Abstract
DNA-dependent protein kinase (DNA-PK) is a key non-homologous end joining (NHEJ) nuclear serine/threonine protein kinase involved in various DNA metabolic and damage signaling pathways contributing to the maintenance of genomic stability and prevention of cancer. In order to examine the role of DNA-PK in processing of non-DSB clustered DNA damage, we have used three different models of DNA-PK deficiency i.e. chemical inactivation of its kinase activity by novel inhibitors IC86621 and NU7026, knock-down and complete absence of the protein in human breast cancer (MCF-7) and glioblastoma cell lines (MO59-J/K). Compromised DNA-PK repair pathway has lead to accumulation of clustered DNA lesions induced by γ-rays. Tumor cells lacking protein expression or with inhibited kinase activity showed a marked decrease in their ability to process oxidatively-induced non-DSB clustered DNA lesions measured using a modified version of pulsed field gel electrophoresis or single cell gel electrophoresis (Comet assay). In all cases, DNA-PK inactivation lead to a higher level of lesion persistence even after 24–72 hrs of repair. We suggest a model in which DNA-PK deficiency affects the processing of these clusters by first compromising base excision repair and second by the presence of catalytically inactive DNA-PK inhibiting the efficient processing of these lesions due to the failure of DNA-PK to disassociate from the DNA ends. The information rendered will be important not only for understating cancer etiology in the presence of a NHEJ deficiency but also lead to a better understanding of cancer treatments based on the induction of oxidative stress and inhibition of cluster repair.
Keywords: Oxidative clustered DNA lesions, DNA-PKcs, cancer, DNA damage, γ-H2AX
Introduction
Oxidative stress resulting from the production of reactive oxygen species (ROS) can induce a plethora of DNA lesions in the form of single and clustered DNA damage [1, 2]. The high density of DNA lesions forming clusters poses a significant challenge to the cellular repair machinery. Persistence of any type of DNA damage can lead to mutations and subsequent transformation and carcinogenesis and other pathophysiological conditions [3, 4]. Clustered DNA lesions are divided in two major groups, double-stranded breaks (DSBs) and non-DSB oxidatively-induced clustered DNA lesions (OCDLs). DSBs are considered highly genotoxic because they potentially lead to chromosomal breakage if unrepaired [5]. DSBs can arise directly after exposure to exogenous sources like ionizing radiation (IR) and endogenously either by free radicals that emerge as byproducts of normal cellular metabolism or as repair intermediates during the processing of OCDLs [6]. Even in the case that DSBs do not form during the attempted repair of clusters, studies have shown a significantly delayed processing of these lesions [7, 8] and enhanced mutation frequency [9]. Two major pathways have been implicated in the processing of DSBs: non-homologous end joining (NHEJ) and homologous recombination repair (HR) [10]. Compromised NHEJ repair favors the induction of HR proteins such as BRCA1 and the processing of DSBs by HR, evidence of interplay between these two repair pathways [11]. A key protein complex in the NHEJ pathway is the DNA-PK holoenzyme consisting of the Ku70/80 heterodimer and the catalytic subunit of DNA-PK (DNA-PKcs) [12]. The pivotal role of DNA-PKcs in processing of DSBs has been shown by several studies that have demonstrated a severe radiosensitivity and decreased DSB repair in cells with compromised DNA-PKcs activity or reduced expression of the protein [13] and telomere dysfunction [14]. In addition, the heterodimeric component of DNA-PK holoenzymee Ku70/80 (Ku) complex has been shown to play a significant role in the repair of closely opposed base lesions in DNA [15].
Polymorphisms of the Prkdc gene (encoding DNA-PKcs) result in decreased activity and/or expression of DNA-PKcs and have been associated with elevated risk for breast [16, 17], lung [18], gastric [19], colon [20] and cervical [21] cancer. Reduced DNA-PKcs levels have been detected in nuclear cortical extracts from brains of Alzheimer’s disease (AD) patients [22]. Hippocampal neurons from severe combined immunodeficient (scid) mice lacking DNA-PK activity have been found extremely susceptible to various damaging agents and oxidative stress [23]. DNA-PKcs has also been suggested to be a potential breast cancer susceptibility gene [24]. Knock-down of DNA-PKcs by siRNA has been shown to parallel the effects of reduced expression of ATM and Artemis [25], two key proteins in the processing of DSBs. Other studies have shown that cells with compromised DNA-PKcs may utilize an alternative (DNA-PKcs independent) but “error-prone” and slower DSB repair pathway involving the Mre11-Rad50-NBS1 complex [26]. Recently DNA-PKcs were shown to interact with traditional BER proteins including XRCC1 [27], APE1 and Polβ [28] implying DNA-PKcs may play a role in the processing of single oxidative DNA lesions. In fact, our preliminary studies showed defective repair of non-DSB clustered lesions in MCF-7 cells with partial DNA-PKcs deficiency [29].
To examine the role of DNA-PKcs in the processing of OCDLs we studied the effect of chemically-induced inactivation of DNA-PKcs or its absence in the repair of OCDLs. Both ‘phenotypes’ (i.e., absent or inactive DNA-PKcs) lead to a significant decrease in clustered lesion processing and enhanced cell death. The specific accumulation of clustered lesions also leads to the persistence of single DNA lesions.
Materials and Methods
Cell culture, irradiation and statistical analysis
Human breast cancer MCF-7 cells were purchased from Tissue Culture Collection (ATCC, Manassas, VA) while isogenic MO59J/K were a kind gift of Dr. Joan Turner (Cross Cancer Institute, Edmonton, AB) and the Alberta Cancer Board. M059-J/K cells were grown in DMEM (Gibco) supplemented with 10% Standard FBS (Atlanta Biologicals) while MCF-7 as previously described [30]. Cells were treated with γ-rays (5 Gy) or 100 µM H2O2 (10 min at room temperature) as previously described [30, 31]. At these moderate H2O2 concentrations [32], negligible induction of clustered DNA lesions is expected and therefore only single (non-clustered) DNA lesions (base damage and SSBs) are induced as also previously showed for NALM-6 cells [31]. Preliminary experiments revealed also for MCF-7 cells a similar trend where the levels of Fpg- and EndoIII-clusters were very close to background levels (data not shown).
Statistical analysis of the data
Paired Student’s t-tests were used to evaluate the differences between averages of the different groups (p<0.05).
siRNA Transfection and drug treatment
Silencing of the Prkdc gene in MCF-7 cells and IC86621 treatment was performed as detailed in the Supplemental Data. In order to eliminate the possibility of off-target effects an additional highly specific DNA-PK inhibitor was used (NU7026) [33]. The optimal concentration of siRNAs was found to be 0.1 µM and, under the specified conditions, a reduction in DNA-PKcs expression of ~85% was achieved [29]. MCF-7 cells were treated with either 100 µM IC86621 (Sigma) or 10 µM NU7026 (Sigma) in the growth medium for 24 h or 1 h respectively. Control (medium containing only DMSO) and drug-treated cells were γ-irradiated and allowed to repair under the drug presence. Cells at indicated post-irradiation repair time points were harvested and either processed for immunofluorescence or for damage measurement using pulsed field gel electrophoresis (PFGE).
Immunofluorescence and immunoblotting
Assays for the detection of DNA-PKcs or γ-H2AX in MCF-7 cells are analytically described in the Supplemental Data. For the γ-H2AX analysis in M059J/K cells, the procedure described in [34] was followed. Detection of XRCC1 was performed using standard Western blotting [29]. Forty (40) µg of whole cell protein were mixed with an equal quantity of 2X SDS buffer (2.5% SDS, 5.0 % β-mercaptoethanol), boiled for 10 minutes and placed on ice. Protein was loaded onto a 4–20% Tris-HCl gradient gel (BioRad) along with 5 µl of Kaleidoscope marker (BioRad, Hercules, CA). Blots were incubated with XRCC1 mouse monoclonal antibody IgG (Abcam, ab1838), secondary goat anti-mouse IgG-HRP (sc-2005) from Santa Cruz Biotech. Inc. Chemiluminescence with SuperSignal West Dura (Pierce) and FluorChem 8900 visualization system (Alpha Innotech).
