Abstract
Prohormone convertase 2 (PC2) functions in the generation of neuropeptides from their precursors. A quantitative peptidomics approach was used to evaluate the role of PC2 in the processing of peptides in a variety of brain regions. Altogether, 115 neuropeptides or other peptides derived from secretory pathway proteins were identified. These peptides arise from 28 distinct secretory pathway proteins, including proenkephalin, proopiomelanocortin, prodynorphin, protachykinin A and B, procholecystokinin, and many others. Forty one of the peptides found in wild type mice were not detectable in any of the brain regions of PC2 knockout mice, and another twenty four peptides were present at levels ranging from 20–79% of wild type levels. Most of the other peptides were not substantially affected by the mutation, with levels ranging from 80–120% of wild type levels, and only three peptides were found to increase in one or more brain regions of PC2 knock-out mice. Taken together, these results are consistent with a broad role for PC2 in neuropeptide processing, but with functional redundancy for many of the cleavages. Comparison of the cleavage sites affected by the absence of PC2 confirms previous suggestions that sequences with a Trp, Tyr and/or Pro in the P1′ or P2′ position are preferentially cleaved by PC2 and not by other enzymes present in the secretory pathway.
Keywords: neuropeptide biosynthesis, proprotein convertase, carboxypeptidase E, proteomics, protease, peptidase
Introduction
Neuropeptides play important roles in a large variety of physiological processes, from feeding and body weight regulation to anxiety, fear, learning and memory, reproduction, and many others. Neuropeptides are generated from precursor molecules by limited proteolysis at specific sites that usually contain multiple basic amino acids such as KR, RR, or RXXR (Zhou et al. 1999;Steiner 1998;Rouille et al. 1995;Seidah et al. 1999). Because the mature forms of the bioactive peptides usually lack N- or C-terminal basic amino acids, these need to be removed from the peptide-processing intermediates. In some cases, additional post-translational modifications are required: these modifications include phosphorylation, acetylation, sulfation, and C-terminal amidation (Prigge et al. 2000). In the traditional scheme for peptide biosynthesis, endoproteolytic cleavage occurs to the C-terminal side of the basic amino acid(s). Then, a carboxypeptidase removes the C-terminal basic residues from the intermediates (Supplemental Figure S1A). An alternative scheme has been proposed which involves initial endopeptidase cleavage to the N-terminal side of the basic residues, and the subsequent removal of these basic residues by an aminopeptidase (Figure S1B).
Enzymes that process peptide precursors for both the traditional and the alternative schemes described above have been identified, and those involved in the traditional scheme are generally well-accepted by most investigators. In the traditional scheme, the key peptide-processing endopeptidases in neuroendocrine tissues are PC1/3 and PC2, and the carboxypeptidases (CP) are CPE and CPD. PC1/3 and PC2 are members of a calcium-dependent, subtilisin-like serine protease family (Rouille et al. 1995;Seidah et al. 1999). Both of these PCs are specifically expressed in neuroendocrine tissues and are enriched in peptide-containing secretory vesicles (Zhou et al. 1999;Zheng et al. 1994;Steiner 1998;Rouille et al. 1995). The enzymatic activities of PC1/3 and PC2 have been extensively studied using a variety of approaches (Zhou et al. 1993;Zhou et al. 1993;Zhou and Mains 1994;Mathis and Lindberg 1992;Breslin et al. 1993;Dupuy et al. 1994;Paquet et al. 1996b;Johanning et al. 1996;Johanning et al. 1998;Day et al. 1998). The substrate specificity has been examined by incubating the purified enzyme with candidate substrates, and then analyzing the products (Zhou et al. 1993;Dupuy et al. 1994;Johanning et al. 1998;Day et al. 1998). In other studies, the enzyme was co-expressed with candidate substrates in neuroendocrine cell lines, and products secreted into the media and/or present inside the cells were characterized (Zhou and Mains 1994;Mathis and Lindberg 1992;Breslin et al. 1993;Paquet et al. 1996b). Conversely, peptide processing in neuroendocrine cell lines endogenously expressing relatively high levels of PC1/3 or PC2 was examined after antisense RNA was used to lower the expression level of the processing enzyme (Dhanvantari and Brubaker 1998;Johanning et al. 1996;Bloomquist et al. 1991). When PC1/3 and PC2 knock-out (KO) mouse models became available, numerous neuropeptide and peptide hormone precursor processing defects were found (Zhu et al. 2002b;Zhu et al. 2002a;Furuta et al. 1997;Allen et al. 2001;Berman et al. 2000;Czyzyk et al. 2003;Dey et al. 2003;Furuta et al. 1997;Johanning et al. 1998;Miller et al. 2003a;Miller et al. 2003b;Taylor et al. 2003;Dey et al. 2004;Pan et al. 2006;Pan et al. 2005;Scamuffa et al. 2006). Similarly, many processing defects were found in mice lacking CPE activity, adding overwhelming support to the traditional peptide processing scheme involving PCs and CPs (Fricker et al. 1996;Rovere et al. 1996;Cain et al. 1997;Udupi et al. 1997;Fricker and Leiter 1999;Lim et al. 2006;Zhang et al. 2008). The strong support for the traditional processing scheme does not rule out the possibility that an alternative processing scheme contributes to peptide biosynthesis in some brain regions, and some evidence in support of a pathway involving cathepsin L and aminopeptidase B has been reported by Hook and co-workers (Hook et al. 2008;Hook 2006;Hwang et al. 2007a;Hwang et al. 2007b;Funkelstein et al. 2008b;Funkelstein et al. 2008a).
In the present study, we have extended our initial analysis (Pan et al. 2006) of neuropeptides in the PC2 KO mouse using a quantitative peptidomics approach. In this approach, extracts from KO and wild type mice are separately labeled with reagents containing different stable isotopes, the samples are pooled, and peptides are purified and analyzed using liquid chromatography and electrospray ionization mass spectrometry (LC/MS). This approach enables the analysis of >100 peptides and also reveals the precise molecular form of each peptide, including post-translational cleavages and other modifications. The finding that a large number of peptides are substantially decreased in the PC2 KO mice, including several suggested to be processed by cathepsin L, supports a broad, required role for PC2 in the biosynthesis of many peptides and argues against a role for cathepsin L in their production. However, some peptides are not affected by the absence of PC2 activity and others are only partially affected, indicating that either PC1/3 or another endopeptidase is involved with neuropeptide production. In addition, this analysis provides insights as to the substrate specificity of the PCs.
Materials and Methods
Reagents
Hydrochloric acid (HCl) (6 N, sequanal grade, constant boiling), trifluoroacetic acid, and formic acid were purchased from Pierce (Rockford, IL, USA). Acetonitrile of HPLC grade was from Fisher Scientific (Fair Lawn, NJ, USA). Sodium phosphate dodecahydrate (Na2HPO4·12H2O), sodium hydroxide (NaOH), glycine, hydroxylamine, and dimethylsulfoxide (DMSO) were obtained from Aldrich Chemicals (St Louis, MO, USA). The isotopic labeling reagents, D0- and D9-forms of 3-(2,5-dioxopyrrolidin-1-yloxycarbonyl)propyl trimethylammonium chloride (abbreviated as D0-TMAB and D9-TMAB) were synthesized as described previously (Che et al. 2005;Che et al. 2005;Zhang et al. 2002). Water was purified with a Milli-Q system (Millipore, Bedford, MA, USA).
Mice
Adult (age range 14~16 weeks) PC2 KO mice and wild-type (WT) littermate mice were obtained from the mating of PC2 +/− mice and genotyping was done by Southern blot analysis as previously described in the initial report describing the generation of the PC2 KO (Furuta et al. 1997). Altogether, 9 male KO mice and 9 male WT mice were used and divided into groups of 3 mice/group, for a total of 3 groups of KO mice and 3 groups of WT mice. Each mouse was sacrificed by decapitation and the head was immediately placed into a microwave oven and irradiated for 8 s at full power to raise the brain temperature to 80°C, as described (Che et al. 2005). After cooling, the brain was removed, cut into coronal sections with a razor blade, and the prefrontal cortex, hypothalamus, striatum, hippocampus, thalamus, and amygdala dissected as previously described (Zhang et al. 2008). Tissues from each group of three KO mice or WT mice were separately pooled, immediately frozen in dry ice, and stored at −70°C until further processing.
