Abstract
Purpose
To analyze human transforming growth factor b-induced (TGFBI) gene mutations in Chinese patients with corneal dystrophies (CDs).
Methods
Twenty-one families with corneal dystrophies were subjected to phenotypic and genotypic characterization. The corneal phenotypes of patients were documented by slit lamp photography. Mutation screening of the coding regions of TGFBI was performed by direct sequencing. An additional 43 families and 3 sporadic patients with TGFBI dystrophies from China reported in the literature were reviewed.
Results
Five mutations of TGFBI were identified in 21 families with CDs, including one novel small deletion mutation, c.△1838–1849 (p.Δ613–616VAEP), responsible for one variant lattice CD (LCD) family and 4 known mutations, R555W mutation for 10 granular cornea dystrophy type I (GCD1) families, R124H for 5 GCD type II (GCD2), R124C for 4 LCD1, and H626R for one variant LCD. In a cohort of Chinese patients (n=355) with TGFBI dystrophies from 64 families and 3 sporadic cases, 19 distinct mutations were found in several different CD subtypes. The 3 most common phenotypes were ranked as follows: GCD1, GCD2, and LCD1. Mutation hot spots at R124 and R555 occurred in >80% of these families.
Conclusions
Our findings extend the mutational spectrum of TFGBI, and this is also the first extensively delineated TGFBI mutation profile associated with the various corneal dystrophies in the Chinese population.
Introduction
Corneal dystrophies (CDs) are characterized by the occurrence of bilateral progressive opacities of the cornea that often arise from the deposition of insoluble material in the corneal stroma and lead to visual impairment. Since 1997, multiple phenotypes have been identified that are related to allelic mutations of the human transforming growth factor b-induced (TGFBI) gene (OMIM 601692) on human chromosome 5q31 [1]. This gene, originally named beta-ig-h3 (BIGH3), comprises 17 exons coding for a unique protein of 683-amino acids, which is produced by both mesenchymal and epithelial cells and denoted as keratoepithelin [1]. At present, more than 50 distinct disease-causing mutations have been identified in TFGBI that are associated with different corneal dystrophies, including granular cornea dystrophy (GCD) type I (GCD1; OMIM 121900), GCD type II (GCD2; OMIM 607541), lattice corneal dystrophy type I(LCD1; OMIM 122200), variant LCD, Thiel-Behnke corneal dystrophy (CDTB; OMIM 602082), Reis-Buckler corneal dystrophy (CDRB; OMIM 608470), and epithelial basement membrane corneal dystrophy (EBMD; OMIM 121820). In the present study, we designated all of the cornea dystrophies associated with TGFBI mutations as TGFBI dystrophies.
Of these mutations, two mutational hot spots have been identified corresponding to R124 and R555 of the TGFBI protein as being the most frequent sites of mutation in various populations [2-7]. Despite TGFBI dystrophies represent a clinically heterogeneous group of disorders, a strong correlation between specific mutations at these two positions and the observed phenotypes has been observed: GCD1/R555W, CDTB/R555Q, LCD1/R124C, GCD2/R124H, and CDRB/R124L. These above CDs were also classified as the classic form of TGFBI dystrophies [8].
The TGFBI mutation spectrum and their clinical consequences have been investigated in patients with CDs in different ethnic populations [2-7]. However, the spectrum of TGFBI mutations and the correlation between genotype and phenotype in the Chinese population has not been studied extensively. In this study, we report our findings on 21 new CD families with TGFBI mutations, and an additional 43 families and 3 sporadic patients with TGFBI dystrophies in a Chinese population collected from the literature [9-31], and delineate extensively the TGFBI mutation profile associated with the various corneal dystrophies in China.
