Abstract
Anisotropic tissue structures provide guidance for navigating neurons in vitro and in vivo. Here we optimized the generation of comparable anisotropic monolayers of astrocytes, endothelial cells, and Schwann cells as a first step toward determining which properties of anisotropic cells are sufficient for nerve guidance. The statistical experimental design method Design of Experiments and the experimental analysis method Response Surface Methodology were applied to improve efficiency and utility. Factors investigated included dimensions of microcontact printed protein patterns, cell density, and culture duration. Protein patterning spacing had the strongest influence. When cells initially aligned at borders and proliferated to fill in spaces, space between stripes was most effective when it was comparable to cell size. Maximizing the area of adhesive molecule coverage was also important for confluence of these types of cells. When cells adhered and aligned over the width of a stripe and broadened to fill spaces, space width about half the cell width was most effective. These findings suggest that if the mechanism of alignment, alignment at borders or over the width of the stripe, is predetermined and the cell size determined, the optimal size of the micropatterning for aligned monolayers of other cell types can be predicted. This study also demonstrates the effective use of DOE and RSM to probe cellular responses to various and multiple factors toward determination of optimal conditions for a desired cellular response.
KEY TERMS: Design of Experiments (DOE), d-optimal, Schwann cell, astrocyte, endothelial cell
INTRODUCTION
The generation of anisotropic tissues is important for a variety of biomedical engineering applications including devising organ replacements, guiding tissue regeneration, designing biomimetic materials, and modeling cellular interactions. The formation of highly organized tissues is also critically important in development i.e. vasculogenesis and angiogenesis1, 2, skeletogenesis3, myogenesis 4, etc. Anisotropic guidance is of particular importance in the nervous system. Oriented glial cells and vasculature guide the formation of precise neuronal connections during development 5–8. Injury to the central nervous system (CNS) results in an inhibitory biochemical barrier to regeneration, but furthermore disrupts the geometric organization of the surrounding tissue 9. The inherent injury response in the peripheral nervous system (PNS) includes reorganization of the injury site with a favorable anisotropic geometry as well as permissive biochemical factors 10. As nerves attempt to regenerate after injury in either the CNS or PNS, oriented tissues are important for effective nerve guidance.
Many studies have established that many cell types have the ability to orient to specific guidance cues in their local environments. Endothelial cells (ECs) have been aligned with shear stress 11, 12, underlying substrate topography 13, and protein patterns 13–16. Astrocytes have been aligned with electric fields 17, 18, mechanical strain 19, and substrate topography 19–22. Schwann cells (SCs) have been aligned with substrate topography 23 and protein patterns 24–26. Oligodendrocytes and fibroblasts have been aligned with topography 20, 21, 27–30. Generally, cell alignment has been demonstrated, but optimization of alignment is less completely understood.
Most of the experimentation in cellular alignment optimization is performed by changing the levels of each variable separately while keeping the remaining variables constant, leading to a large number of experiments. One-factor-at-a-time investigations also ignore interactions between factors and do not elucidate which factors are most important and thus often do not provide information about the overall optimum. One of the most well-utilized experimental analysis methods for examination of factors is Response Surface Methodology (RSM)31–34. RSM is a set of statistical methods which consists of using a data set from designed experiments to determine conditions by which and a desired response can be obtained. Design of Experiments (DOE)has been demonstrated to be efficient and satisfactory for the acquisition of information to correlate independent factors with response in formulation composition and/or manufacturing processing parameters. While the power of DOE and RSM has been well demonstrated in chemical and process engineering, these tools have not been applied extensively in cellular and molecular bioengineering.
In the particular case of the nervous system, previous work has shown that neurons are guided by anisotropic tissue structures in vitro and in vivo. Neurons can align to tissues such as nerve 35, 36, white matter 9, muscle 37–39, and blood vessels 40–42; to transplanted olfactory ensheathing cells 43, marrow stromal cells 44, and tracks of SCs 45; and in co-cultures with aligned astrocytes 17, 19, 20, 46, SCs 23, 47, 48, meningeal cells 49, olfactory ensheathing cells 20, and fibroblasts 49, 50. In motivating strategies to promote nerve regeneration, this work gives rise to several questions: Is anisotropy of any tissue or cell type sufficient for nerve guidance? Which cellular properties are important for guidance?
In this study, we generated comparable confluent and aligned monolayers of SCs, astrocytes, and ECs. Investigation of the influence of protein patterning parameters on cellular confluence and alignment was optimized by DOE and statistically modeled and analyzed quantitatively by RSM, and conditions for desired responses were predicted and confirmed for each cell type. The systematic optimization of biochemical conditions to enhance cellular alignment and confluence was investigated as a first step toward generating anisotropic tissues and biomimetic substrates for studying neuron guidance. Successful application of the statistical experimental design method DOE, the experimental analysis method RSM, and statistical optimization to surface engineering and cell culture parameters was also demonstrated.
MATERIALS AND METHODS
Experimental design and statistical analysis
Statistical software package Design-Expert (Version 7, Stat-Ease, Minneapolis, MN, USA) was used to design and analyze the experiment. A four-factor d-optimal design with a full set of replicates was calculated to model alignment and confluence. The same experimental design was used for the three cell types investigated: astrocytes (A7s), human umbilical vein ECs (HUVECs), and SCs. The four factors investigated for effects on alignment and confluence were width of space between protein stripes, width of protein stripes, starting cell density, and time in culture (Table 1). The first and second factors had five levels each: 10, 20, 30, 40, and 50 μm. The third factor had three levels (determined from confluence experiments): 17K cells cm−2, 26K cells cm−2, and 49K cells cm−2. The fourth factor had two levels: 24h and 48h for A7s and HUVECs, and 48h and 72h for SCs. The number of cells cm−2 and the mean vector length (MVL) of the clustering of nuclear alignment at the end of each run were measured with the analysis described below(Table 2).
Table 1. Factors investigated for effects on cell alignment and confluence.
| Factor | Low Value | High Value |
|---|---|---|
| A-space width | 10 μm | 50 μm |
| B-stripe width | 10 μm | 50 μm |
| C-plating density | 17K cells cm−2 | 49K cells cm−2 |
| D-culture time | 24h (48h for SCs) | 48h (72h for SCs) |
Table 2. Responses recorded in 118 experimental runs for each cell types.
| Response | Low Value | High Value | Average |
|---|---|---|---|
| A7 | |||
| mean vector length | 0.095 | 0.693 | 0.443 |
| terminal cell density | 7,979 cells cm−2 | 106,252 cells cm−2 | 45,395 cells cm−2 |
| HUVEC | |||
| mean vector length | 0.127 | 0.418 | 0.282 |
| terminal cell density | 4,663 cells cm−2 | 54,974 cells cm−2 | 26,776 cells cm−2 |
| SC | |||
| mean vector length | 0.532 | 0.806 | 0.705 |
| terminal cell density | 15,310 cells cm−2 | 100,933 cells cm−2 | 50,771 cells cm−2 |
Cell culture
Cell culture reagents were from Invitrogen Life Technologies (Carlsbad, CA, USA) unless otherwise indicated. Cultures were incubated at 37°C with 5% CO2 in a humidified environment.
SCs from rat sciatic nerve, a generous gift of Dr. Mary Bunge (University of Miami, Coral Gables, FL, USA), were cultured on tissue culture plastic dishes coated with 100 μg ml−1 poly(L-lysine) (PLL; Sigma, St. Louis, MO, USA) in Dulbecco’s modified eagle’s medium (DMEM) with 10% fetal bovine serum (FBS), 4 mM L-glutamine, 100 U ml−1 penicillin and 100 μg ml−1 streptomycin supplemented with 2 μM forskolin (Sigma), 10 μg ml−1 bovine pituitary extract (Sigma) and 2 μM here gulin (generous gift from Genentech, Vacaville, CA, USA) (SC media). Cells used in experiments were between passages 5 and 9.
A7s, a generous gift of Dr. Herbert Geller (Developmental Neurobiology Section, NHLBI, NIH, Bethesda, MD, USA), were cultured on tissue-culture plastic dishes in DMEM with 10% FBS, 4 mM L-glutamine, 100 U ml−1 penicillin and 100 μg ml−1 streptomycin supplemented with 5 μg ml−1 insulin from bovine pancreas. Cells used in experiments were between passages 9 and 13.
HUVECs (PCS-100-010, ATCC, Manassas, VA, USA) were cultured on tissue-culture plastic dishes in endothelial growth media-2 (EGM-2) (Cambrex, Rockland, ME, USA). Cells used in experiments were between passages 5 and 11.
Determination of cell density at confluence
Cells were allowed to grow to confluence in T25 culture flasks. Samples were fixed with 2% paraformaldehyde (Sigma) in 0.1 M phosphate-buffered saline (PBS). Cells were then stained with 4′,6′-diamidino-2-phenylindole (DAPI, Sigma) and analyzed for cell density as described below.
Alignment study
Substrates were prepared as previously described 51. For SCs and A7s, poly(dimethyl siloxane) (PDMS) microstamps with plateaus of 10 mm length, 10–50 μm width and 10–50 μm pitch were coated with laminin (LN) and stamped onto glass coverslips via micro-contact printing. For HUVECs, coverslips were microstamped with human fibronectin (FN) stripes. SCs, A7s, and HUVECs were plated on the appropriate substrates at 17K, 26K, or 49K cells cm−2 and incubated for 24–72 h. Cells were fixed with 2% paraformaldehyde (Sigma) in 0.1 M phosphate-buffered saline (PBS), stained with DAPI and analyzed for alignment as described below. Control coverslips coated uniformly with LN or FN were also plated at these cell densities, fixed at appropriate time points, and analyzed.