Peptide extraction and the differential isotopic labeling
The extraction of peptides from brain regions was carried out as described previously (Pan et al. 2006). Briefly, tissue was sonicated in ~5 volumes of water, heated to 70°C for 20 minutes, cooled 15 minutes in ice, acidified to a final concentration of 10 mM HCl, and centrifuged in a microfuge for 15 minutes at 4°C. The supernatant was removed and stored at −70°C until labeling. Extracted peptides from PC2 KO mice or WT mice were labeled by D0- or D9-TMAB isotopic labeling reagents according to the labeling strategy shown in Supplemental Figure S2. For all brain regions except hypothalamus, each of the three groups of PC2 KO mice was paired with a WT group and the extracted peptides were labeled with the isotopic reagents (Figure S2; note the opposite orientation of the labeling of LC/MS Run 2, relative to Runs 1 and 3). For hypothalamus, two of the replicate groups were labeled as for Runs 1 and 2 (Figure S2). However, the third group was split and half was labeled as shown for Run 3, and the other half in the reverse orientation (Run 4). This strategy has been previously described in more detail (Pan et al. 2006). The labeling was performed as described preciously (Pan et al. 2006;Zhang et al. 2008). The combined D0- and D9-TMAB labeled samples were stored at −70°C until subsequent MS analyses.
LC-MS and data analysis
To detect and quantify the peptides from PC2 KO and WT mouse tissues, liquid chromatography and tandem mass spectrometry were performed on an ABI Q-star Pulsar-i™ quadrupole time-of-flight mass spectrometer (Applied Biosystems/MDS Sciex) equipped with a nanoelectrospray ionization source. The instrument setting and detection parameters were identical to those described previously (Pan et al. 2006). Database searching and peptide identification procedures are the same as described previously (Pan et al. 2006;Zhang et al. 2008). In brief, peptide identification was performed using a combination of Mascot searches followed by manual interpretation of the mass spectra to reduce false positives. Criteria for accepting or rejecting a peptide sequence have been described in several previous publications (Che et al. 2007;Zhang et al. 2008;Morano et al. 2008).
Results
To examine the relative levels of a number of peptides in PC2 KO mouse brain regions, a quantitative peptidomics method was used in which extracts were labeled with either light (D0-TMAB) or heavy (D9-TMAB) isotopic tags according to the scheme in Figure S2. The labeling strategy of the hypothalamus, and some of the hypothalamic data, were previously reported (Pan et al. 2006); additional analyses of the hypothalamic data were performed and included with the new data for the additional brain regions in the present study. These additional brain regions were analyzed in triplicate (Figure S2), with each replicate representing pools of tissue from distinct animals and involving a comparison of both KO and WT mice in the same LC/MS run. In addition, for the analysis of some peptides (described below) we included previously published data that involved the comparison of multiple groups of WT animals with other WT animals; this provides an estimate of the natural variation of the peptide levels among groups of animals with the same genetic background (Zhang et al. 2008).
Altogether, 115 distinct peptides that arose from secretory pathway proteins were detected in the 19 different LC/MS analyses. Most of these peptides were found in multiple runs from the same region and/or in different regions, for a total of 1082 separate data points. A small number were detected in just a single run, and a few peptides were detected in every LC/MS run from every brain region. In considering each peptide and each brain region separately, but combining data for the multiple LC/MS runs and/or charge states, the 1082 separate data points were reduced to 409 distinct entries in Table S1 (Supplemental information). In addition to these secretory pathway peptides, other peptides were detected which arose from cytosolic, nuclear, or mitochondrial proteins; these were not included in the present analysis. In general, the number of peptides derived from cytosolic, nuclear, or mitochondrial proteins was comparable to the number of neuropeptides/secretory pathway proteins.
A frequency plot of the primary data of each of the 1082 separate data points shows that approximately 1/3 of all detected secretory pathway peptides are present in the PC2 KO mouse brain at levels ≤20% of their levels in WT mouse brain (i.e. PC2 KO: WT ratio ≤0.20) (Figure 1, filled symbols and solid line). In contrast, a similar analysis comparing multiple groups of WT animals with other WT animals shows a ratio close to the theoretical 1.00 (Figure 1, open symbols and dashed line). Approximately 40–50% of the secretory pathway peptides shows a ratio between the PC2 KO and WT mice that is within the range of the WT:WT comparison (i.e. 0.80 to 1.2), and ~20% of the peptides show a partial decrease with PC2 KO: WT ratios of 0.20 to 0.80 (Figure 1). These three groups are discussed further below. In most cases, a similar PC2 KO:WT ratio was found in each brain region in which a peptide was detected. However, there were some exceptions and several peptides showed a difference between brain regions; these are also discussed below. Representative spectra of peptides that are altered in one or more brain regions of PC2 KO mice but not all other brain regions are shown in Figures S3, S4, and S5 (Supplemental information).
Figure 1.
Frequency plot showing the distribution of the number of peptides observed with each relative abundance ratio (in ranges of 0.05 increments). X axis: the relative ratio between each peptide in two different groups of samples. Filled circles represent the ratio between PC2 KO mice and WT mice. Open circles represent the ratio between separate groups of WT mice, and has been previously reported (Zhang et al. 2008).
Of the 115 distinct peptides, 41 were found only in the WT-labeled peak of each data set, and were not detectable in the PC2 KO mice (Table 1 and Supplementary Tables S1 and S2). Based on the detection limit (estimated from the noise in the MS spectrum), these peptides are present in the mutant mice at levels ≤10% of their levels in WT mice. Three of the peptides in this group are fragments of PC2 itself, and therefore should not be present in the PC2 KO mice. One peptide from the PC2 chaperone protein 7B2 was found to be absent in PC2 KO tissue. In addition, a peptide derived from the C-terminal region of PC1/3 that was found in the hypothalamus of WT mice is not detectable in the PC2 KO hypothalamus, suggesting that PC2 is involved in the generation of this peptide in the hypothalamus. Other notable members of the group of peptides which are not detectable in the PC2 KO mice include α-melanocyte-stimulating hormone, corticotropin-like intermediate lobe peptide, and other proopiomelanocortin fragments. Several peptides derived from proenkephalin, prodynorphin, chromogranin A and B, and procholecystokinin are also in this group (Table 1).
Table 1.
Neuropeptides and other secretory pathway peptides detected in wild type mouse brain and either not detectable in PC2 KO mouse brain (left), present in PC2 KO mouse brain at reduced levels (middle), or unaffected by the PC2 mutation (right).