Single DNA lesion detection using alkaline single cell gel electrophoresis (SCGE)
For the measurement of total number of single DNA lesions using single cell gel electrophoresis (Comet assay) we have used a novel adaptation of the method originally described by Visvardis et al. [35]. This adaptation involves measurement of DNA damage in human DNA agarose plugs similar to the PFGE assay. Single DNA damage was induced by 100 µM H2O2. An analytical description of the procedure is presented under Supplementary Data.
OCDL detection using Pulsed Field Gel Electrophoresis (PFGE)
Cells at each time point were harvested by immersion in liquid nitrogen, retrieved and embedded into low melting point agarose (Biorad, Hercules CA) plugs (~250,000 cells/plug) as analytically described in Tsao et al. [36]. Lysis and preparations of human DNA was performed as previously described [29, 30]. For the detection of DSBs and OCDL an adaptation of PFGE was used with different E. coli repair enzymes as damage probes (Fpg, Endo III and Endo IV). Calculation of DSBs and OCDL was performed using number average length analysis (NALA) [36].
Apoptosis detection
Detection of apoptosis was performed as previously described using the Annexin V FITC Apoptosis detection Kit (Calbioshem, San Diego, CA) and fluorescence microscopy [29]. Apoptosis is expressed as the percentage (%) of Annexin V positive cells.
Results
Measurement of DSB repair in three types of DNA-PKcs deficient cells
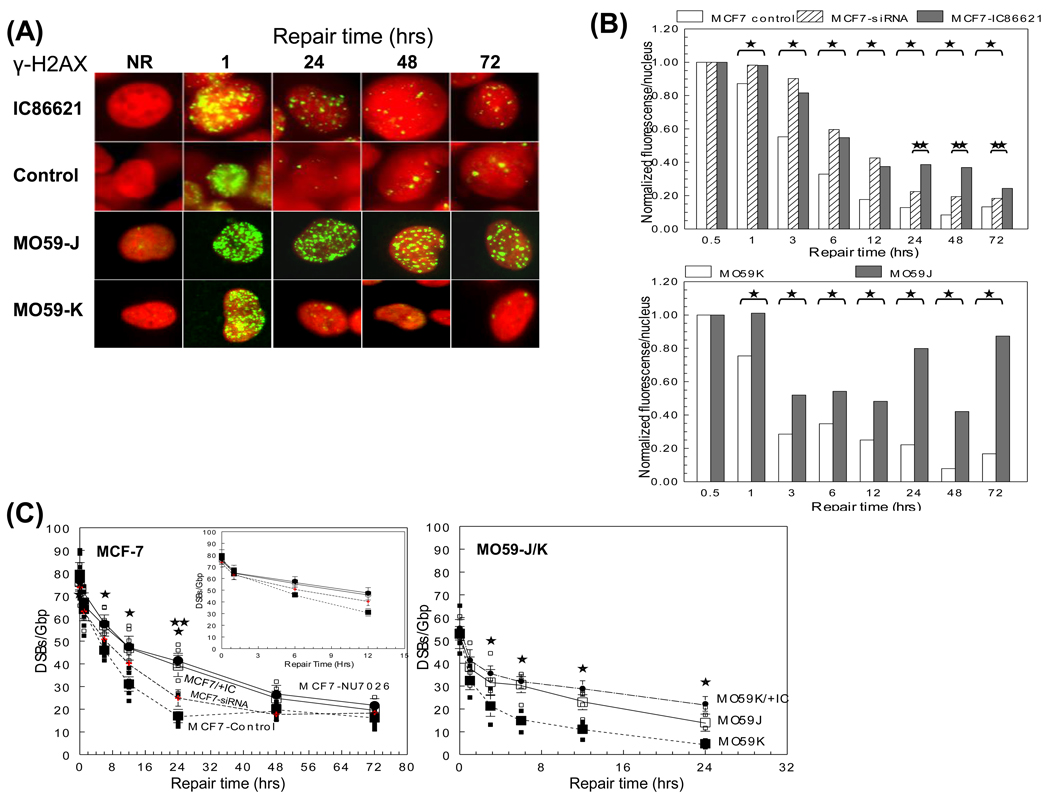
DNA-PKcs is a major component of NHEJ and DSB repair. In order to determine if any qualitative or quantitative differences arise as a result of DNA-PKcs deficiency, three different repair deficient cell models were used. MCF-7 or MO59-K cancer cells were examined in the presence or absence of the specific DNA-PKcs kinase inhibitors IC86621/NU7026 or after siRNA knock-down of DNA-PKcs. MO59-J cells completely lack DNA-PKcs expression and therefore are significantly less efficient in DSB repair, in contrast with their isogenic MO59-K, due to a frameshift mutation [37]. Two independent assays were utilized to compare these model systems, the γ-H2AX focus formation assay and an adaptation of pulsed field gel electrophoresis (PFGE) (Fig. 1). MCF-7 or MO59-K cells exposed to 5 Gy IR in the presence of the drug (IC86621 or NU7026) showed as expected significant DSB persistence compared to control cells over time. Measurements made indirectly using γ-H2AX foci total intensity (Fig. 1A–B) or directly using PFGE (Fig. 1C) indicated a serious DNA repair defect. Extended repair times up to 72 hrs also revealed a significant persistence of γ-H2AX foci and dramatically delayed repair kinetics for M059-J cells compared to controls (p<0.05). In control MCF-7 and M059-K cells, γ-H2AX foci levels returned to background values after 48 hrs. However, both drug-treated and DNA-PKcs deficient cells retained ~40% of initial DNA damage. Comparison with siRNA-MCF-7 cells showed differences only at 24–72 hrs. Fig. 1C depicts the DSBs remaining after exposure to 5 Gy as a function of post-irradiation time for all cell lines assessed using PFGE. For repair times up to 24 hrs, the remaining number of DSBs was always higher for NU7026-treated MCF-7 and IC86621-treated MO59-K (MO59K/+IC) cells compared to controls (significantly different only at 3, 6, 12 and 24 hrs post-IR, p<0.05). Although no statistical difference was found between MO59J and MO59K/+IC cells, a higher accumulation of DSBs was always found for the drug-treated cells. Comparison between NU7026 and IC86621 drugs (only for MCF-7) showed no real difference while comparison of both drugs to siRNA showed no significant differences with the exception of 24 hrs. The residual DSBs present 24 hrs post irradiation in all experimental cell lines indicate incomplete rejoining of DNA damage in the absence of DNA-PKcs. Quantitative comparison between the two assay methods employed showed some differences which can be attributed to the increased sensitivity of the γ-H2AX assay compared to PFGE [38].
Fig. 1.
Processing of DSBs in DNA-PKcs proficient and deficient cell lines (IC86621-, NU7026-, siRNA-treated MCF-7 and MO59-K, MO59-J and MO59-K) as a function of post-irradiation time. (A) Representative microscopic fluorescence images of γ-H2AX foci accumulation after 5 Gy IR at times (1, 24, 48 and 72 hrs). Non-irradiated samples (NR) are also shown. Nuclei were stained using either propidium iodide PI (MCF-7-top) or DAP1 (MO59-J/K-btm) (B) Quantitation of the normalized total mean γ-H2AX fluorescence intensity expressed as intensity/nucleus (% of maximum value at 1 hr). (C) Detection of DSB repair using PFGE analysis. Values for DNA-PK siRNA cells (up to 24 hrs) are taken from Peddi et al. [29] to allow direct comparison while for 48 and 72 hrs are from this study (crosses). Values are averages from three (MCF-7) or two (MO59-J/K) independent experiments. Inset: Average values for MCF-7 with repair times up to 12 hr. Small symbols, individual data points from independent irradiation experiments; Large symbols, averages. Error bars, SEM; in some cases are smaller than the corresponding symbol. Closed symbols, control MCF-7 or MO59-K cells. Closed large circles, NU7026-treated MCF-7 or IC86621-treated MO59-K (MO59K/+IC). Open symbols, IC86621-treated MCF-7 cells or M059-J. Statistically significant differences between IC86621-/NU7026-MCF-7/MO59-K and controls (*) and IC86621-/NU7026-MCF-7 and siRNA-MCF-7 (**) at p<0.05 are shown.
