Abstract
We previously reported that a single DNA double-strand break (DSB) near a telomere in mouse embryonic stem cells can result in chromosome instability. We have observed this same type of instability as a result of spontaneous telomere loss in human tumor cell lines, suggesting that a deficiency in the repair of DSBs near telomeres has a role in chromosome instability in human cancer. We have now investigated the frequency of the chromosome instability resulting from DSBs near telomeres in the EJ-30 human bladder carcinoma cell line to determine whether subtelomeric regions are sensitive to DSBs, as previously reported in yeast. These studies involved determining the frequency of large deletions, chromosome rearrangements, and chromosome instability resulting from I-SceI endonuclease-induced DSBs at interstitial and telomeric sites. As an internal control, we also analyzed the frequency of small deletions, which have been shown to be the most common type of mutation resulting from I-SceI-induced DSBs at interstitial sites. The results demonstrate that although the frequency of small deletions is similar at interstitial and telomeric DSBs, the frequency of large deletions and chromosome rearrangements is much greater at telomeric DSBs. DSB-induced chromosome rearrangements at telomeric sites also resulted in prolonged periods of chromosome instability. Telomeric regions in mammalian cells are therefore highly sensitive to DSBs, suggesting that spontaneous or ionizing radiation-induced DSBs at these locations may be responsible for many of the chromosome rearrangements that are associated with human cancer.
Keywords: Telomeres, gross chromosomal rearrangements, sister chromatid fusion, chromosomal instability, double-strand break
1. Introduction
Telomeres are DNA-protein complexes containing short TTAGGG repeat sequences that are added on to the ends of chromosomes by the enzyme telomerase [1–3]. Telomeres serve a critical function in preventing the ends of chromosomes from appearing as DNA double-strand breaks (DSBs) and preventing fusion. In mammals, telomeres are maintained in germ line cells but undergo age-related shortening in somatic cells due to insufficient telomerase activity, which is associated with cell senescence [4]. Cell senescence is initiated when a telomere shortens to the point that it is recognized as a DSB [5]. Human cells that lose the ability to senesce continue to show telomere shortening and eventually enter crisis, which involves increased chromosome fusion, aneuploidy, and cell death [6,7]. Thus, it has been proposed that an important step in carcinogenesis is for cells to regain the ability to maintain telomeres, not only to avoid senescence, but also to avoid the extensive chromosome fusion during crisis [4,6]. Consistent with this hypothesis, most tumors demonstrate telomerase activity [8,9], although tumors maintaining telomeres through an alternative mechanism are also observed [10].
In addition to gradual shortening in aging cells, loss of telomere function can occur as a result of altered expression of proteins involved in telomere maintenance or as a result of DNA damage within telomeres or subtelomeric regions [11]. Telomere loss combined with defects in cell cycle checkpoints can result in continued growth of cells with gross chromosomal rearrangements (GCRs) and chromosome instability, which can occur through DNA degradation or breakage/fusion/bridge (B/F/B) cycles [12]. B/F/B cycles are initiated when the broken ends of chromosomes or sister chromatids fuse, form a bridge during anaphase, and then break when the centromeres are pulled in opposite directions during cytokinesis. Because the broken chromosomes in the daughter cells lack a telomere, they will again fuse and continue to undergo B/F/B cycles in subsequent generations, leading to additional DNA rearrangements. B/F/B cycles continue until the chromosomes acquire a new telomere, which can occur through a variety of mechanisms [13]. One of the most common mechanisms for telomere acquisition is through nonreciprocal translocation, which can transfer the instability onto the chromosome donating the translocation. As a result, the loss of a single telomere can result in the instability of multiple chromosomes [12].
Telomere loss has been demonstrated to play a role in the chromosomal rearrangements associated with cancer [14]. This relationship between telomere loss and cancer is illustrated by the fact that mice deficient in both telomerase and p53 have a high incidence of human-like cancers, with the tumor cells from these mice showing chromosome rearrangements typical of B/F/B cycles [15]. In preneoplastic lesions of colon cancer, telomere shortening has been observed and is correlated with DNA damage response and telomeric protein down-regulation [16]. In addition, early passage human tumor cell lines and tumors have also been shown to have high rates of spontaneous telomere loss and chromosome fusions [17–20]. The telomere loss in cancer cells can occur during crisis when telomeres become critically short in cells that do not express telomerase. However, we have also demonstrated that tumor cell lines that express telomerase can continue to have high rates of spontaneous telomere loss and prolonged instability associated with B/F/B cycles [21]. One possible mechanism by which spontaneous telomere loss in cancer cells could occur is through oncogene-driven replication stress, which results in stalled replication forks and DSBs at regions that pose problems for DNA replication [22,23]. DSBs occurring near telomeres may be especially sensitive to replication stress, because studies in yeast have shown that replication forks stall near telomeres [24]. Moreover, subtelomeric regions in yeast have also been shown to be defective in repair of I-SceI-induced DSBs by nonhomologous end joining (NHEJ), which leads to GCRs [25]. Thus, although replication stress would result in DSBs at multiple sites, the DSBs occurring at telomeres would be especially prone to generating chromosome instability. [13]
To investigate the role of sensitivity of subtelomeric regions to DSBs in telomere loss and chromosome instability in cancer cells, we have studied the consequences of DSBs near telomeres in the EJ-30 human tumor cell line. These studies utilize clones of EJ-30 that contain plasmid sequences containing selectable marker genes and an 18 base pair recognition site for the I-SceI endonuclease that are integrated adjacent to a telomere. Because the I-SceI recognition site is not found in the mammalian genome, it has been used extensively to study mechanisms of recombination and repair in mammalian cells [26–32]. EJ-30 is ideal for these studies, because we have previously demonstrated that spontaneous telomere loss in this cell line results in both chromosome healing and sister chromatid fusions [13,21], which have not been observed at DSBs at interstitial sites [Honma, 2007 #2802; Rebuzzini, 2005 #2793; Sargent, 1997 #1658; Varga, 2005 #2963]. The relationship between DSB-induced chromosome rearrangements and spontaneous rearrangements in cancer cells can therefore be established. The analysis of I-SceI-induced events has the advantage that unlike spontaneous events, the location of the DSB is known and therefore the extent of DNA degradation relative to the GCRs can be determined. We previously reported that I-SceI-induced DSBs near telomeres can lead to chromosome instability in mouse embryonic stem (ES) cells [33]. However, we were unable to determine the relative frequency of the DSB-induced chromosome instability because of the low frequency of DSBs that were produced using transient expression of I-SceI endonuclease. To increase the efficiency of generating DSBs with I-SceI, in the present study we have constitutively expressed the I-SceI endonuclease in the EJ-30 human tumor cell line. The relative frequency of large deletions and GCRs at I-SceI sites was then compared to the frequency of small deletions at both interstitial and telomeric I-SceI sites. Small deletions were used as an internal control to monitor the efficiency of generating DSBs at the different locations, because they are the most common I-SceI-induced DNA rearrangement at interstitial sites in mammalian cells, while GCR are relatively rare [29–32]. The results provide definitive evidence for the sensitivity of subtelomeric regions to DSBs in the EJ-30 human tumor cell line.
2. Materials and Methods
2.1. Cell lines and culture conditions
All cell clones used in this study were derived from the EJ-30 human bladder cell carcinoma cell line (obtained from Dr. William Dewey, UCSF), which was subcloned from the EJ cell line that is also called MGH-U1 [34]. The cell clones were grown in αMEM (UCSF Cell Culture Facility) supplemented with 5% fetal calf serum (Invitrogen-Gibco), 5% newborn calf serum with iron (Invitrogen-Gibco), 1 mM L-glutamine (Invitrogen-Gibco), and Gentamicin (Invitrogen-Gibco). Cells were propagated at 37°C in humidified incubators.
2.2. Plasmids
The pNCT-tel plasmid has been described previously [21]. pNCT-tel contains an ampicillin-resistance gene (amp) for selection in bacterial cells, a neomycin-resistance gene (neo), the Herpes Simplex Virus Thymidine Kinase (HSV-tk) gene under the control of a cytomegalo virus (CMV) promotor, and 0.8 kb of telomeric repeats. pNCT-tel also contains an I-Sce1 endonuclease recognition site between the neo and the HSV-tk genes for the generations of DSBs.
The pNTIL-tel plasmid is identical to pNCT-tel except for the fact that the I-SceI site is located within the 5' end of the HSV-tk coding sequence. pNTIL-tel was made by first removing the I-SceI site from pNCT-tel by digesting with I-SceI endonuclease, removing the 4 bp overhang with DNA polymerase, and re-ligation. The new I-SceI site was then inserted in frame by site-directed mutagenesis using the Gene Tailor kit (Invitrogen) at a location 18 amino acids from downstream from the first methionine. The oligonucleotide linker inserted at this site consisted of annealed primers 5'- CGTTCGACCAGGCTGCGCGTAGGGATAACAGGGTAATTATTCGACGTACGGC-3' and 5'-ACGCGCAGCCTGGTCGAACGCAGACGCGTG-3' bearing the new I-SceI site (bold). In addition to the I-SceI recognition site, a thymidine was removed just prior to the I-SceI site to avoid in-frame stop codons in the I-SceI site, and the addition of a thymidine was added immediately after the I-SceI site to restore the reading frame.
The insertion of the I-SceI site in the HSV-tk gene in pNTIL-tel was meant to enable us to detect both small deletions and GCRs using ganciclovir selection for the loss of HSV-tk function. Because the first 33 amino acids of HSV-tk are not required for its activity [35] it was thought that the insertion of the I-SceI site would not affect HSV-tk activity. However, this was not the case, as cells that contain the pNTIL-tel plasmid show only a limited sensitivity to ganciclovir. In addition, small deletions were not detectable using pNTIL-tel, apparently because truncated forms of HSV-tk that initiate translation from the 2nd or 3rd methionine are also functional [36,37]. As a result, pNTIL-tel was used in our studies only to confirm the results obtained with pNCT-tel.
The pQCXIP-ISce1 plasmid was used for the expression of the I-Sce1 endonuclease by viral infection. pQCXIP-ISce1 was prepared by cloning an EcoRI fragment containing the I-Sce1 gene from the pCBASce expression vector [28] into an EcoRI site in the pQCXIP retrovirus vector (Invitrogen). The plasmid contains an internal ribosome entry site to allow for expression of the genes for puromycin (puro) resistance and I-Sce1 endonuclease from the same promoter.
2.3. Virus preparation
Packaging of the pQCXIP control and pQCXIP-ISceI retroviral vectors was performed using Amphoteric 293 (A293) cells (Invitrogen). Packaging involved plating 5×106 293A cells the day before on collagen-coated 100 mm tissue culture plates (BD Biocoat). For each plate, 20 μg of plasmid was added to 1.25 ml of OptiMEM (Invitrogen), and 75 μl of Lipfectamine 2000 (Invitrogen) was added to 1.25 ml of OptiMEM. The two solutions were then mixed together, allowed to sit at room temperature for 20 minutes, and the mixture was added to the 10 ml of culture medium without antibiotics that was already on the plate. After 8 hours, the transfection medium was removed and replaced with EJ-30 growth medium. The medium was harvested from the infected A293 cells after 40, 48, 64, and 72 hours, and was filtered using a non-protein binding Puradisc filter (Whatman) and frozen at −80°C.