| Peptides greatly decreased in PC2 KO mice (PC2KO:WT ratio <0.10) | Peptides partially decreased in PC2 KO mice (PC2KO:WT ratio 0.10 to 0.80) | Peptides present in PC2 KO mice at levels similar to WT mice (PC2KO:WT ratio 0.80–1.20) | |||
|---|---|---|---|---|---|
| Precursor | Peptide Sequence | Precursor | Precursor | Precursor | Peptide Sequence |
| 7B2 | (LLYEKMKGGQ) | Chromogranin B | LLDEGHYPV | Cerebellin 1 | GSAKVAFSAIRSTN |
| CART | APGAMLQIEALQEVLKKL | Chromogranin B | SFARAPQLDL | Cerebellin 1 | SGSAKVAFSAIRSTN |
| CART | APGAMLQIEALQEVLKKLKS | Chromogranin B | LGALFNPYFDPLQWKNSDFE | Cerebellin 1 | GSAKVAFSAIRSTNH |
| Chromogranin A | AYGFRDPGPQL | Procholecystokinin | AVLRTDGEPRARLGALLA | Cerebellin 1 | SGSAKVAFSAIRSTNH |
| Chromogranin A | WSRMDQLAKELTAE | Procholecystokinin | KAPSGRMSVLKNLQSLDPSHRISD | Cerebellin 2 | GSAKVAFSATRSTN |
| Chromogranin A | LEGEDDPDRSMKLSFRT | Prodynorphin | YGGFLRKYPK | Cerebellin 2 | (SGSAKVAFSATRSTN) |
| Chromogranin B | YPQSKWQEQE | Proenkephalin | YGGFMRF | Cerebellin 2 | (GSAKVAFSATRSTNH) |
| Chromogranin B | QYDGVAELDQLLHY | Proenkephalin | SPQLEDEAKELQ | Cerebellin 2 | (SGSAKVAFSATRSTNH) |
| Chromogranin B | LLDEGHYPVRESPIDT | Proenkephalin | FAESLPSDEEGENYSKEVPEIE | Cerebellin 4 | SKVAFSAVRSTN |
| Chromogranin B | LLDEGHYPVRESPIDTA | Proenkephalin | FAESLP-phosphoS-DEEGENYSKEVPEIE | Cerebellin 4 | ANSKVAFSAVRSTN |
| Chromogranin B | PSPKESKEADVATVRLGE | Proneuropeptide Y | (YPSKPDNPGEDAPAEDMARYYSALRHYIN LITRQRY-amide) | Cerebellin 4 | AANSKVAFSAVRSTN |
| Chromogranin B | SGKEVKGEEKGENQNSKFEVRLL | Proneurotensin | KIPYIL | Chromogranin B | GLQYRGRGSEEDRAPRPR |
| Procholecystokinin | ARLGALLA | Proopiomelanocortin | AEEEAVWGDGSPEPSPRE-amide | Chromogranin B | GLQYRGRG-phosphoS-EEDRAPRPR |
| Procholecystokinin | (QLRAVLRTDGEPRARLGA) | Proopiomelanocortin | (ELEGERPLGLEQVLESDAEKDDGPY) | Chromogranin B | (SGKEVKGEEKGENQNSKFEV) |
| Procholecystokinin | (APSGRMSVLKNLQSLDPSHRIS) | ProSAAS | (LENPSPQAPARRLLP) | Chromogranin B | (EERRPSPKESKEADVATVRLGE) |
| Procholecystokinin | APSGRMSVLKNLQSLDPSHRISD | ProSAAS | ARPVKEPRSLSAASAPLVETSTPL | Procholecystokinin | (GEPRARLGALL) |
| Procholecystokinin | (AVLRTDGEPRARLGALLARYIQQV) | ProSAAS | AVPRGEAAGAVQELARALAHLLEAERQE | Procholecystokinin | (AVLRTDGEPRARL-amide) |
| Prodynorphin | YGGFLRRI | Protachykinin A | HKTDSFVGLM-amide | Procholecystokinin | AVLRTDGEPRARLGA |
| Prodynorphin | PKLKWDNQ | Protachykinin A | RPKPQQFFGLM-amide | Procholecystokinin | AVLRTDGEPRARLGAL |
| Prodynorphin | (YGGFLRRQFKVVT) | Protachykinin A | ALNSVAYERSAMQNYE | Procholecystokinin | AVLRTDGEPRARLGALL |
| Prodynorphin | (YGGFLRRIRPKLKWDNQ) | Secretogranin II | IPVGSLKNEDTPNRQYLDEDMLLKVLEYLNQ EQAEQGREHLA | Prodynorphin Progastrin Releasing | YGGFLRKYP |
| Proenkephalin | YGGFL | VGF | NAPPEPVPPPRAAPAPTHV | Peptide | (APVSTGAGGGTVLAKMYP) |
| Proenkephalin | YGGFMRSL | VGF | LEGSFLGGSEAGERLLQQGLAQVEA-(amide) | PC1/3 | LSDDDRVTWAEQQYEKERS |
| Proenkephalin | YGGFMRRV-amide | VGF | (AQEEADAEERRLQEQEELENYIEHVLLHRP) | Propeptidyl-amidating-monooxygenase | FRSPLSVF |
| Proenkephalin | YGGFMRRVGR | ProSAAS | ASAPLVETSTPLRL | ||
| Proenkephalin Progastrin Releasing | VGRPEWWMDYQ | ProSAAS | SLSAASAPLVETSTPL | ||
| Peptide | (GSHWAVGHLM-amide) | ProSAAS | SVDQDLGPEVPPENVLGALL | ||
| PC1/3 | GVEKMVNVVE | ProSAAS | SVDQDLGPEVPPENVLGALLRV | ||
| PC2 | SLQSILRKN | ProSAAS | GEAAGAVQELARALAHLLEAERQE | ||
| PC2 | IKMALQQEGFD | ProSAAS | (AVPRGEAAGAVQELARALAHLLEAERQERA) | ||
| PC2 | pyroE-ELEEELDEAVERSLQSILRKN | Protachykinin B | DMHDFFVGLM-amide | ||
| ProMHC | EIGDEENSAKFPI-amide | ProTRH | SFPWMESDVT | ||
| Proopiomelanocortin | SYSMEHFRWGKPV-amide | ProTRH | DLQRVRGDLGAALDSWIT | ||
| Proopiomelanocortin | Ac-SYSMEHFRWGKPV-amide | ProTRH | FIDPELQRSWEETEGEEGGLMPE | ||
| Proopiomelanocortin | Ac-SYS-Mox-EHFRWGKPV-amide | ProTRH | LLEAAQEEGAVTPDLPGLEKVQVRPE | ||
| Proopiomelanocortin | RPVKVYPNVAENESAEAFPLEF | Provasopressin | VQLAGTRESVDSAKP | ||
| Proopiomelanocortin | RPVKVYPNVAENE-phosphoS-AEAFPLEF | Provasopressin | (AGTRESVDSAKPRVY) | ||
| Protachykinin A Provasoactive | DADSSVEKQVALLKALYGHGQISH | Provasopressin | VQLAGTRESVDSAKPRVY | ||
| Intestinal Peptide | ISSSISEDPVPI | Secretogranin II | ESKDQLSEDASKVITYL | ||
| VGF | KKNAPPEPVPPPRAAPAPTHV | Secretogranin III | FPKPEGSQDKSLHN | ||
| VGF | (QQETAAAETETRTHTLTRVNLESPGPERVW) | ||||
Precursor: name of protein precursor from which peptide is derived. Abbreviations: CART, cocaine- and amphetamine-regulated transcript; proMHC, promelanin concentrating hormone; proTRH, prothyrotropin releasing hormone. Note that 7B2 and VGF are names, not abbreviations. Peptide name: common name of peptide, or sequence position within precursor. CLIP, corticotropin-like intermediate lobe peptide; MSH, melanocyte-stimulating hormone. N-term flanking: the 5 amino acids in the precursor adjacent to the N-terminus of the peptide are indicated. Peptide sequence: The peptide sequences bracketed by parentheses represent tentatively identified peptides (i.e. based on mass, charge state, and number of isotopic tags incorporated). All other peptides were identified unambiguously by sequencing. Mox, oxidized Met; phosphoS, phosphorylated Ser. C-term flanking: the 5 amino acids in the precursor adjacent to the C-terminus of the peptide are indicated. * in this column represent the C-terminal end of the precursor protein. Theor. mass: theoretical monoisotopic mass (in Da) of the unprotonated peptide (after subtracting the mass of the tags). Region: A, amygdala; Hi, hippocampus; Hy, hypothalamus; PC, prefrontal cortex; S, striatum; T, thalamus. PC2:WT Avg: the average ratio of peptide levels in extracts of PC2 KO mouse brain regions relative to the levels in extracts from wild type mice. n: number of times the peptide was detected in different samples.
A total of 24 peptides were detectable in PC2 mouse extracts at levels lower than those in WT extracts, with average PC2 KO: WT ratios ranging from 0.20 to 0.79 (Table 1 and Supplementary Tables S1 and S3). Data from each brain region was pooled to obtain a sufficient sample size for statistical analysis, comparing the results with previously analyzed data from WT:WT comparisons of the same peptides and brain regions (Zhang et al. 2008). Nineteen of the peptides showed a statistically significant difference between the PC2 KO:WT ratio and the WT:WT ratio (Supplementary Table S3). The other 5 peptides showed a tendency for a decrease in the PC2 KO mice, but due to the variation in the data for either the PC2 KO:WT or WT:WT ratios, statistical significance could not be established.
A third group of peptides showed generally similar levels in the PC2 KO and WT groups, with ratios between 0.80 and 1.20 (Table 1 and Supplementary Tables S1 and S4). A total of 40 peptides fell into this group. Many of the peptides in this group do not contain a PC consensus site at either side. Examples include peptides derived from the precursor proteins for cerebellin 1, cerebellin 2, and cerebellin 4, and peptides derived from procholecystokinin and provasopressin (Supplementary Table S4).