Measurement of OCDLs repair in three types of DNA-PKcs deficient cells
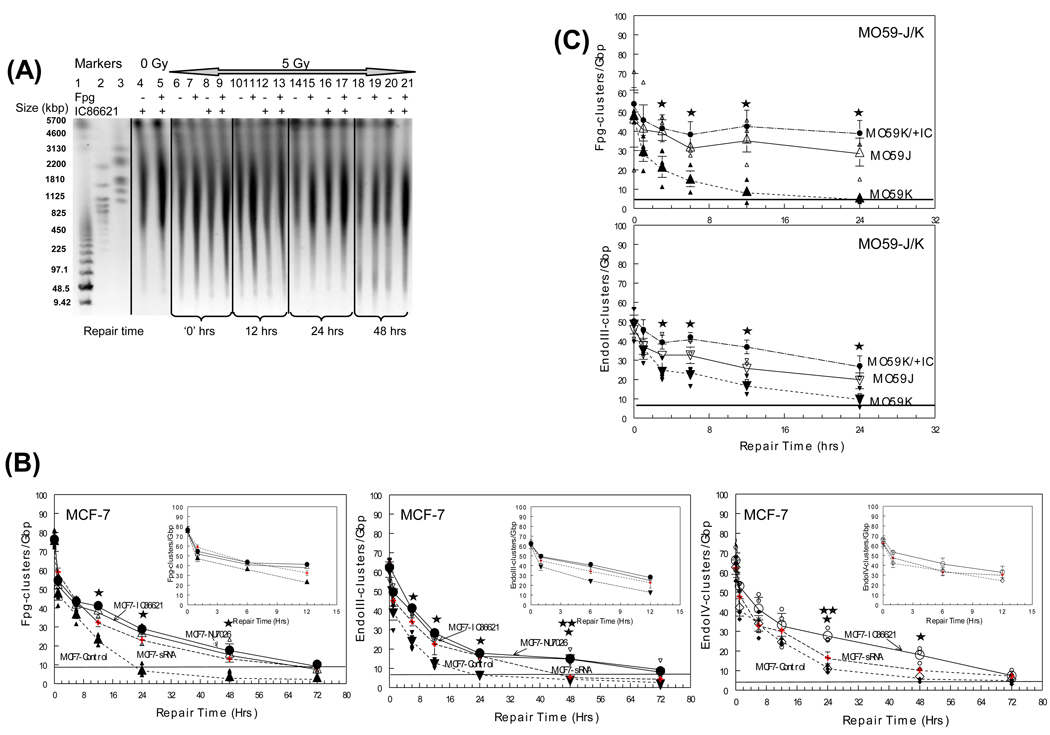
As DNA-PKcs seems to play an important role in the processing of DNA DSBs, we next examined the role of DNA-PKcs in the repair of OCDLs. In Fig. 2A, segments of a PFGE gel containing DNA isolated from sham-irradiated IC86621-treated cells (lanes 4–5), as well as from cells exposed to 5 Gy (+/− IC86621) and then incubated at 37°C to allow repair are shown. The 0, 12, 24 and 48 hr time points are shown (lanes 6–21). DSB rejoining is measured by electrophoresing DNA without the enzyme (–, lanes). Cells incubated at 37°C resealed DSBs, observed here as an increase in DNA size from 0 to 48 hrs (lanes 6, 10, 14 and 18). The incubation of the non-irradiated samples with enzymes alone results in a very minimum increase in DNA fragmentation and reduction of DNA size (lanes 4–5). This correlates with the expected low level of endogenous damage clusters in the absence of IR. Treatment of the irradiated samples (5 Gy) with the enzyme results in a significant increase in small DNA fragments, particularly in the Mbp size range, that persists even after 48 hrs (lanes 18–21). This is indicative of the higher levels of oxidative lesions upon DNA irradiation. Quantitation for all cluster types and cell lines are presented in Figures 2B–C. Significant differences were detected for drug-treated MCF-7, MO59K/+IC and M059-J cells similar to results obtained when DSB repair was monitored (6–48 hrs, p<0.05). Again and as for DSBs, no significant difference was found between MO59-J and drug-treated MO59-K cells but cluster accumulation was always higher for drug-treated cells compared to MO59-J cells. Comparison of IC86621- or NU7026-treated MCF-7 cells with siRNA-treated cells did not reveal any significant differences with the exception of abasic clusters detected by Endo IV at 24 hrs and oxypyrimidine clusters detected by EndoIII at 48 hrs. In all cases OCDL levels for drug treated cells did not return to background levels before 72 hrs of repair. MO59J cells showed a similar trend with high levels of residual (~40% of initial values) clustered lesion damage even after 24 hrs of repair.
Fig. 2.
Processing of non-DSB oxidative DNA clusters in DNA-PKcs proficient and deficient cell lines (IC86621-, NU7026-, siRNA-treated MCF-7 and MO59-K, MO59-J and MO59-K) as a function of post-irradiation time. Different types of damage clusters were detected using PFGE analysis and E. coli repair enzymes Fpg and EndoIII as damage probes: (A) Representative image of a neutral gel containing DNA from MCF-7 cells unirradiated (0 Gy) and exposed to 5 Gy of γ rays, and harvested as a function of repair time post-exposure. Molecular size markers, lanes 1–3: S. pombe and low range PFG marker, lane 1; H. wingei, lane 3; S. cerevisae, lane 3. The molecular sizes of these DNAs are shown in kbp at the left of the gel image; 0 Gy, lanes 4–5; 5 Gy at increasing repair times (0–72 hrs), lanes 6–21. For each time point four samples are shown: enzyme or IC86621 untreated, (−); Fpg- or IC86621-treated, (+). (B–C) Cluster repair detected in MCF-7 and MO59-J/K cells. Fpg-detected oxypurine clusters, upright triangles. Endo III-detected oxypyrimidine clusters, inverted triangles. Endo IV detected abasic sites, circles. Values for DNA-PK siRNA cells (up to 24 hrs) are taken from Peddi et al. [29] to allow direct comparison while for 48 and 72 hrs are from this study (crosses). Values are averages from three (MCF-7) or two (MO59-J/K) independent experiments. Insets: Average values for MCF-7 with repair times up to 12 hr. Small symbols, individual data points from independent irradiation experiments; Large symbols, averages. Error bars, SEM; in some cases are smaller than the corresponding symbol. Closed symbols, control MCF-7 or MO59-K cells. Closed large circles, NU7026-MCF-7 or IC86621-MO59K (MO59K/+IC). Open symbols, IC86621-treated MCF-7 cells or M059-J. Statistically significant differences between IC86621-/NU7026-treated MCF-7 or MO59-K (MO59K/+IC) and controls (*) and between IC86621-/NU7026-MCF-7 and siRNA-MCF-7 (**) at p<0.05 are shown. Straight lines correspond to sham-irradiated (0 Gy) average values for each type of cluster.