Methods
Patients and subjects
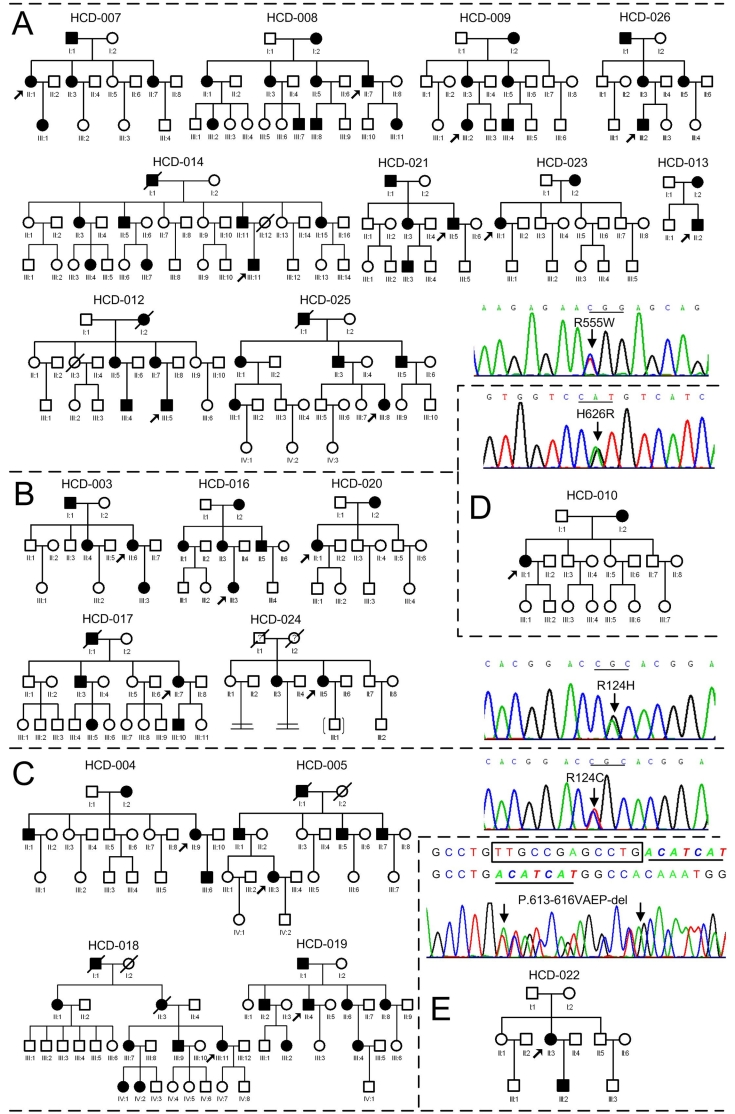
As part of a genetic screening program for genetic eye disorders, we collected materials from 26 families diagnosed with corneal dystrophies from China. A slit-lamp examination was performed for all participating individuals to determine if they were affected or unaffected with corneal dystrophies and to determine the disease phenotype. Of these families, 21 were further classified into 3 diagnostic categories: GCD1 in 10 families, LCD in 6 families, and GCD2 in 5 families. The pedigrees of these 21 families are shown in Figure 1, and the slit-lamp photographs of the representative patients with CDs are shown in Figure 2. After informed consent conforming to the tenets of the Declaration of Helsinki and following the guidance of sample collection of National Infrastructure Program of Chinese Genetic Resources (NICGR), 112 blood samples (58 samples from patients and 54 from normal members of families) from the 21 families and other 50 samples from ethnically matched control individuals were obtained before the study.
Figure 1.
Pedigrees of 21 Chinese families with corneal dystrophies and sequence chromatograms of their corresponding mutations TGFBI. GCD 1/R555W mutation (A), GCD2/R124H mutation (B), LCD1/R124C mutation (C), variant LCD/H626R mutation (D), and Variant LCD/Δ613–616VAEP mutation (E). Squares and circles symbolize males and females, respectively. The open and closed symbols indicate unaffected and affected individuals, respectively. The proband is marked with an arrow.
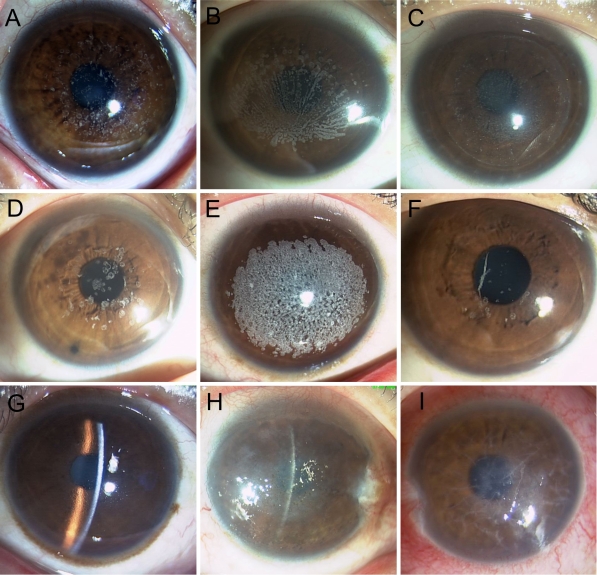
Figure 2.