In addition to the peptides described above which consistently fell into one of the three groups, another 10 peptides either increased in one or more brain regions, or showed changes in some brain regions that were different from those in another region (Supplementary Tables S1 and S5). The procholecystokinin-derived peptide (region 65–94) was found to be elevated ~45% in the prefrontal cortex of PC2 KO mice, relative to WT mice (Supplementary Table S5). This peptide contains a PC consensus site near the N-terminus, and the shorter peptide (region 72–94) was only detected in WT mice and not the PC2 KO mice (Supplementary Table S2). Thus, the longer peptide appears to be a PC2 substrate and the shorter one a PC2 product. Two other peptides also show an increase in some brain regions of the PC2 KO mice, relative to WT mice: chromogranin B 357–373 with and without a phosphate group on Ser365 (Supplementary Table S5). However, shorter peptides corresponding to products could not be found in our data, and there were no clear PC consensus sites (although several potential single basic cleavage sites). Instead, it is possible that the levels of these peptides are increased in the PC2 KO mice due to the action of another enzyme; CPE or CPD. Both of these chromogranin B-derived peptides require CP cleavage of a Pro-Arg bond, and neither CPE nor CPD are efficient with substrates containing a penultimate Pro (Smyth et al. 1989;Novikova et al. 1999). The Arg-extended peptides (Chromogranin B 357–374, with and without phosphoserine) were also detected in mouse brain in the present study (Supplementary Table S4). Therefore, the increase in the relative levels of the shorter des-Arg form of the chromogranin peptides in some brain regions of the PC2 KO mice could reflect increased activity of one of the CPs, possibly due to less competition with other peptides as a result of the decreased processing of many peptides in the PC2 KO mouse brain.
Other peptides showed variability in the extent of decrease in PC2 KO mice, relative to WT mice, among various brain regions (Supplementary Table S5). For example, the C-terminal region of neuropeptide Y is present in PC2 KO mouse prefrontal cortex at 44% of the level in WT brain but present in all other brain regions at ~95% of the level in WT brain (Supplementary Table S5 and Figure S4). Similarly, the proSAAS peptide “big SAAS” was present in PC2 KO mouse amygdala, hippocampus, and prefrontal cortex at levels close to WT mouse, but present in PC2 KO mouse hypothalamus, striatum, and thalamus at levels ~58% those of the WT mouse (Supplementary Table S5 and Figure S3). Several other peptides showed smaller differences between regions (Supplementary Tables S1 and S5).
Analysis of the cleavages site of the 41 peptides substantially affected by the absence of PC2 activity (i.e. those peptides in Table S2) was performed in order to examine whether specific amino acids were over- or under-represented in various positions nearby the cleavage site. For this analysis, both the N- and C-terminal sites were considered together because it was not possible to know if the processing defect was due to the lack of cleavage of one or both of these sites (even if only one cleavage site is used exclusively by PC2, the enzyme knockout will lead to the disappearance of this peptide). All residues in the P1 site are basic (Figure 2), which agrees with numerous previous studies on the activity of purified PC2 (Smeekens et al. 1992;Steiner 1998;Johanning et al. 1998;Lamango et al. 1996). Arg is favored over Lys in this P1 position approximately 6-fold (Figure 2). The P2 site is frequently also a basic residue, with Lys favored over Arg, but many other residues are found in this position. Preferences in the P3 to P5 positions are less pronounced, although there is a slight preference for Arg or an acidic residue in the P4 and P5 positions (Figure 2). Some residues are completely absent in one or more positions, although it is difficult to interpret this due to the small sample size (relative to the number of amino acids). However, the absence of Cys in the vicinity of the cleavage site (P5 through P5′) is consistent with previous studies analyzing PC1/3 and 2 cleavages (Cameron et al. 2001;Duckert et al. 2004;Pan et al. 2005;Pan et al. 2006). The P1′ site is frequently Tyr, Ala, or Ser, the P2′ site is frequently Tyr, Pro, or Gly, and the P3′ site is frequently Gly.
Figure 2.
Frequency of amino acids within the cleavage site of the PC2-selective cleavages. Only those peptides that are present in wild-type mice but not detected in PC2 KO mice were considered for this analysis (i.e. peptides in Table S2). Both up-stream and down-stream cleavage sites needed to produce the peptide were included. For the cleavage site on the N-terminus of the observed peptide, P1 was defined as the residue immediately upstream of the observed peptide and P1′ as the N-terminal residue of the observed peptide. For the cleavage site on the C-terminus of the observed peptide, the analysis was more complex and needed to take into account the activity of a carboxypeptidase and in some cases, the amidating enzyme that converts C-terminal Gly to an amide group. Thus, the P1 residue was defined as the basic residue that would produce an intermediate that would be further cleaved by a carboxypeptidase and amidating enzyme to generate the observed peptide.
A comparison of the cleavage sites found in the peptides grouped into Tables S2, S3, and S4 was performed to test if the amino acid frequencies noted in Figure 2 are predictors of whether a peptide is an exclusive product of PC2 or not. Those peptides that decreased substantially in the PC2 KO mice (Table S2) represent peptides that depend on PC2 activity and cannot be produced in sufficient levels without this enzyme. Peptides that show a partial decrease in the PC2 KO mice (Table S3) represent peptides that need PC2 but other enzymes can partially compensate and produce some of the peptide. Those peptides that are not affected by the absence of PC2 (Table S4) may be cleaved by this enzyme in part, but one or more additional enzymes are clearly involved in their production such that the absence of PC2 has no major effect on peptide levels. The presence of a Lys-Arg in the cleavage site was more frequently found in the peptides that require PC2 activity than in those that do not (Table 2). Arg-Arg in the cleavage site was less frequently found than Lys-Arg and showed the reverse trend, with more Arg-Arg sites in the peptides that do not require PC2 (Table 2). Cleavages at single basic Arg that lack another basic in the P2 position were relatively common among peptides that do not require PC2 and less frequently found in peptides that require PC2. For this analysis, only cleavage sites involving basic residues were considered; cleavages by signal peptidases or other non-basic cleavages were excluded because these steps are mediated by non-PC enzymes.
Table 2. Analysis of cleavage site residues.
Only cleavage sites at basic residues were considered; non-basic cleavage sites such as signal peptide cleavages were excluded from this analysis.
| P2 |
P1 |
P1′ |
P2′ |
Undetectable in PC2 KOa |
Partial decrease in PC2 KOb |
No change in PC2 KOc |
|---|---|---|---|---|---|---|
| K | R | 54% | 60% | 27% | ||
| R | R | 10% | 14% | 20% | ||
| K | K | 5% | 2% | 2% | ||
| R | K | 4% | 0% | 2% | ||
| other | R | 24% | 23% | 48% | ||
| other | K | 3% | 0% | 0% | ||
| P in P1′ or P2′ | 28% | 12% | 5% | |||
| Y in P1′or P2′ | 30% | 24% | 5% | |||
| W in P1′ or P2′ | 4% | 0% | 0% | |||
| F in P1′ or P2′ | 3% | 7% | 7% | |||
| H in P1′ or P2′ | 4% | 2% | 9% | |||
| G in P1′ or P2′ | 20% | 17% | 25% | |||
| A in P1′ or P2′ | 18% | 26% | 30% | |||
| S in P1′ or P2′ | 22% | 19% | 36% | |||
The fraction of cleavage sites for the peptides listed in Table S2 containing the indicated P2, P1, P1′, or P2′ residues is indicated. A total of 74 cleavage sites were considered.
The fraction of cleavage sites for the peptides listed in Table S3 containing the indicated P2, P1, P1′, or P2′ residues is indicated. A total of 42 cleavage sites were considered.
The fraction of cleavage sites for the peptides listed in Table S4 containing the indicated P2, P1, P1′, or P2′ residues is indicated. A total of 44 cleavage sites were considered.
Analysis of residues found in either the P1′ and/or P2′ position was also performed for the three groups of peptides. For this, all residues found to show higher frequency in these positions in Figure 2 as well as related residues were examined and compared among groups. The presence of Pro or Tyr in the P1′ or P2′ positions highly correlates with whether the peptide is cleaved primarily by PC2 or is cleaved by additional enzymes (Table 2). Other large aromatic residues such as Trp also showed this same trend, although there were fewer examples of cleavages containing this residue. On the other hand, Phe and His did not show the same trend (Table 2). As a side point, although His is often considered a basic residue because it can become positively charged at acidic pH values, at neutral pH it is usually uncharged and is cleaved efficiently by carboxypeptidase A type exopeptidases (which cleave aromatic residues) but not by carboxypeptidases with a specificity for basic residues (Smyth et al. 1989;Lyons et al. 2008), and therefore appears to some enzymes as an aromatic residue rather than a basic residue. Although Gly, Ala, and Ser appeared in the P1′ and P2′ positions of the cleavage sites of the PC2 products more frequently than most other amino acids, these residues were also commonly found in comparable positions in the peptides cleaved by other enzymes (Table 2), and therefore are not good predictors of whether a peptide will be dependent on PC2 for its production.