Confirmation of protein patterning
Fixed samples were blocked for 1 h at room temperature with 10% goat serum (Jackson Immuno Research Laboratories, West Grove, PA, USA) and 1% bovine serum albumin (Sigma) in 0.1 M PBS (blocking buffer) with the addition of 0.1% Triton X-100 (VWR, West Chester, PA, USA) for permeabilization. Samples were incubated overnight at 4°C with a rabbit polyclonal anti-LN primary antibody (Biomedical Technologies Inc., Stoughton, MA, USA) or monoclonal anti-cellular FN primary antibody (Sigma), diluted 1:200 in blocking buffer. Samples were rinsed in PBS, incubated for 1 h at room temperature with Cy2 or Cy3-conjugated goat anti-rabbit secondary antibody or Cy2 or Cy3-conjugated goat anti-mouse secondary antibody (Jackson Immuno Research Laboratories) diluted 1:200 in blocking buffer, rinsed in PBS and viewed under fluorescence microscopy.
Microscopy
12-bit fluorescent images were obtained at 100× magnification with a 30-ms exposure for imaging of DAPI, 100-ms exposure for imaging of immunohistochemistry, and 10-ms exposure for imaging of phase contrast and analyzed as described below.
To visualize alignment and proliferation of support cells on micropatterned substrates, ten 664 μm × 872 μm fields of view (FOVs) were acquired at 100× magnification on a Nikon Eclipse TE2000-S microscope equipped with phase-contrast and epifluorescence optics and software-controlled motorized stage. Samples were oriented so that substrate pattern direction was uniform between samples. Corresponding phase and epifluorescence images were captured using a Hamamatsu Orca-ER camera, an Orbit shutter controller, and a Ludl stage controller, outputting to OpenLab v4.0.4 (Improvision, Lexington, MA, USA). A custom stage automation was written which allows the user to select the area of protein patterning, choose 10 points at random within this area, and obtain corresponding images of phase, DAPI, and stained micropattern.
DAPI images for adhesion and cell density analysis were converted to 8-bit greyscale with Adobe Photoshop CS2, inverted, and the contrast was adjusted using the level and curve functions. Nuclei number were assessed with ImageJ v1.36, using the Otsu function of the multithresholder plugin and the particle analyzer plugin with size filter set to 50–850 pixels to exclude image artifacts and cell clusters. For alignment and confluence analysis, DAPI images were processed as described, and nuclei number and angle of orientation were assessed with ImageJ. This data was analyzed with Oriana v2.02c (Kovach Computing Services, Pentraeth, UK).
Statistical analysis
Circular analysis was used to analyze the angular distributions of the major axes of the nuclei (Li et al 2008). Circular mean vectors were calculated, where the length of the mean vector corresponds to the degree of clustering of the data, and the direction of the mean vector corresponds to the mean direction of the data. Uniformity of the distribution of nuclei was assessed using a Rao’s spacing test and alignment of nuclei in the direction of the substrate pattern was assessed with a V-test. Oriana 2.02c software was used for circular analysis. For all cases, a P value of 0.05 was taken to be significant. Design results were analyzed by standard analysis of variance (ANOVA) and fitted to a second-order polynomial equation.
RESULTS
Design of Experiment
In this study, we investigated the effects of protein pattern dimensions, cell plating density, and time in culture on the generation of aligned, confluent monolayers of three cell types. In order to optimize the experimental efficiency, DOE was utilized. A D-optimal design, which is generated by an iterative search algorithm that minimizes the covariance of the parameter estimates for a specified model, was selected and reduced the number of required experiments from 150 to 59 for each cell type. We hypothesized that several factors would interact to affect the monolayers, i.e. that plating density and culture time would interact to affect the terminal cell density. Therefore, a second order interaction model was chosen so that both the main effects of each individual factor as well as any dual factor interactions could be investigated. The following model was proposed to describe the monolayers.
Where Y is the response (either mean vector length or terminal cell density), A is the space between protein stripes, B is the width of protein stripes, C is the initial plating density, D is the culture time, and xi and xij are the coefficients.
Factors and Responses Analyzed
The space between protein stripes and the width of protein stripes were varied from 10–50 μm in 10 μm intervals. The initial plating densities and culture times were determined from pilot experiments that established the density of each cell type at confluence (Figure 1A). At confluence, the density of A7s was 105,000 ± 20,000 cells cm−2 (Figure 1B), the density of HUVECs was 41,000 ± 9,000 cells cm−2 (Figure 1C), and the density of SCs was 55,000 ± 14,000 cells cm−2 (Figure 1D). Initial plating densities for the experiments of the study were chosen to correspond to approximately half of the cell density of each cell type at confluence, i.e. 49K, 17K, and 26K cells cm−2. In pilot experiments plating cells on LN coated coverslips at these densities, most samples were confluent within 48h (A7s, HUVECs) or 72h (SCs). Thus culture times of 24h, 48h, and 72h were chosen for investigation. Mean vector length of the distribution of nuclear angles was used as a measure of overall alignment (Figure 2), and the number of cells stained by DAPI and counted was used as a measure of the final confluence.
Figure 1. Cell density at confluence varied among A7s, HUVECs, and SCs.
(A) Graph of the cell density of A7s, HUVECs, and SCs grown to confluence and counted (n=3). Phase contrast micrographs of A7s (B), HUVECs (C), and SCs (D) at confluence. * Indicates difference from other cell types with p<0.05. Scale bar, 200 μm.
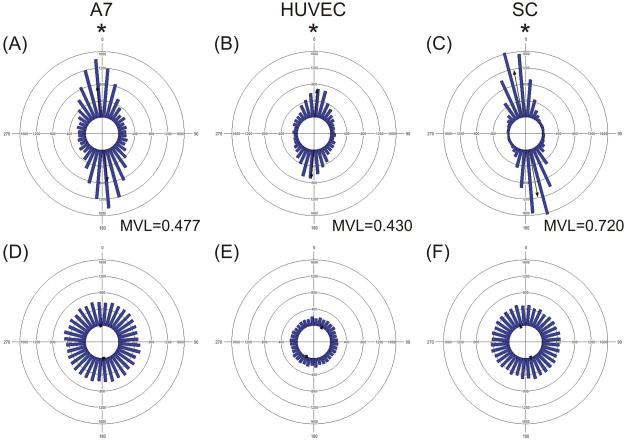
Figure 2. Cellular alignment on micropatterned substrates was clustered in the direction of the micropatterns.
Circular histograms of A7s (A,D), HUVECs (B,E), and SCs (C,F) plated at 49K cm−2 and cultured for 48h on optimal protein micropatterned substrates (A–C) or on uniformly protein coated substrates (D–F). * Indicates difference from a uniform distribution with p<0.05 by Rao’s spacing test.
The D-optimal mixture design and response results for each run can be found in supplementary materials. From the 150 candidate points for each cell type, 59 experimental points were chosen by Design Expert software to establish the model and assess the fit of the model. These experimental points included vertices, centers of the edges, blends of the inner experimental space, and overall centroid. Each run was replicated for a total of 118 runs for each cell type. All runs were carried out in the order given by the software. Of the 354 runs in the study, 21 runs were repeated at the end of all trials to replace runs that contained sub-optimal microstamping or cell health as revealed by immunohistochemistry. The high, low, and average responses recorded for each cell type are presented in Table 2.
The RSM models for all cell types are presented in Tables 3 and 4 and Figures 3–6. The models are presented in two ways: (1) with coefficients that correspond to the actual experimental variables (i.e. protein stripe width in μm); and (2) with coded coefficients that are transformed so the high value of the variable becomes +1 and the low value becomes −1 (i.e. transformation of the equation such that protein stripe width of 50 μm would equal +1 and protein stripe width of 10 μm would equal −1). Coded coefficients allowed for comparison across conditions. In each case tested, the model explained the response well, as the lack of fit was not significant (p>0.05). Stepwise regression was performed; where the coefficients with the largest p-value were consecutively deleted until only significant coefficients remained. The coefficients and contributions before elimination of insignificant variables can be found in supplementary materials.
Table 3. Effect estimates for alignment from the result of Design Expert analysis.
Coded coefficients are transformed so the high value of the factor becomes +1 and the low value becomes −1 (i.e. for A, 50μm becomes +1 and 10 μm becomes −1).
| Factor | Coefficient (Actual Factor) | Coefficient (Coded Factor) | Standard Error (Coded) | Percent Contribution | P-value | |
|---|---|---|---|---|---|---|
| A7 | Model | <0.0001* | ||||
| Intercept | 0.643 | 0.440 | 0.009 | |||
| A-space width | 2.09 × 10−3 | −0.031 | 0.012 | 14.9% | 0.0109* | |
| B-stripe width | −2.85 × 10−3 | −0.057 | 0.012 | 47.7% | <0.0001* | |
| C-plating density | −6.56 × 10−6 | −0.015 | 0.01 | 4.5% | 0.1621 | |
| D-culture time | −1.16 × 10−3 | 0.012 | 0.009 | 4.2% | 0.1728 | |
| AD | −1.01 × 10−4 | −0.024 | 0.012 | 9.0% | 0.0468* | |
| CD | 1.57 × 10−7 | 0.03 | 0.01 | 19.6% | 0.0037* | |
| Lack of fit | 0.9172** | |||||
| HUVEC | Model | <0.0001* | ||||
| Intercept | 0.338 | 0.290 | 0.006 | |||
| A-space width | −1.34 × 10−3 | −0.027 | 0.009 | 32.6% | 0.0032* | |
| B-stripe width | 1.64 × 10−3 | −0.033 | 0.009 | 47.3% | 0.0005* | |
| C-plating density | 1.13 × 10−6 | 0.018 | 0.008 | 20.1% | 0.019* | |
| Lack of fit | 0.7534** | |||||
| SC | Model | <0.0001* | ||||
| Intercept | 0.633 | 0.710 | 0.005 | |||
| A-space width | 3.42 × 10−3 | −0.013 | 0.006 | 11.2% | 0.0432* | |
| C-plating density | 1.34 × 10−6 | 0.022 | 0.006 | 42.7% | <0.0002* | |
| D-culture time | 8.22 × 10−4 | −0.015 | 0.005 | 28.1% | 0.0019* | |
| AD | −6.81 × 10−5 | −0.016 | 0.007 | 18.0% | 0.0140* | |
| Lack of fit | 0.5838** | |||||
indicates the factor as a significant contributor to the model with p<0.05,
indicates an insignificant lack of fit with p>0.05
Table 4. Effect estimates for terminal density from the result of Design Expert analysis.