Measurement of single DNA lesions, phospho-Thr2609 DNA-PKcs foci and apoptosis
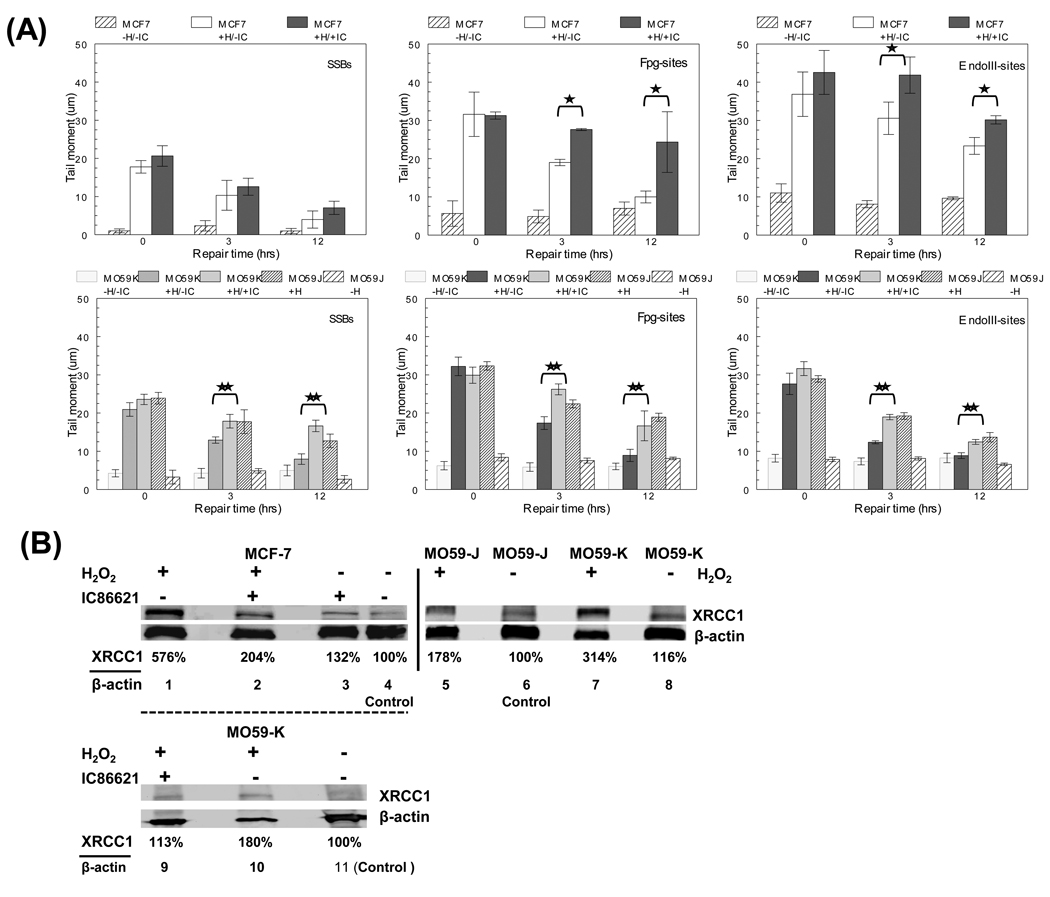
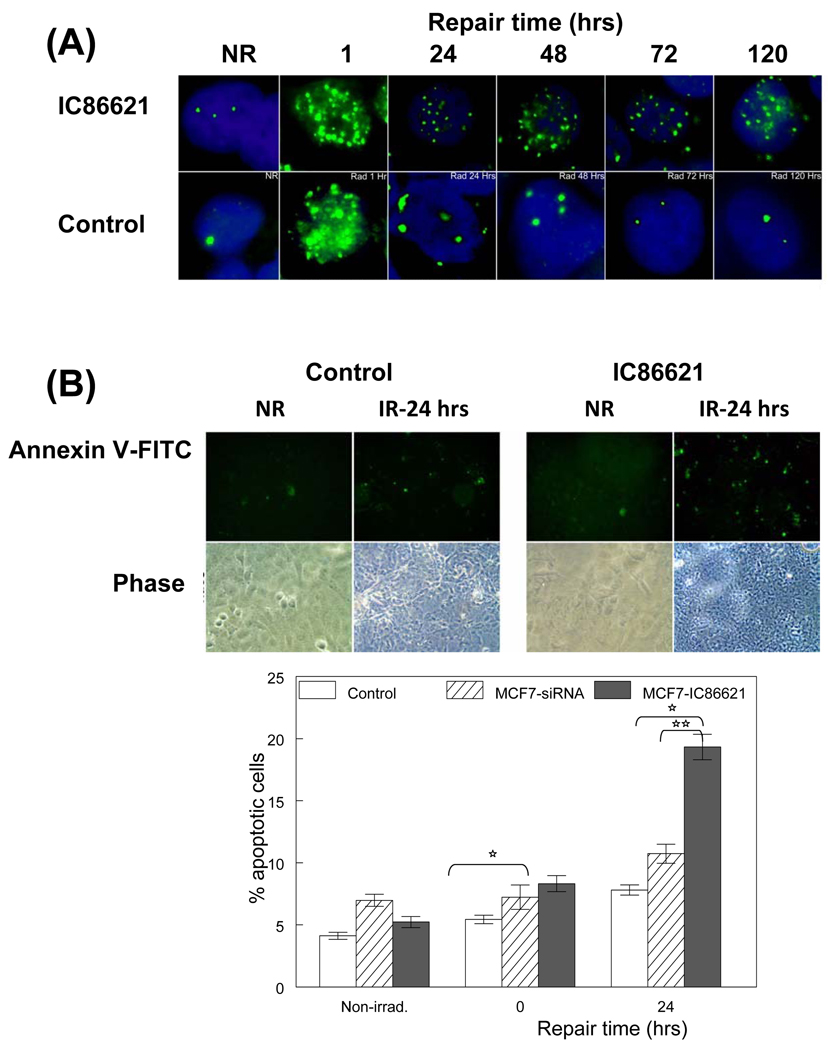
In order to assess the role of DNA-PKcs in the processing of single (not clustered) DNA lesions in MCF-7 and MO59-J/K cells after exposure to H2O2, we utilized alkaline single cell gel electrophoresis and examined the resulting tail moment (TM) in cells with or without IC88621 treatment. In addition, we performed parallel repair experiments for the MO59J/K cells after H2O2 and IC86621 treatment (for MO59-K). As shown in Figure 3A (top), drug treatment did not affect for MCF-7 the processing of simple DNA SSBs (compare white bars to dark grey bars). On the other hand, MO59J cells and MO59K/+IC were found to have a significant deficiency of processing SSBs compared to MO59K cells (**, p<0.05). However, treatment of MCF-7 or MO59-K cells with the enzymes Fpg or EndoIII resulted in an increased TM upon exposure to H2O2 and the repair of these oxidative lesions was significantly decreased in the presence of the DNA-PKcs inhibitor. MO59-J cells showed also a deficiency in the processing of Fpg- or EndoIII-sites but less pronounced compared to SSBs. Comparison between Fpg- and EndoIII-cleavage sites revealed a prevalence of EndoIII-sites at a ratio (EndoIII-sites/Fpg-sites) ~ 1.1–1.4 (‘0’ hrs) for all cell lines. These results indicate that oxypyrimidines were likely the predominant oxidative lesion induced by H2O2 treatment and in addition a possible impairment of BER. For this reason we measured the expression level of a key BER repair protein XRCC1 in MCF-7 and MO59-K cells after drug treatment (Fig. 3B). Inhibition of DNA-PKcs activity by drug treatment resulted in a significant reduction in the expression level of XRCC1 compared to no drug-treated cells after H2O2 induction suggesting a significant defect in the specific repair pathway consistent with the deficient processing of single DNA lesions (compare lanes 1 to 2 and 10 to 9). In addition, also in MO59J cells, XRCC1 expression was found reduced but in a less pronounced way (~44%) compared to MO59-K cells. The XRCC1 levels of basal expression (lanes 6 and 8) were also found reduced for MO59-J compared to MO59-K cells (Fig. 3A) as also suggested by Toulany et al. [39]. On the other hand the ‘basal levels for IC86621-treated MCF-7 or MO59-K cells were found slightly increased compared to controls (lanes 3–4 and 9–11). Treatment of MCF-7 cells with IC86621 not only resulted in deficient repair of OCDLs and DSBs but also in persistence of phosphorylated (Thr2609) DNA-PKcs foci over 120 hrs (5 days) of repair after 5 Gy IR (Fig. 4A and Figure S2). This is a strong indication of persistent DNA damage (DSBs or OCDLs). Finally, in order to investigate the cytotoxicity of residual DNA damage caused by DNA-PKcs inhibition, apoptosis was evaluated through Annexin V staining (Fig. 4B). Cell division and cell death was previously found to contribute to the disappearance of repair resistant oxidative DNA clustered lesions [7]. Since cell growth of IC86621-treated cells was found to be reduced ~10% compared to controls (72 hrs, data not shown), we measured apoptosis after exposure to 5 Gy in controls and drug-treated cells. A 2-fold greater incidence of apoptosis was detected for IC8861-treated cells 24 hrs post-irradiation. Cells treated with DNA-PKcs siRNA on the other hand did not display a significant increase in cell death. These results indicate that catalytic inhibition of DNA-PKcs is more deleterious to cells than partial knock-down of protein expression. Taken together, these data indicate that DNA-PKcs plays a fundamental and important role in the processing of a variety of oxidative DNA lesions including DSBs, abasic sites, and oxidized base damage.