2.4. Selection of cells with constitutive expression of I-Sce1
The infection of cells with the pQCXIP-ISceI retrovirus or control pQCXIP retrovirus was performed using T-75 flasks that were 50% confluent. The cells were incubated with a mixture of 5 ml of virus-containing medium, 5 ml of EJ-30 growth medium, and 8 μg/ml polybrene (Sigma). After 24 hours, the medium was removed and replaced with EJ-30 growth medium, and the cells were incubated for another 24 hours. The selection for infected cells was then achieved through addition of growth medium containing 2 μg/ml puromycin (Sigma). The cells were then cultured for 10 days in medium containing puromycin, with medium changes every 2 days, to allow for expression of I-SceI endonuclease and the generation of DSBs. After 10 days, the cells were trypsinized, pooled together, and replated into T-75 tissue culture flasks. The pooled cells were then either used for preparation of genomic DNA for analysis of small deletions by PCR, or replated again as single cells in medium with or without selection for analysis of large deletions or GCRs. Replating was necessary for analysis of large deletions or GCRs, because the initial puror colonies contain mixtures of rearrangements as a result of the relatively slow rate of cutting of the I-SceI endonuclease in the infected cells. The analysis of the consequences of I-SceI-induced DSBs in our studies was performed 10 days after viral infection, because our preliminary studies using PCR and ganciclovir selection demonstrated that the percentage of cells with small deletions and ganciclovir resistance remained constant 6 to 10 days after infection (data not shown).
2.5. Selection of cells with GCRs and large deletions
The frequency of cells with loss of function of the HSV-tk gene was performed by plating the virally-infected puror cell populations in medium containing 50 μM ganciclovir. Selection was performed by plating 100 mm tissue culture dishes with 100 cells per dish and 1000 cells per dish in triplicate. As controls, the cells were also plated in dishes containing growth medium without ganciclovir. In some instances, selection was also performed in medium containing both 50 μM ganciclovir and 400 μg/ml G418 to select for cells that had inactivated the HSV-tk gene, but retained the neo gene. After 2 weeks, the colonies were fixed, stained, and counted. The percentage of Ganr cells was then determined by dividing the number of colonies in the selective medium by the number of colonies in the regular growth medium, and multiplying by 100. Standard deviations were obtained from a total of three independent experiments.
2.6. Analysis of small deletions by digestion of PCR products with I-SceI
Small deletions were analyzed by digesting PCR products spanning the I-SceI site with the I-SceI endonuclease. Long PCR was performed on genomic DNA isolated from either the pooled virally-infected puror cell cultures, or individual subclones selected at random from the pooled puror cultures. Long PCR was performed using the Elongase PCR kit (Invitrogen) as described by the manufacturer. PCR involved 94°C for 30 seconds, then 35 cycles of 94°C for 30 seconds, 57°C for 30 seconds, and 68°C for 105 seconds. Long PCR for EJ-30 clones containing the pNCT-tel plasmid was performed using primers OZNEO1 AAAAGCGGCCATTTTCCACCA and OZCMV1 TGCCTCACGACCAACTTCTGC. Long PCR for EJ-30 clones containing the pNTIL-tel plasmid was performed using primers CMV1-F TATATGGAGTTCCGCGTTACA and TK1-R GTTTGGCCAAGACGTCCAAG. 25 μl of the PCR product was then digested with 20 units of I-SceI endonuclease at 37°C overnight, and the products were run on 1% agarose gels. After staining with ethidium bromide, digital images were analyzed using free ImageJ software (http://www.versiontracker.com/dyn/moreinfo/macosx/37303) to calculate the intensity of the bands. The percent of cells containing small deletions at the I-SceI site was determined by dividing the area of the uncut band by the combined total area of the cut and uncut bands.
PCR for chromosome healing in EJ-30 clones A4 and B3 containing the pNCT-tel plasmid was performed as previously described, using one primer specific for the plasmid sequences and one primer specific for telomeric repeat sequences [38]. Direct DNA sequencing of the PCR product containing the site of chromosome healing using the NEOZ-F primer AGACAAGCGGCTGTCTGATG was performed by MCLAB (South San Francisco).
2.7. Selection of individual subclones for analysis of DNA rearrangements
Individual subclones were isolated to analyze the nature of the I-SceI-induced DNA rearrangements. Individual colonies generated from the puror populations of cells infected with pQCXIP or pQCXIP-ISceI were isolated by ring cloning. Ring cloning was performed by first washing the plates with Saline A buffer (1.4 M NaCL, 0.05 M KCl), placing an 8×8 mm cloning cylinder (Fisher Scientific) with silicone grease on its lower edge over a colony, and addition of trypsin to the cylinder (UCSF Cell Culture Facility). After 4 minutes in trypsin, the cells in each colony were pipeted up and down, and transferred to a 12 well tissue culture dish containing growth medium. The resulting subclones were later expanded into T-75 cell culture flasks for preparation of genomic DNA.
2.8. Southern blot analysis and plasmid rescue for analysis of large deletions and GCRs
Southern blot analysis of genomic DNA from the individual Ganr or randomly-selected subclones was performed following digestion with HindIII as previously described [38]. Plasmid rescue for analysis of DNA rearrangements in A4 subclones was performed as previously described [33,38] following digestion of genomic DNA with the XbaI and AccI restriction enzymes for A4 and B3, respectively. Briefly, the genomic DNA was digested with the restriction enzyme, the DNA was circularized by ligation at a dilute concentration of 1 μg/ml overnight at 16°C, re-concentrated to 50 μl using sequential Ultra-4/Microcon 30,000 MWCO spin filters (Millipore), and then used to transform STBL2 bacteria (Invitrogen). The selection for cells containing the plasmid was performed using ampicillin, and the rescued plasmids were mapped with restriction enzymes and sequenced using various primers specific for the plasmid DNA.
2.9. Cytogenetic analysis by FISH
The preparation of chromosomes and cytogenetic analysis by FISH was performed as previously described [13]. FISH was performed in 5 sequential steps, consisting of 1) the protein nucleic acid (PNA) telomeric probe, 2), the subtelomeric probes for chromosome 16, 3) the cosmid probes located adjacent to the integration site, 4) the M-FISH chromosome painting probe, and 5) the subtelomeric probes for other chromosomes. Telomere analysis using PNA probes labeled with Cy3 (Perseptive Biosystems) was performed as previously described [39]. M-FISH was performed using multi-FISH probes (MetaSystems, GmbH) according to the manufacturer's recommendations. The RT99 (Genebank accession No. AC004653) and 317H7 (Genebank accession No. AC005569) cosmid clones that were used as probes have been mapped to the end of chromosome 16p [40]. These cosmids were isolated from partially-digested DNA libraries made from flow-sorted human chromosome 16 [41]. The subtelomeric BAC probes (Cytocell) were hybridized according to manufacturer's protocol. The subtelomeric BAC clones for chromosome16 consisted of GS-121-I4, which is located a maximum of 160 kb from the telomere on the short arm, and GS-240-G10, which is located a maximum of 200 kb from the telomere on 16q [42]. Images of hybridized metaphases were captured with a CCD camera (Zeiss) coupled to a Zeiss Axioplan microscope and were processed with ISIS software (MetaSystems, GmbH). Probes for each subtelomeric region were obtained from Cytocell, and hybridizations were performed according to the manufacturer's recommendations.
3. Results
3.1. Generation of cell clones containing telomeric and interstitial I-SceI sites
The EJ-30 human tumor cell clones used in these studies contain the pNCT-tel or pNTIL-tel plasmids that are integrated at either interstitial or telomeric locations. Both plasmids contain a neo for positive selection in G418, an HSV-tk gene for negative selection in ganciclovir, and an I-SceI recognition site for introducing DSBs (Fig. 1). The I-SceI site in pNTC-tel is located between the neo and HSV-tk genes, while the I-SceI site in pNTIL-tel is located at the 5' end of the HSV-tk coding sequence. Prior to transfection, the plasmids were linearized with NotI to place the telomeric repeat sequences on one end. Following transfection, colonies resistant to G418 were selected at random and analyzed by Southern blot analysis. At some integration sites, the telomeric repeat sequences seeded the formation of a new telomere, positioning the plasmid at the new end of a chromosome [43,44]. However, the plasmids also commonly undergo rearrangements that result in the loss of the telomeric plasmid sequences, and then integrate at interstitial sites within chromosomes [45]. The presence of a plasmid integrated at a telomere is easily identified by Southern blot analysis by the presence of a diffuse band containing the telomeric repeat sequences, which vary in length in different cells in the population [21,46,47]. Telomeric integration sites were then confirmed by sensitivity to BAL31 exonuclease, which is a well-established method for detection of terminal restriction fragments located at the ends of chromosomes [21,44].
FIG. 1.
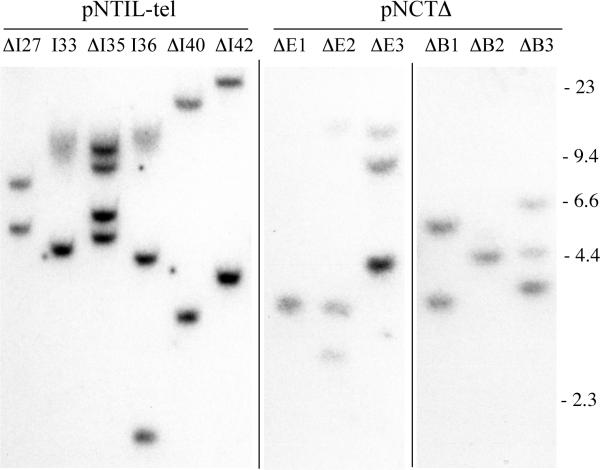
Characterization of integrated plasmid sequences in clones with telomeric or interstitial integration sites. Similar to our earlier isolation and characterization of clones B3 and A4 transfected with the pNCT-tel plasmid, cell line EJ-30 was transfected with the linearized pNTIL-tel plasmid with telomeric repeat sequences on one end. Following transfection and selection with G418, individual colonies were then selected and their genomic DNA was analyzed by Southern blot analysis following digestion with XbaI, which cuts once in the center of the plasmid. Clones in which the linearized pNTIL-tel plasmid seeded the formation of a new telomere upon integration demonstrate a diffuse band (I33 and I36) as a result of variability in the length of the telomere in different cells in the population. All other clones contain the plasmid integrated at interstitial sites (ΔI27, ΔI35, ΔI40, ΔI42). Additional cell clones with telomeric integration sites (ΔE1, ΔE2, ΔE3, ΔB1, ΔB2, ΔB3) were also isolated following transfection with the linearized pNCTΔ plasmid that does not contain telomeric repeat sequences.