Coded coefficients are transformed so the high value of the factor becomes +1 and the low value becomes −1 (i.e. for A, 50μm becomes +1 and 10 μm becomes −1).
| Factor | Coefficient (Actual Factor) | Coefficient (Coded Factor) | Standard Error (Coded) | Percent Contribution | P-value | |
|---|---|---|---|---|---|---|
| A7 | Model | <0.0001* | ||||
| Intercept | −41900 | 49700 | 1430 | |||
| B-stripe width | 538 | 6880 | 2060 | 4.7% | 0.0011* | |
| C-plating density | 0.698 | 20200 | 1680 | 61.1% | <0.0001* | |
| D-culture time | 1620 | 11200 | 1380 | 28.3% | <0.0001* | |
| BC | 0.0189 | 6030 | 2380 | 2.7% | 0.0127* | |
| BD | −22.7 | −5450 | 1980 | 3.2% | 0.0068* | |
| Lack of fit | 0.1557** | |||||
| HUVEC | Model | <0.0001* | ||||
| Intercept | −1960 | 28000 | 811 | |||
| C-plating density | 0.445 | 7120 | 951 | 57.7 | <0.0001* | |
| D-culture time | 424 | 5090 | 795 | 42.3 | <0.0001* | |
| Lack of fit | 0.9799** | |||||
| SC | Model | <0.0001* | ||||
| Intercept | 1330 | 53300 | 1030 | |||
| A-space width | 248 | −5930 | 1480 | 6.4% | 0.0001* | |
| B-stripe width | −666 | 7580 | 1490 | 10.3% | <0.0001* | |
| C-plating density | 1.43 | 15000 | 1230 | 59.3% | <0.0001* | |
| D-culture time | 38.7 | 6730 | 1020 | 17.3% | <0.0001* | |
| AC | −0.0165 | −5280 | 1790 | 3.5% | 0.004* | |
| BD | 17.4 | 4180 | 1480 | 3.2% | 0.0057* | |
| Lack of fit | 0.1925** | |||||
indicates the factor as a significant contributor to the model with p<0.05,
indicates an insignificant lack of fit with p>0.05
Figure 3. Micropattern dimensions affected alignment of cells.
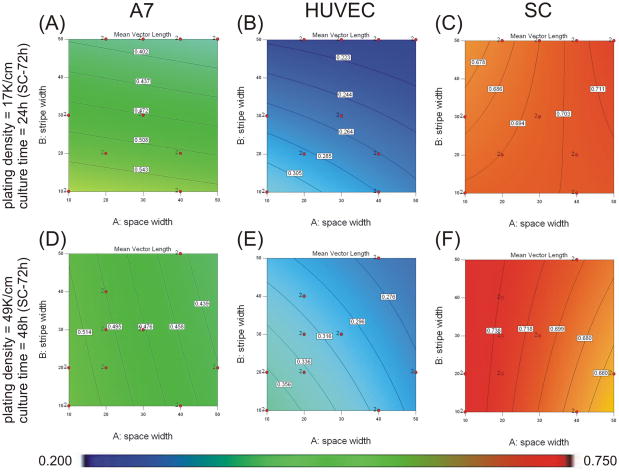
Contour graphs showing the effects of protein stripe and space width on mean vector length when plated at 17K cells cm−2 for 24h (48h for SCs) (A–C) or plated at 49K cells cm−2 for 48h (72h for SCs) (D–F). Experimental conditions were determined and analyzed with Design Expert 7 software. An ANOVA model of mean vector length was calculated and graphed for A7s (A,D), HUVECs (B, E), and SCs (C, F). Mean vector magnitudes are indicated by contour lines and colorimetric scale, blue-red, 0.20–0.75. Design points carried out experimentally are indicated with red dots.
Figure 6. Protein stripe width affected the confluence of A7s and SCs.
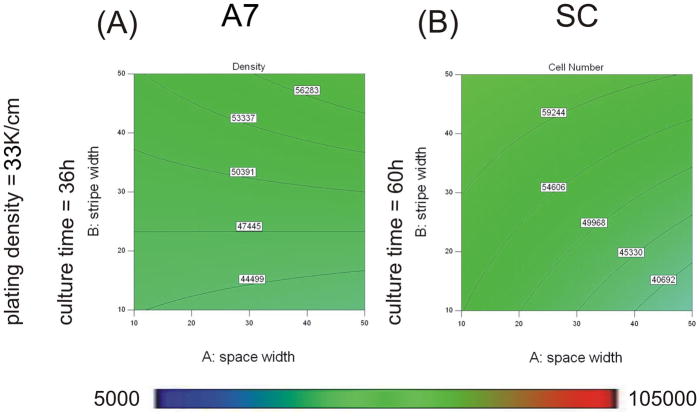
Contour graphs showing the effect of protein stripe and space width on cell number when plated at 33K/cm2 for 36h for A7s (A) and 60h for SCs (B). Experimental conditions were determined and analyzed with Design Expert 7 software. An ANOVA model of cell number was calculated and graphed for A7s (A) and SCs (B). Cell number magnitudes are indicated by contour lines and colormetric scale, blue-red, 5K–105K cells cm−2. Design points carried out experimentally are indicated with red dots.
Protein pattern dimensions influenced A7 alignment
The width of protein patterning and spacing had significant effects on the alignment of A7s (Table 3). Interactions between spacing and culture time and interactions between starting density and culture time were also significant (Table 3). The following model for A7 alignment was obtained, with coefficients corresponding to actual factors:
Where A is the space between protein stripes (μm), B is the width of protein stripes (μm), C is the initial plating density (cells cm−2), and D is the culture time (h). Contour plots based on this equation are shown in Figure 3. The resulting model suggests that at low starting cell density and shorter culture time, alignment increased with decreasing stripe width (Figure 3A). At higher starting density and longer culture time, alignment increased with decreasing space width (Figure 3D). The contribution of the influence of stripe width on A7 alignment was stronger than the contribution of space width in the range of conditions tested (contribution of 47.7% for stripe width vs. 14.9% for space width). The interaction terms of AD (9.0%) and CD (19.6%) also significantly contributed to the model.
Protein stripe pattern, plating density, and culture time influenced terminal density of A7s
The width of protein stripes, starting cell density, and culture time had significant effects on the terminal density of A7s (Table 4). The interaction between stripe width and plating density was also significant. The following model for A7 terminal density was obtained, with coefficients corresponding to actual factors:
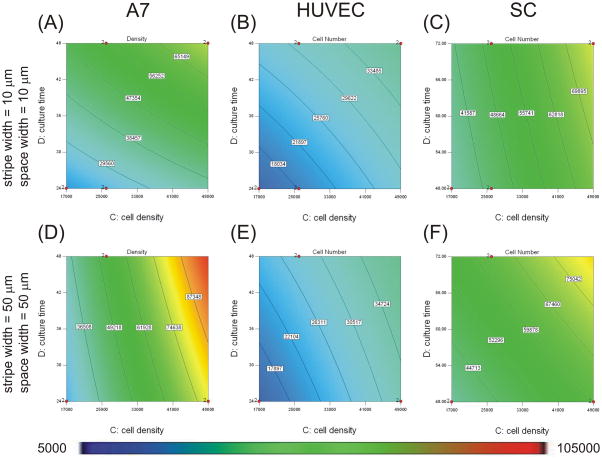
Where B is the width of protein stripes (μm), C is the initial plating density (cells cm−2), and D is the culture time (h). Contour plots based on this equation are shown in Figures 5 and 6. Final cell density increased as plating density and culture time increased (Figure 5A, D). As stripe width increased, the final cell density increased (Figure 6A). The influence of plating density on A7 confluence was stronger than the influence of culture time in the range tested (contributions of 61.1% for plating density vs. 28.3% for culture time), and both were stronger than the influence of stripe width (contribution of 4.7%). The interaction terms of BC (2.7%) and BD (3.2%) also contributed to the model.
Figure 5. Plating density and culture time affected terminal density of cells.
Contour graphs showing the effects of protein stripe and space width on cell number when plated on micropatterns with 10 μm protein stripe and space width (A–C) or micropatterns with 50 μm protein stripe and space width (D–F). Experimental conditions were determined and analyzed with Design Expert 7 software. An ANOVA model of final cell number was calculated and graphed for A7s (A,D), HUVECs (B, E), and SCs (C, F). Cell number magnitudes are indicated by contour lines and colorimetric scale, blue-red, 5K–105K cells cm−2. Design points carried out experimentally are indicated with red dots.
Protein pattern dimensions and plating density influenced HUVEC alignment
The width of protein patterning and spacing had significant effects on the alignment of HUVECs (Table 3). The effect of the starting cell density was also significant. The following model for HUVEC alignment was obtained, with coefficients corresponding to actual factors:
Where A is the space between protein stripes (μm), B is the width of protein stripes (μm), and C is the initial plating density (cells cm−2). Contour plots based on this equation are shown in Figures 3 and 4. The resulting model shows that alignment increased with decreasing protein stripe width and decreasing spacing independent of the starting density and culture time (Figure 3B and 3E). Alignment also increased as the starting cell density increased (Figure 4A). The contribution of the influence of stripe width was stronger than the contribution of space width to HUVEC alignment in the range tested (contributions of 47.3% for stripe width vs. 32.6% for space width), and both were stronger than the influence of cell density (20.1%).