Fig. 3.
Effect of DNA-PKcs inhibition in the processing of single oxidative DNA lesions and base excision repair (BER) pathway (A) Processing of total single DNA lesions in IC86621-treated MCF-7 or MO59-K (+H/+IC: H2O2 and IC86621-treated) and regular MO59J/K cells and controls after exposure for 15 min to 100 µM H2O2 (+H). Detection of SSBs as well as Fpg- and EndoIII-sites using alkaline single cell gel electrophoresis at three post-treatment repair times (‘0’, 3 and 12 hrs). The additional general control no H2O2/IC86621-treated samples has also been included (−H/−IC). Values are averages from two independent experiments. (B) Detection of XRCC1 in IC86621-treated MCF-7 cells 30 min after exposure to 100 µM H2O2 using Western blotting. Forty (40) µg of total protein were loaded/lane. Lanes 1–4, MCF-7 cells; lanes 5–8, M059J/K cells; lanes 9–11 MO59K cells. Lane 4, densitometry control for MCF-7 (DMSO); lanes 6 and 11, MO59J or MO59K (DMSO) with no H2O2 or drug treatment: densitometry controls for MO59J/K. β-actin used as load control. Densitometry values presented as the ratio of XRCC1/actin normalized to control samples. Values averages from two independent experiments. Statistically significant differences between IC86621-treated MCF-7 or MO59-K (MO59K/+IC) and controls (*,** respectively) at p<0.05 are shown.
Fig. 4.
Persistence of DNA-PKcs phosphorylated form and apoptosis in MCF-7 cells exposed to γ-rays. (A) phospho-Thr2609 DNA-PKcs foci in IC86621-treated MCF-7 and control cells after exposure to 5 Gy of γ rays. Cells were fixed at different post-irradiation repair times. Both control samples and IC86621-treated were immunostained with rabbit polyclonal to phospho-(Thr2609) DNA-PKcs antibody (green foci). DAPI was used to stain nucleus in all cells. The non-irradiated samples have been included (NR). The results shown here are representative of two independent experiments. (B) Detection of apoptosis using the Annexin V binding assay and fluorescence microscopy at 0 and 24 hrs post-irradiation. Fluorescent and phase pictures for irradiated IC86621-MCF-7 and control cells (IR) as well as non-irradiated (NR) are shown. Annexin V positive cells are green. Quantitation of 200 cells expressed as the percentage of positively stained cells is shown below the representative images. Values are averages from two independent experiments. Values for DNA-PK siRNA cells are taken from Peddi et al. [29] to allow direct comparison. Statistically significant differences between MCF7-IC86621 and controls (*) at p<0.05 are shown.
Discussion
Many cancer cells and tumors exhibit elevated levels of DSBs and OCDLs [40, 41]. OCDLs have the potential to be used as a cancer biomarker and predictive marker for treatment efficacy especially in the case of DSB-deficient tumors. Preliminary evidence from our laboratory suggested a possible involvement of DNA-PKcs in the repair of this type of non-DSB complex DNA damage [29]. Different laboratories have implicated NHEJ components in the repair of non-DSB oxidative clustered lesions [15, 42, 43]. DNA-PK activation has been shown by SSBs induced in plasmid DNA [44] or by the radiomimetic drug bleomycin [45] which induces both DSBs as well as non-DSB clustered DNA damage [46]. We used three different models of DNA-PKcs disruption in human cancer cells exposed to IR. These were the pharmacological (by using the highly specific drugs IC86621 and NU7026) and siRNA-targeted inhibition of DNA-PKcs, simulating the conditions of catalytically inactive (but present DNA-PKcs) and reduced DNA-PKcs expression respectively. In order to simulate of absence of DNA-PKcs we have used MO59-J cells and comparison was performed to MO59-K cells. In all cases deficient cells displayed a compromised (DNA-PKcs dependent) DSB repair pathway (Fig. 2), and this delay of DNA repair is in agreement with previous studies suggesting a slower processing using DNA-PK independent DSB repair pathways [47–49].
In addition to delayed repair kinetics of DSBs for DNA-PKcs deficient cells, a parallel persistence of OCDLs has been detected in all cases as also suggested by in vitro studies [50]. Related to the repair of OCDLs is the idea of ‘abortive excision repair’ i.e., non-DSB clusters converted to DSBs [6]. Although in our study such an effect has not been detected, the measured relative increase in DSBs for MO59-J cells at extended repair time points (Fig. 1B: 24–72 hrs) maybe due to this phenomenon. The DSB repair deficiency of MO59-J would certainly favor the detection of these indirect DSBs which under normal repair conditions may be processed very fast for our techniques to detect. Taken together with the role of DNA-PK in the NHEJ repair pathway, our present results suggest a model for the role of DNA-PKcs in the processing of non-DSB clusters (Fig. 5). In the case of a normally functioning DNA-PKcs (Pathway I), repair of DSBs will be performed quickly and efficiently within 12–24 hrs. This fast processing of SSBs and DSBs in normal cells decreases the possibility of compromised non-DSB clusters’ repair [50]. The persistence of DSBs due to a malfunctioning DSB repair system (Pathway II or III) enhances the possibility of repair inhibition or retardation (indirect action). An increased lifetime of SSBs and DSBs can result in the inhibition or delay of proper processing of non-DSB clusters. Our data definitely supports this idea since after extended repair times, the numbers of all types of clusters returned to background levels. In the case of chemical inactivation of DNA-PKcs the overall effect is even more pronounced. IC86621 or NU7026 unlike other inhibitors of the PIKK family like wortmannin (Figure S1) are expected to be very specific against DNA-PKcs. According to our model, DNA-PKcs is normally induced (Figure S3) but fails to dissociate properly from DNA ends due to the inhibition of its autophosphorylation activity on any of the ‘ABCDE’ cluster sites as suggested by Meek et al. [51, 52]. Since the DNA-PK complex stays at the DNA ends, access to broken ends by repair factors like HR or BER proteins decreases [11]. Of course since DSBs are eventually processed, roles for other NHEJ or HR independent pathways cannot be excluded. The prevention of DNA-PKcs autophosphorylation by drug treatment initiates ATM action. We and others believe that ATM phosphorylates Thr2609 and since the DSBs are not being repaired, the Thr2609 signal persists (Fig. 4A) [53]. The compromise, by drug treatment, of the efficient processing of DNA lesions induced by H2O2 supports an additional role of DNA-PKcs in the processing of DNA lesions traditionally repaired by BER (Fig. 3A). Inactive or reduced levels of DNA-PKcs maybe compromising the processing of base damage through BER (direct action). DNA-PKcs has been shown recently to interact with many BER proteins suggesting a repairsome formation [28]. Alternatively, DNA-PKcs could activate pre-existing XRCC1 through phosphorylation of serine 371 in its BRCT1 domain [27] and regulate its expression/stabilization upon exposure to an oxidizing agent favoring a recruitment process at the damage sites rather new synthesis [39]. The more pronounced inhibitory effects of catalytically inactive DNA-PK compared to its absence (MO59-J) in the repair of Fpg- or EndoIII-sites (Fig. 2C and 3A) suggest a combination of a direct and indirect effect as suggested in Fig. 5 (Pathway III). The significantly compromised repair of SSBs in MO59-J compared to MO59K cells cannot exclude an additional deficiency in a protein involved in the processing of SSBs. The results presented in Figure 3A certainly support a compromised BER through at least XRCC1 reduced expression. Previous studies have shown an important role for XRCC1 in the processing of an abasic site within a damage cluster containing an 8-oxodG [54]. Of course we cannot exclude a negative effect of inactive DNA-PKcs not only on BER but also on nucleotide excision repair (NER), mismatch repair (MMR), transcription coupled repair (TCR) or global genome repair (GGR), since all these pathways have been implicated in the processing of oxidatively-induced DNA lesions [55].