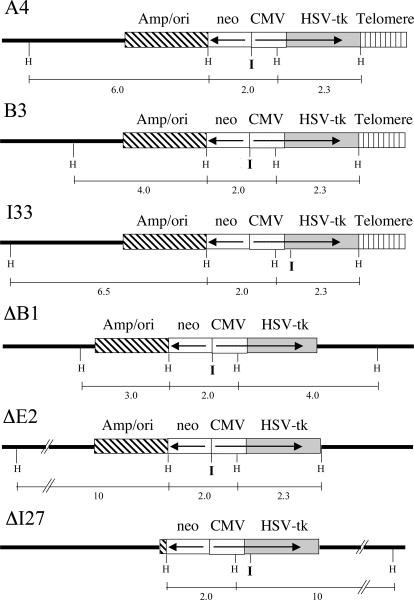
Three EJ-30 clones containing telomeric plasmid integration sites were used in this study, A4, B3 and I33. Two of these clones, A4 and B3, contain a single unrearranged copy of the pNCT-tel plasmid and have been previously described [21]. The telomeric plasmid in clone B3 has been shown to be located near the original end of chromosome 16p, both by FISH and by rescue of the plasmid and adjacent cellular DNA [21]. The third clone with a telomeric integration site, I33, which was isolated for the present study, was demonstrated by Southern blot analysis to contain a single copy of the pNTIL-tel plasmid located at a telomere (Fig. 2).
FIG. 2.
The structure of the integrated plasmid sequences used to investigate the consequences of I-SceI-induced DSBs at telomeric and interstitial locations. EJ-30 clones A4 and B3 contain a single copy of the pNCT-tel plasmid integrated on the end of a chromosome, while EJ-30 clones ΔB1 and ΔE2 contain single rearranged copies of pNCT-tel integrated at interstitial sites. EJ-30 clone I33 contains a single copy of the pNTIL-tel plasmid integrated at the end of a chromosome, while clone ΔI27 contains a single rearranged copy of pNTIL-tel integrated at an interstitial site. pNCT-tel and pNTIL-tel both contain an HSV-tk gene with a CMV promoter, a neo gene with an HSV-tk promoter, vector sequences (amp/ori), and telomeric repeat sequences for seeding new telomeres. The only difference between pNCT-tel and pNTIL-tel is the location of an 18 bp recognition site for introducing DSBs with the I-SceI endonuclease (I), which is located between the neo and HSV-tk genes in pNCT-tel and at the 5' end of the HSV-tk gene in pNTIL-tel. The location of the HindII sites (H) and the size of the HindIII restriction fragments used for Southern blot analysis are shown.
EJ-30 clones containing the same plasmids integrated at random interstitial sites were used as controls, including ΔB1 and ΔE2, which were transfected with the pNCTΔ plasmid, and ΔI27, which was transfected with the pNTIL-tel plasmid. The pNCTΔ plasmid is the same as pNCT-tel, except that it lacks the telomeric repeat sequences. Each of the clones was screened for sensitivity to ganciclovir to insure that the HSV-tk gene was intact, and Southern blot analysis was performed to eliminate clones containing multiple copies of the plasmid (Fig. 2). The number of plasmid integration sites was determined by digestion of the genomic DNA with XbaI, which cuts once in the center of these plasmids, and will therefore produce two bands for each integration site.
3.2. Selection of cells with constitutive expression of I-SceI endonuclease
Constitutive expression of I-SceI endonuclease was used to maximize the efficiency of cutting at the I-SceI site. A retroviral vector approach was chosen because it avoids the harsh conditions associated with DNA transfection and the introduction of large amounts of transfected DNA into cells. The retrovirus vector containing the I-SceI gene used in our studies, pQCXIP-ISceI, contains the puro gene, and therefore selection of cells with the integrated I-SceI gene was performed using puromycin. Virtually all of the puromycin-resistant (puror) cells that contain the pQCXIP-ISceI retrovirus should also express the I-SceI gene, because both the puro and I-SceI genes are in this vector expressed from the same promoter.
3.3. The analysis of the frequency of large deletions and GCRs using ganciclovir selection
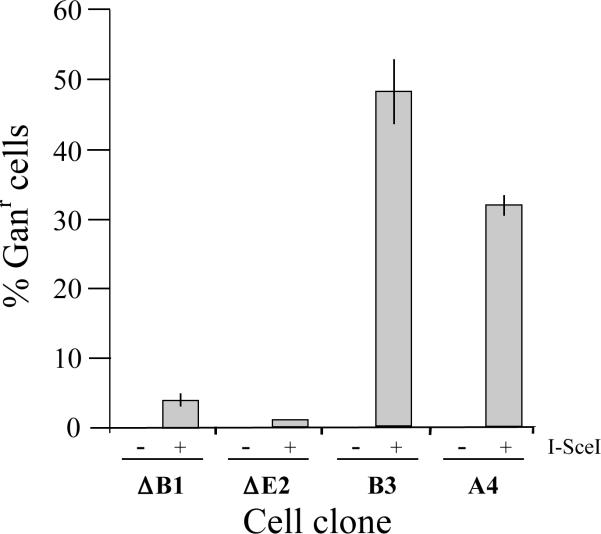
The influence of DSBs on large deletions and GCRs at the interstitial and telomeric I-SceI sites was analyzed by plating the pooled puror populations of cells infected with the pQCXIP-ISceI retrovirus in medium containing 50 μM ganciclovir. Growth in medium containing ganciclovir selects for cells that have lost the function of the HSV-tk gene [48,49]. The generation of ganciclovir-resistant (Ganr) cells from clones containing the pNCT-tel plasmid requires the deletion of at least 226 bps in one direction, which is the distance to the enhancer sequences in CMV promoter for the HSV-tk gene [50]. As seen in Fig. 3, following infection with the pQCXIP-ISceI retrovirus, clones A4 and B3 with telomeric I-SceI sites had a much higher frequency of Ganr colonies than clones ΔE2 and ΔB1 with interstitial I-SceI sites, demonstrating that DSBs at telomeric sites are far more likely to result in large deletions or GCRs than DSBs at interstitial sites. Control cultures consisting of pooled puror populations of these clones infected with the pQCXIP retrovirus had a much lower frequency of Ganr colonies (<1%), consistent with our earlier studies demonstrating that spontaneous Ganr cells in EJ-30 clones occur at a rate of 10−4 events/cell/generation [21].
FIG. 3.
Increased frequency of inactivation of the HSV-tk gene as a result of I-SceI-induced DSBs near telomeres. The EJ-30 clones ΔB1 and ΔE2 containing interstitial I-SceI sites, and clones A4 and B3 containing telomeric I-SceI sites were infected with the control pQCXIP retrovirus (−) and the pQCXIP-ISceI retrovirus containing the I-SceI gene (+). The pooled puror cultures of infected cells were then plated into medium containing 50 μM ganciclovir to determine the percentage of Ganr cells in the population. The standard deviations are shown for the 3 separate experiments.
Inactivation of the HSV-tk gene in cells containing the pNTIL-tel plasmid would require deletions that extend at least 76 bps from the I-SceI site, which is the distance to the alternative methionine start site within the HSV-tk gene [36,37]. However, quantitative studies on the frequency of inactivation of the HSV-tk gene in the I33 and ΔI27 clones using ganciclovir resistance was not possible, because the insertion of the I-SceI site at the 5' end of the HSV-tk gene severely reduced the sensitivity of the cells to ganciclovir. Selection for cells containing large deletions and GCRs for qualitative analysis could be achieved at a much higher concentration of 200 μM ganciclovir (see below), however, at this higher concentration most cells that do not contain the HSV-tk gene also die.
3.4. The analysis of DSB-induced DNA rearrangements using ganciclovir selection
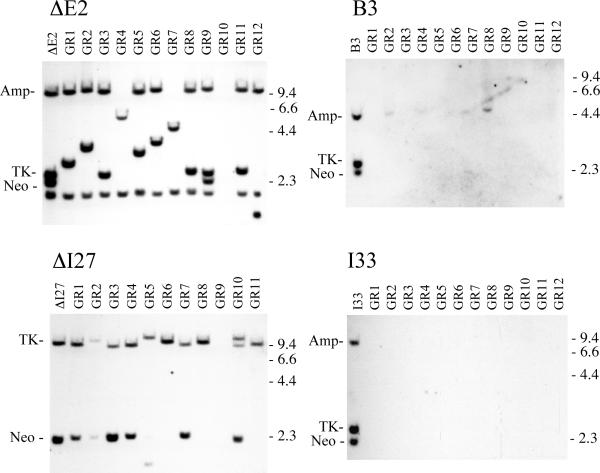
We next compared the types of rearrangements resulting from telomeric and interstitial DSBs in the individual Ganr subclones by Southern blot analysis. The Ganr subclones isolated from clone ΔE2 with an interstitial I-SceI site nearly all show loss of the original band containing the neo gene, which is the fragment containing the I-SceI site (Fig. 4). In its place, most clones show a new band that varies in size, demonstrating that rearrangements had occurred. Only a few Ganr ΔE2 subclones showed the complete loss of the integrated plasmid sequences, i.e., showed no plasmid specific bands. Similar results were observed with the Ganr subclones isolated from clone ΔI27, which contains the pNTIL-tel plasmid integrated at an interstitial site. Although not quantitative due to selection with 200 μM ganciclovir, the Ganr subclones of ΔI27 that survived all show changes in the size of the band containing the HSV-tk gene, which is the fragment containing the I-SceI site (Fig. 4). As with clone ΔE2, few of the Ganr ΔI27 subclones showed complete loss of the integrated plasmid sequences.
FIG. 4.
Southern blot analysis demonstrated that inactivation of the HSV-tk at telomeric sites most often results from large deletions involving loss of the integrated plasmid sequences. Genomic DNA from parental clones B3 and I33 with telomeric I-SceI sites, and ΔE2, ΔI27 with interstitial I-SceI sites, is compared with genomic DNA from 12 of their Ganr subclones infected with the pQCXIP-ISceI retrovirus (GR1 – GR12). The genomic DNA was digested with HindIII, and hybridization was performed using the pNTPΔ plasmid probe. pNTPΔ contains the amp, neo and HSV-tk genes, but lacks the telomeric repeat sequences. pNTPΔ also lacks the CMV promoter, which cross hybridizes with the CMV promoter in the pQCXIP retrovirus. The location of the bands containing the neo and HSV-tk genes, and the location of the lambda bacteriophage HindIII restriction fragments are shown.
In contrast to the clones with interstitial I-SceI sites, the most common event in the Ganr subclones of clone B3 with a telomeric I-SceI site was the complete loss of the integrated plasmid sequences (Fig. 4). Approximately 9 in 10 Ganr subclones isolated from clone B3 contained rearrangements that involved the complete loss of the plasmid-specific bands. Similar results were observed with the Ganr subclones isolated from clone I33, which contains the pNTIL-tel plasmid integrated at a telomere (Fig. 4). Therefore, in addition to a quantitative difference in the frequency of Ganr cells, there is also a qualitative difference in the types of rearrangements that occur at DSBs at interstitial and telomeric sites. As a result, the frequency of cells in the population that experience complete loss of the plasmid sequences is approximately 100-fold higher as a result of DSBs within subtelomeric regions. This difference is not limited to I-SceI-induced DSBs, because the complete loss of the integrated plasmid sequences is also the most frequent event observed in spontaneous Ganr subclones isolated from clone B3 [21].