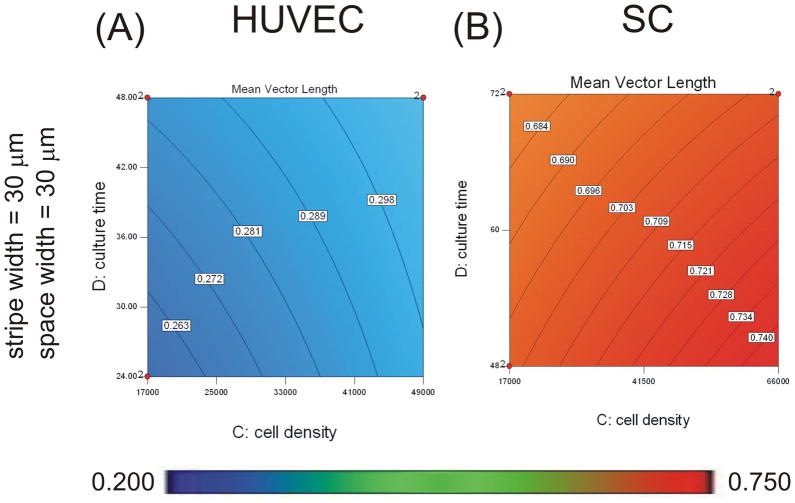
Figure 4. Plating density affected alignment of HUVECs and SCs.
Contour graphs showing the effects of plating density and culture time on mean vector length when plated on micropatterns with 30 μm protein stripe and space width. Experimental conditions were determined and analyzed with Design Expert 7 software. An ANOVA model of mean vector length was calculated and graphed for HUVECs (A), and SCs (B). Mean vector magnitudes are indicated by contour lines and colorimetric scale, blue-red, 0.20–0.75. Design points carried out experimentally are indicated with red dots.
Plating density and culture time influenced HUVEC terminal density
Starting cell density and culture time had significant effects on the confluence of HUVECs (Table 4). The following model for HUVEC terminal density was obtained, with coefficients corresponding to actual factors:
Where C is the initial plating density (cells cm−2) and D is the culture time (h). Contour plots based on this equation are shown in Figure 5. Final cell density increased as plating density and culture time increased (Figure 5B, E). The influence of plating density on HUVEC confluence was stronger than the influence of culture time in the range tested (contributions of 57.7% for plating density vs. 42.3% for culture time).
Plating density and culture time influenced SC alignment
Starting cell density and culture time had significant effects on the alignment of SCs (Table 3). The space between protein stripes was also significant. The following model for SC alignment was obtained, with coefficients corresponding to actual factors:
Where A is the space between protein stripes (μm), C is the initial plating density (cells cm−2), and D is the culture time (h). Contour plots based on this equation are shown in Figures 3 and 4. The resulting model shows that at low starting cell density and shorter culture time, alignment increased with increasing space width (Figure 3C). At higher starting density and longer culture time, alignment increased with decreasing space width (Figure 3F). Alignment increased with increased starting cell density and culture time (Figure 4B). The contribution of plating density was stronger than the contributions of culture time (contributions of 42.7% for plating density vs. 28.1% for culture time), and both were stronger than the contribution of space width to SC alignment (11.2%). The interaction term AD also contributed (18.0%).
Pattern dimensions, plating density, and culture time influenced SC terminal cell density
All four factors had significant effects on the terminal density of SCs (Table 3). The interaction between space width and plating density and between stripe width and culture time were also significant (Table 4). The following model for SC confluence was obtained, with coefficients corresponding to actual factors:
Where A is the space between protein stripes (μm), B is the width of protein stripes (μm), C is the initial plating density (cells cm−2), and D is the culture time (h). Contour plots based on this equation are shown in Figures 5 and 6. Final cell density increased as plating density and culture time increased (Figure 5C, F). As stripe width increased, the terminal cell density increased, and as space width increased, cell density decreased (Figure 6B). The influence of cell density had the strongest influence on SC confluence (contribution of 59.3%). The influences of stripe width and culture time in the range tested were weaker (contributions of 10.3% for stripe width and 17.3% for culture time) with the smallest influence coming from space width (6.4%). The interaction terms of AC and BD also contributed (3.5% for AC and 3.2% for BD).
Contributions of factors varied across cell types
Presentation of coded coefficients allowed us to compare the relative contributions of experimental parameters to cell responses across cell types. Cellular alignment of all three cell types was affected by pattern spacing. The contribution of pattern spacing to alignment was similar for A7s and HUVECs (coded coefficients of −0.031 for A7s and −0.027 for HUVECs), but reduced by half for SCs (coded coefficient of −0.013). The contribution of stripe width to the alignment of A7s was greater than that for HUVECs (coded coefficients of −0.057 for A7s vs. −0.033 for HUVECs). The contribution of plating density to the alignment of HUVECs and SCs was similar (coded coefficients of 0.018 for HUVECs and 0.022 for SCs).
Monolayer terminal cell density of all three cell types was affected by plating density and culture time. The contribution of plating density to terminal cell density was smallest for HUVECs (coded coefficient of 7120), intermediate for SCs (coded coefficient of 15000), and strongest for A7s (coded coefficient of 20200). The contribution of culture time to the terminal cell density was weakest for HUVECs (coded coefficient of 5090), intermediate for SCs (coded coefficient of 6730), and strongest for A7s (coded coefficient of 11200). The contribution of stripe width to the terminal cell density of SCs was stronger than the contribution to the terminal cell density of A7s (coded coefficients of 7580 for SCs vs 6880 for A7s).
Optimal conditions for culture of confluent, aligned monolayer varied among cell types
To determine the optimal conditions for confluent and/or aligned monolayers, a series of rules was applied systematically. To determine the conditions by which confluence would be achieved for monolayers of each cell type, the following criteria were applied: the space and stripe width must be between 10 and 50 μm, the starting cell density must be between 17K and 49K cells cm−2, the culture time must be between 24 and 48h for A7s and HUVECs and be between 48 and 72h for SCs, and the cell density must be between 85–125K cells cm−2 for A7s, 32–50K cells cm−2 for HUVECs, and 41–69K cells cm−2 for SCs. The required conditions for each cell type are presented in Table 5.1. For A7 confluence, there were 60 answers with equal desirability. These combinations can be found in supplementary materials. Space widths ranged from 17–50 μm, stripe widths ranged from 17–50 μm, plating density ranged from 45,000–49,000 cells cm−2, and culture time ranged from 33–48h. For HUVEC confluence, there were 80 answers with equal desirability. These combinations can be found in supplementary materials. Space widths ranged from 10–49 μm, stripe widths ranged from 10–50 μm, plating density ranged from 29,000–49,000 cells cm−2, and culture time ranged from 27–48h. For SC confluence, there were 80 answers with equal desirability. These combinations can be found in supplementary materials. Space widths ranged from 10–50 μm, stripe widths ranged from 10–50 μm, plating density ranged from 17,000–49,000 cells cm−2, and culture time ranged from 48–72h.
Table 5. Numerical optimizations of culture conditions with Design Expert.
| Factor | Input Bounds | Model Output | ||
|---|---|---|---|---|
| A7 | HUVEC | SC | ||
| 5.1 CONFLUENT | ||||
| space width | 10–50 μm | 17–50 μm | 10–49 μm | 10–50 μm |
| stripe width | 10–50 μm | 17–50 μm | 10–50 μm | 10–50 μm |
| plating density | 17–49K cm −2 | 45–49K cm−2 | 29–49K cm−2 | 17K–49K cm−2 |
| culture time | 24–48h 48–72h (SC) |
33–48h | 27–48h | 48–72h |
| terminal density | 85–125K cm−2 (A7) 32–50K cm−2 (HUVEC) 41–69K cm−2 (SC) |
85–96K cm−2 | 32–41K cm−2 | 41–68K cm−2 |
| Desirability | 1.000 (60 answers) | 1.000 (80 answers) | 1.000 (80 answers) | |
| 5.2 OPTIMAL ALIGNMENT | ||||
| space width | 10–50μm | 10 μm | 10 μm | 10 μm |
| stripe width | 10–50 μm | 10 μm | 10 μm | 50 μm |
| plating density | 17–49K cm−2 | 17K cm−2 | 49K cm−2 | 49K cm−2 |
| culture time | 24–48h 48–72h (SC) |
24h | 48h | 48h |
| mean vector length | maximize | 0.578 ± 0.035 | 0.373 ± 0.017 | 0.763 ± 0.014 |
| desirability | 0.808 | 0.752 | 0.844 | |
| 5.3 OPTIMAL ALIGNMENT AT CONFLUENCE (IN RANGE OF EXPERIMENTS) | ||||
| space width | 10–50 μm | 16 μm | 10 μm | 10 μm |
| stripe width | 10–50 μm | 50 μm | 10 μm | 49 μm |
| plating density | 17–49K cm−2 | 49K cm−2 | 49K cm−2 | 46K cm−2 |
| culture time | 24–48h 48–72h (SC) |
48h | 48h | 48h |
| mean vector length | maximize | 0.498 ± 0.046 | 0.373 ± 0.017 | 0.759 ± 0.014 |
| terminal density | 85–125K cm−2 (A7) 32–50K cm −2 (HUVEC) 41–69K cm−2 (SC) |
85K ± 8K cm−2 | 32K ± 2K cm−2 | 69K ± 6K cm−2 |
| desirability | 0.674 | 0.752 | 0.829 | |
| 5.4 MATCHED ALIGNMENT AT CONFLUENCE | ||||
| space width | 10–50 μm | 50 μm | 10 μm | 50 μm |
| stripe width | 10–50 μm | 50 μm | 10 μm | 28 μm |
| plating density | 17–49K cm−2 | 49K cm−2 | 49K cm−2 | 17K cm−2 |
| culture time | 24–48h 48–72h (SC) |
31h | 48h | 72h |
| mean vector length | 0.373 minimize (SC) |
0.376 ± 0.036 | 0.373 ± 0.017 | 0.659 ± 0.011 |
| terminal density | 85–125K cm−2 (A7) 32–50K cm−2 (HUVEC) 41–69K cm−2 (SC) |
85K ± 6K cm−2 | 32K ± 2K cm−2 | 41K ± 3K cm−2 |
| Desirability | 1.000 | 0.752 | 0.533 | |
| 5.5 OPTIMAL ALIGNMENT AT CONFLUENCE (LARGER RANGE) | ||||
| space width | 1–100 μm | 1–31 μm | 1 μm | 1–84 μm |
| stripe width | 1–100 μm | 1–100 μm | 1 μm | 1–98 μm |
| plating density | 0–100K cm−2 | 50–81K cm−2 | 100K cm−2 | 45–68K cm−2 |
| culture time | 0–72h 0–96h (SC) |
58–72h | 18h | <1–36h |
| mean vector length | maximize | 0.693–0.784 | 0.435 ± 0.037 | 0.806–0.863 |
| terminal density | 85–125K cm−2 (A7) 32–50K cm−2 (HUVEC) 41–69K cm−2 (SC) |
95–125K cm−2 | 50K ± 5K cm−2 | 42–68K cm−2 |
| Desirability | 1.000 (75 answers) | 0.941 | 1.000 (77 answers) | |
To determine the best alignment under the conditions tested (sub-confluent or confluent), the same bounds were applied for the factors, no bound was placed on the terminal density, and the mean vector length had to be maximized. The strongest alignment conditions for each cell type are presented in Table 5.2. SC optimal alignment (MVL = 0.763 ± 0.014) was notably higher than A7 optimal alignment (MVL = 0.578 ± 0.035) and HUVEC optimal alignment (MVL = 0.373± 0.017).