Representative corneal phenotypes as shown by slit lamp examination. The proband of GCD1 family HCD-007 with R555W mutation had crumb-shaped and round white opacities (A). The proband of GCD1 family HCD-023 with R555W mutation showed confluent gray white opacities in a fan-like distribution in the central cornea (B) and recurrence corneal deposits after penetrating keratoplasty (C). The proband of GCD2 family HCD-016 with R124H mutation showed that the corneal opacities resemble rings, disks or snowflakes (D). The patient with the severe form of corneal dystrophy had confluent round white opacities in a coralloid shape in the superficial stromal layer (E), whereas his brother had fewer round opacities and thick spicular opacities (F) in a consanguineous marriage Chinese family clinically diagnosed as GCD2. The proband of LCD1 family HCD-005 with R124C mutation showed numerous fine, branching refractile lattice lines in subepithelial and stromal layers of the cornea (G). The proband of LCD family HCD-010 with H626R mutation showed recurrent diffuse stromal haze (H). The proband of LCD family HCD-022 with Δ613–616VAEP mutation showed thick, stellate lattice lines with intervening opacities and extending more to the periphery (I).
Genetic analysis
Total genomic DNA was extracted from whole blood using the Wizard Genomic DNA Purification Kit (Promega, Beijing, China) according to the manufacturer’s instructions. Amplification of the coding regions of TGFBI from genomic DNA was performed with the primers as described previously [1,2,22]. The sequences of the primers are listed in Table 1. Each PCR reaction was carried out in a 50 µl reaction mixture containing 100-200 ng of genomic DNA, 0.25 µl of TaKaRa Ex TaqTM (5U/µl; TaKaRa Biotechnology Co., Ltd., Dalian, China), 4.0 µl of dNTPs (2.5 mM each), 1.0 µl of each primer (10 pmol), 5 µl of 10× Ex Taq Buffer (Mg2+ plus). PCR conditions were as follows: 94 °C for 3 min; 30 cycles of 94 °C for 30 s, 52 °C or 55 °C or 58 °C for 30 s, 72 °C for 1 min; and further extension step at 72 °C for 5 min. PCR products were purified and sequenced on an ABI 3730XL Automated Sequencer (PE Biosystems, Foster City, CA), using the same PCR forward primers.
Table 1. Summary of the primers used for the amplification of the 17 exons of the human TGFBI gene.
| Exon | Primersequnce(5’→3') | Annealing temperature (°C) | Amplicon length (bp) |
|---|---|---|---|
| 1 |
F: GCGCTCTCACTTCCCTGGAG |
52 |
252 |
| |
R: GACTACCTGACCTTCCGCAG |
|
|
| 2 |
F: GGTGGACGTGCTGATCATCT |
58 |
194 |
| |
R: AGCCAGCGTGCATACAGCTT |
|
|
| 3 |
F: ACCTGTGAGGAACAGTGAAG |
58 |
200 |
| |
R: GCCTTTTATGTGGGTACTCC |
|
|
| 4 |
F: CCCCAGAGGCCATCCCTCCT |
58 |
225 |
| |
R: CCGGGCAGACGGAGGTCATC |
|
|
| 5 |
F: TAAACACAGAGTCTGCAGCC |
58 |
260 |
| |
R: TTCATTATGCACCAAGGGCC |
|
|
| 6 |
F: TGTGTTGACTGCTCATCCTT |
58 |
317 |
| |
R: CATTCAGGGGAACCTGCTCT |
|
|
| 7 |
F: TTCAGGGAGCACTCCATCTT |
55 |
224 |
| |
R: ATCTAGCTGCACAAATGAGG |
|
|
| 8 |
F: CTTGACCTGAGTCTGTTTGG |
58 |
324 |
| |
R: GAAGTCGCCCAAAGATCTCT |
|
|
| 9 |
F: ACTTTTGAACCCACTTTCTC |
58 |
200 |
| |
R: CAATCTAACAGGGATGCCTT |
|
|
| 10 |
F: TCTGGACCTAACCATCACCC |
58 |
206 |
| |
R: CAGGAGCATGATTTAGGACC |
|
|
| 11 |
F: CTCGTGGAAGTATAACCAGT |
58 |
223 |
| |
R: TGGGCAGAAGCTCCACCCGG |
|
|
| 12 |
F: CATTCCAGTGGCCTGGACTCTACTATC |
58 |
318 |
| |
R: GGGGCCCTGAGGGATCACTACTT |
|
|
| 13 |
F: GGGATTAACTCTATCTCCTT |
58 |
249 |
| |
R: TGTGTATAATTCCATCCTGG |
|
|
| 14 |
F: CTGTTCAGTAAACACTTGCT |
58 |
262 |
| |
R: CTCTCCACCAACTGCCACAT |
|
|
| 15 |
F: CACTCTGGTCAAACCTGCCT |
58 |
147 |
| |
R: AGGCTAGGCGCAAACCTAGC |
|
|
| 16 |
F: CAGTTGCAGGTATAACTTTC |
58 |
120 |
| |
R: TAAACAGGTCTGCAATGACT |
|
|
| 17 |
F: GGGAGATCTGCACCTATTTG |
58 |
113 |
| R: TGGTGCATTCCTCCTGTAGT |
Publications selection
To evaluate the incidence of TFGBI mutations in Chinese patients with TGFBI dystrophies, we also reviewed published articles obtained by an electronic search on PubMed/MEDLINE, VIP Information, CNKI, WANGFANG, data and Google using the keywords “corneal dystrophy” and “TGFBI” or “BIGH3.” Articles were limited to full papers in the English or Chinese language literature, and the search ended in February, 2010. When the same author reported results obtained on a complete or partial similar patient population in several publications, only the most recent or the most complete report was included in the analysis to avoid overlap. According to these criteria, 23 eligible trials published between 2002 and 2010 were selected [9-31]. The articles reported mutations of the TGFBI/BIGH3 gene associated with corneal dystrophies in the Chinese population. All of these TGFBI dystrophies were reclassified as classic or variant form according to Aldave and Sonmez’s [8] criterion.