3.5. The analysis of small deletions at the I-SceI site using PCR
We next compared the frequency of small deletions at the I-SceI site in the pooled puror populations of the various clones infected with the pQCXIP-ISceI retrovirus. This approach involves the amplification of a 1.8 kb region spanning the I-SceI site by long PCR, followed by digestion of the PCR product with I-SceI endonuclease. Using this approach, small deletions result in a full-sized PCR product that is not cut by the I-SceI endonuclease. Large deletions and large insertions would generate detectably smaller or larger PCR products, respectively, or no PCR product if one or both primer sites are lost. Similarly, cells with GCRs at the I-SceI site would also not generate a PCR product. The percent of cells in the population that have lost the I-SceI site can therefore be calculated from the fraction of DNA in the uncut band compared to the total DNA in the cut and uncut bands.
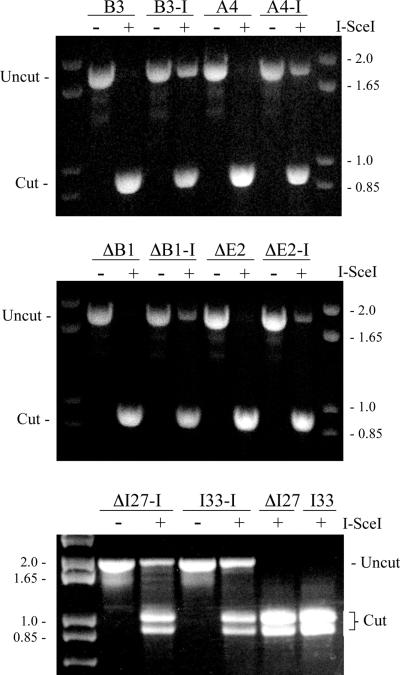
Digestion of the PCR products with I-SceI showed that all of the clones contained a significant fraction of uncut full-length PCR product, demonstrating the presence of small deletions at the I-SceI site (Fig. 5). In contrast, all of PCR product generated from the pooled puror populations of the same clones infected with the control pQCXIP retrovirus was cut by I-SceI. A comparison of the percentage of uncut PCR product generated from the pooled puror populations showed that there was consistently more uncut PCR product in the A4, B3 and I33 clones containing the telomeric I-SceI sites than in the ΔB1, ΔE2 and ΔI27 clones containing interstitial I-SceI sites (Fig. 5, Table 1). Based on these results, the frequency of small deletions would at first appear to be somewhat greater in the clones with telomeric I-SceI sites. However, these results must first be corrected for the frequency of cells in the population that are resistant to ganciclovir, since nearly all Ganr cells have large deletions that result in complete loss of the integrated plasmid sequences. The lack of a PCR product from cells with large deletions or GCRs would result in a corresponding overestimation of the frequency of small deletions in the cell population. The correction of the percent of PCR product digested with I-SceI for the fraction of Ganr cells in the pooled puror populations (1 – fraction of Ganr cells) demonstrates that the percentage of cells with small deletions is similar in the clones with telomeric and interstitial I-SceI sites (Table 1). We were not able to estimate the fraction of Ganr cells in the ΔI27 and I33 clones containing the pNTIL-tel plasmid due to their limited sensitivity to ganciclovir. The correction for the fraction of cells that have lost the plasmid sequences in these clones was therefore based on PCR analysis of individual subclones selected from the puror population (see below). The corrected values for the percentage of small deletions in these clones is similar to the percentage of small deletions in the clones containing the pNCT-tel plasmid (Table 1).
FIG. 5.
The analysis of small deletions at the I-SceI site in EJ-30 clones with interstitial and telomeric integration sites. Small deletions were detected by monitoring the ability of the I-SceI endonuclease to cut a 1.8 kb PCR product spanning the I-SceI site. Genomic DNA was isolated from pooled puror cultures of pQCXIP (A4, B3 and I33) or pQCXIP-ISceI (A4-I, B3-I and I33-I) infected cultures of clones with telomeric integration sites, or pQCXIP (ΔB1, ΔE2 and ΔI27) or pQCXIP-ISceI (ΔB1-I, ΔE2-I and ΔI27-I) infected cultures of clones with interstitial integration sites. The genomic DNA from the pooled puror cells was isolated 10 days after infection with the retroviruses. The percentage of cells that had small deletions was determined by comparing the intensity of the undigested band with the combined intensity of the digested and undigested bands.
Table 1.
Percent of cells in the population that contain small deletions at the I-SceI site.
| Clone | Percent uncut DNA | Fraction Ganr/PCR− | Percent small del* |
|---|---|---|---|
| Interstitial | |||
| ΔB1 | 20.5 | 0.034 | 19.8 |
| ΔE2 | 18.2 | 0.008 | 17.9 |
| ΔI27 | 20.9 | 0.066 | 19.5 |
| Telomeric | |||
| A4 | 34.9 | 0.30 | 24.4 |
| B3 | 35.9 | 0.48 | 18.7 |
| I33 | 31.2 | 0.52 | 15.0 |
The percent of cells with small deletions is determined by multiplying the fraction of uncut PCR product by (1- Fraction of Ganr or PCR− cells).
3.6. The analysis DSB-induced rearrangements in subclones isolated without ganciclovir selection
We next analyzed the types of rearrangements in individual subclones picked at random without ganciclovir selection after replating puror populations of clones ΔI27 and I33 infected with either the pQCXIP or pQCXIP-ISceI retroviruses. This approach allowed us to extend our analysis to determine the presence of large deletions or GCRs in puror populations of clones ΔI27 and I33, which we were not able to determine by ganciclovir selection due to their limited sensitivity. In addition, the analysis of the types of rearrangements in these random subclones allowed us to validate our assays, which are based on the digestion of the PCR product with I-SceI for small deletions and ganciclovir selection for large deletions/GCRs. To select random subclones, the pooled puror cells infected with either pQCXIP or pQCXIP-ISceI were replated in medium without ganciclovir. Individual colonies were then selected and the subclones were analyzed for the presence of the integrated plasmid sequences by PCR using the same primers used previously to analyze the pooled puror populations. Subclones with no PCR product were considered to have large deletions or GCR, as was confirmed by Southern blot analysis (see below). Subclones with PCR products similar in size to untreated controls were considered to be unrearranged or contain small deletions or insertions, while those with PCR fragments that were smaller or larger than controls were considered to have intermediate-sized deletions or insertions, respectively. To identify small deletions, the PCR products were tested for the presence of the intact I-SceI site by digestion with I-SceI endonuclease, as performed earlier with the pooled puror populations (see Fig. 5). Sequence analysis of PCR fragments that did not digest with I-SceI confirmed the presence of small deletions (data not shown), typical of those previously observed at I-SceI sites in mammalian cells [29–32].
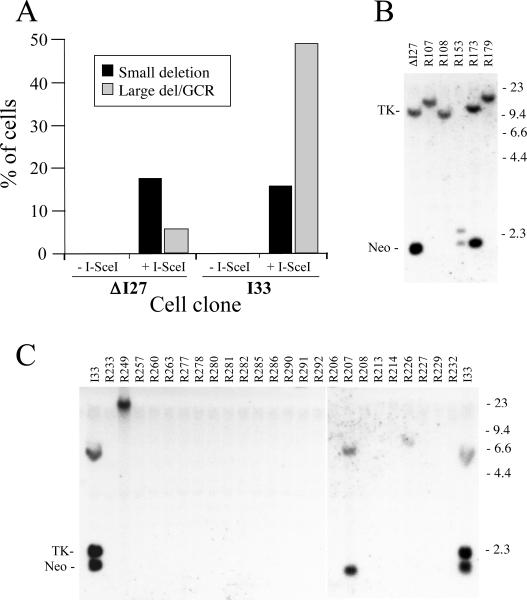
The combined results demonstrate that the percentage of small deletions (PCR products that do not cut with I-SceI) generated by I-SceI is similar in the random ΔI27 and I33 subclones, while the percentage of large deletions/GCRs (no PCR product) is much higher in the random I33 subclones (Fig. 6A). The increased percentage of large deletions/GCRs in the random I33 subclones is consistent with the increased percentage of Ganr cells in the pooled puror populations of the EJ-30 clones with telomeric I-SceI sites compared to the clones with interstitial I-SceI sites (see Fig. 3). The similar percentage of small deletions in the random ΔI27 and I33 subclones is also consistent with the results from the PCR analysis of the pooled puror populations of the EJ-30 clones with telomeric and interstitial I-SceI sites (Table 1). The analysis of the individual puror subclones selected at random without ganciclovir selection therefore confirms that the analysis of the pooled puror cells by PCR/I-SceI digestion accurately reflects the frequency of cells in the population with small deletions when an adjustment is made for the frequency of Ganr cells. Similarly, the analysis of the random puror subclones confirms that the frequency of Ganr cells in the pooled puror populations accurately reflects the frequency of cells with large deletions and/or GCRs.
FIG. 6.
Comparison of the frequency of small deletions, large deletions, and GCRs resulting from DSBs at telomeric and interstitial sites. (A) The types of rearrangements were characterized in subclones of EJ-30 clones ΔI27 and I33 that were selected at random without ganciclovir selection from pooled puror populations infected with the pQCXIP and pQCXIPISceI retroviruses. The puror colonies were pooled 10 days after infection and were replated as single cells in growth medium without ganciclovir. Individual colonies were then selected and the subclones (+I-SceI, n=81 for ΔI27, n=101 for I33) were analyzed for the presence of small deletions, large deletions or GCRs. As controls, subclones selected at random from puror populations of ΔI27 and I33 (−I-SceI, n=25) infected with the pQCXIP retrovirus were also analyzed. The analysis of small deletions in the random subclones involved amplification of the region containing the I-SceI site by PCR, followed by digestion of the PCR product with I-SceI, as performed for the pooled populations in Fig. 5. Subclones in which the PCR fragment was not cut by I-SceI were scored as having small deletions. The subclones that did not produce a PCR product were scored as having large deletions or GCRs, and were subsequently analyzed by Southern blot analysis to determine the type of rearrangement that had occurred. The results are shown for (B) the parental ΔI27 clone and all 5 PCR-negative ΔI27 subclones (R107–R179), and (C) the parental I33 clone and 24 PCR-negative I33 subclones (R206–R292). Southern blot analysis was performed using HindIII digestion and the pNPTΔ probe as described in Fig. 4. The data for I33 subclones R206–R232 were taken from a separate hybridization than the other I33 subclones.
Southern blot analysis was performed on genomic DNA from the random ΔI27 and I33 subclones that did not produce a PCR product to determine the nature of the rearrangements that were present. The results are consistent with the analysis of rearrangements in the Ganr subclones isolated from the pooled puror populations of these clones (see Fig. 4). All 5 of the random subclones of ΔI27 that did not generate a PCR product contained new plasmid-specific bands that varied in size (Fig. 6B), while nearly all of the random subclones of I33 that did not generate PCR bands showed no plasmid-specific bands demonstrating the loss of the plasmid sequences (Fig. 6C). Selection with ganciclovir therefore accurately reflects the frequency of large deletions and GCRs in the pooled puror populations of the EJ-30 clones infected with the pQCXIP-ISceI retrovirus.