For optimization of a confluent, aligned monolayer of each cell type, the same rules were applied for the factors, the mean vector length of nuclear angles had to be maximized, and the cell number had to be between 85–125K cells cm−2 for A7s, 32–50K cells cm−2 for HUVECs, and 41–69K cells cm−2 for SCs. The optimal conditions for each cell type are presented in Table 5.3. Within the bounds applied to the factors, SC optimal alignment (MVL = 0.759 ± 0.014) was higher than A7 optimal alignment (MVL = 0.498 ± 0.046) and HUVEC optimal alignment (MVL = 0.373 ± 0.017). Alignment at confluence for HUVECs and SCs did not vary from alignment at any terminal density.
Monolayers of all three cell types were cultured under these conditions (Figure 2, 7). A7 alignment (MVL = 0.477) and confluence (87K cells cm−2) were in the range of the predicted results (0.498 ±0.046 and 85,000±7,531). HUVEC alignment (MVL = 0.430) and confluence (48K cells cm−2) were higher than the predicted results (0.373±0.017 and 32,000±2,180). SC alignment (MVL = 0.720) was lower than the predicted result (0.759±0.014), while the confluence (67K cell cm−2) was in the range of the predicted result(69,000±5,632).
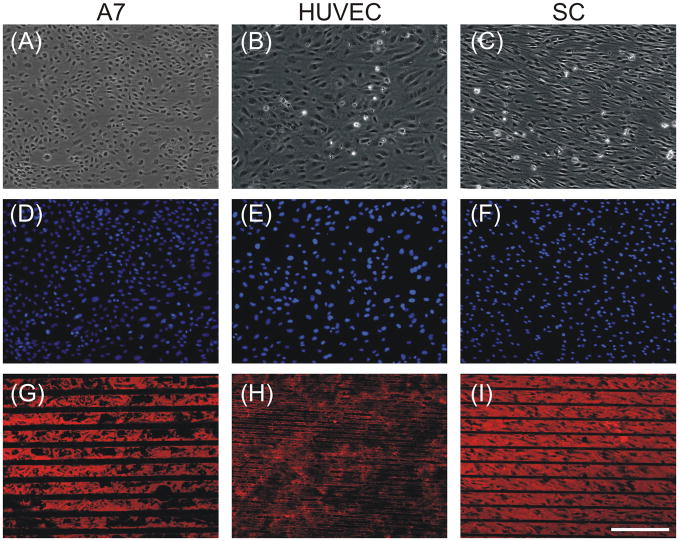
Figure 7. Experimental runs supported model predictions.
Phase contrast (A–C) and epifluorescent images (D–I) of A7s plated at 49K cells cm−2 for 48h on LN stripes of 50 μm with 20 μm spaces (A, D, G), HUVECs plated at 49K cells cm−2 for 48h on FN stripes of 10μm with 10 μm spaces (B, E, H), and SCs plated at 49K cells cm−2 for 48h on LN stripes of 50 μm with 10 μm spaces(C, F, I). Cultures were stained with DAPI (D–F), with anti-LN (G,I) or anti-FN (H). Scale bar, 200 μm.
Cellular dynamics underlying alignment varied among cell types
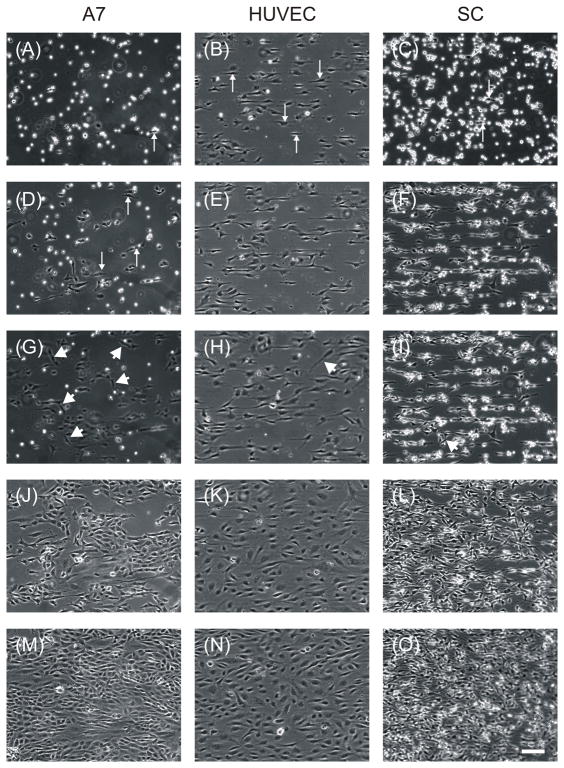
Time lapse microscopy under optimal conditions for each cell type revealed further differences in cellular alignment among the cell types. Timelapse movies can be found in supplementary materials. At one hour after plating, A7s had just begun to adhere to the substrate (Figure 8A) while SCs had adhered and a few cells had begun to orient in the direction of the pattern (Figure 8C). The SCs initially appeared to be aligning along the boundaries of the stripes as they were in single file lines that were spaced 50 μm apart. By one hour, most of the HUVECs had adhered and elongated in the direction of the pattern, over the width of the stripe (Figure 8B). By four hours, SCs and HUVECs had similar elongation and confluence (Figure 8F and 8E), but elongation of A7s had just begun (Figure 8D). While most of the HUVECs and SCs at these time points appeared to move only parallel to the direction of the pattern, there were a number of astrocytes that moved in other directions, even perpendicular to the pattern and crossing several stripes. SCs rarely crossed over more than one stripe, and HUVECs remained confined to the 10 μm stripes. Between 4h and 10h, little change in alignment or confluence was seen in any of the cell types (Figure 8G–I), but the cells remained motile, moving mostly parallel to the pattern direction. HUVEC cell bodies were no longer as confined to the 10 μm stripes with their cell bodies broadened into the spaces between stripes. Between 10h and 24h, there was marked proliferation of all three cell types (Figure 8J–L). By 24h, A7s were 75% confluent, SCs were 90% confluent, and HUVECs were 95% confluent. At 48h, the cultures were confluent and remained aligned in the direction of the underlying protein pattern (Figure 8M–O).
Figure 8. Timelapse microscopy of aligned monolayer generation revealed differences among cells types.
Phase contrast images obtained by timelapse microscopy of A7s plated at 49K cells cm−2 after 1h (A), 4h (D), 10h (G), 24h (J), and 48h (M) on LN stripes of 50 μm with 20 μm spaces, HUVECs plated at 49K cells/cm2 after 1h (B), 4h (E), 10h (H), 24h (K), and 48h (N) on FN stripes of 10μm with 10 μm spaces, and SCs plated at 49K cells/cm2 after 1h (C), 4h (F), 10h (I), 24h (L), and 48h (O) on LN stripes of 50 μm with 10 μm spaces. Arrows indicate cells beginning elongation along micropattern. Arrow heads indicate cells moving in directions different from the pattern. Scale bar, 100 μm.
DISCUSSION
The central goal of this study was to generate confluent and aligned monolayers of support cells for studying neuron guidance. A broader goal of the study was to demonstrate the utility of DOE and RSM in cellular bioengineering experiments. Toward this end, the influence of protein patterning and culture parameters on cellular confluence and alignment were statistically modeled and analyzed quantitatively, and optimal conditions were determined from these models for each cell type. Confluent and aligned monolayers of A7s, HUVECs, and SCs were achieved through culture on micropatterned proteins, and the predicted optimal conditions by which these monolayers can be achieved varied among the cell types tested.