Discussion
A major finding of the present study is that PC2 plays a dominant role in the production of many neuropeptides and other secretory pathway peptides. Altogether, approximately 1/3 of the secretory pathway peptides detected in 6 different brain regions of WT mice are undetectable in the PC2 KO mice, suggesting that these peptides require PC2 for cleavage at either the N- and/or C-terminal site and this cleavage cannot be performed by a related enzyme such as PC1/3. Furthermore, this finding argues against a role for the cathepsin L/aminopeptidase pathway (Figure S1B) in the production of these peptides. Several recent studies have suggested that cathepsin L and aminopeptidase B contribute to the production of many peptides, including enkephalin, neuropeptide Y, and α-melanocyte stimulating hormone (αMSH) (Funkelstein et al. 2008b;Funkelstein et al. 2008a;Hook et al. 2008;Hook et al. 2009;Hwang et al. 2007b;Yasothornsrikul et al. 2003). Based on the present data, there is no evidence for an alternative pathway to PC2 for the production of Leu-enkephalin, other proenkephalin-derived peptides (octapeptide, metorphamide), or αMSH in the mouse brain regions examined; in the absence of PC2 activity these peptides are not detected (Tables S1 and S2). On the other hand, the proneuropeptide Y C-terminal peptide was only partially affected by the absence of PC2 activity in the prefrontal cortex, and was not affected at all in the other 5 brain regions examined (Table S5); this implies an alternative endopeptidase is involved in the cleavage of proneuropeptide Y into the bioactive N-terminal peptide and the C-terminal peptide. PC1/3 has been shown to efficiently cleave proneuropeptide Y at the dibasic site in several cell lines (Paquet et al. 1996b;Paquet et al. 1996a;Brakch et al. 1997). Further studies on PC1/3 knock-out mice are required to address the role of this enzyme in vivo.
Previous studies with purified enzymes have revealed considerable overlap in the specificity of PC1/3 and PC2, and therefore it is not possible to know whether PC2 participates in the processing of peptides unaffected by the disruption of the PC2 gene (i.e. peptides in Table S4). However, it is clear that an enzyme other than PC2 contributes to their processing, either redundantly with PC2 or alone; otherwise their levels would be affected by the PC2 KO. Those peptides that partially decreased in the PC2 KO mice but which were still detectable in the mutants presumably are cleaved by both PC2 and other endopeptidases, while those peptides showing a very large decrease in the PC2 KO mice represent cleavage products unique to PC2 that another enzyme cannot perform. Analysis of the cleavage sites of these three groups of peptides is generally consistent with previous studies testing purified PC2 with synthetic substrates (Smeekens et al. 1992;Steiner 1998;Johanning et al. 1998;Lamango et al. 1996;Kacprzak et al. 2005). In a comparison of tri-peptides with a flurogenic group in the P1′ position, Arg-Arg and Lys-Arg sequences were found to be cleaved by PC2 with comparable kinetics (Johanning et al. 1998). In the present study, Lys-Arg was more commonly found in peptides affected by the PC2 KO than in the peptides not affected by the mutation (54–60% versus 27%), while Arg-Arg was less prevalent in all groups and showed the reverse trend (10% versus 20%; see Table 2). Cleavages at single basic residues were more frequently found in the group of peptides unaffected by the PC2 gene disruption than in the groups of peptides that decreased in the PC2 KO mice; this finding fits with some of the previous studies on purified PC2 and synthetic substrates (Johanning et al. 1998). The presence of Pro, Tyr, or Trp in the P1′ or P2′ position was the most important predictor of whether a peptide was exclusively cleaved by PC2 (i.e. undetectable in PC2 KO mice), partially cleaved by PC2, or cleaved by other endopeptidases (possibly in addition to PC2). This tendency was noted in a previous study focused only on hypothalamus of the PC2 KO mice (Pan et al. 2006), and holds for the 5 other brain regions and many additional peptides examined in the present study. However, previous tendencies noted by Pan et al, such as the presence of a basic residue in the P3 position of preferred PC2 substrates, did not hold up when the larger number of peptides and brain regions was considered. Previous studies on purified enzyme also support the importance of Pro in the P1′ and P2′ positions in conferring exclusive or preferred processing of a site by PC2 and not the other PCs (Day et al. 1998;Kacprzak et al. 2005). However, other trends noted from the studies using purified PC2 and synthetic substrates, such as charged residues in the P2′ position or positively charged residues in the P1′ or P3′ positions (Kacprzak et al. 2005), do not appear to predict whether peptides require PC2 in vivo (Figure 2).
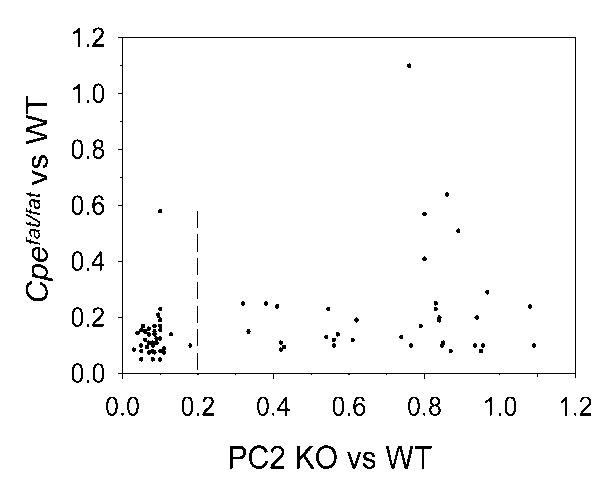
Recently, we used our quantitative peptidomics approach to compare levels of peptides in WT versus Cpefat/fat mice (Lim et al. 2006;Zhang et al. 2008); these mice lack CPE activity due to a point mutation in the coding region of the gene that causes the enzyme to be inactive and unstable (Naggert et al. 1995;Varlamov et al. 1996). Many peptides were found to be greatly reduced in the Cpefat/fat mice while only a small number of peptides that required removal of basic C-terminal residues were not affected, or partially affected, by the absence of CPE (Lim et al. 2006;Zhang et al. 2008). Thus, while PC1/3 and PC2 are both located within the mature secretory vesicle (Cameron et al. 2001;Zhou et al. 1999;Muller et al. 1998), CPE and CPD have overlapping substrate specificities but are localized to distinct compartments; CPE is present in mature secretory vesicles while CPD is present in Golgi, trans Golgi network, and immature vesicles but not detectable in mature secretory vesicles (Novikova et al. 1999;Chen et al. 2001;Varlamov et al. 1999a;Varlamov et al. 1999b;Guest et al. 1991). Thus, the peptides most dramatically affected in the Cpefat/fat mice are presumably those cleaved by endopeptidases late in the secretory pathway while those unaffected by the absence of CPE are cleaved by endopeptidases earlier in the Golgi, trans Golgi network, or immature vesicles where CPD is present and able to remove the basic residues from the endopeptidase reaction product. Therefore, we compared the results from the study on Cpefat/fat mice with the present results for PC2 KO mice and examined only those peptides which underwent both endopeptidase and carboxypeptidase cleavages and which were observed in both studies. For this analysis, mutant:WT ratios were assigned a value based on the detection limit, which varied from <0.03 to <0.13 due to the signal to noise ratio for each peptide (Figure 3). Although there is not a linear correlation between the relative levels of peptides in Cpefat/fat mice and the relative level in PC2 KO, there is considerable overlap among those peptides that are cleaved by both PC2 and CPE (Figure 3). Specifically, of the 74 peptides found in both studies that require endopeptidase and carboxypeptidase cleavages, half of these have a PC2 KO: WT ratio of <0.20. Of this group, the vast majority (>90%) have a Cpefat/fat: WT ratio of <0.20, indicating that they are cleaved late in the secretory pathway (Figure 3). In contrast, those peptides which had PC2 KO: WT ratios >0.70, showed a more equal distribution in their Cpefat/fat: WT ratio (Figure 3). This is consistent with both CPD and PC1/3 acting earlier in secretory granule maturation than their related peptidases, CPE and PC2, as previously proposed based on analysis of the enzymatic properties such as pH optima and activation by Ca2+ (Seidah and Chretien 2004b;Seidah and Chretien 2004a;Steiner 1998;Zhou et al. 1999;Fricker 2004a;Fricker 2004b). Thus, a combination of cleavage site preferences (Figure 2 and Table 2) and other factors such as secretory granule maturation and pH appear to contribute to the processing specificity of PC2 and explain why some peptides and not others are altered in the PC2 KO mice.