3.7. The analysis of DSB-induced DNA rearrangements in Ganr/G418r subclones
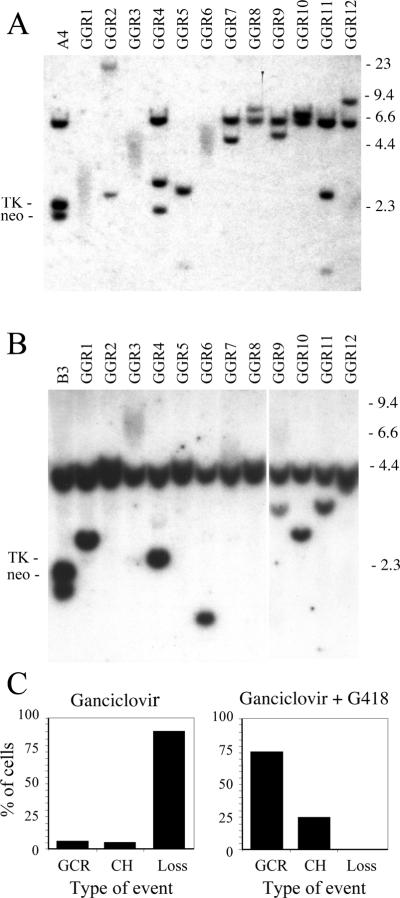
The types of DNA rearrangements resulting from DSBs near telomeres are difficult to analyze in subclones selected with ganciclovir, because most Ganr subclones have completely lost the telomeric plasmid sequences. We therefore performed selection with both ganciclovir and G418 to isolate subclones that have retained the portion of the plasmid containing the neo gene. Including G418 during selection results in far fewer colonies, since only approximately 1 in 20 Ganr cells is also resistant to G418, i.e., Ganr/G418r. The retention of some portion of the integrated plasmid in these subclones allows for analysis of the rearrangements by Southern blot analysis and plasmid rescue. Southern blot analysis of Ganr/G418r subclones of clones A4 and B3 demonstrated a variety of new bands of different sizes (Fig. 7). In some subclones, the new bands were highly diffuse (A4 subclones GGR1, GGR3, and GGR6; B3 subclone GGR3), typical of the new bands that resulted from chromosome healing following spontaneous telomere loss in EJ-30 clone B3 [21]. As in our earlier studies with mouse ES cells [38,51], confirmation of chromosome healing in these subclones was performed by PCR, using one primer with homology to the plasmid sequences adjacent to the I-SceI site and one primer with homology to the telomeric repeat sequences (data not shown). DNA sequence analysis of the resulting PCR products demonstrated that the telomeric repeat sequences were added directly at the site of the break (data not shown), as previously reported for mouse ES cells [38,51]. The frequency of chromosome healing in the Ganr/G418r subclones was 9 of 35 for A4 (26%), and 12 of 43 for B3 (28%). Chromosome healing in the human EJ-30 tumor cell line is therefore less frequent than in mouse ES cells, in which chromosome healing was observed in 60% of the Ganr/G418r subclones [38].
FIG. 7.
Southern blot analysis of the types of DNA rearrangements resulting from DSBs near telomeres in Ganr/G418r subclones. Genomic DNA from (A) the parental clone A4 and 12 of its Ganr/G418r subclones, GGR1 through 12, and (B) the parental clone B3 and 12 of its Ganr/G418r subclones infected with pQCXIP-ISceI, GGR1 through 12, was analyzed following digestion with HindIII and hybridization with the pNPTΔ plasmid probe as described in Fig. 4. The data for B3 subclones GGR9–12 were taken from a separate hybridization. Subclones with new highly diffuse bands have undergone chromosome healing, while subclones with new discrete bands have undergone GCRs. (C) The combined percent of cells in clones A4 and B3 with GCRs, chromosome healing (CH), or complete loss (Loss) of the plasmid sequences, when selecting with ganciclovir alone (left), or with ganciclovir and G418 together (right).
3.8. The characterization of recombination junctions involved in the formation of GCRs
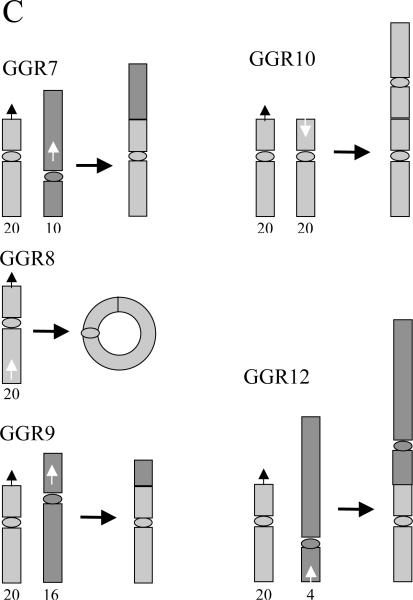
We next analyzed the DNA found at the recombination junctions in 5 Ganr/G418r subclones of clone A4 to determine the nature of the rearrangements involved. DNA sequence analysis of the genomic DNA adjacent to the rescued plasmid demonstrated that the new telomere seeded by the pNCT-tel plasmid in clone A4 was located on the long arm of chromosome 20 (20q11.23), which would result in the loss of a large portion of the original arm. Our earlier analysis of DNA rearrangements resulting from spontaneous telomere loss in 4 Ganr/G418r subclones of clone B3 demonstrated that all 4 contained inverted repeats [33], which we have shown result from sister chromatid fusion [52]. However, DNA sequence analysis of the rescued plasmid DNA from A4 subclones demonstrated that only 1 of the 5 DSB-induced Ganr/G418r subclones contained an inverted repeat, while the other 4 had a variety of other GCRs, including translocations, ring chromosome formation, and fusion at a telomeric site to form a dicentric chromosome (Fig. 8). DNA sequence analysis of the recombination site in subclone A4-GGR7 demonstrated that DNA from chromosome 10 was joined on at the I-SceI site without the loss of a single nucleotide from the ATAA overhang generated by I-SceI. There were 2 bps of microhomology at the recombination junction. The orientation of the chromosome 10 sequence is towards the telomere on the long arm at 10q21, indicating that a translocation had occurred.
FIG. 8.
Characterization of the types of rearrangements resulting from DSBs near telomeres using plasmid rescue and DNA sequence analysis. Genomic DNA from Ganr/G418r subclones isolated from clone A4 was used for rescue of the plasmid sequences and adjacent cellular DNA. (A) DNA sequence analysis was performed using a primer adjacent to the I-SceI site to analyze the recombination junction and the identity of the DNA involved in rearrangements in the A4 Ganr/G418r subclones GGR7, GGR8, GGR9, GGR10 and GGR12. The sequence of the pNCT-tel plasmid ending in the 4 bp overhang generated by I-SceI (ATAA) is compared with the sequence of the recombination site it the A4 subclones and of the sequence of the other DNA involved in the rearrangement. Nucleotides with homology (asterisks), microhomology at the recombination junction (bold), and inserted nucleotides (italics) are shown. (B) DNA sequence analysis of the recombination site involved in the integration of the pNCT-tel plasmid on chromosome 20 in clone A4. The DNA from clone A4 is compared with the NotI-digested end of pNCT-tel and the sequence of chromosomeo 20q. The NotI site used to linearize the pNCT-tel plasmid prior to transfection is indicated (italics). (C) The structure of chromosome rearrangements predicted from the sequence analysis of the DNA found at the site of recombination. The location and orientation of the sequences at the site of recombination at the I-SceI site (white arrows) and other chromosomes involved in the recombination events (black arrows) are shown. The positions of centromeres are indicated by ovals.
DNA sequence analysis of the recombination site in subclone A4-GGR8 demonstrated that DNA from chromosome 20 was located at the I-SceI site, with the loss of 7 nucleotides from the site of the DSB, including the ATAA overhang generated by I-SceI. There was 1 bp inserted at the recombination junction. The orientation of the chromosome 20 sequence is towards the centromere on the long arm at the 20p12/20p13 junction, indicating the formation of a ring chromosome.
DNA sequence analysis of the recombination site in subclone A4-GGR9 demonstrated the presence of 124 bps of DNA from chromosome 4, followed by DNA from chromosome 16. There were 64 nucleotides lost at the I-SceI site, with 2 bps of microhomology at the recombination junction. Recombination between chromosome 4 and 16 involved 4 bps of microhomology. The orientation of the chromosome 16 sequence is towards the telomere on the short arm at 16p13, indicating that a translocation had occurred.
DNA sequence analysis of the recombination site in subclone A4-GGR10 demonstrated an inverted repeat at the I-SceI site, with the loss of 7 nucleotides including the ATAA overhang from one inverted repeat, and the loss of 1.9 kb of DNA from the other inverted repeat. There were 4 bps of microhomology at the recombination junction. The substantial degradation of DNA from one of the inverted repeats is typical of what we have observed in mouse ES cells [33], since selection in G418 requires only that one neo gene be intact.
DNA sequence analysis of the recombination site in subclone A4-GGR12 demonstrated that DNA from chromosome 4 was located at the I-SceI site, without the loss of any nucleotides from the ATAA overhang generated by I-SceI. There was no microhomology at the recombination junction. The site of recombination was 2002 bps from the canonical telomeric repeat sequences on the short arm of chromosome 4, with the sequence directed towards the centromere, indicating that a chromosome fusion involving the formation of a dicentric chromosome had occurred. The site of fusion so close to the telomere suggests that the fusion with chromosome 4 resulted from spontaneous telomere loss, which occurs at a rate of 10−4 events/cell/generation in EJ-30 [21].
3.9. The analysis of DSB-induced chromosome instability in Ganr/G418r subclones of clone B3
We have previously demonstrated that spontaneous telomere loss in clone B3 results in chromosome instability involving B/F/B cycles [13,21]. To demonstrate whether DSBs near telomeres also cause chromosome instability, we analyzed whether three Ganr/G418r subclones of B3 showed instability of chromosome 16. One of the subclones, B3-GGR62, contains a duplication from chromosome 5 at the end of chromosome 16p in 98% of the cells in the population, demonstrating that the marker chromosome 16 had undergone a stable DNA rearrangement (Table 2). However, in the other two subclones, B3-GGR70 and B3-GGR76, the marker chromosome 16 was highly unstable and appeared to still be undergoing B/F/B cycles (Fig. 9, Table 2). This instability was evident by the fact that the arm containing the telomeric plasmid was missing a telomere in many cells in the population, which we have demonstrated is associated with anaphase bridges and amplification of subtelomeric DNA [52]. Moreover, in the great majority of cells in these subclones, chromosome 16 has acquired a new telomere by one of a variety of mechanisms that we have previously observed after spontaneous telomere loss [13]. Some of these mechanisms of telomere acquisition stabilize the chromosome, as demonstrated by the homogeneity of the structure of the marker chromosome in secondary subclones [21]. However, other mechanisms of telomere acquisition can lead to further chromosome instability, both on the chromosome that acquired the telomere and on other chromosomes. These include the formation of nonreciprocal translocations [13], dicentric chromosomes that result from the fusion with other chromosomes, and ring chromosomes resulting from the fusion with the other end of chromosome 16. Interestingly, most of the events observed during B/F/B cycles were also observed as the initial rearrangements occurring after I-SceI-induced DSBs in Ganr/G418r subclones of clone A4 (see Fig. 8).
Table 2.