Results
Mutational analysis
We screened the coding regions of TGFBI using PCR-based amplification followed by direct sequencing. Mutations were identified in all patients from the 21 unrelated Chinese families (Figure 1). Five distinct mutations of TGFBI were identified, including one novel small deletion mutation, c.Δ1838–1849 (p.Δ613–616VAEP), in exon 14 responsible for the HCD-022 family with variant LCD (Figure 1E and Figure 2I). This mutation was found in the proband and her son, but not in her parents and 50 normal controls. The other four known mutations were R555W mutation for 10 GCD1 families (Figure 1A and Figure 2A-C), R124H for 5 GCD2 families (Figure 1B and Figure 2D), R124C for 4 LCD1 families (Figure 1C and Figure 2G) and H626R for one variant LCD family (Figure 1D and Figure 2H).
Profile of TGFBI dystrophies
To delineate the TGFBI mutation profile associated with the various corneal dystrophies in the Chinese population, an additional 43 families and 3 sporadic cases with TGFBI dystrophies collected from the literature were also included in the investigation. Thus, a total of 355 patients from 64 families and 3 sporadic patients were included in the statistical analysis. We reclassified these TGFBI dystrophies as classic or variant form, with GCD1/R555W, GCD2/R124H, LCD1/R124C, CDRB/R124L, and CDTB/R555Q as classic forms and the others as corresponding variants [8].
In this cohort of Chinese patients, 19 distinct TGFBI mutations were identified in several different CD subtypes (Table 2). The most common phenotypes were the GCD1 associated with the R555W mutation (26.6%, 17/64 families), GCD2 with the R124H mutation (26.6%,17/64families), and LCD1 with the R124C mutation (18.8%,12/64 families). The mutational hot spots at positions R124 and R555 occurred in 53.1% (34/64) and 28.1% (18/64) of the families, respectively. Our observation also showed that the phenotypes correlated strongly with specific mutations in TGFBI, including that between GCD1 and R555W, GCD2 and R124H, and LCD1 and R124C. The exception is two CDTB families associated with the R124C mutation [26]. In addition, more than half (11/19) of these mutations were observed in Chinese patients with variant LCDs (Table 2).
Table 2. TGFBI mutations and phenotypes of patients in 61 families and 3 sporadic cases from Chinese origin.
| Corneal phenotype | Mutation | Exon | Number of families | Number of patients (Male/Female) | Reference |
|---|---|---|---|---|---|
| GCD1 |
R555W |
12 |
17 |
92 (37/65) |
[10-12,17,18,23,28], This study |
| |
A546D |
12 |
1 |
10 (6/4) |
[23] |
| GCD2 |
R124H |
4 |
17 |
65 (32/33) |
[10-12,17,24,25], This study |
| |
R124H+c.Δ307–308CT |
4 |
|
1 |
[9] |
| LCD1 |
R124C |
4 |
12 |
89 (43/46) |
[15,16,21,25,31], This study |
| Variant LCD |
H626R |
14 |
3 |
14 (7/7) |
[15,25], This study |
| |
V505D |
11 |
1 |
10 (4/6) |
[14] |
| |
T538P |
12 |
1 |
3 (1/2) |
[16] |
| |
V625D |
14 |
1 |
2 (1/1) |
[20] |
| |
Δ613–616VAEP |
14 |
1 |
2 (1/1) |
This study |
| |
I522N |
12 |
1 |
4 (1/3) |
[29] |
| |
P501T |
11 |
|
1 |
[16] |
| |
R514P+F515L |
11 |
1 |
5 (4/1) |
[31] |
| |
A546T |
12 |
|
1 |
[30] |
| |
H572R |
13 |
1 |
3 (2/1) |
[31] |
| CDRB |
R124L |
4 |
3 |
14 (3/11) |
[10,13] |
| |
G623D |
14 |
1 |
7 (3/4) |
[22,27] |
| CDTB |
R555Q |
12 |
1 |
14 (6/8) |
[17,19] |
| |
R124C |
4 |
2 |
18 (9/9) |
[26] |
| Total | 19 | 64 | 355 |
Discussion