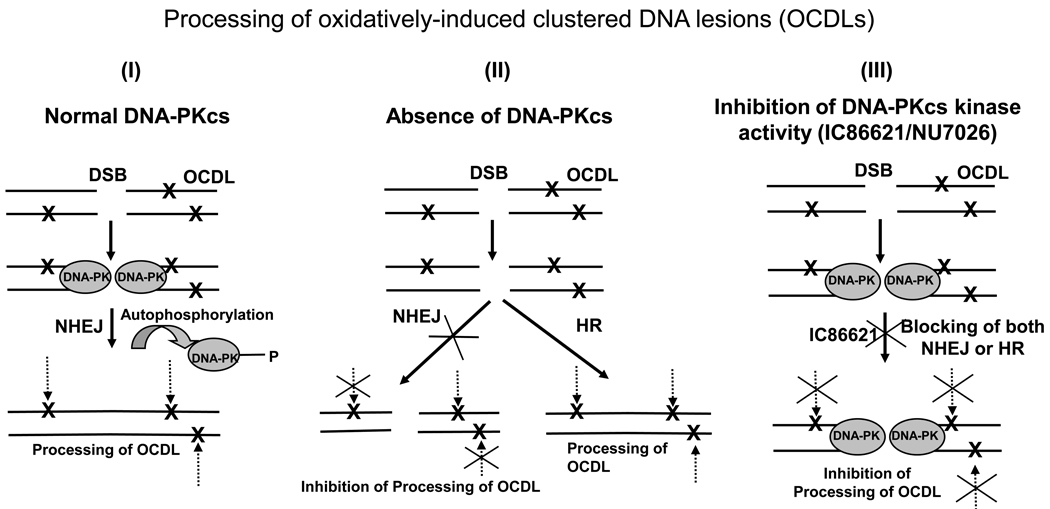
Fig. 5.
Schematic representing the alternative pathways of OCDL processing in the presence, absence, or inhibition of DNA-PKcs activity. Here OCDLs are shown flanking a DSB based on current theoretical and experimental evidence [2]. In DNA-PK normal cells the autophosphorylation of DNA-PK leads to its disassociation from the DNA ends allowing NHEJ or HR to proceed in DSB repair (Pathway I). As soon as the DSB is repaired all the other neighboring clustered DNA lesions can be processed. In the case of DNA-Pkcs absence, broken DNA ends cannot be repaired by NHEJ, and HR or other slower pathways repair the DSB. The presence of a SSB or DSB can be a significant inhibitory factor for the processing of other neighboring damage (Pathway II). Treatment of cells with drug is expected to lead to the inactivation of DNA-PKcs due to inhibition of its autophosphorylation. Therefore DNA-PKcs is expected, according to current status of knowledge, to be unable to dissociate from the DNA ends, physically blocking both NHEJ and HR (Pathway III). The presence of the DSB and the DNA-PK molecule is expected to have a profound inhibitory effect on the processing of all neighboring clustered DNA lesions. Additionally, the inactivation of DNA-PK can lead to the compromise of other repair pathways associated with its kinase activity including base excision repair (BER) inhibiting directly the processing of base lesions.
Here, we provide the first evidence of the actual involvement of DNA-PKcs in the processing of non-DSB oxidatively-induced clustered DNA lesions. The compromised repair of single and complex DNA lesions is primarily associated with the persistence of strand breaks which act as an inhibitory factor for the processing of neighboring base lesions in the cluster (indirect action) [1]. Complete absence or inactivation of DNA-PKcs significantly inhibits OCDL repair efficiency compared to partial deficiency induced by siRNA targeting. Drug induced inactivation of DNA-PKcs leads to a failure of DNA-PKcs to dissociate properly from the DNA ends and this can act as a further compromising factor for efficient lesion processing. In support of this idea, DNA-PK inhibition by wortmannin has been shown to lead to accumulation of DNA damage (expressed as micronuclei) induced by the chemotherapeutic drug bleomycin [56] which a well known inducer of clustered DNA damage [46].
Over the last decade a great emphasis has been placed on DNA-PK inhibition as a potential pharmacological target in radiotherapy and chemotherapy. In several studies drug-induced inhibition of DNA-PKcs has radio- or chemo-sensitized resistant tumor cells [57, 58]. Combined use of novel inhibitors taking advantage of interactions between DNA-PK and BER proteins, like PARP-1, has resulted in significant radiosensitization effects [33]. In addition, ROS have been shown to modulate (decrease) DNA-PK activity [59, 60] in vitro. Our present studies in human cancer cells appear to support an additional role of DNA-PKcs in the processing of oxidative DNA lesions and emphasize the potential use of DNA-PK inhibition or suppression as a potential tool for anticancer therapeutic intervention. In addition suggest and in conjunction with previous studies [61, 62], that genetic variation in DSB repair pathways may have a significant effect on the in vivo response of organisms to ionizing radiation, oxidative stress and tumorigenesis.
Supplementary Material
Acknowledgements
The authors would like to thank Drs. Jac Nickoloff for helpful discussions. We would also like to thank Angela Cecil, Peter Kalogerinis and Chris Richardson for their help on the data analysis. This work was supported by funds provided Dr. Georgakilas by a 2009/2010 Research/Creative Activity Grant from East Carolina University and partly by the Intramural Research Program of the National Cancer Institute, NIH (JSD, WMB, and OAS).