Mechanisms of telomere acquisition and rearrangement of chromosome 16 in subclones of EJ-30 clone B3 that contain inverted repeats following I-SceI-induced DSB near the telomere.
| Clone | No Tel | Tel Acq Tel lost | Tel Acq No tel lost | NRT | Transl Dup | Dup 16q | Loss 16p | Ring | Cascade | Dicent | TDL |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GGR62 | 0 | 0 | 0 | 2 | 98 | 0 | 0 | 0 | 0 | 0 | 0 |
| GGR70 | 9 | 11 | 11 | 13 | 19 | 8 | 24 | 0 | 1 | 3 | 1 |
| GGR76 | 8 | 21 | 21 | 12 | 15 | 5 | 0 | 2 | 5 | 2 | 9 |
Results given as percent of mitoses. Number of mitoses analyzed: 46 for GGR62, 75 for GGR70, 58 for GGR76. No Tel - no telomere on 16p. Tel Acq/Tel lost - Telomere acquired at 16p with telomere missing on another chromosome. Tel Acq/No tel lost - Telomere acquired at 16p without a telomere missing on another chromosome. NRT - telomere acquired by nonreciprocal translocation. Transl/Dup - telomere acquired without the loss of arm from other chromosome. Dup 16q - telomere acquired by duplication of the other end of chromosome 16. Loss 16p – loss of all or part of 16p. Ring - chromosome 16 as a ring chromosome. Cascade - translocations on chromosomes donating NRT to chromosome 16. Dicentric - chromosome 16 as a dicentric with another chromosome. TDL – nonreciprocal translocation with loss of donor chromosome.
FIG. 9.
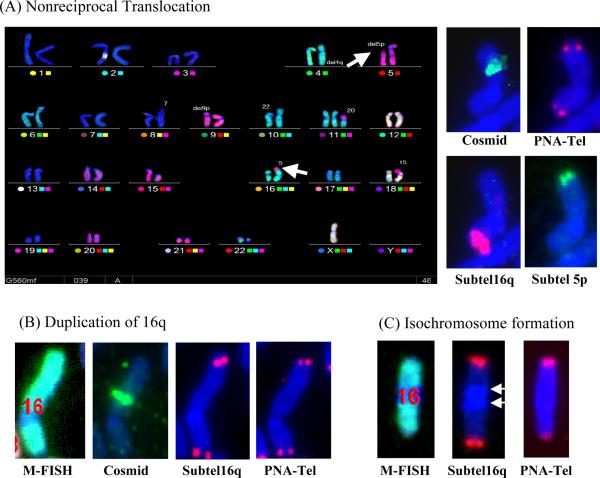
Instability of chromosome 16 in the Ganr/G418r subclone GGR70 isolated from clone B3. FISH analysis demonstrated that a variety of different types of rearrangements on the end of chromosome 16 containing the I-SceI-induced DSB, demonstrating an ongoing instability (Table 2). These rearrangements include (A) nonreciprocal translocations, (B) duplications originating from the other arm of chromosome 16, and (C) isochromosome formation. Chromosome analysis involved sequential hybridization using (1) M-FISH to identify all chromosomes, (2) cosmid RT99 adjacent to the integrated plasmid at 16p to identify the location of the site of sister chromatid fusion, (3) a PNA telomere-specific probe to identify telomeres, and (4) subtelomeric DNA for 16q, the arm that is opposite the plasmid integration site, or subtelomeric regions of chromosome 5p, to demonstrate the presence of portions of these chromosome arms on the end of chromosome 16. (A) M-FISH demonstrated that the nonreciprocal translocation involved the loss of the short (p) arm from chromosome 5 (arrow) and its appearance on chromosome 16p (arrow). The presence of the translocation near the original end of chromosome 16p is demonstrated by hybridization with the cosmid probe. Hybridization with subtelomeric probes demonstrate that one end of the chromosome is composed of chromosome 16q, while the other end is from 5p. Hybridization with the PNA telomeric probe demonstrates that the chromosome has telomeres on both ends. (B) The duplication of the other end of chromosome 16 in this cell is demonstrated by hybridization of a probe for the subtelomeric region of the long arm (16q) to both ends of the chromosome. The short arm is still present, as demonstrated by hybridization with the cosmid probe adjacent to the integrated plasmid, and telomeres are present on both ends. (C) The formation of an isochromosome by chromosome 16 in this subclone resulted in the loss of the 16p arm and the plasmid, and the duplication of the entire 16q arm, including the centromere (arrows) and telomere.
4. Discussion
The results presented here clearly establish that DSBs near telomeres have severe consequences for chromosome instability in the EJ-30 human tumor cell line. Consistent with earlier studies [29–32], we found that I-SceI-induced DSBs at interstitial sites most often resulted in small deletions, while large deletions and GCRs were relatively rare. We also found that the frequency of small deletions resulting from I-SceI-induced DSBs near telomeres was similar to that observed for interstitial DSBs (see Figs. 5 and 6A, Table 1), however, the frequency of large deletions and GCRs was greatly increased near telomeres (see Figs. 4 and 6A). In fact, large deletions and GCRs are by far the most common types of DNA rearrangements that are observed in cells experiencing DSBs near telomeres. This appears to be a general feature of mammalian cells, because we have also observed large deletions and GCRs as a result of I-SceI-induced DSBs near telomeres in mouse ES cells, although the frequency of these events was not determined [33,38]. Our results are consistent with the increased sister chromatid exchanges and dynamic evolution that is observed in human subtelomeric regions [53]. This increase in large deletions and GCRs near telomeres strongly indicates a high degree of sensitivity of subtelomeric regions to DSBs. An alternative explanation, that large deletions and GCRs also occur at a high frequency at interstitial sites, but are selected against because they are lethal, is highly unlikely in view of the excess number of large deletions and GCRs that are observed relative to small deletions. While large deletions and GCRs may be more likely to be lethal at some interstitial sites, they have been uniformly found to be rare events at a large number of different interstitial I-SceI sites in this and other studies [29–32]. Moreover, a study in human cells that detected chromosome translocations by PCR 48 hours after the introduction of two DSBs, and therefore was not dependent upon cell survival, demonstrated that translocations are several orders of magnitude less frequent than simple rejoining of ends at a single DSB [54]. The increased sensitivity of telomeric regions to DSBs is also consistent with studies in yeast, which were performed with a nonessential chromosome so that increased cell death at interstitial GCRs was not a factor [25].
The increased frequency of large deletions and GCRs as a result of DSBs near telomeres could result from a deficiency in one or more DSB repair pathways in telomeric regions. Not all DSB repair pathways would appear to be deficient, because the frequency of small deletions is similar at telomeric and interstitial sites. There are multiple mechanisms for repair of DSBs, including NHEJ [55], alternative NHEJ (Alt-NHEJ) [56], and homologous recombination repair (HRR) [57]. NHEJ and Alt-NHEJ involve the direct rejoining of broken DNA ends, while HRR in mammalian cells involves the utilization of the sister chromatid as a template for repair involving homologous recombination. A deficiency in NHEJ is one possible explanation for the high frequency of large deletions and GCRs resulting from DSBs near telomeres. I-SceI-induced DSBS at interstitial sites are faithfully repaired by NHEJ without deletions, while small deletions result from repair by Alt-NHEJ [58–60]. As a result, a deficiency in repair by NHEJ results in more extensive deletions [58–60]. Therefore, if NHEJ were defective near telomeres, DSBs at interstitial sites would go undetected due to faithful repair by NHEJ, while DSBs at telomeric sites would result in large deletions and GCRs. On the other hand, small deletions resulting from Alt-NHEJ would occur at the same frequency at both locations. Consistent with this model, studies in S. cerevisiae have reported deficient repair of I-SceI-induced DSBs by NHEJ near telomeres, which results in an increase in GCRs [25]. The rejoining of DSBs near telomeres by Alt-NHEJ might also be involved in the generation of large deletions and GCRs, since Alt-NHEJ is commonly associated with extensive degradation and chromosome rearrangements [56]. Alternatively, a deficiency in HRR near telomeres might also explain the high frequency of large deletions and GCRs resulting from DSBs near telomeres. The HRR process involves the resection of DNA at DSBs to generate large single-stranded regions that are used to mediate DSB repair through homologous recombination [57]. This resection can also involve the other strand, resulting in deletions at the site of the DSB [61], which could account for the extensive degradation observed at DSBs near telomeres.
The most common type of rearrangement we observed as a result of I-SceI-induced DSBs near telomeres is a large deletion that results in the complete loss of the plasmid sequences (see Figs. 4 and 6). Consistent with this observation, large deletions are also an important mechanism for chromosome instability following telomere loss in yeast [62]. Large deletions resulting in the complete loss of the telomeric plasmid sequences are also the most common rearrangement resulting from spontaneous telomere loss in the EJ-30 clone B3 [21], demonstrating that large deletions near telomeres are not limited to I-SceI-induced DSBs. The complete loss of the telomeric plasmid sequences could occur through several different mechanisms, all of which we have observed in our studies. The loss of the telomeric plasmid sequences could occur as a result of extensive degradation at the site of the DSB. Degradation at the I-SceI site is evident in the analysis of DNA rearrangements in our studies. Selection with G418 selects against cells that have extensive degradation that results in the loss of the neo gene. However, because only one copy of the neo gene is required for resistance to G418, there is no selection against degradation on one of the inverted repeats. Extensive degradation of one of the inverted repeats is seen in subclone B3-GGR10 (Fig. 8A), where one of the inverted repeats was joined at the I-SceI site and the other was degraded by 1.9 kb. The analysis I-SceI-induced inverted repeats in mouse ES cells also demonstrated extensive degradation at DSBs near telomeres, by as much as 30 kb in one Ganr/G418r subclone [33,38]. The complete loss of the telomeric plasmid sequences could also occur through the breakage of the chromosome following sister chromatid fusions or fusions to other chromosomes. Chromosome fusions result in the formation of chromosome bridges during anaphase, which results in the random breakage of the fused chromosomes [12,52]. As a result, some daughter cells will acquire additional copies of portions of the fused chromosomes, while the other daughter cells will acquire the corresponding chromosomes deletions. Finally, the complete loss of the telomeric plasmid sequences could occur through the loss of the entire chromosome or chromosome arm following DSB-induced GCRs. In fact, we have observed both an increase in the rate of chromosome loss and loss of the chromosome arm containing the telomeric plasmid sequences (isochromosome formation) in a subclone undergoing B/F/B cycles [13]. Consistent with this mechanism, dicentric and ring chromosomes have been demonstrated to have an increased likelihood of being lost with time in dividing cell populations [63].
The results presented here demonstrate for the first time that DSBs near telomeres in mammalian cells result in an increased frequency of propagated GCRs and chromosome instability compared to DSBs at interstitial sites. These GCRs and chromosome instability are likely to have severe consequences in that they result in many of the types of rearrangements associated with human cancer [64–66]. Moreover, the frequency of GCRs resulting from DSBs near telomeres is almost certainly underestimated in our study, because most subclones experiencing complete loss of the plasmid are also likely to experience GCRs. As mentioned above, complete loss of the plasmid sequences can result directly from GCRs as a result of the breakage or loss of rearranged chromosomes. We have also shown that GCRs are typically accompanied by extensive degradation [33,38], which is evident in subclone A4-GGR10 (Fig. 8) and ES cell subclones in which one sister chromatid was degradation up to 30 kb prior to sister chromatid fusion. Therefore, complete loss of the plasmid is likely to often result from a similar extent of degradation prior to formation of GCRs. However, although extensive degradation often precedes GCRs, it is not the sole reason for the GCRs that result from DSBs near telomeres, since we sometimes observe GCRs with minimal degradation at the site of the DSB in both EJ-30 human tumor cells (see Fig. 8) and in mouse ES cells [33,38].
