaques infected with a CCR5-tropic simian/human immunodeficiency virus (SHIV) develop broadly reactive anti-HIV neutralizing antibodies. J Virol. 2007;81:6402–6411. doi: 10.1128/JVI.00424-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beaumont T, van Nuenen A, Broersen S, Blattner WA, Lukashov VV, Schuitemaker H. Reversal of human immunodeficiency virus type 1 IIIB to a neutralization-resistant phenotype in an accidentally infected laboratory worker with a progressive clinical course. J Virol. 2001;75:2246–2252. doi: 10.1128/JVI.75.5.2246-2252.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choisy M, Woelk CH, Guegan JF, Robertson DL. Comparative study of adaptive molecular evolution in different human immunodeficiency virus groups and subtypes. J Virol. 2004;78:1962–1970. doi: 10.1128/JVI.78.4.1962-1970.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korber BT, MacInnes K, Smith RF, Myers G. Mutational trends in V3 loop protein sequences observed in different genetic lineages of human immunodeficiency virus type 1. J Virol. 1994;68:6730–6744. doi: 10.1128/jvi.68.10.6730-6744.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, Hoffmann F, He J, He X, Kankasa C, Ruprecht R, et al. Evolution of subtype C HIV-1 Env in a slowly progressing Zambian infant. Retrovirology. 2005;2:67. doi: 10.1186/1742-4690-2-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Humbert M, Rasmussen RA, Song R, Ong H, Sharma P, Chenine AL, et al. SHIV-1157i and passaged progeny viruses encoding R5 HIV-1 clade C env cause AIDS in rhesus monkeys. Retrovirology. 2008;5:94. doi: 10.1186/1742-4690-5-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song RJ, Chenine AL, Rasmussen RA, Ruprecht CR, Mirshahidi S, Grisson RD, et al. Molecularly cloned SHIV-1157ipd3N4: a highly replication- competent, mucosally transmissible R5 simian-human immunodeficiency virus encoding HIV clade C Env. J Virol. 2006;80:8729–8738. doi: 10.1128/JVI.00558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hofmann-Lehmann R, Swenerton RK, Liska V, Leutenegger CM, Lutz H, McClure HM, Ruprecht RM. Sensitive and robust one-tube real-time reverse transcriptase-polymerase chain reaction to quantify SIV RNA load: comparison of one- versus two-enzyme systems. AIDS Res Hum Retroviruses. 2000;16:1247–1257. doi: 10.1089/08892220050117014. [DOI] [PubMed] [Google Scholar]
- 31.Zhang H, Hoffmann F, He J, He X, Kankasa C, West JT, et al. Characterization of HIV-1 subtype C envelope glycoproteins from perinatally infected children with different courses of disease. Retrovirology. 2006;3:73. doi: 10.1186/1742-4690-3-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kosakovsky Pond SL, Frost SD. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 34.Grenfell BT, Pybus OG, Gog JR, Wood JL, Daly JM, Mumford JA, Holmes EC. Unifying the epidemiological and evolutionary dynamics of pathogens. Science. 2004;303:327–332. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- 35.Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ross HA, Rodrigo AG. Immune-mediated positive selection drives human immunodeficiency virus type 1 molecular variation and predicts disease duration. J Virol. 2002;76:11715–11720. doi: 10.1128/JVI.76.22.11715-11720.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williamson S. Adaptation in the env gene of HIV-1 and evolutionary theories of disease progression. Mol Biol Evol. 2003;20:1318–1325. doi: 10.1093/molbev/msg144. [DOI] [PubMed] [Google Scholar]
- 38.Markham RB, Wang WC, Weisstein AE, Wang Z, Munoz A, Templeton A, et al. Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proc Natl Acad Sci U S A. 1998;95:12568–12573. doi: 10.1073/pnas.95.21.12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salvatori F, Masiero S, Giaquinto C, Wade CM, Brown AJ, Chieco-Bianchi L, De Rossi A. Evolution of human immunodeficiency virus type 1 in perinatally infected infants with rapid and slow progression to disease. J Virol. 1997;71:4694–4706. doi: 10.1128/jvi.71.6.4694-4706.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Troyer RM, Collins KR, Abraha A, Fraundorf E, Moore DM, Krizan RW, et al. Changes in human immunodeficiency virus type 1 fitness and genetic diversity during disease progression. J Virol. 2005;79:9006–9018. doi: 10.1128/JVI.79.14.9006-9018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore PL, Gray ES, Choge IA, Ranchobe N, Mlisana K, Abdool Karim SS, et al. The c3-v4 region is a major target of autologous neutralizing antibodies in human immunodeficiency virus type 1 subtype C infection. J Virol. 2008;82:1860–1869. doi: 10.1128/JVI.02187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morrison HG, Kirchhoff F, Desrosiers RC. Effects of mutations in constant regions 3 and 4 of envelope of simian immunodeficiency virus. Virology. 1995;210:448–455. doi: 10.1006/viro.1995.1361. [DOI] [PubMed] [Google Scholar]