We searched for mutations of TGFBI in 21 unrelated Chinese families affected with GCDs or LCDs. Five distinct mutations of TGFBI were identified in all of the families (Figure 1), and there have been perfect correlations between GCD1 and R555W, between GCD2 (ACD) and R124H, and between LCD1 and R124C. Among of these mutations, one novel 12 bp deletion mutation, c.Δ1838–1849 (p.Δ613–616VAEP), in exon 14 was found in the HCD-022 family (Figure 1E) with variant LCD (Figure 2I). Compare with the five small deletion mutations (1~6 bp deleted) in TGFBI reported previously by other groups [2,4,9,32,33], this mutation was the longest deletion mutation in TGFBI. It was also identified as a spontaneous inheritable TGFBI mutation because only the proband and her affected son carried the mutation, and not her parents. Spontaneous mutations in this gene are rare, but have been reported. Tanhehco et al. [34] reported a spontaneous R124L mutation in TGFBI in 2 patients with CDRB, and Zhao et al. [35] reported a spontaneous R555Q mutation in TGFBI in two unrelated families with Bowman's layer corneal dystrophies (CDTB and CDRB). Our finding not only extends the mutational spectrum of TFGBI, but also has historic value as a possible explanation for the presence of this autosomal dominant disorder in the population.
Through extensive investigation of the clinical and genetic findings in TGFBI dystrophies in the Chinese population, we observed that GCD1 in association with the R555W mutation (17 families) and GCD2 in association with the R124H mutation (17 families) accounted for more than half (53.1%, 34/64) of the families and were the most prevalent type of this disease. Our observations also confirm the presence of mutation hot spots at positions R124 and R555, which occur in more than 80% (52/64) of the families with TGFBI dystrophies in China. There were strong genotype-phenotype correlations between R555W and GCD1, between R124H and GCD2, and between R124C and LCD1 in our collection of 64 families, which is consistent with previous reports in patients from other ethnic origins [2-7]. The exception is 2 Chinese typical CDTB families associated with the R124C mutation [26]. Although these two mutations have been identified as major causes of LCDs or GCDs in different ethnic groups [2-7], population-specific variations in the prevalence of different mutations in TGFBI have been documented. The classic form of GCD1 with the R555W mutation and LCD1 with the R124C mutation are more common in different ethnic groups in Europe, the United States, and India [2,4,6,7], but in other populations, such as in Japan and Korea, GCD1 is found much less frequently than GCD2 associated with the R124H mutation [3,5], and H626R may be more frequent than R124C in Vietnamese patients with LCDs [36].
The second common type of TGFBI dystrophies is the LCDs, which accounted for more than one third of all of the families (34.4%, 22/64) in the Chinese population. Of these LCDs families, the major subtype is the classic form of LCD (LCD1) associated with the R124C mutation (54.5%,12/22). The other different forms of lattice dystrophy were reclassified as variant LCDs, which were the most mutationally heterogeneous and accounted for half of the mutations identified. In comparison to these common types, CDRB and CDTB were rare (about 5% of each subtype) in the Chinese population.
Most of the TGFBI mutations associated with corneal dystrophies are heterozygous. However, homozygous mutations are also found in some patients with severe form of TGFBI dystrophies [37,38]. Only two patients bearing homozygous R124H mutations from a consanguineous marriage family with GCD2 have been reported in China [24]. This disease is also described as a semidominant condition in which the heterozygous mutant is less severely affected than the homozygote. We also found a patient with severe form of corneal dystrophy in a consanguineous marriage family clinically diagnosed as GCD2. He had confluent round opacities in a coralloid shape in both of his eyes (Figure 2E), which is similar in phenotype to the patients with GCD2 bearing a homozygous R124H mutation reported previously [38]. His parents and brother had fewer round opacities and spicular opacities (Figure 2F). Unfortunately, we could not obtain blood samples for gene examination.