Figure 3.
Comparison of Cpefat/fat mice and PC2 KO mice, relative to WT mice. The analysis was performed on all peptides detected in both the current report and a previous study (Zhang et al. 2008) which required both processing by an endopeptidase and by a carboxypeptidase. The relative ratios of the peptides from Cpefat/fat versus WT mice were plotted against the ratio for the same peptide from PC2 KO versus WT mice. The identities of the peptides used for this figure are provided in Table S6 (supplemental information).
In summary, the quantitative peptidomics approach allows for the efficient analysis of the relative levels of peptides in two groups of mice, and provides information on the exact form of a peptide being measured. The application of this technique to the analysis of multiple brain regions of the PC2 KO mouse has greatly extended previous studies examining peptides in these mice, and collectively these studies provide a more complete understanding of the important role of PC2 in the production of bioactive peptides in vivo.
Supplementary Material
Acknowledgments
This work was supported primarily by grant DA04494, and also by grant DK51271 (L.D.F.), by grant DA015237 (J.E.P.), and by grant DK13914 and the Howard Hughes Medical Institute (D.F.S.). Special thanks to Ben Segall for creating a program for de novo sequencing of isotopic tagged peptides, which was used to determine the sequence of several of the peptides described in this paper. Mass spectrometry was performed in the Laboratory for Macromolecular Analysis and Proteomics of the Albert Einstein College of Medicine.
Abbreviations
- CP
carboxypeptidase
- KO
knock-out
- LC/MS
liquid chromatography mass spectrometry
- MSH
melanocyte stimulating hormone
- PC
prohormone convertase
- TMAB
3-(2,5-dioxopyrrolidin-1-yloxycarbonyl)propyl trimethylammonium chloride
- WT
wild-type
References
- Allen RG, Peng B, Pellegrino MJ, Miller ED, Grandy DK, Lundblad JR, Washburn CL, Pintar JE. Altered processing of pro-orphanin FQ/nociceptin and pro-opiomelanocortin-derived peptides in the brains of mice expressing defective prohormone convertase 2. J Neurosci. 2001;21:5864–5870. doi: 10.1523/JNEUROSCI.21-16-05864.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman Y, Mzhavia N, Polonskaia A, Furuta M, Steiner DF, Pintar JE, Devi LA. Defective prodynorphin processing in mice lacking prohormone convertase PC2. J Neurochem. 2000;75:1763–1770. doi: 10.1046/j.1471-4159.2000.0751763.x. [DOI] [PubMed] [Google Scholar]
- Bloomquist BT, Eipper BA, Mains RE. Prohormone-converting enzymes: Regulation and evaluation of function using antisense RNA. Mol Endocrinol. 1991;5:2014–1024. doi: 10.1210/mend-5-12-2014. [DOI] [PubMed] [Google Scholar]
- Brakch N, Rist B, Beck-Sickinger AG, Goenaga J, Wittek R, Burger E, Brunner HR, Grouzmann E. Role of prohormone convertases in pro-neuropeptide Y processing: coexpression and in vitro kinetic investigations. Biochemistry. 1997;36:16309–16320. doi: 10.1021/bi9714767. [DOI] [PubMed] [Google Scholar]
- Breslin MB, Lindberg I, Benjannet S, Mathis JP, Lazure C, Seidah NG. Differential processing of proenkephalin by prohormone convertases 1(3) and 2 and furin. J Biol Chem. 1993;268:27084–27093. [PubMed] [Google Scholar]
- Cain BM, Wang W, Beinfeld MC. Cholecystokinin (CCK) levels are greatly reduced in the brains but not the duodenums of Cpefat/Cpefat mice: A regional difference in the involvement of carboxypeptidase E (Cpe) in pro-CCK processing. Endocrinol. 1997;138:4034–4037. doi: 10.1210/endo.138.9.5490. [DOI] [PubMed] [Google Scholar]
- Cameron A, Apletalina EV, Lindberg I. The Enzymology of PC1 and PC2. Enzymes. 2001;22:291–332. [Google Scholar]
- Che FY, Lim J, Biswas R, Pan H, Fricker LD. Quantitative neuropeptidomics of microwave-irradiated mouse brain and pituitary. Mol Cell Proteomics. 2005;4:1391–1405. doi: 10.1074/mcp.T500010-MCP200. [DOI] [PubMed] [Google Scholar]
- Che FY, Zhang X, Berezniuk I, Callaway M, Lim J, Fricker LD. Optimization of neuropeptide extraction from the mouse hypothalamus. J Proteome Res. 2007;6:4667–4676. doi: 10.1021/pr060690r. [DOI] [PubMed] [Google Scholar]
- Chen H, Jawahar S, Qian Y, Duong Q, Chan G, Parker A, Meyer JM, Moore KJ, Chayen S, Gross DJ, Glasser B, Permutt MA, Fricker LD. A missense polymorphism in the human carboxypeptidase E gene alters its enzymatic activity: Possible implications in type 2 diabetes mellitus. Hum Mut. 2001;18:120–131. doi: 10.1002/humu.1161. [DOI] [PubMed] [Google Scholar]
- Czyzyk TA, Morgan DJ, Peng B, Zhang J, Karantzas A, Arai M, Pintar JE. Targeted mutagenesis of processing enzymes and regulators: implications for development and physiology. J Neurosci Res. 2003;74:446–455. doi: 10.1002/jnr.10792. [DOI] [PubMed] [Google Scholar]
- Day R, Lazure C, Basak A, Boudreault A, Limperis P, Dong W, Lindberg I. Prodynorphin processing by proprotein convertase 2: Cleavage at single basic residues and enhanced processing in the presence of carboxypeptidase activity. J Biol Chem. 1998;273:829–836. doi: 10.1074/jbc.273.2.829. [DOI] [PubMed] [Google Scholar]
- Dey A, Norrbom C, Zhu X, Stein J, Zhang C, Ueda K, Steiner DF. Furin and prohormone convertase 1/3 are major convertases in the processing of mouse pro-growth hormone-releasing hormone. Endocrinology. 2004;145:1961–1971. doi: 10.1210/en.2003-1472. [DOI] [PubMed] [Google Scholar]
- Dey A, Xhu X, Carroll R, Turck CW, Stein J, Steiner DF. Biological processing of the cocaine and amphetamine-regulated transcript precursors by prohormone convertases, PC2 and PC1/3. J Biol Chem. 2003;278:15007–15014. doi: 10.1074/jbc.M212128200. [DOI] [PubMed] [Google Scholar]
- Dhanvantari S, Brubaker PL. Proglucagon processing in an islet cell line: effects of PC1 overexpression and PC2 depletion. Endocrinology. 1998;139:1630–1637. doi: 10.1210/endo.139.4.5936. [DOI] [PubMed] [Google Scholar]
- Duckert P, Brunak S, Blom N. Prediction of proprotein convertase cleavage sites. Protein Eng Des Sel. 2004;17:107–112. doi: 10.1093/protein/gzh013. [DOI] [PubMed] [Google Scholar]
- Dupuy A, Lindberg I, Zhou Y, Akil H, Lazure C, Chretien M, Seidah NG, Day R. Processing of prodynorphin by the prohormone convertase PC1 results in high molecular weight intermediate forms. Cleavage at a single arginine residue. FEBS Lett. 1994;337:60–65. doi: 10.1016/0014-5793(94)80630-6. [DOI] [PubMed] [Google Scholar]
- Fricker LD. Carboxypeptidase E. In: Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of Proteolytic Enzymes. Academic Press; San Diego: 2004a. pp. 840–844. [Google Scholar]
- Fricker LD. Metallocarboxypeptidase D. In: Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of Proteolytic Enzymes. Academic Press; San Diego: 2004b. pp. 848–851. [Google Scholar]
- Fricker LD, Berman YL, Leiter EH, Devi LA. Carboxypeptidase E activity is deficient in mice with the fat mutation: Effect on peptide processing. J Biol Chem. 1996;271:30619–30624. doi: 10.1074/jbc.271.48.30619. [DOI] [PubMed] [Google Scholar]