Our results demonstrate that the initial GCRs resulting from DSBs near telomere can take several different forms, including translocations, sister chromatid fusions (inverted repeats), rings, and dicentric chromosomes, all of which can be unstable and result in additional rearrangements or chromosome loss. Similar types of GCRs are also subsequently observed during B/F/B cycles that result from spontaneous telomere loss [13] or DSBs near telomeres (Fig. 9, Table 2) in the EJ-30 cell line. This is not surprising, because these rearrangements result from the absence of a telomere on the end of the chromosomes in subsequent cell generations as a result of breakage of the fused sister chromatids during anaphase. However, this variety of GCRs differs from the initial GCRs that we have observed as a result of DSBs near telomeres in 6 subclones of mouse ES cells [33,38], all of which involved inverted repeats. Similarly, the initial GCRs that we observed following spontaneous telomere loss in 4 subclones of EJ-30 also involved inverted repeats [33], which were demonstrated by cytogenetic analysis to result from sister chromatid fusions [52]. We have previously proposed that the difference between the initial GCRs resulting from spontaneous telomere loss and the subsequent GCRs occurring during B/F/B cycles is due to the fact that the initial events occur during DNA replication [12]. During DNA replication, the ends of the sister chromatids would be in close proximity and therefore sister chromatid fusion would be the most likely GCR to occur. In contrast, the breakage of the sister chromatids during anaphase would allow ample time for the telomere-deficient chromosomes to be rejoined with a variety of other DSBs prior to DNA replication in the next cell cycle when sister chromatid fusion could again occur. This model would predict that spontaneous telomere loss in cancer cells occurs during DNA replication. Cancer cells have been demonstrated to have increased rates of chromosome breaks at fragile sites due to replication stress [22,23], which would be likely to occur near telomeres because subtelomeric regions, like fragile sites, have been shown in yeast to cause replication forks to stall [24]. This model would also predict that the timing of I-SceI-induced DSBs in human cancer cells is different from mouse ES cells, because all 6 of the I-SceI-induced GCRs analyzed in mouse ES cells consisted of inverted repeats. This difference in timing of formation of I-SceI-induced DSBs could be due to the limited access of the I-SceI endonuclease to the DNA due to chromatin structure, since the telomeric plasmid sequences in mouse ES cells [45], but not human tumor cells (unpublished observation, [67]), undergo DNA methylation and heterochromatin formation upon passage in culture.
The results presented here also confirm that DSBs near telomeres in human cells can result in chromosome healing. However, unlike many of the other events resulting from DSBs near telomeres, chromosome healing occurs at the site of the break and can prevent DNA degradation and chromosome instability [38]. Chromosome healing may therefore serve as an alternative mechanism for repair of DSBs near telomeres, which otherwise would lead to chromosome instability. Although chromosome healing results in a terminal deletion, this would have little consequence for DSBs near telomeres. An important observation in the current study is that the frequency of chromosome healing in the EJ-30 human tumor cell line is much lower than in mouse ES cells. As shown in Fig. 7, chromosome healing is rarely seen in EJ-30 subclones selected with ganciclovir alone, accounting for only 4 % of the rearrangements that were observed. In contrast, chromosome healing in mouse ES cells accounts for 25% of the I-SceI-induced events in subclones selected with ganciclovir. This low frequency of chromosome healing in EJ-30 has important implications for chromosome instability for DSB-induced or spontaneous telomere loss, since it may mean that in addition to an increased rate of telomere loss, human tumor cells are less capable of preventing chromosome instability involving B/F/B cycles. A deficiency in chromosome healing could therefore contribute to the chromosome instability resulting from telomere loss in human cancer cells.
The sensitivity of telomeric regions to DSBs provides new insights into how DSBs may contribute to telomere loss and the chromosome instability in human cancer cells. In some instances, DSBs near telomeres could result from exogenous sources, such as DSBs produced by ionizing radiation. Ionizing radiation is now known to initiate chromosome instability, although the mechanisms involved have yet to be elucidated [68–71]. However, as discussed above, endogenous DSBs also occur at an increased frequency in cancer cells due to replication stress [22,23], and may contribute to the spontaneous telomere loss that we, and others, have demonstrated as an important mechanism for chromosome instability in human cancer cell lines [17,21]. Although replication stress-induced DSBs may occur at many locations, our results demonstrate that those occurring near telomeres would be especially prone to initiating chromosome instability. A more complete understanding of the mechanisms responsible for the sensitivity of subtelomeric regions to DSBs and the role of chromosome healing in preventing chromosome instability should therefore provide important insights into the mechanisms of chromosome rearrangement in cancer.
Acknowledgement
The work in the J.P.M. laboratory was supported by National Institutes of Health grant CA12025. The work in the L.S. laboratory was supported by RISC-RAD contract number FI6RCT2003-508842.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- [2].Chan SR, Blackburn EH. Telomeres and telomerase. Philos Trans R Soc Lond B Biol Sci. 2004;359:109–121. doi: 10.1098/rstb.2003.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Vega LR, Mateyak MK, Zakian VA. Getting to the end: telomerase access in yeast and humans. Nat Rev Mol Cell Biol. 2003;4:948–959. doi: 10.1038/nrm1256. [DOI] [PubMed] [Google Scholar]
- [4].Harley CB. Telomeres and aging. In: Blackburn EH, Greider CW, editors. Telomeres. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1995. pp. 247–263. [Google Scholar]
- [5].Deng Y, Chan SS, Chang S. Telomere dysfunction and tumour suppression: the senescence connection. Nat Rev Cancer. 2008;8:450–458. doi: 10.1038/nrc2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB, Bacchetti S. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ducray C, Pommier J-P, Martins L, Boussin F, Sabatier L. Telomere dynamics, end-to-end fusions and telomerase activation during the human fibroblast immortalization process. Oncogene. 1999;18:4211–4223. doi: 10.1038/sj.onc.1202797. [DOI] [PubMed] [Google Scholar]
- [8].Shay JW, Wright WE. Telomerase activity in human cancer. Curr. Opin. Oncol. 1996;8:66–71. doi: 10.1097/00001622-199601000-00012. [DOI] [PubMed] [Google Scholar]
- [9].Raynaud CM, Sabatier L, Philipot O, Olaussen KA, Soria JC. Telomere length, telomeric proteins and genomic instability during the multistep carcinogenic process. Crit Rev Oncol Hematol. 2008;66:99–117. doi: 10.1016/j.critrevonc.2007.11.006. [DOI] [PubMed] [Google Scholar]
- [10].Reddel RR, Bryan TM. Alternative lengthening of telomeres: dangerous road less travelled. Lancet. 2003;361:1840–1841. doi: 10.1016/S0140-6736(03)13538-6. [DOI] [PubMed] [Google Scholar]
- [11].Bailey SM, Murnane JP. Telomeres, chromosome instability and cancer. Nucleic Acids Res. 2006;34:2408–2417. doi: 10.1093/nar/gkl303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Murnane JP. Telomeres and chromosome instability. DNA Repair (Amst) 2006;5:1082–1092. doi: 10.1016/j.dnarep.2006.05.030. [DOI] [PubMed] [Google Scholar]
- [13].Sabatier L, Ricoul M, Pottier G, Murnane JP. The loss of a single telomere can result in genomic instability involving multiple chromosomes in a human tumor cell line. Mol. Cancer Res. 2005;3:139–150. doi: 10.1158/1541-7786.MCR-04-0194. [DOI] [PubMed] [Google Scholar]
- [14].De Lange T. Telomere-related genome instability in cancer. Cold Spring Harb Symp Quant Biol. 2005;70:197–204. doi: 10.1101/sqb.2005.70.032. [DOI] [PubMed] [Google Scholar]
- [15].Artandi SE, Chang S, Lee S-L, Alson S, Gottlieb GJ, Chin L, DePinho RA. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- [16].Raynaud CM, Jang SJ, Nuciforo P, Lantuejoul S, Brambilla E, Mounier N, Olaussen KA, Andre F, Morat L, Sabatier L, Soria JC. Telomere shortening is correlated with the DNA damage response and telomeric protein down-regulation in colorectal preneoplastic lesions. Ann Oncol. 2008;19:1875–1881. doi: 10.1093/annonc/mdn405. [DOI] [PubMed] [Google Scholar]
- [17].Gisselsson D, Jonson T, Petersen A, Strombeck B, Dal Cin P, Hoglund M, Mitelman F, Mertens F, Mandahl N. Telomere dysfunction triggers extensive DNA fragmentation and evolution of complex chromosome abnormalities in human malignant tumors. Proc. Natl. Acad. Sci. USA. 2001;98:12683–12688. doi: 10.1073/pnas.211357798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gisselsson D, Pettersson L, Hoglund M, Heidenblad M, Gorunova L, Wiegant J, Mertens F, Dal Cin P, Mitelman F, Mandahl N. Chromosomal breakage-fusion-bridge events cause genetic intratumor heterogeneity. Proc. Natl. Acad. Sci. USA. 2000;97:5357–5362. doi: 10.1073/pnas.090013497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Montgomery E, Wilentz RE, Argani P. Analysis of anaphase figures in routine histologic sections distinguishes chromosomally unstable from chromosomally stable malignancies. Cancer Biol. Ther. 2003;2:248–252. doi: 10.4161/cbt.2.3.362. [DOI] [PubMed] [Google Scholar]
- [20].Meeker AK, Hicks JL, Iacoluzio-Donahue CA, Montgomery EA, Westra WH, Chan TY, Ronnett BM, De Marzo AM. Telomere length abnormalities occur early in the initiation of epithelial carcinogenesis. Clin. Can. Res. 2004;10:3317–3326. doi: 10.1158/1078-0432.CCR-0984-03. [DOI] [PubMed] [Google Scholar]
- [21].Fouladi B, Miller D, Sabatier L, Murnane JP. The relationship between spontaneous telomere loss and chromosome instability in a human tumor cell line. Neoplasia. 2000;2:540–554. doi: 10.1038/sj.neo.7900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Orntoft T, Lukas J, Bartek J. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- [23].Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr., Kastrinakis NG, Levy B, Kletsas D, Yoneta A, Herlyn M, Kittas C, Halazonetis TD. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- [24].Ivessa AS, Zhou J-Q, Schulz VP, Monson EK, Zakian VA. Saccharomyces Rrm3p, a 5' to 3' DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 2002;16:1383–1396. doi: 10.1101/gad.982902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ricchetti M, Dujon B, Fairhead C. Distance from the chromosome end determines the efficiency of double-strand break repair in subtelomeres of haploid yeast. J. Mol. Biol. 2003;328:847–862. doi: 10.1016/s0022-2836(03)00315-2. [DOI] [PubMed] [Google Scholar]