References
- 1.Georgakilas AG. Processing of DNA damage clusters in human cells: Current status of knowledge. Mol. Biosyst. 2008;4:30–35. doi: 10.1039/b713178j. [DOI] [PubMed] [Google Scholar]
- 2.Hada M, Georgakilas AG. Formation of clustered DNA Damage after High-LET Irradiation: A review. J. Radiat. Res. 2008;49:203–210. doi: 10.1269/jrr.07123. [DOI] [PubMed] [Google Scholar]
- 3.Sedelnikova OA, Redon CE, Dickey JS, Nakamura AJ, Georgakilas AG, Bonner WM. Role of oxidatively induced DNA lesions in human pathogenesis. Mutat. Res. Reviews. 2009 doi: 10.1016/j.mrrev.2009.12.005. In press; doi:10.1016/j.mrrev.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franco R, Schoneveld O, Georgakilas AG, Panayiotidis MI. Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett. 2008;266:6–12. doi: 10.1016/j.canlet.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 5.Burma S, Chen BPC, Chen DJ. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair. 2006;5:1042–1048. doi: 10.1016/j.dnarep.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 6.Blaisdell JO, Wallace S. Abortive base-excision repair of radiation-induced clustered DNA lesions in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2001;98:7426–7430. doi: 10.1073/pnas.131077798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Georgakilas AG, Bennett PV, Wilson DM, III, Sutherland BM. Processing of bistranded abasic DNA clusters in gamma-irradiated human hematopoietic cells. Nucleic Acids Res. 2004;32:5609–5620. doi: 10.1093/nar/gkh871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozmin SG, Sedletska Y, Reynaud-Angelin A, Gasparutto D, Sage E. The formation of double-strand breaks at multiply damaged sites is driven by the kinetics of excision/incision at base damage in eukaryotic cells. Nucl. Acids Res. 2009;37:1767–1777. doi: 10.1093/nar/gkp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bellon S, Shikazono N, Cunniffe S, Lomax M, O'Neill P. Processing of thymine glycol in a clustered DNA damage site: mutagenic or cytotoxic. Nucl. Acids Res. 2009:gkp422. doi: 10.1093/nar/gkp422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Couedel C, Mills KD, Barchi M, Shen L, Olshen A, Johnson RD, Nussenzweig A, Essers J, Kanaar R, Li GC, Alt FW, Jasin M. Collaboration of homologous recombination and nonhomologous end-joining factors for the survival and integrity of mice and cells. Genes Dev. 2004;18:1293–1304. doi: 10.1101/gad.1209204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 12.Meek K, Gupta S, Ramsden DA, Lees-Miller SP. The DNA-dependent protein kinase: the director at the end. Immunol. Rev. 2004;200:132–141. doi: 10.1111/j.0105-2896.2004.00162.x. [DOI] [PubMed] [Google Scholar]
- 13.Lees-Miller SP, Godbout R, Chan DW, Weinfeld M, Day RS, 3rd, Barron GM, Allalunis-Turner J. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science. 1995;267:1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- 14.Williams ES, Klingler R, Ponnaiya B, Hardt T, Schrock E, Lees-Miller SP, Meek K, Ullrich RL, Bailey SM. Telomere Dysfunction and DNA-PKcs Deficiency: Characterization and Consequence. Cancer Res. 2009;69:2100–2107. doi: 10.1158/0008-5472.CAN-08-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hashimoto M, Donald CD, Yannone SM, Chen DJ, Roy R, Kow YW. A possible role of Ku in mediating sequential repair of closely opposed lesions. J. Biol. Chem. 2001;276:12827–12831. doi: 10.1074/jbc.M010906200. [DOI] [PubMed] [Google Scholar]
- 16.Yu Y, Okayasu R, Weil MM, Silver A, McCarthy M, Zabriskie R, Long S, Cox R, Ullrich RL. Elevated breast cancer risk in irradiated BALB/c mice associates with unique functional polymorphism of the Prkdc (DNA-dependent protein kinase catalytic subunit) gene. Cancer Res. 2001;61:1820–1824. [PubMed] [Google Scholar]
- 17.Fu YP, Yu JC, Cheng TC, Lou MA, Hsu GC, Wu CY, Chen ST, Wu HS, Wu PE, Shen CY. Breast cancer risk associated with genotypic polymorphism of the nonhomologous end-joining genes: a multigenic study on cancer susceptibility. Cancer Res. 2003;63:2440–2446. [PubMed] [Google Scholar]
- 18.Auckley DH, Crowell RE, Heaphy ER, Stidley CA, Lechner JF, Gilliland FD, Belinsky SA. Reduced DNA-dependent protein kinase activity is associated with lung cancer. Carcinogenesis. 2001;22:723–727. doi: 10.1093/carcin/22.5.723. [DOI] [PubMed] [Google Scholar]
- 19.Lee HS, Choe G, Park KU, Park dJ, Yang HK, Lee BL, Kim WH. Altered expression of DNA-dependent protein kinase catalytic subunit (DNA-PKcs) during gastric carcinogenesis and its clinical implications on gastric cancer. Int. J. Oncol. 2007;31:859–866. [PubMed] [Google Scholar]
- 20.Kurimasa A, Ouyang H, Dong L-j, Wang S, Li X, Cordon-Cardo C, Chen DJ, Li GC. Catalytic subunit of DNA-dependent protein kinase: Impact on lymphocyte development and tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:1403–1408. doi: 10.1073/pnas.96.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson-Bennett BL, Deford J, Diaz-Arrastia C, Levine L, Wang HQ, Hannigan EV, Papaconstantinou J. Implications of tyrosine phosphoproteomics in cervical carcinogenesis. J. Carcinog. 2008;7:1–10. doi: 10.1186/1477-3163-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shackelford DA. DNA end joining activity is reduced in Alzheimer's disease. Neurobiol. Aging. 2006;27:596–605. doi: 10.1016/j.neurobiolaging.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 23.Culmsee C, Bondada S, Mattson MP. Hippocampal neurons of mice deficient in DNA-dependent protein kinase exhibit increased vulnerability to DNA damage, oxidative stress and excitotoxicity. Mol. Brain Res. 2001;87:257–262. doi: 10.1016/s0169-328x(01)00008-0. [DOI] [PubMed] [Google Scholar]
- 24.Someya M, Sakata K, Matsumoto Y, Yamamoto H, Monobe M, Ikeda H, Ando K, Hosoi Y, Suzuki N, Hareyama M. The association of DNA-dependent protein kinase activity with chromosomal instability and risk of cancer. Carcinogenesis. 2006;27:117–122. doi: 10.1093/carcin/bgi175. [DOI] [PubMed] [Google Scholar]
- 25.Peng Y, Woods RG, Beamish H, Ye R, Lees-Miller SP, Lavin MF, Bedford JS. Deficiency in the catalytic subunit of DNA-dependent protein kinase causes down-regulation of ATM. Cancer Res. 2005;65:1670–1676. doi: 10.1158/0008-5472.CAN-04-3451. [DOI] [PubMed] [Google Scholar]
- 26.Udayakumar D, Bladen CL, Hudson FZ, Dynan WS. Distinct pathways of nonhomologous end joining that are differentially regulated by DNA-dependent protein kinase-mediated phosphorylation. J. Biol. Chem. 2003;278:41631–41635. doi: 10.1074/jbc.M306470200. [DOI] [PubMed] [Google Scholar]
- 27.Lévy, N.; Martz A, Bresson A, Spenlehauer C, de Murcia G, Ménissier-de Murcia J. XRCC1 is phosphorylated by DNA-dependent protein kinase in response to DNA damage. Nucleic Acids Res. 2006;34:32–41. doi: 10.1093/nar/gkj409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parlanti E, Locatelli G, Maga G, Dogliotti E. Human base excision repair complex is physically associated to DNA replication and cell cycle regulatory proteins. Nucleic Acids Res. 2007;35:1569–1577. doi: 10.1093/nar/gkl1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peddi P, Francisco DC, Cecil A, Hair JM, Panayiotidis MI, Georgakilas AG. Deficient processing of clustered DNA damage in human breast cancer cells MCF-7 with silenced DNA-PKcs expression. Cancer Lett. 2008;269:174–183. doi: 10.1016/j.canlet.2008.04.049. [DOI] [PubMed] [Google Scholar]
- 30.Francisco DC, Peddi P, Hair JM, Flood BA, Cecil AM, Kalogerinis PT, Sigounas G, Georgakilas AG. Induction and processing of complex DNA damage in human breast cancer cells MCF-7 and non-malignant MCF-10A cells. Free Radic. Biol. Med. 2008;44:558–569. doi: 10.1016/j.freeradbiomed.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 31.Holt SM, Georgakilas AG. Detection of complex DNA damage in γ-irradiated acute lymphoblastic leukemia pre-B NALM-6 cells. Radiat. Res. 2007;168:527–534. doi: 10.1667/RR0974.1. [DOI] [PubMed] [Google Scholar]