- Fricker LD, Leiter EH. Peptides, enzymes, and obesity: new insights from a “dead” enzyme. Trends Biochem Sci. 1999;24:390–393. doi: 10.1016/s0968-0004(99)01448-6. [DOI] [PubMed] [Google Scholar]
- Funkelstein L, Toneff T, Hwang SR, Reinheckel T, Peters C, Hook V. Cathepsin L participates in the production of neuropeptide Y in secretory vesicles, demonstrated by protease gene knockout and expression. J Neurochem. 2008a;106:384–391. doi: 10.1111/j.1471-4159.2008.05408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funkelstein L, Toneff T, Mosier C, Hwang SR, Beuschlein F, Lichtenauer UD, Reinheckel T, Peters C, Hook V. Major role of cathepsin L for producing the peptide hormones ACTH, beta-endorphin, and alpha-MSH, illustrated by protease gene knockout and expression. J Biol Chem. 2008b;283:35652–35659. doi: 10.1074/jbc.M709010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta M, Yano H, Zhou A, Rouille Y, Holst JJ, Carroll R, Ravazzola M, Orci L, Furata H, Steiner DF. Defective prohormone processing and altered pancreatic islet morphology in mice lacking active SPC2. Proc Natl Acad Sci USA. 1997;94:6646–6651. doi: 10.1073/pnas.94.13.6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guest PC, Ravazzola M, Davidson HW, Orci L, Hutton JC. Molecular heterogeneity and cellular localization of carboxypeptidase H in the islets of langerhans. Endocrinol. 1991;129:734–740. doi: 10.1210/endo-129-2-734. [DOI] [PubMed] [Google Scholar]
- Hook V, Funkelstein L, Lu D, Bark S, Wegrzyn J, Hwang SR. Proteases for processing proneuropeptides into Peptide neurotransmitters and hormones. Annu Rev Pharmacol Toxicol. 2008;48:393–423. doi: 10.1146/annurev.pharmtox.48.113006.094812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook V, Funkelstein L, Toneff T, Mosier C, Hwang SR. Human pituitary contains dual cathepsin L and prohormone convertase processing pathway components involved in converting POMC into the peptide hormones ACTH, alpha-MSH, and beta-endorphin. Endocrine. 2009 doi: 10.1007/s12020-009-9163-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook VY. Protease pathways in peptide neurotransmission and neurodegenerative diseases. Cell Mol Neurobiol. 2006;26:449–469. doi: 10.1007/s10571-006-9047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SR, Garza C, Mosier C, Toneff T, Wunderlich E, Goldsmith P, Hook V. Cathepsin L expression is directed to secretory vesicles for enkephalin neuropeptide biosynthesis and secretion. J Biol Chem. 2007a;282:9556–9563. doi: 10.1074/jbc.M605510200. [DOI] [PubMed] [Google Scholar]
- Hwang SR, O’Neill A, Bark S, Foulon T, Hook V. Secretory vesicle aminopeptidase B related to neuropeptide processing: molecular identification and subcellular localization to enkephalin- and NPY-containing chromaffin granules. J Neurochem. 2007b;100:1340–1350. doi: 10.1111/j.1471-4159.2006.04325.x. [DOI] [PubMed] [Google Scholar]
- Johanning K, Juliano MA, Juliano L, Lazure C, Lamango NS, Steiner DF, Lindberg I. Specificity of prohormone convertase 2 on proenkephalin and proenkephalin-related substrates. J Biol Chem. 1998;273:22672–22680. doi: 10.1074/jbc.273.35.22672. [DOI] [PubMed] [Google Scholar]
- Johanning K, Mathis JP, Lindberg I. Role of PC2 in proenkephalin processing: antisense and overexpression studies. J Neurochem. 1996;66:898–907. doi: 10.1046/j.1471-4159.1996.66030898.x. [DOI] [PubMed] [Google Scholar]
- Kacprzak MM, Than ME, Juliano L, Juliano MA, Bode W, Lindberg I. Mutations of the PC2 substrate binding pocket alter enzyme specificity. J Biol Chem. 2005;280:31850–31858. doi: 10.1074/jbc.M505567200. [DOI] [PubMed] [Google Scholar]
- Lamango NS, Zhu X, Lindberg I. Purification and enzymatic characterization of recombinant prohormone convertase 2: Stabilization of activity by 21 kDa 7B2. Arch Biochem Biophys. 1996;330:238–250. doi: 10.1006/abbi.1996.0249. [DOI] [PubMed] [Google Scholar]
- Lim J, Berezniuk I, Che FY, Parikh R, Biswas R, Pan H, Fricker LD. Altered neuropeptide processing in prefrontal cortex of Cpefat/fat mice: Implications for neuropeptide discovery. J Neurochem. 2006;96:1169–1181. doi: 10.1111/j.1471-4159.2005.03614.x. [DOI] [PubMed] [Google Scholar]
- Lyons PJ, Callaway MB, Fricker LD. Characterization of carboxypeptidase A6, an extracellular-matrix peptidase. J Biol Chem. 2008 doi: 10.1074/jbc.M707680200. [DOI] [PubMed] [Google Scholar]
- Mathis JP, Lindberg I. Posttranslational processing of proenkephalin in AtT-20 cells: Evidence for cleavage at Lys-Lys site. Endocrinol. 1992;131:2287–2296. doi: 10.1210/endo.131.5.1425427. [DOI] [PubMed] [Google Scholar]
- Miller R, Aaron W, Toneff T, Vishnuvardhan D, Beinfeld MC, Hook VY. Obliteration of alpha-melanocyte-stimulating hormone derived from POMC in pituitary and brains of PC2-deficient mice. J Neurochem. 2003a;86:556–563. doi: 10.1046/j.1471-4159.2003.01856.x. [DOI] [PubMed] [Google Scholar]
- Miller R, Toneff T, Vishnuvardhan D, Beinfeld M, Hook VY. Selective roles for the PC2 processing enzyme in the regulation of peptide neurotransmitter levels in brain and peripheral neuroendocrine tissues of PC2 deficient mice. Neuropeptides. 2003b;37:140–148. doi: 10.1016/s0143-4179(03)00027-1. [DOI] [PubMed] [Google Scholar]
- Morano C, Zhang X, Fricker LD. Multiple Isotopic Labels for Quantitative Mass Spectrometry. Anal Chem. 2008;80:9298–9309. doi: 10.1021/ac801654h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller L, Picart R, Barret A, Seidah NG, Tougard C. Immunocytochemical localization of the prohormone convertases PC1 and PC2 in rat prolactin cells. J Histochem Cytochem. 1998;46:101–108. doi: 10.1177/002215549804600113. [DOI] [PubMed] [Google Scholar]
- Naggert JK, Fricker LD, Varlamov O, Nishina PM, Rouille Y, Steiner DF, Carroll RJ, Paigen BJ, Leiter EH. Hyperproinsulinemia in obese fat/fat mice associated with a point mutation in the carboxypeptidase E gene and reduced carboxypeptidase E activity in the pancreatic islets. Nature Genetics. 1995;10:135–142. doi: 10.1038/ng0695-135. [DOI] [PubMed] [Google Scholar]
- Novikova EG, Eng FJ, Yan L, Qian Y, Fricker LD. Characterization of the enzymatic properties of the first and second domains of metallocarboxypeptidase D. J Biol Chem. 1999;274:28887–28892. doi: 10.1074/jbc.274.41.28887. [DOI] [PubMed] [Google Scholar]
- Pan H, Che FY, Peng B, Steiner DF, Pintar JE, Fricker LD. The role of prohormone convertase-2 in hypothalamic neuropeptide processing: a quantitative neuropeptidomic study. J Neurochem. 2006;98:1763–1777. doi: 10.1111/j.1471-4159.2006.04067.x. [DOI] [PubMed] [Google Scholar]
- Pan H, Nanno D, Che FY, Zhu X, Salton SR, Steiner DF, Fricker LD, Devi LA. Neuropeptide Processing Profile in Mice Lacking Prohormone Convertase-1. Biochemistry. 2005;44:4939–4948. doi: 10.1021/bi047852m. [DOI] [PubMed] [Google Scholar]