In addition to allelic heterogeneity, TGFBI dystrophies also exhibited extensive intrafamily and interfamily variations in clinical phenotypes, even when the same single point mutation in TGFBI is involved. Perhaps the most extreme example of the phenotypic variability is unaffected carriers who had the disease-causing mutations in TGFBI. In our collection of 64 Chinese families, 11 these carriers from 4 families were reported [21,23,24]. Of these cases, 2 unaffected relatives bearing the A546D mutation from a GCD1 family [23] and 2 from 2 LCD1 families carrying the R124C mutation [21] were too young to rule out inheritance of these two diseases by clinical examination. However, 7 individuals carrying a heterogeneous R124H mutation showed no clinical manifestations in a reduced penetrance GCD2 Chinese family, and an age-related factor was excluded in the contribution to these carriers [24]. All of this clinical evidence suggests that other factors such as age, environmental factors, and modifier genes may influence the expressivity and the penetrance of this disease, but further studies are needed for confirmation.
In summary, this study reported our findings in 21 new CD families with TGFBI mutations, and delineated extensively the TGFBI mutation profile associated with the various corneal dystrophies in the Chinese population: both GCD1 associated with the R555W mutation and GCD2 with the R124H mutation are the most common forms of this disease. We also confirmed the presence of mutational hotspots at positions R124 and R555, and a strong correlation between these two mutations and their phenotypes.
Acknowledgments
The authors thank all patients and their family members for taking part in this study. This work was supported by the National Infrastructure Program of Chinese Genetic Resources (2006DKA21301), National Science & Technology Pillar Program of China (2008BAH24B05), National Natural science Foundation of China (30872843), Natural Science Foundation of Fujian Province (C0810018), the Key Program of Scientific Research of FuJian Medical University(09ZD016-1), and Program for New Century Excellent Talents in Fujian Province University (Juhua Yang). Professor Xu Ma (genetic@263.net.cn) and Dr. Juhua Yang (julian_yang@mail.fjmu.edu.cn) contributed equally to the research presented and can be considered as co-corresponding authors.
References
- 1.Munier FL, Korvatska E, Djemaï A, Le Paslier D, Zografos L, Pescia G, Schorderet DF. Kerato-epithelin mutations in four 5q31-linked corneal dystrophies. Nat Genet. 1997;15:247–51. doi: 10.1038/ng0397-247. [DOI] [PubMed] [Google Scholar]
- 2.Munier FL, Frueh BE, Othenin-Girard P, Uffer S, Cousin P, Wang MX, Héon E, Black GC, Blasi MA, Balestrazzi E, Lorenz B, Escoto R, Barraquer R, Hoeltzenbein M, Gloor B, Fossarello M, Singh AD, Arsenijevic Y, Zografos L, Schorderet DF. BIGH3 mutation spectrum in corneal dystrophies. Invest Ophthalmol Vis Sci. 2002;43:949–54. [PubMed] [Google Scholar]
- 3.Mashima Y, Yamamoto S, Inoue Y, Yamada M, Konishi M, Watanabe H, Maeda N, Shimomura Y, Kinoshita S. Association of autosomal dominantly inherited corneal dystrophies with BIGH3 gene mutations in Japan. Am J Ophthalmol. 2000;130:516–7. doi: 10.1016/s0002-9394(00)00571-7. [DOI] [PubMed] [Google Scholar]
- 4.Chakravarthi SV, Kannabiran C, Sridhar MS, Vemuganti GK. TGFBI gene mutations causing lattice and granular corneal dystrophies in Indian patients. Invest Ophthalmol Vis Sci. 2005;46:121–5. doi: 10.1167/iovs.04-0440. [DOI] [PubMed] [Google Scholar]
- 5.Kim HS, Yoon SK, Cho BJ, Kim EK, Joo CK. BIGH3 gene mutations and rapid detection in Korean patients with corneal dystrophy. Cornea. 2001;20:844–9. doi: 10.1097/00003226-200111000-00013. [DOI] [PubMed] [Google Scholar]