Design of Experiments
Statistical experimental design was applied to improve the experimental efficiency of modeling confluence and alignment of these cell types. DOE has been demonstrated to be efficient and satisfactory for the acquisition of information to correlate independent factors with response in formulation composition and/or manufacturing processing parameters 31, 32. For RSM analysis, experimental factors are modeled as a continuous function to approximate the optima of individual or multiple responses. Contour plots are often used to represent the response to two variables. Points on these plots are generated from a number of experimental trials and intermediate values between them are determined by mathematical interpolation. Results that can be analyzed by RSM can be designed D-optimally. The reasons for using optimal design instead of standard classical design, i.e the simplex design, are that standard factorial designs require numbers of runs that are prohibitively large for reasonable allocation of resources and time, and/or the design space is constrained i.e. the process space contains factor settings that are not feasible or are impossible to run 52. A D-optimal design is generated by an iterative search algorithm and seeks to minimize the covariance of the parameter estimates for a specified model. This is equivalent to maximizing the determinant D = |XTX|, where X is the design matrix of model terms (the columns i.e. space width, stripe width, etc.) evaluated at specific treatments in the design space (the rows i.e. 10 μm, 20 μm, etc.). When the responses of interest are expressed in a model as a continuous function of the factors involved, the model could reveal, graphically and mathematically, regions of desirable formulation compositions that satisfy the criteria imposed by the experimenter 52.
A7s and SCs were chosen for modeling and generation of anisotropic cultures because while it has been well recognized that the different chemical cues presented by SCs and astrocytes after injury contribute to the varied capacities for regeneration in the PNS and CNS, the contributions of the physical features of these distinct glia to the promotion of axon regrowth and guidance have not been studied together. We were also interested in determining if guidance by anisotropic tissues was limited to the features of glial cells, so HUVEC was chosen for characterization as a non-glial cell type. Astrocytes, ECs, and SCs have been cultured to aligned confluence previously, but not under similar conditions so that their influence on neuron growth can be compared in the same experiments. Generation of confluent layers is important so that the influence of the cellular cue can be evaluated. Micropatterning was chosen as a common alignment method since previous work with SCs and ECs has shown it effective, and because of its ease and flexibility 53. Protein stripe and space width were varied at 10 um intervals from 10–50 um as these dimensions are on the scale of cellular dimensions and are in the range of dimensions that have been employed to align these and other cell types in our lab as well as other labs. Cell plating density was chosen as the number of cells that would be required to cover half of the substrate for each cell type. Culture time was chosen based on pilot experiments where it was qualitatively determined that the cells had reached confluence.
In order to study these cell types as anisotropic substrates, we were interested in achieving cultures that were both aligned and confluent. Circular statistical analysis and mean vector length of nuclear angles were chosen to measure alignment because circular orientation data such as alignment is better described by a resultant vector rather than an arithmetic mean 54 and the mean vector length corresponds to the clustering of the angles of the nuclei 51. Changes in cellular shape have been shown to affect the shape of the nucleus 30, 55, and previous work has shown that the angle of the major axis of the nucleus is an effective measure of cellular orientation 48, 56. Other studies have utilized the nuclear form factor (NFF) as a measure of cellular alignment 13. Because compared alignment across cell types, this method was not used as nuclear shape of the three cell types varied.
Contributions of Factors and Interpretation of Responses
Our models and analysis of cellular alignment and confluence revealed different contributions of factors among the cell types tested (Table 6). SCs had the highest degree of alignment of the cell types, and plating density had the largest contribution to both their alignment and their confluence. Culture time and space width each contributed to SC alignment and confluence as well. Our multi-factorial approach also revealed that the effect of space width changed when influenced by other factors; for short cultures, increased space width increased alignment, but for longer cultures, increased space width decreased alignment. As the aligned SC monolayers were forming, SCs showed a high capacity for alignment to protein patterns. The size of protein stripe was not a significant factor to SC alignment as SCs initially aligned to stripe borders and demonstrated a high affinity for alignment to each other as they filled in. The optimal conditions for SC alignment at confluence were the largest stripe width tested (50 μm), the smallest space width tested (10 μm), the highest plating density tested (49Kcells cm−2), and the shortest culture time tested (48h).
Table 6. Summary of key findings.
| A7 | HUVEC | SC | |
|---|---|---|---|
| Most important factor for aligned monolayer: | Protein stripe width | Cell density | Cell density |
| Other key factors: | Protein space width Cell density Culture time |
Protein stripe width | Culture time Protein space width |
| How well optimized: (MVL) | 0.498 model 0.443 experimental |
0.373 model* 0.418 experimental* |
0.759 model 0.705 experimental |
| Suggested mechanism: |
|
|
|
Note: optimal conditions may be outside range tested.
HUVECs had the lowest alignment of the cell types tested. The most important factor in achieving anisotropic HUVEC monolayers was plating density as it was the strongest contributing factor to confluence of HUVECs and also played a role in alignment. Stripe width was also an important factor for HUVEC alignment, as the cells’ initial confinement to small stripes was imperative for achieving aligned monolayers. The optimal conditions for HUVEC alignment and confluence were small stripe and space width (10 μm) and high cell density (49K cells cm−2) and culture time (48h).
A7s’ alignment was intermediate between the HUVECs and the SCs. The most important factor in achieving confluent, aligned monolayers of A7s was protein stripe width as it was the strongest contributing factor to mean vector length and also significantly influenced terminal cell density. Our multi-factorial approach revealed several interesting interactions of factors that influenced this cell type. At low starting cell density and shorter culture time, alignment increased with decreasing stripe width, whereas at higher starting density and longer culture time, alignment increased with decreasing space width. Space width played an important role at longer culture times, suggesting that the way cells fill in the spaces is critical for the maintenance of alignment at confluence. The optimal conditions for confluent, aligned A7s were the largest stripe width tested (50 μm), space width of approximately the width of A7s (17 μm), and high plating density (49Kcells cm−2), and culture time(48h).
Potential Mechanisms of Aligned Monolayer Formation
With our combinatorial approach, we observed and analyzed the complexity of cellular interactions with micropatterned materials under a range of conditions; taking these observations together points toward possible mechanisms that underlie the formation of confluent, aligned monolayers of cells. SCs achieved their highly aligned monolayers by aligning first to the protein stripe edges, then proliferating and aligning to each other. Thus initial plating density strongly affected SC alignment and confluence. Increased width of the protein stripes increased terminal cell density by providing larger areas of adhesive substrate for initial adhesion and proliferation, but was in consequential for alignment in these studies. Further, when space widths between the protein stripes were on the order of the size of a single SC, cells could fill in and align within the spaces more efficiently and overall alignment was enhanced.
The relatively low alignment of HUVECs achieved in this study may indicate a lower capacity for alignment to protein tracks by these cells, or may suggest that the optimal conditions for aligning HUVECs were outside the range tested. For example, timelapse microscopy revealed that HUVECs aligned to stripes much earlier than A7s and SCs, so their optimal alignment may be achieved at an earlier timepoint than those studied here. While SCs initially aligned to the edges of stripes, optimally aligned HUVECs adhered and aligned onto the entire 10μm wide stripes and initially remained confined to the stripes. HUVECs then broadened by 24h to fill the spaces between the stripes.
The intermediately aligned A7 cultures exhibited the widest range of responses among the cell types to the range of factors tested, and had clear optimal conditions for achieving aligned, confluent monolayers. A7s adhered and aligned to the 50μm protein stripes, then filled in the 20 μm spaces between the stripes, taking the longest time of the three cell types to adhere and spread. Steric hindrance and/or cell-cell interactions likely allowed the A7s to fill in most effectively when the spaces were on the scale of cellular size. A7s differed from SCs and HUVECs in their dynamics, with a number of A7s moving in directions non-parallel to the protein patterns, most likely because they were larger than 20μm and could interact with more than one stripe at a time. Of note, A7s were the most closely packed together cell type at confluence, and required the highest plating density and the longest time in culture to achieve their confluence.
Our study of the generation of anisotropic monolayers of three cell types revealed general trends of importance across cell types. The spacing of the protein patterning was the most important. When cells initially adhered and aligned at borders and proliferated to fill in spaces (A7s or SCs), space between the protein stripes was most effective when it was on the order of cell width (20 μm for A7s and 10 μm for SCs). When cells adhered and aligned over the width of a stripe and broadened to fill spaces (HUVECs), space width about half the size of the cell was most effective. Some cell types appeared to have an aligned morphology that was not dependent on stripe width (A7s and SCs), but the aligned shape of other cells was strongly influenced by the pattern on which they were aligned (HUVECs). Maximizing the area of adhesive molecule coverage was also important for confluence unless cells were large enough to broaden to fill spaces between the stripes. Taken together, these results suggest that if the mechanism of alignment is predetermined and the cell size determined, the optimal size of the micropatterning for aligned monolayers of other cell types can be predicted.
Comparisons to Single Factor Approaches
Several of these culture parameters have also been studied previously, but typically in one-at-a-time approaches. Vartanian et al. studied the effect of culture time on baboon carotid artery ECs plated at 50,000 cells cm−2 on 25 μm collagen I stripes with 100 μm spaces and found that alignment increased from 0 to 24h, but was not changed between 24 and 48h 14. While culture time was not a significant contributing factor to HUVEC alignment in the present study, our choice of FN may have provided a stronger molecule for alignment of these cells. Feinberg et al. also studied the effect of pattern dimension on the alignment of primary porcine vascular ECs and found that while ECs did not align to 5 μm stripes of FN with 5 μm spaces, they did align to 50 μm stripes and spaces 13. The smaller stripe and space sizes may have been too small to be detected by ECs which are larger than 5 μm. In comparison, HUVECs aligned to stripes tested in our study, where the smallest features tested were larger than this threshold. Wu et al. studied the effect of FN stripe width on the alignment of HUVECs and found a similar trend to our results; as width decreased from 60 to 15 μm, alignment increased15. They also found that cell area decreased, the length of the short axis of the cell decreased, and apoptosis increased. While we did not test for apoptosis directly, HUVECs were plated at confluence for many of our conditions, and it was clear from timelapse microscopy that not all of the cells survived. Similar to the Wu study, our cells were also qualitatively less broad on micropatterns than on flat substrates.