- 32.Ward JF, Blakely WF, Joner EI. Mammialian cells are not killed by DNA single-strand breaks caused by hydroxyl radicals from hydrogen peroxide. Radiat. Res. 1985;103:383–392. [PubMed] [Google Scholar]
- 33.Veuger SJ, Curtin NJ, Richardson CJ, Smith GCM, Durkacz BW. Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent Protein kinase and poly(ADP-Ribose) polymerase-1. Cancer Res. 2003;63:6008–6015. [PubMed] [Google Scholar]
- 34.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Visvardis EE, Haveles KS, Pataryas TA, Margaritis LH, V S, Sideris EG. Diversity of peripheral blood mononuclear cells as revealed by a novel multiple microgel ldquocomet assayrdquo. Environmental and Molecular Mutagenesis. 2000;36:32–39. doi: 10.1002/1098-2280(2000)36:1<32::aid-em5>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 36.Tsao D, Kalogerinis P, Tabrizi I, Dingfelder M, Stewart RD, Georgakilas AG. Induction and processing of clustered DNA lesions in human monocytes exposed to low doses of HZE 56Fe particles. Radiat. Res. 2007;168:87–97. doi: 10.1667/RR0865.1. [DOI] [PubMed] [Google Scholar]
- 37.Anderson CW, Dunn JJ, Freimuth PI, Galloway AM, Allalunis-Turner MJ. Frameshift mutation in PRKDC, the gene for DNA-PKcs, in the DNA repair-defective, human, glioma-derived cell line M059J. Radiat. Res. 2001;156:2–9. doi: 10.1667/0033-7587(2001)156[0002:fmiptg]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 38.Kinner A, Wu W, Staudt C, Iliakis G. {gamma}-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucl. Acids Res. 2008;36:5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toulany M, Dittmann K, Fehrenbacher B, Schaller M, Baumann M, Rodemann HP. PI3K-Akt signaling regulates basal, but MAP-kinase signaling regulates radiation-induced XRCC1 expression in human tumor cells in vitro. DNA Repair. 2008;7:1746–1756. doi: 10.1016/j.dnarep.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 40.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. [gamma]H2AX and cancer. Nat. Rev. Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nowsheen S, Wukovich RL, Aziz K, Kalogerinis PT, Richardson CC, Panayiotidis MI, Bonner WM, Sedelnikova OA, Georgakilas AG. Accumulation of oxidatively-induced clustered DNA lesions in human tumor tissues. Mutat. Res. 2009;674:131–136. doi: 10.1016/j.mrgentox.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gulston M, C dL, Jenner T, Davis E, O'Neill P. Processing of clustered DNA damage generates additional double-strand breaks in mammalian cells post-irradiation. Nucleic Acids Res. 2004;32:1602–1609. doi: 10.1093/nar/gkh306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malyarchuk S, Castore R, Harrison L. DNA repair of clustered lesions in mammalian cells: involvement of non-homologous end-joining. Nucleic Acids Res. 2008;36:4872–4882. doi: 10.1093/nar/gkn450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plumb MA, Smith GCM, Cunniffe SMT, O'neill P. DNA-PK activation by ionizing radiation-induced DNA single-strand breaks. Int. J. Radiat. Biol. 1999;75:553–561. doi: 10.1080/095530099140195. [DOI] [PubMed] [Google Scholar]
- 45.Mårtensson S, Nygren J, Osheroff N, Hammarsten O. Activation of the DNA-dependent protein kinase by drug-induced and radiation-induced DNA strand breaks. Radiat. Res. 2003;160:291–301. doi: 10.1667/0033-7587(2003)160[0291:aotdpk]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 46.Regulus P, Duroux B, Bayle PA, Favier A, Cadet J, Ravanat JL. Oxidation of the sugar moiety of DNA by ionizing radiation or bleomycin could induce the formation of a cluster DNA lesion. Proc. Natl. Acad. Sci. USA. 2007;104:14032–14037. doi: 10.1073/pnas.0706044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karlsson KH, Stenerlow B. Focus formation of DNA repair proteins in normal and repair-deficient cells irradiated with high-LET ions. Radiat Res. 2004;161:517–527. doi: 10.1667/rr3171. [DOI] [PubMed] [Google Scholar]
- 48.Perrault R, Wang H, Wang M, Rosidi B, Iliakis G. Backup pathways of NHEJ are suppressed by DNA-PK. J. Cell. Biochem. 2004;92:781–794. doi: 10.1002/jcb.20104. [DOI] [PubMed] [Google Scholar]
- 49.Riballo E, Kuhne M, Rief N, Doherty A, Smith GCM, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, Parker AR, Jackson SP, Gennery A, Jeggo PA, Lobrich M. A pathway of double-strand break rejoining dependent upon ATM, Artemis and proteins locating to H2AX foci. Mol. Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 50.Budworth H, Dianova II, Podust VN, Dianov GL. Repair of clustered DNA lesions. J. Biol. Chem. 2002;277:21300–21305. doi: 10.1074/jbc.M201918200. [DOI] [PubMed] [Google Scholar]
- 51.Meek K, Douglas P, Cui X, Ding Q, Lees-Miller SP. trans Autophosphorylation at DNA-dependent protein kinase's two major autophosphorylation site clusters facilitates end processing but not end joining. Mol. Cell. Biol. 2007;27:3881–3890. doi: 10.1128/MCB.02366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meek K, Dang V, Lees-Miller SP, Frederick WA. Adv. Immunol. Academic Press; 2008. Chapter 2 DNA-PK: The means to justify the ends? pp. 33–58. [DOI] [PubMed] [Google Scholar]
- 53.Chen BPC, Uematsu N, Kobayashi J, Lerenthal Y, Krempler A, Yajima H, Lobrich M, Shiloh Y, Chen DJ. Ataxia telangiectasia mutated (ATM) Is essential for DNA-PKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. J. Biol. Chem. 2007;282:6582–6587. doi: 10.1074/jbc.M611605200. [DOI] [PubMed] [Google Scholar]
- 54.Mourgues S, Lomax ME, O'Neill P. Base excision repair processing of abasic site/single-strand break lesions within clustered damage sites associated with XRCC1 deficiency. Nucl. Acids Res. 2007;35:7676–7687. doi: 10.1093/nar/gkm947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holt SM, Scemama JL, Panayiotidis MI, Georgakilas AG. Compromised repair of clustered DNA damage in the human acute lymphoblastic leukemia MSH2-deficient NALM-6 cells. Mutat. Res. 2009;674:123–130. doi: 10.1016/j.mrgentox.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 56.Nuno GO, Matilde C, António SR, Octávia MG, José MT-R, José R. DNA-PK inhibitor wortmannin enhances DNA damage induced by bleomycin in V79 Chinese hamster cells. Teratogenesis Carcinog. Mutagen. 2002;22:343–351. doi: 10.1002/tcm.10029. [DOI] [PubMed] [Google Scholar]
- 57.Kashishian A, Douangpanya H, Clark D, Schlachter ST, Eary CT, Schiro JG, Huang H, Burgess LE, Kesicki EA, Halbrook J. DNA-dependent protein kinase inhibitors as drug candidates for the treatment of cancer. Mol Cancer Ther. 2003;2:1257–1264. [PubMed] [Google Scholar]
- 58.Westhoff MA, Kandenwein JA, Karl S, Vellanki SHK, Braun V, Eramo A, Antoniadis G, Debatin KM, Fulda S. The pyridinylfuranopyrimidine inhibitor, PI-103, chemosensitizes glioblastoma cells for apoptosis by inhibiting DNA repair. Oncogene. 2009 doi: 10.1038/onc.2009.215. [DOI] [PubMed] [Google Scholar]
- 59.Lu H-R, Zhu H, Huang M, Chen Y, Cai Y-J, Miao Z-H, Zhang J-S, Ding J. Reactive oxygen species elicit apoptosis by concurrently disrupting topoisomerase II and DNA-dependent protein kinase. Mol Pharmacol. 2005;68:983–994. doi: 10.1124/mol.105.011544. [DOI] [PubMed] [Google Scholar]
- 60.Boldogh I, Roy G, Lee M-S, Bacsi A, Hazra TK, Bhakat KK, Das GC, Mitra S. Reduced DNA double strand breaks in chlorambucil resistant cells are related to high DNA-PKcs activity and low oxidative stress. Toxicology. 2003;193:137–152. doi: 10.1016/j.tox.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 61.Vaganay-Juery S, Muller C, Marangoni E, Abdulkarim B, Deutsch E, Lambin P, Calsou P, Eschwege F, Salles B, Joiner M, Bourhis J. Decreased DNA-PK activity in human cancer cells exhibiting hypersensitivity to low-dose irradiation. Br. J. Cancer. 2000;83:514–518. doi: 10.1054/bjoc.2000.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Okayasu R, Suetomi K, Yu Y, Silver A, Bedford JS, Cox R, Ullrich RL. A deficiency in DNA repair and DNA-PKcs expression in the radiosensitive BALB/c Mouse. Cancer Res. 2000;60:4342–4345. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.