- [26].Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- [27].Richardson C, Jasin M. Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells. Mol. Cell. Biol. 2000;20:9068–9075. doi: 10.1128/mcb.20.23.9068-9075.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Honma M, Sakuraba M, Koizumi T, Takashima Y, Sakamoto H, Hayashi M. Non-homologous end-joining for repairing I-SceI-induced DNA double strand breaks in human cells. DNA Repair (Amst) 2007;6:781–788. doi: 10.1016/j.dnarep.2007.01.004. [DOI] [PubMed] [Google Scholar]
- [30].Rebuzzini P, Khoriauli L, Azzalin CM, Magnani E, Mondello C, Giulotto E. New mammalian cellular systems to study mutations introduced at the break site by non-homologous end-joining. DNA Repair (Amst) 2005;4:546–555. doi: 10.1016/j.dnarep.2004.12.011. [DOI] [PubMed] [Google Scholar]
- [31].Sargent RG, Brenneman MA, Wilson JH. Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol. Cell. Biol. 1997;17:267–277. doi: 10.1128/mcb.17.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Varga T, Aplan PD. Chromosomal aberrations induced by double strand DNA breaks. DNA Repair (Amst) 2005;4:1038–1046. doi: 10.1016/j.dnarep.2005.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lo AWI, Sprung CN, Fouladi B, Pedram M, Sabatier L, Ricoul M, Reynolds GE, Murnane JP. Chromosome instability as a result of double-strand breaks near telomeres in mouse embryonic stem cells. Mol. Cell. Biol. 2002;22:4836–4850. doi: 10.1128/MCB.22.13.4836-4850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].O'Toole CM, Povey S, Hepburn P, Franks LM. Identity of some human bladder cancer cell lines. Nature. 1983;301:429–430. doi: 10.1038/301429a0. [DOI] [PubMed] [Google Scholar]
- [35].Wild K, Bohner T, Aubry A, Folkers G, Schulz GE. The three-dimensional structure of thymidine kinase from Herpes simplex virus type 1. FEBS. 1995;368:289–292. doi: 10.1016/0014-5793(95)00680-8. [DOI] [PubMed] [Google Scholar]
- [36].Haarr L, Marsden HS, Preston CM, Smiley JR, Summers WC, Summers WP. Utilization of internal AUG codons for initiation of protein synthesis directed by mRNAs from normal and mutant genes encoding herpes simplex virus-specified thymidine kinase. J Virol. 1985;56:512–519. doi: 10.1128/jvi.56.2.512-519.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Irmiere AF, Manos MM, Jacobson JG, Gibbs JS, Coen DM. Effect of an amber mutation in the herpes simplex virus thymidine kinase gene on polypeptide synthesis and stability. Virology. 1989;168:210–220. doi: 10.1016/0042-6822(89)90260-2. [DOI] [PubMed] [Google Scholar]
- [38].Gao Q, Reynolds GE, Wilcox A, Miller D, Cheung P, Artandi SE, Murnane JP. Telomerase-dependent and -independent chromosome healing in mouse embryonic stem cells. DNA Repair. 2008;7:1233–1249. doi: 10.1016/j.dnarep.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lansdorp PM, Verwoerd NP, van de Rijke FM, Dragowska V, Little M-T, Dirks RW, Raap AK, Tanke HJ. Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet. 1996;5:685–691. doi: 10.1093/hmg/5.5.685. [DOI] [PubMed] [Google Scholar]
- [40].Doggett NA, Goodwin LA, Tesmer JG, Meincke LJ, Bruce DC, Clark LM, Altherr MR, Ford AA, Chi H-C, Marrone BL, Longmire JL, Lane SA, Whitmore SA, Lowenstein MG, Sutherland RD, Mundt MO, Knill EH, Bruno WJ, Macken CA, Torney DC, Wu JR, Griffith J, Sutherland GR, Deaven LL, Callen DF, Moyzis RK. An integrated physical map of human chromosome 16. Nature. 1995;377(Suppl.):335–365. doi: 10.1038/377335a0. [DOI] [PubMed] [Google Scholar]
- [41].Longmire JL, Brown NC, Meincke LJ, Campbell ML, Albright KL, Fawcett JJ, Campbell EW, Moyzis RK, Hildebrand CE, Evans GA, Deaven LL. Construction and characterization of partial digest DNA. Genet. Anal. Tech. Appl. 1993;10:69–76. doi: 10.1016/1050-3862(93)90037-j. [DOI] [PubMed] [Google Scholar]
- [42].Knight SJL, Lese CM, Precht KS, Kuc J, Ning Y, Lucas S, Regan R, Brenan M, Nicod A, Lawrie NM, Cardy DLN, Nguyen H, Hudson TJ, Riethman HC, Ledbetter DH, Flint J. An optimized set of human telomere clones for studying telomere integrity and architecture. Am. J. Hum. Genet. 2000;67:320–332. doi: 10.1086/302998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Barnett MA, Buckle J, Evans EP, Porter ACG, Rout D, Smith AG, Brown WRA. Telomere directed fragmentation of mammalian chromosomes. Nucl. Acids Res. 1993;21:27–36. doi: 10.1093/nar/21.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hanish JP, Yanowitz JL, De Lange T. Stringent sequence requirements for the formation of human telomeres. Proc. Natl. Acad. Sci. USA. 1994;91:8861–8865. doi: 10.1073/pnas.91.19.8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Pedram M, Sprung CN, Gao Q, Lo AWI, Reynolds G, Murnane JP. Telomere position effect and silencing of transgenes near telomeres in the mouse. Mol. Cell Biol. 2006;26:1865–1878. doi: 10.1128/MCB.26.5.1865-1878.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sprung CN, Afshar G, Chavez EA, Lansdorp P, Sabatier L, Murnane JP. Telomere instability in a human cancer cell line. Mutat. Res. 1999;429:209–223. doi: 10.1016/s0027-5107(99)00115-3. [DOI] [PubMed] [Google Scholar]
- [47].Sprung CN, Sabatier L, Murnane JP. Telomere dynamics in a human cancer cell line. Exp. Cell Res. 1999;247:29–37. doi: 10.1006/excr.1998.4293. [DOI] [PubMed] [Google Scholar]
- [48].Brisebois JJ, DuBow MS. Selection for spontaneous null mutations in a chromosomally-integrated HSV-1 thymidine kinase gene yields deletions and a mutation caused by intragenic illegitimate recombination. Mutat. Res. 1993;287:191–205. doi: 10.1016/0027-5107(93)90012-5. [DOI] [PubMed] [Google Scholar]
- [49].Murata S, Matsuzaki T, Takai S, Yaoita H, Noda M. A new retroviral vector for detecting mutations and chromosomal instability in mammalian cells. Muatat. Res. 1995;334:375–383. doi: 10.1016/0165-1161(95)90075-6. [DOI] [PubMed] [Google Scholar]
- [50].Meier JL, Stinski MF. Effect of a modulator deletion on transcription of the human cytomegalovirus major immediate-early genes in infected undifferentiated and differentiated cells. J Virol. 1997;71:1246–1255. doi: 10.1128/jvi.71.2.1246-1255.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Sprung CN, Reynolds GE, Jasin M, Murnane JP. Chromosome healing in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA. 1999;96:6781–6786. doi: 10.1073/pnas.96.12.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lo AWI, Sabatier L, Fouladi B, Pottier G, Ricoul M, Murnane JP. DNA amplification by breakage/fusion/bridge cycles initiated by spontaneous telomere loss in a human cancer cell line. Neoplasia. 2002;6:531–538. doi: 10.1038/sj.neo.7900267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Mefford HC, Trask BJ. The complex structure and dynamic evolution of human subtelomeres. Nat Rev Genet. 2002;3:91–102. doi: 10.1038/nrg727. [DOI] [PubMed] [Google Scholar]
- [54].Weinstock DM, Brunet E, Jasin M. Induction of chromosomal translocations in mouse and human cells using site-specific endonucleases. J Natl Cancer Inst Monogr. 2008:20–24. doi: 10.1093/jncimonographs/lgn009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- [56].Nussenzweig A, Nussenzweig MC. A backup DNA repair pathway moves to the forefront. Cell. 2007;131:223–225. doi: 10.1016/j.cell.2007.10.005. [DOI] [PubMed] [Google Scholar]
- [57].Thompson LH, Schild D. Recombinational DNA repair and human disease. Mutat Res. 2002;509:49–78. doi: 10.1016/s0027-5107(02)00224-5. [DOI] [PubMed] [Google Scholar]
- [58].Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, Sabatier L, Lopez BS. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- [59].Guirouilh-Barbat J, Rass E, Plo I, Bertrand P, Lopez BS. Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc Natl Acad Sci U S A. 2007;104:20902–20907. doi: 10.1073/pnas.0708541104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zierhut C, Diffley JF. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. Embo J. 2008;27:1875–1885. doi: 10.1038/emboj.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Hackett JA, Greider CW. End resection initiates genomic instability in the absence of telomerase. Mol. Cell. Biol. 2003;23:8450–8461. doi: 10.1128/MCB.23.23.8450-8461.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Tucker JD, Cofield J, Matsumoto K, Ramsey MJ, Freeman DC. Persistence of chromosome aberrations following acute radiation: I, PAINT translocations, dicentrics, rings, fragments, and insertions. Environ. Mol. Mutagen. 2005;45:229–248. doi: 10.1002/em.20090. [DOI] [PubMed] [Google Scholar]
- [64].Brodeur GM, Hogarety MD. Gene amplification in human cancers: biological and clinical significance. In: Vogelstein B, Kinzler KW, editors. The genetic basis of human cancer. McGraw-Hill; New York: 1998. pp. 161–172. [Google Scholar]
- [65].Schwab M. Oncogene amplification in solid tumors. Sem. Cancer Biol. 1999;9:319–325. doi: 10.1006/scbi.1999.0126. [DOI] [PubMed] [Google Scholar]
- [66].Gollin SM. Chromosomal alterations in squamous cell carcinomas of the head and neck: window to the biology of disease. Head Neck. 2001;23:238–253. doi: 10.1002/1097-0347(200103)23:3<238::aid-hed1025>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- [67].Koering CE, Pollice A, Zibella MP, Bauwens S, Puisieux A, Brunori M, Brun C, Martins L, Sabatier L, Pulitzer JF, Gilson E. Human telomeric position effect is determined by chromosomal context and telomeric chromatin integrity. EMBO Rep. 2002;3:1055–1061. doi: 10.1093/embo-reports/kvf215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Sabatier L, Dutrillaux B, Martin MB. Chromosomal instability (Scientific Correspondence) Nature. 1992;357:548. doi: 10.1038/357548a0. [DOI] [PubMed] [Google Scholar]
- [69].Sabatier L, Lebeau J, Dutrillaux B. Chromosomal instability and alterations of telomeric repeats in irradiated human fibroblasts. Int. J. Radiat. Biol. 1994;66:611–613. doi: 10.1080/09553009414551701. [DOI] [PubMed] [Google Scholar]
- [70].Ayouaz A, Raynaud C, Heride C, Revaud D, Sabatier L. Telomeres: hallmarks of radiosensitivity. Biochimie. 2008;90:60–72. doi: 10.1016/j.biochi.2007.09.011. [DOI] [PubMed] [Google Scholar]
- [71].Little JB. Genomic instability and radiation. J. Radiol. Prot. 2003;23:173–181. doi: 10.1088/0952-4746/23/2/304. [DOI] [PubMed] [Google Scholar]