Schmalenberg and Uhrich studied the effects of pattern dimensions and culture time on SC alignment and found that there was no difference in the alignment of SCs plated at 100K cellscm−2 at 4h and at confluence and that pattern spacing of 40 μm did not direct alignment as well as spacing of 20 and 30 μm 26. This trend also agrees with our finding that decreased space width increases alignment at confluence. Thompson and Buettner also studied the effects of pattern dimension, plating density, and culture time on SC alignment, but not in a multi-factorial approach. They employed micropatterns with LN stripe width ranging from 20–30 μm and space width ranging from 2–20 μm and found no significant effect on the degree of orientation 25. Our study supports these findings as we also showed that stripe width was not a significantly contributing factor to SC alignment. Our study also extends this work since by testing a wider range of space widths we were able to determine an optimal space width (10 μm). They found that seeding density had no effect on alignment at 6 and 24 h 24, while in longer cultures, alignment dipped at 48h but increased by 120h 25. Of note, though our starting densities were higher, we also found that 48h was our optimal culture time for SC alignment. With time lapse microscopy, Thompson and Buettner found that most SC elongation occurred in the first 12h of culture, and cells were motile throughout the experiment, continuing to move up and down the pattern even after they were completely aligned. Crossing to neighboring stripes was also noted, although crossing behavior diminished following alignment. Our timelapse results for SCs were similar despite the differences in plating densities.
Versatility of Response Surface Methodology
RSM of confluence and alignment of the three cell types also allows the possibility for the alignment and/or terminal cell density of the cell types investigated to be matched. To determine the conditions by which cultures could be generated in which the three cell types have similar alignments, the same input parameters were applied, but rather than maximizing the mean vector for each cell type, the mean vector length was set to 0.373, the optimal value for HUVECs. The predicted conditions are shown in Table 5.4. The optimal HUVEC value is outside of the range of possible SC values predicted by the model in the range of conditions tested. Minimization of the alignment of SCs in the range of conditions tested toward matching the optimal alignment of the HUVECs predicts a mean vector length of 0.659 ± 0.011. Of note, because SCs align more strongly than HUVECs to the tested micropatterns, matching mean vector lengths cannot be achieved in this range of conditions.
This model can also be used to predict the alignment and confluence of cells cultured outside the range of the conditions tested. For example the bounds can be expanded such that the space and stripe width must be between 1 and 100 μm, the starting cell density must be between 0K and 100K cells cm−2, the culture time must be between 0 and 72h for A7s and HUVECs and be between 0 and 96h for SCs, the mean vector length of nuclear angles must be maximized, and the cell number must be between 85–125K cells cm−2 for A7s, 32–50K cells cm−2 for HUVECs, and 41–69K cells cm−2 for SCs. The predictions under these conditions are presented in Table 5.5. For A7 and SC, there were multiple answers with equal desirability. These combinations can be found in supplementary materials.
Conclusion
In conclusion, the formation of aligned monolayers of three cell types was optimized by response surface methodology as a first step toward generation of anisotropic tissues and biomimetic substrates for the study of nerve guidance. A strength of biomedical engineering approaches is the combination of design and problem-solving techniques of engineers with the multifaceted experiments of biological sciences. The problem-solving power of DOE and RSM tools has been well recognized in chemical and pharmaceutical engineering; here we have demonstrated the benefits of applying these powerful tools to biomedical engineering areas such as tissue engineering and biomaterials. Current approaches to biological problems that require large number of variables can suffer from the expense of large amounts of time and resources, ignore interactions between inputs, and importantly, are not always well equipped to determine the relative contributions of inputs. The approaches utilized in this study offer a more efficient, economical, informative, and rigorous way to probe cellular responses to various and multiple factors. Finally, this study has also provided insights into the general mechanisms of cellular adhesion, alignment, and monolayer formation–key events that underlie the development of cell-and biomaterial-based therapeutics.
Supplementary Material
Acknowledgments
The authors would like to thank Christina Johnson and Grace Li for helpful discussion of the initial experimental set-up. They would also like to thank Carmichael Ong, Liane Livi, Cameron Rementer, and Jesse Thon for assistance with imaging and analysis. Si wafers were fabricated at the BioMEMS Resource Center with the generous assistance of Octavio Hurtado. This work was funded by an NSF Career Award and NIH R01 EB005722-01A2 to DHK and an AAUW Selected Professions dissertation fellowship to CK.
References
- 1.Schmidt A, Brixius K, Bloch W. Endothelial precursor cell migration during vasculogenesis. Circ Res. 2007;101:125–36. doi: 10.1161/CIRCRESAHA.107.148932. [DOI] [PubMed] [Google Scholar]
- 2.Tomanek RJ. Formation of the coronary vasculature during development. Angiogenesis. 2005;8:273–84. doi: 10.1007/s10456-005-9014-9. [DOI] [PubMed] [Google Scholar]
- 3.Wood A, Thorogood P. An ultrastructural and morphometric analysis of an in vivo contact guidance system. Development. 1987;101:363–381. [Google Scholar]
- 4.Sabourin LA, Rudnicki MA. The molecular regulation of myogenesis. Clinical genetics. 2000;57:16. doi: 10.1034/j.1399-0004.2000.570103.x. [DOI] [PubMed] [Google Scholar]
- 5.Singer M, Nordlander RH, Egar M. Axonal guidance during embryogenesis and regeneration in the spinal cord of the newt: The blueprint hypothesis of neuronal pathway patterning. J Comp Neurol. 1979;185:1–21. doi: 10.1002/cne.901850102. [DOI] [PubMed] [Google Scholar]
- 6.Gomes FC, Spohr TC, Martinez R, Moura Neto V. Cross-talk between neurons and glia: Highlights on soluble factors. Braz J Med Biol Res. 2001;34:611–20. doi: 10.1590/s0100-879x2001000500008. [DOI] [PubMed] [Google Scholar]
- 7.Silver J, Edwards MA, Levitt P. Immunocytochemical demonstration of early appearing astroglial structures that form boundaries and pathways along axon tracts in the fetal brain. The Journal of Comparative Neurology. 1993;328:415–436. doi: 10.1002/cne.903280308. [DOI] [PubMed] [Google Scholar]
- 8.Makita T, Sucov HM, Gariepy CE, Yanagisawa M, Ginty DD. Endothelins are vascular-derived axonal guidance cues for developing sympathetic neurons. Nature. 2008;452:759–63. doi: 10.1038/nature06859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pettigrew DB, Crutcher KA. White matter of the cns supports or inhibits neurite outgrowth in vitro depending on geometry. J Neurosci. 1999;19:8358–66. doi: 10.1523/JNEUROSCI.19-19-08358.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bunge RP, Fernandez-Valle C. The biology of schwann cells. In: Kettenmann H, Ransom BR, editors. Neuroglia. Vol. 44. Oxford University Press; Oxford: 1995. p. 57. [Google Scholar]
- 11.Nerem RM, Levesque MJ, Cornhill JF. Vascular endothelial morphology as an indicator of the pattern of blood flow. J Biomech Eng. 1981;103:172–6. doi: 10.1115/1.3138275. [DOI] [PubMed] [Google Scholar]
- 12.Malek AM, Izumo S. Mechanism of endothelial cell shape change and cytoskeletal remodeling in response to fluid shear stress. J Cell Sci. 1996;109(Pt 4):713–26. doi: 10.1242/jcs.109.4.713. [DOI] [PubMed] [Google Scholar]
- 13.Feinberg AW, Wilkerson WR, Seegert CA, Gibson AL, Hoipkemeier-Wilson L, Brennan AB. Systematic variation of microtopography, surface chemistry and elastic modulus and the state dependent effect on endothelial cell alignment. J Biomed Mater Res A. 2007 doi: 10.1002/jbm.a.31626. [DOI] [PubMed] [Google Scholar]
- 14.Vartanian KB, Kirkpatrick SJ, Hanson SR, Hinds MT. Endothelial cell cytoskeletal alignment independent of fluid shear stress on micropatterned surfaces. Biochem Biophys Res Commun. 2008;371:787–92. doi: 10.1016/j.bbrc.2008.04.167. [DOI] [PubMed] [Google Scholar]
- 15.Wu CC, Li YS, Haga JH, Kaunas R, Chiu JJ, Su FC, Usami S, Chien S. Directional shear flow and rho activation prevent the endothelial cell apoptosis induced by micropatterned anisotropic geometry. Proc Natl Acad Sci U S A. 2007;104:1254–9. doi: 10.1073/pnas.0609806104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276:1425–8. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 17.Alexander JK, Fuss B, Colello RJ. Electric field-induced astrocyte alignment directs neurite outgrowth. Neuron Glia Biol. 2006;2:93–103. doi: 10.1017/S1740925X0600010X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borgens RB, Shi R, Mohr TJ, Jaeger CB. Mammalian cortical astrocytes align themselves in a physiological voltage gradient. Exp Neurol. 1994;128:41–9. doi: 10.1006/exnr.1994.1111. [DOI] [PubMed] [Google Scholar]