- 6.Korvatska E, Munier FL, Djemaï A, Wang MX, Frueh B, Chiou AG, Uffer S, Ballestrazzi E, Braunstein RE, Forster RK, Culbertson WW, Boman H, Zografos L, Schorderet DF. Mutation hot spots in 5q31-linked corneal dystrophies. Am J Hum Genet. 1998;62:320–4. doi: 10.1086/301720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El-Ashry MF, Abd El-Aziz MM, Hardcastle AJ, Bhattacharya SS, Ebenezer ND. A Clinical and Molecular Genetic Study of Autosomal-Dominant Stromal Corneal Dystrophy in British Population. Ophthalmic Res. 2005;37:310–7. doi: 10.1159/000087791. [DOI] [PubMed] [Google Scholar]
- 8.Aldave AJ, Sonmez B. Elucidating the Molecular Genetic Basis of the Corneal Dystrophies: Are We There Yet? Arch Ophthalmol. 2007;125:177–86. doi: 10.1001/archopht.125.2.177. [DOI] [PubMed] [Google Scholar]
- 9.Pang CP, Lam DS. Differential occurrence of mutations causative of eye diseases in the Chinese population. Hum Mutat. 2002;19:189–208. doi: 10.1002/humu.10053. [DOI] [PubMed] [Google Scholar]
- 10.Yu J, Zou LH, He JC, Liu NP, Zhang W, Lu L, Sun XG, Dong DS, Wu YY, Yin XT. Analysis of mutation of BIGH3 gene in Chinese patients with corneal dystrophies. Zhonghua Yan Ke Za Zhi. 2003;39:582–6. [PubMed] [Google Scholar]
- 11.Jin T, Zou LH, Yang L, Dong WL, Yu J, Lu L. Identification of BIGH3 gene mutations in the patients with two types of corneal dystrophies. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2004;21:32–4. [PubMed] [Google Scholar]
- 12.Li Y, Sun XG, Ren HY, Dong B, Wang ZQ, Sun XY. Analysis of human transforming growth factor beta-induced gene mutation in corneal dystrophy. Chin Med J (Engl) 2004;117:1418–21. [PubMed] [Google Scholar]
- 13.Tian X, Liu ZG, Li Q, Li B, Wang W, Xie PY, Fujiki K, Murakami A, Kanai A. Analysis of gene mutation in Chinese patients with Reis-Bücklers corneal dystrophy. Zhonghua Yan Ke Za Zhi. 2005;41:239–42. [PubMed] [Google Scholar]
- 14.Tian X, Fujiki K, Wang W, Murakami A, Xie P, Kanai A, Liu Z. Novel mutation (V505D) of the TGFBI gene found in a Chinese family with lattice corneal dystrophy, type I. Jpn J Ophthalmol. 2005;49:84–8. doi: 10.1007/s10384-004-0167-7. [DOI] [PubMed] [Google Scholar]
- 15.Dong WL, Zou LH, Pan ZQ, Jin T, Yu J. Molecular genetic study on patients with lattice corneal dystrophy in China. Zhonghua Yan Ke Za Zhi. 2005;41:523–6. [PubMed] [Google Scholar]
- 16.Yu P, Gu Y, Yang Y, Yan X, Chen L, Ge Z, Qi M, Si J, Guo L. A clinical and molecular-genetic analysis of Chinese patients with lattice corneal dystrophy and novel Thr538Pro mutation in the TGFBI (BIGH3) gene. J Genet. 2006;85:73–6. doi: 10.1007/BF02728974. [DOI] [PubMed] [Google Scholar]
- 17.Qi YH, He HD, Li Y, Lin H, Gu JZ, Su H, Huang SZ. A research on TGFBI gene mutations in Chinese families with corneal dystrophies. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2006;23:310–2. [PubMed] [Google Scholar]
- 18.Yang J, Tong Y, Zhu Y, Xiao J, Chen Y, Lin J. Exon12 of BIGH3 (R555W) mutation in granular corneal dystrophy in a Chinese kindred. J Fujian Medical University. 2007;41:14–6. [Google Scholar]
- 19.Qi YH, He HD, Li Y, Wang L, Lin H, Su H, Gu JZ, Huang SZ. TGFBI gene mutation analysis in a Chinese family with Thiel-Behnke corneal dystrophy. Zhonghua Yan Ke Za Zhi. 2007;43:718–21. [PubMed] [Google Scholar]
- 20.Tian X, Fujiki K, Zhang Y, Murakami A, Li Q, Kanai A, Wang W, Hao Y, Ma Z. A novel variant lattice corneal dystrophy caused by association of mutation (V625D) in TGFBI gene. Am J Ophthalmol. 2007;144:473–5. doi: 10.1016/j.ajo.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 21.Liu Z, Wang YQ, Gong QH, Xie LX. An R124C mutation in TGFBI caused lattice corneal dystrophy type I with a variable phenotype in three Chinese families. Mol Vis. 2008;14:1234–9. [PMC free article] [PubMed] [Google Scholar]
- 22.Li D, Qi Y, Wang L, Lin H, Zhou N, Zhao L. An atypical phenotype of Reis-Bücklers corneal dystrophy caused by the G623D mutation in TGFBI. Mol Vis. 2008;14:1298–302. [PMC free article] [PubMed] [Google Scholar]
- 23.Yu P, Gu Y, Jin F, Hu R, Chen L, Yan X, Yang Y, Qi M. p.Ala546 > Asp and p.Arg555 > Trp mutations of TGFBI gene and their clinical manifestations in two large Chinese families with granular corneal dystrophy type I. Genet Test. 2008;12:421–5. doi: 10.1089/gte.2008.0005. [DOI] [PubMed] [Google Scholar]