- Paquet L, Massie B, Mains RE. Proneuropeptide Y processing in large dense-core vesicles: manipulation of prohormone convertase expression in sympathetic neurons using adenoviruses. J Neurosci. 1996a;16:964–973. doi: 10.1523/JNEUROSCI.16-03-00964.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet L, Zhou A, Chang EY, Mains RE. Peptide biosynthetic processing: distinguishing prohormone convertases PC1 and PC2. Mol Cell Endocrinol. 1996b;120:161–168. doi: 10.1016/0303-7207(96)03834-8. [DOI] [PubMed] [Google Scholar]
- Prigge ST, Mains RE, Eipper BA, Amzel LM. New insights into copper monooxygenases and peptide amidation: structure, mechanism and function. Cell Mol Life Sci. 2000;57:1236–1259. doi: 10.1007/PL00000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouille Y, Duguay SJ, Lund K, Furuta M, Gong Q, Lipkind G, Oliva AA, Jr, Chan SJ, Steiner DF. Proteolytic processing mechanisms in the biosynthesis of neuroendocrine peptides: the subtilisin-like proprotein convertases. Front Neuroendocrinol. 1995;16:322–361. doi: 10.1006/frne.1995.1012. [DOI] [PubMed] [Google Scholar]
- Rovere C, Viale A, Nahon J, Kitabgi P. Impaired processing of brain proneurotensin and promelanin-concentrating hormone in obese fat/fat mice. Endocrinol. 1996;137:2954–2958. doi: 10.1210/endo.137.7.8770919. [DOI] [PubMed] [Google Scholar]
- Scamuffa N, Calvo F, Chretien M, Seidah NG, Khatib AM. Proprotein convertases: lessons from knockouts. FASEB J. 2006;20:1954–1963. doi: 10.1096/fj.05-5491rev. [DOI] [PubMed] [Google Scholar]
- Seidah NG, Benjannet S, Hamelin J, Mamarbachi AM, Basak A, Marcinkiewicz J, Mbikay M, Chretien M, Marcinkiewicz M. The subtilisin/kexin family of precursor convertases. Emphasis on PC1, PC2/7B2, POMC and the novel enzyme SKI-1. Ann N Y Acad Sci. 1999;885:57–74. doi: 10.1111/j.1749-6632.1999.tb08665.x. [DOI] [PubMed] [Google Scholar]
- Seidah NG, Chretien M. Proprotein convertase 2. In: Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of Proteolytic Enzymes. Academic Press; San Diego: 2004a. pp. 1865–1868. [Google Scholar]
- Seidah NG, Chretien M. Proprotein convertase I. In: Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of Proteolytic Enzymes. Academic Press; San Diego: 2004b. pp. 1861–1864. [Google Scholar]
- Smeekens SP, Montag AG, Thomas G, Albiges-Rizo C, Carroll R, Benig M, Phillips LA, Martin S, Ohag S, Gardner P, Swift HH, Steiner DF. Proinsulin processing by the subtilisin-related proprotein convertases, PC2 and PC3. Proc Natl Acad Sci USA. 1992;89:8822–8826. doi: 10.1073/pnas.89.18.8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DG, Maruthainar K, Darby NJ, Fricker LD. C-terminal processing of neuropeptides: involvement of carboxypeptidase H. J Neurochem. 1989;53:489–493. doi: 10.1111/j.1471-4159.1989.tb07360.x. [DOI] [PubMed] [Google Scholar]
- Steiner DF. The proprotein convertases. Curr Opin Chem Biol. 1998;2:31–39. doi: 10.1016/s1367-5931(98)80033-1. [DOI] [PubMed] [Google Scholar]
- Taylor NA, Van de Ven WJ, Creemers JW. Curbing activation: proprotein convertases in homeostasis and pathology. FASEB J. 2003;17:1215–1227. doi: 10.1096/fj.02-0831rev. [DOI] [PubMed] [Google Scholar]
- Udupi V, Gomez P, Song L, Varlamov O, Reed JT, Leiter EH, Fricker LD, Greeley GHJ. Effect of carboxypeptidase E deficiency on progastrin processing and gastrin mRNA expression in mice with the fat mutation. Endocrinol. 1997;138:1959–1963. doi: 10.1210/endo.138.5.5113. [DOI] [PubMed] [Google Scholar]
- Varlamov O, Eng FJ, Novikova EG, Fricker LD. Localization of metallocarboxypeptidase D in AtT-20 cells: Potential role in prohormone processing. J Biol Chem. 1999a;274:14759–14767. doi: 10.1074/jbc.274.21.14759. [DOI] [PubMed] [Google Scholar]
- Varlamov O, Leiter EH, Fricker LD. Induced and spontaneous mutations at Ser202 of carboxypeptidase E: Effect on enzyme expression, activity, and intracellular routing. J Biol Chem. 1996;271:13981–13986. doi: 10.1074/jbc.271.24.13981. [DOI] [PubMed] [Google Scholar]
- Varlamov O, Wu F, Shields D, Fricker LD. Biosynthesis and packaging of carboxypeptidase D into nascent secretory vesicles in pituitary cell lines. J Biol Chem. 1999b;274:14040–14045. doi: 10.1074/jbc.274.20.14040. [DOI] [PubMed] [Google Scholar]
- Yasothornsrikul S, Greenbaum D, Medzihradszky KF, Toneff T, Bundey R, Miller R, Schilling B, Petermann I, Dehnert J, Logvinova A, Goldsmith P, Neveu JM, Lane WS, Gibson B, Reinheckel T, Peters C, Bogyo M, Hook V. Cathepsin L in secretory vesicles functions as a prohormone-processing enzyme for production of the enkephalin peptide neurotransmitter. Proc Natl Acad Sci U S A. 2003;100:9590–9595. doi: 10.1073/pnas.1531542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Sioma CS, Thompson RA, Xiong L, Regnier FE. Controlling deuterium isotope effects in comparative proteomics. Anal Chem. 2002;74:3662–3669. doi: 10.1021/ac025614w. [DOI] [PubMed] [Google Scholar]
- Zhang X, Che FY, Berezniuk I, Sonmez K, Toll L, Fricker LD. Peptidomics of Cpe(fat/fat) mouse brain regions: implications for neuropeptide processing. J Neurochem. 2008;107:1596–1613. doi: 10.1111/j.1471-4159.2008.05722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Streck RD, Scott REM, Seidah NG, Pintar JE. The developmental expression in rat of proteases furin, PC1, PC2, and carboxypeptidase E: Implications for early maturation of proteolytic processing capacity. J Neurosci. 1994;14:4656–4673. doi: 10.1523/JNEUROSCI.14-08-04656.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Bloomquist BT, Mains RE. The prohormone convertases PC1 and PC2 mediate distinct endoproteolytic cleavages in a strict temporal order during proopiomelanocortin biosynthetic processing. J Biol Chem. 1993;268:1763–1769. [PubMed] [Google Scholar]
- Zhou A, Mains RE. Endoproteolytic processing of proopiomelanocortin and prohormone convertases 1 and 2 in neuroendocrine cells overexpressing prohormone convertases 1 or 2. J Biol Chem. 1994;269:17440–17447. [PubMed] [Google Scholar]
- Zhou A, Webb G, Zhu X, Steiner DF. Proteolytic processing in the secretory pathway. J Biol Chem. 1999;274:20745–20748. doi: 10.1074/jbc.274.30.20745. [DOI] [PubMed] [Google Scholar]
- Zhu X, Orci L, Carroll R, Norrbom C, Ravazzola M, Steiner DF. Severe block in processing of proinsulin to insulin accompanied by elevation of des-64,65 proinsulin intermediates in islets of mice lacking prohormone convertase 1/3. Proc Natl Acad Sci USA. 2002a;99:10299–10304. doi: 10.1073/pnas.162352799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Zhou A, Dey A, Norrbom C, Carroll R, Zhang C, Laurent V, Lindberg I, Ugleholdt R, Holst JJ, Steiner DF. Disruption of PC1/3 expression in mice causes dwarfism and multiple neuroendocrine peptide processing defects. Proc Natl Acad Sci USA. 2002b;99:10293–10298. doi: 10.1073/pnas.162352599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.