- 19.Biran R, Noble MD, Tresco PA. Directed nerve outgrowth is enhanced by engineered glial substrates. Exp Neurol. 2003;184:141–52. doi: 10.1016/s0014-4886(03)00253-x. [DOI] [PubMed] [Google Scholar]
- 20.Deumens R, Koopmans GC, Den Bakker CG, Maquet V, Blacher S, Honig WM, Jerome R, Pirard JP, Steinbusch HW, Joosten EA. Alignment of glial cells stimulates directional neurite growth of cns neurons in vitro. Neuroscience. 2004;125:591–604. doi: 10.1016/j.neuroscience.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 21.Webb A, Clark P, Skepper J, Compston A, Wood A. Guidance of oligodendrocytes and their progenitors by substratum topography. J Cell Sci. 1995;108(Pt 8):2747–60. doi: 10.1242/jcs.108.8.2747. [DOI] [PubMed] [Google Scholar]
- 22.Recknor JB, Recknor JC, Sakaguchi DS, Mallapragada SK. Oriented astroglial cell growth on micropatterned polystyrene substrates. Biomaterials. 2004;25:2753–67. doi: 10.1016/j.biomaterials.2003.11.045. [DOI] [PubMed] [Google Scholar]
- 23.Miller C, Shanks H, Witt A, Rutkowski G, Mallapragada S. Oriented schwann cell growth on micropatterned biodegradable polymer substrates. Biomaterials. 2001;22:1263–9. doi: 10.1016/s0142-9612(00)00278-7. [DOI] [PubMed] [Google Scholar]
- 24.Thompson DM, Buettner HM. Schwann cell response to micropatterned laminin surfaces. Tissue Eng. 2001;7:247–65. doi: 10.1089/10763270152044125. [DOI] [PubMed] [Google Scholar]
- 25.Thompson DM, Buettner HM. Oriented schwann cell monolayers for directed neurite outgrowth. Ann Biomed Eng. 2004;32:1120–30. doi: 10.1114/b:abme.0000036648.68804.e7. [DOI] [PubMed] [Google Scholar]
- 26.Schmalenberg KE, Uhrich KE. Micropatterned polymer substrates control alignment of proliferating schwann cells to direct neuronal regeneration. Biomaterials. 2005;26:1423–30. doi: 10.1016/j.biomaterials.2004.04.046. [DOI] [PubMed] [Google Scholar]
- 27.Clark P, Connolly P, Curtis AS, Dow JA, Wilkinson CD. Topographical control of cell behaviour. I. Simple step cues. Development. 1987;99:439–48. doi: 10.1242/dev.99.3.439. [DOI] [PubMed] [Google Scholar]
- 28.den Braber ET, de Ruijter JE, Ginsel LA, von Recum AF, Jansen JA. Quantitative analysis of fibroblast morphology on microgrooved surfaces with various groove and ridge dimensions. Biomaterials. 1996;17:2037–44. doi: 10.1016/0142-9612(96)00032-4. [DOI] [PubMed] [Google Scholar]
- 29.Walboomers XF, Croes HJ, Ginsel LA, Jansen JA. Contact guidance of rat fibroblasts on various implant materials. J Biomed Mater Res. 1999;47:204–12. doi: 10.1002/(sici)1097-4636(199911)47:2<204::aid-jbm10>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 30.Dalby MJ, Riehle MO, Yarwood SJ, Wilkinson CD, Curtis AS. Nucleus alignment and cell signaling in fibroblasts: Response to a micro-grooved topography. Exp Cell Res. 2003;284:274–82. doi: 10.1016/s0014-4827(02)00053-8. [DOI] [PubMed] [Google Scholar]
- 31.El-Malah Y, Nazzal S, Khanfar NM. D-optimal mixture design: Optimization of ternary matrix blends for controlled zero-order drug release from oral dosage forms. Drug Development and Industrial Pharmacy. 2006;32:1207–1218. doi: 10.1080/03639040600685167. [DOI] [PubMed] [Google Scholar]
- 32.Kalil SJ, Maugeri F, Rodrigues MI. Response surface analysis and simulation as a tool for bioprocess design and optimization. Process Biochemistry. 2000;35:539–50. [Google Scholar]
- 33.Li X, Xu T, Ma X, Guo K, Kai L, Zhao Y, Jia X, Ma Y. Optimization of culture conditions for production of cis-epoxysuccinic acid hydrolase using response surface methodology. Bioresource Technology. 2008;99:5391–5396. doi: 10.1016/j.biortech.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 34.Lumor SE, Akoh CC. Enzymatic incorporation of stearic acid into a blend of palm olein and palm kernel oil: Optimization by response surface methodology. Journal of the American Oil Chemists’ Society. 2005;82:421–426. [Google Scholar]
- 35.David S, Aguayo AJ. Axonal elongation into peripheral nervous system “Bridges” After central nervous system injury in adult rats. Science. 1981;214:931–3. doi: 10.1126/science.6171034. [DOI] [PubMed] [Google Scholar]
- 36.Richardson PM, McGuinness UM, Aguayo AJ. Axons from cns neurons regenerate into pns grafts. Nature. 1980;284:264–5. doi: 10.1038/284264a0. [DOI] [PubMed] [Google Scholar]
- 37.Glasby MA, Gschmeissner SG, Hitchcock RJI, Huang CLH. The dependence of nerve regeneration through muscle grafts in the rat on the availability and orientation of basement membrane. Journal of Neurocytology. 1986;15:497–510. doi: 10.1007/BF01611732. [DOI] [PubMed] [Google Scholar]
- 38.Keynes RJ, Hopkins WG, Huang LH. Regeneration of mouse peripheral nerves in degenerating skeletal muscle: Guidance by residual muscle fibre basement membrane. Brain Res. 1984;295:275–81. doi: 10.1016/0006-8993(84)90976-4. [DOI] [PubMed] [Google Scholar]
- 39.Hall S. Axonal regeneration through a cellular muscle grafts. Journal of Anatomy. 1997;190:57–71. doi: 10.1046/j.1469-7580.1997.19010057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson PN, Turmaine M. Axonal regeneration through arterial grafts. Journal of Anatomy. 1986;147:73. [PMC free article] [PubMed] [Google Scholar]
- 41.Chiu DT, Janecka I, Krizek TJ, Wolff M, Lovelace RE. Autogenous vein graft as a conduit for nerve regeneration. Surgery. 1982;91:226–33. [PubMed] [Google Scholar]
- 42.Foidart-Dessalle M, Dubuisson A, Lejeune A, Severyns A, Manassis Y, Delree P, Crielaard JM, Bassleer R, Lejeune G. Sciatic nerve regeneration through venous or nervous grafts in the rat. Experimental Neurology. 1997;148:236–246. doi: 10.1006/exnr.1997.6542. [DOI] [PubMed] [Google Scholar]
- 43.Li Y, Sauve Y, Li D, Lund RD, Raisman G. Transplanted olfactory ensheathing cells promote regeneration of cut adult rat optic nerve axons. Journal of Neuroscience. 2003;23:7783–7788. doi: 10.1523/JNEUROSCI.23-21-07783.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hofstetter CP, Schwarz EJ, Hess D, Widenfalk J, El Manira A, Prockop DJ, Olson L. Marrow stromal cells form guiding strands in the injured spinal cord and promote recovery. Proc Natl Acad Sci U S A. 2002;99:2199–204. doi: 10.1073/pnas.042678299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brook GA, Lawrence JM, Shah B, Raisman G. Extrusion transplantation of schwann cells into the adult rat thalamus induces directional host axon growth. Exp Neurol. 1994;126:31–43. doi: 10.1006/exnr.1994.1040. [DOI] [PubMed] [Google Scholar]
- 46.Recknor JB, Sakaguchi DS, Mallapragada SK. Growth and differentiation of astrocytes and neural progenitor cells on micropatterned polymer films. Ann N Y Acad Sci. 2005;1049:24–7. doi: 10.1196/annals.1334.004. [DOI] [PubMed] [Google Scholar]
- 47.Dubey N, Letourneau PC, Tranquillo RT. Guided neurite elongation and schwann cell invasion into magnetically aligned collagen in simulated peripheral nerve regeneration. Experimental Neurology. 1999;158:338–350. doi: 10.1006/exnr.1999.7095. [DOI] [PubMed] [Google Scholar]
- 48.Thompson DM, Buettner HM. Neurite outgrowth is directed by schwann cell alignment in the absence of other guidance cues. Ann Biomed Eng. 2006;34:161–8. doi: 10.1007/s10439-005-9013-4. [DOI] [PubMed] [Google Scholar]
- 49.Walsh JF, Manwaring ME, Tresco PA. Directional neurite outgrowth is enhanced by engineered meningeal cell-coated substrates. Tissue Eng. 2005;11:1085–94. doi: 10.1089/ten.2005.11.1085. [DOI] [PubMed] [Google Scholar]
- 50.Stepien E, Stanisz J, Korohoda W. Contact guidance of chick embryo neurons on single scratches in glass and on underlying aligned human skin fibroblasts. Cell Biol Int. 1999;23:105–16. doi: 10.1006/cbir.1998.0256. [DOI] [PubMed] [Google Scholar]
- 51.Bruder JM, Lee AP, Hoffman-Kim D. Biomimetic materials replicating schwann cell topography enhance neuronal adhesion and neurite alignment in vitro. Journal of Biomaterials Science. (Polymer) 2007;18:967–982. doi: 10.1163/156856207781494412. [DOI] [PubMed] [Google Scholar]
- 52.Croarkin C, Tobias P, Zey C S. International, S. National Institute of and Technology. Engineering statistics handbook. The Institute; International SEMATECH; 2001. [Google Scholar]
- 53.Falconnet D, Csucs G, Michelle Grandin H, Textor M. Surface engineering approaches to micropattern surfaces for cell-based assays. Biomaterials. 2006;27:3044–3063. doi: 10.1016/j.biomaterials.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 54.Li GNY, Hoffman-Kim D. Evaluation of neurite outgrowth anisotropy using a novel application of circular analysis. Journal of Neuroscience Methods. 2008 doi: 10.1016/j.jneumeth.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci U S A. 1997;94:849–54. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yim EK, Reano RM, Pang SW, Yee AF, Chen CS, Leong KW. Nanopattern-induced changes in morphology and motility of smooth muscle cells. Biomaterials. 2005;26:5405–13. doi: 10.1016/j.biomaterials.2005.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.