- 24.Cao W, Ge H, Cui X, Zhang L, Bai J, Fu S, Liu P. Reduced penetrance in familial Avellino corneal dystrophy associated with TGFBI mutations. Mol Vis. 2009;15:70–5. [PMC free article] [PubMed] [Google Scholar]
- 25.Wang LM, Wang YC, Qiu DL, Ying M, Li ND. TGFBI gene mutations in three Chinese families with autosomal dominant corneal dystrophy. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2009;26:179–82. doi: 10.3760/cma.j.issn.1003-9406.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 26.Chang L, Zhiqun W, Shijing D, Chen Z, Qingfeng L, Li L, Xuguang S. Arg124Cys mutation of the TGFBI gene in 2 Chinese families with Thiel-Behnke corneal dystrophy. Arch Ophthalmol. 2009;127:641–4. doi: 10.1001/archophthalmol.2009.71. [DOI] [PubMed] [Google Scholar]
- 27.Li DD, Qi YH, Han Q, Lin H, Zhao LM, Zhang CM. Analysis of TGFBI gene mutation in a Chinese family with atypical Reis-Buckler corneal dystrophy. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2009;26:245–8. doi: 10.3760/cma.j.issn.1003-9406.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Hu L, Xu F, Ma WJ, Zhang H, Sui RF. R555W mutation of TGFbetaI related to granular corneal dystrophy in Chinese patients. Chin Med J (Engl) 2009;122:2691–4. [PubMed] [Google Scholar]
- 29.Zhang C, Zeng G, Lin H, Li D, Zhao L, Zhou N, Qi Y. A novel mutation I522N within the TGFBI gene caused lattice corneal dystrophy I. Mol Vis. 2009;15:2498–502. [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L, Gu Y, Yu P. Application of molecular genetic technique in classification of corneal dystrophy. Chin J Pract Ophthalmol. 2006;24:678–80. [Google Scholar]
- 31.Zhong X, Chen S, Huang W, Yang J, Chen X, Zhou Y, Zhou Q, Wang Y. Novel and known mutations of TGFBI, their genotype-phenotype correlation and structural modeling in 3 Chinese families with lattice corneal dystrophy. Mol Vis. 2010;16:224–30. [PMC free article] [PubMed] [Google Scholar]
- 32.Aldave AJ, Rayner SA, Kim BT, Prechanond A, Yellore VS. Unilateral lattice corneal dystrophy associated with the novel His572del mutation in the TGFBI gene. Mol Vis. 2006;12:142–6. [PubMed] [Google Scholar]
- 33.Rozzo C, Fossarello M, Galleri G, Sole G, Serru A, Orzalesi N, Serra A, Pirastu M. A common beta ig-h3 gene mutation (delta f540) in a large cohort of Sardinian Reis Bücklers corneal dystrophy patients. Mutations in brief no. 180. Online. Hum Mutat. 1998;12:215–6. [PubMed] [Google Scholar]
- 34.Tanhehco TY, Eifrig DE, Jr, Schwab IR, Rapuano CJ, Klintworth GK. Two cases of Reis-Bucklers corneal dystrophy (granular corneal dystrophy type III) caused by spontaneous mutations in the TGFBI gene. Arch Ophthalmol. 2006;124:589–93. doi: 10.1001/archopht.124.4.589. [DOI] [PubMed] [Google Scholar]
- 35.Zhao XC, Nakamura H, Subramanyam S, Stock LE, Gillette TE, Yoshikawa S, Ma X, Yee RW. Spontaneous and inheritable R555Q mutation in the TGFBI/BIGH3 gene in two unrelated families exhibiting Bowman's layer corneal dystrophy. Ophthalmology. 2007;114:e39–46. doi: 10.1016/j.ophtha.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 36.Chau HM, Ha NT, Cung LX, Thanh TK, Fujiki K, Murakami A, Kanai A. H626R and R124C mutations of the TGFBI (BIGH3) gene caused lattice corneal dystrophy in Vietnamese people. Br J Ophthalmol. 2003;87:686–9. doi: 10.1136/bjo.87.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okada M, Yamamoto S, Watanabe H, Shimomura Y, Tano Y. Granular corneal dystrophy with homozygous mutations in the kerato-epithelin gene. Am J Ophthalmol. 2000;129:411–2. doi: 10.1016/s0002-9394(00)00348-2. [DOI] [PubMed] [Google Scholar]
- 38.Okada M, Yamamoto S, Inoue Y, Watanabe H, Maeda N, Shimomura Y, Ishii Y, Tano Y. Severe corneal dystrophy phenotype caused by homozygous R124H keratoepithelin mutations. Invest Ophthalmol Vis Sci. 1998;39:1947–53. [PubMed] [Google Scholar]