- 43.Rong R, Gnanakaran S, Decker JM, Bibollet-Ruche F, Taylor J, Sfakianos JN, et al. Unique mutational patterns in the envelope alpha 2 amphipathic helix and acquisition of length in gp120 hypervariable domains are associated with resistance to autologous neutralization of subtype C human immunodeficiency virus type 1. J Virol. 2007;81:5658–5668. doi: 10.1128/JVI.00257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanders RW, Venturi M, Schiffner L, Kalyanaraman R, Katinger H, Lloyd KO, et al. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J Virol. 2002;76:7293–7305. doi: 10.1128/JVI.76.14.7293-7305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, Wilson IA, et al. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1-->2 mannose residues on the outer face of gp120. J Virol. 2002;76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gnanakaran S, Lang D, Daniels M, Bhattacharya T, Derdeyn CA, Korber B. Clade-specific differences between human immunodeficiency virus type 1 clades B and C: diversity and correlations in C3-V4 regions of gp120. J Virol. 2007;81:4886–4891. doi: 10.1128/JVI.01954-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buckley KA, Li PL, Khimani AH, Hofmann-Lehmann R, Liska V, Anderson DC, et al. Convergent evolution of SIV env after independent inoculation of rhesus macaques with infectious proviral DNA. Virology. 2003;312:470–480. doi: 10.1016/s0042-6822(03)00262-9. [DOI] [PubMed] [Google Scholar]
- 48.Chohan B, Lang D, Sagar M, Korber B, Lavreys L, Richardson B, Overbaugh J. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1-V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J Virol. 2005;79:6528–6531. doi: 10.1128/JVI.79.10.6528-6531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, et al. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science. 2004;303:2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 50.Haggerty S, Dempsey MP, Bukrinsky MI, Guo L, Stevenson M. Posttranslational modifications within the HIV-1 envelope glycoprotein which restrict virus assembly and CD4-dependent infection. AIDS Res Hum Retroviruses. 1991;7:501–510. doi: 10.1089/aid.1991.7.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohgimoto S, Shioda T, Mori K, Nakayama EE, Hu H, Nagai Y. Location- specific, unequal contribution of the N glycans in simian immunodeficiency virus gp120 to viral infectivity and removal of multiple glycans without disturbing infectivity. J Virol. 1998;72:8365–8370. doi: 10.1128/jvi.72.10.8365-8370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Willey RL, Smith DH, Lasky LA, Theodore TS, Earl PL, Moss B, et al. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol. 1988;62:139–147. doi: 10.1128/jvi.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu Z, Kayman SC, Honnen W, Revesz K, Chen H, Vijh-Warrier S, et al. Characterization of neutralization epitopes in the V2 region of human immunodeficiency virus type 1 gp120: role of glycosylation in the correct folding of the V1/V2 domain. J Virol. 1995;69:2271–2278. doi: 10.1128/jvi.69.4.2271-2278.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luciw PA, Mandell CP, Himathongkham S, Li J, Low TA, Schmidt KA, et al. Fatal immunopathogenesis by SIV/HIV-1 (SHIV) containing a variant form of the HIV-1SF33 env gene in juvenile and newborn rhesus macaques. Virology. 1999;263:112–127. doi: 10.1006/viro.1999.9908. [DOI] [PubMed] [Google Scholar]
- 55.Li Y, Cleveland B, Klots I, Travis B, Richardson BA, Anderson D, et al. Removal of a single N-linked glycan in human immunodeficiency virus type 1 gp120 results in an enhanced ability to induce neutralizing antibody responses. J Virol. 2008;82:638–651. doi: 10.1128/JVI.01691-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fung MS, Sun CR, Gordon WL, Liou RS, Chang TW, Sun WN, et al. Identification and characterization of a neutralization site within the second variable region of human immunodeficiency virus type 1 gp120. J Virol. 1992;66:848–856. doi: 10.1128/jvi.66.2.848-856.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sullivan N, Thali M, Furman C, Ho DD, Sodroski J. Effect of amino acid changes in the V1/V2 region of the human immunodeficiency virus type 1 gp120 glycoprotein on subunit association, syncytium formation, and recognition by a neutralizing antibody. J Virol. 1993;67:3674–3679. doi: 10.1128/jvi.67.6.3674-3679.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCaffrey RA, Saunders C, Hensel M, Stamatatos L. N-linked glycosylation of the V3 loop and the immunologically silent face of gp120 protects human immunodeficiency virus type 1 SF162 from neutralization by anti-gp120 and anti-gp41 antibodies. J Virol. 2004;78:3279–3295. doi: 10.1128/JVI.78.7.3279-3295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]