Abstract
Background
In the fission yeast Schizosaccharomyces pombe, the TOR (target of rapamycin) and PKA (protein kinase A) signaling transduction pathways regulate the expression of genes required for cell growth and sexual differentiation in response to the nutritional environment. Inhibition of Tor2 signaling results in the induction of genes involved in sexual differentiation, and the cells undergo mating and meiosis, even under good nutritional conditions. The same phenotype is observed in mutants in which the PKA pathway is inactive. By contrast, Tor2 overexpression or mutations that hyperactivate PKA signaling impair sexual differentiation, even under poor nutritional conditions. Accordingly, a very important question is to understand the molecular mechanism by which these two pathways coordinately regulate gene expression in response to nutrients.
Methodology/Principal Findings
Here we demonstrate that TOR and PKA pathways operate coordinately to negatively regulate sexual differentiation by inhibiting the nuclear accumulation of the Ste11 transcription factor. However, the Tor2 pathway is unable to block the nuclear localization of Ste11 under good nutritional conditions when the PKA pathway is inactive. Using microarray analyses, we found that both pathways inhibit sexual differentiation by blocking ste11-dependent gene expression.
Conclusions/Significance
We conclude that both the PKA and the TOR pathways inhibit Ste11 nuclear accumulation to repress Ste11-dependent gene expression. However, the PKA pathway plays a quantitatively more important role than the TOR pathway in this process.
Introduction
In the presence of nutrients, the fission yeast Schizosaccharomyces pombe reproduces asexually by means of the mitotic cell cycle. Nitrogen depletion promotes cell cycle arrest in G1 and cells undergo sexual differentiation and meiosis to produce four resistant haploid spores that remain dormant until they encounter favorable growth conditions. TOR and PKA are two prominent, evolutionarily conserved signal transduction pathways that couple nitrogen and carbon source availability, respectively, to regulate diverse cell responses that ultimately drive cell growth and proliferation and inhibit sexual differentiation.
TOR is a serine/threonine protein kinase that is structurally and functionally conserved from yeasts to mammals. In mammalian cells, mTOR is a critical player in the TSC1-TSC2/Rheb/mTOR signaling pathway, which regulates cell growth in response to growth factors, nutrients, and energy conditions. TOR is activated by the GTPase Rheb, which is negatively regulated by the TSC1-TSC2 tuberous sclerosis complex [1], [2]. In the cell, TOR forms two types of multiprotein complexes: namely, TOR complex 1 (TORC1) and TOR complex 2 (TORC2) [3], [4]. TORC1 contains Raptor, is sensitive to rapamycin, and mediates the effects on protein synthesis and cell growth. In contrast, TORC2, which contains Rictor, regulates Akt and also affects the actin cytoskeleton [5], [6]. Unlike higher eukaryotes, which contain a single TOR protein, S. pombe and Saccharomyces cerevisiae have two: Tor1 and Tor2. Contrary to S. cerevisiae, the TSC1-TSC2/Rheb/TOR pathway is conserved in S. pombe, providing an excellent model to study the TOR pathway. In S. pombe, Tor2 forms part of the TORC1 complex and is essential for cell growth and the repression of sexual differentiation, meiosis and sporulation [7], [8], [9], [10], whereas Tor1 is not essential for growth and is included in the TORC2 complex [7], [9], [11].
Cyclic AMP (cAMP) also plays an important role in the regulation of cellular growth and sexual development in S. pombe. In several organisms, the level of cAMP changes in accordance to environmental conditions. In eukaryotic cells, the major target of cAMP is Protein Kinase A (PKA or cAMP-dependent protein kinase), which is a serine/threonine protein kinase that responds to glucose and is involved in the regulation of a broad diversity of biological processes, including cell growth and differentiation, nutrient sensing, and physiological stress responses [12], [13], [14]. In fission yeast, the intracellular level of cAMP is kept high during cell growth and division. Conditions unfavorable for growth cause a down-regulation of PKA, exit from the cell cycle, and sexual differentiation. S. pombe cells defective in cgs1, the gene for the regulatory subunit of cAMP-dependent protein kinase, constitutively generates active PKA and cells scarcely mate and sporulate [15]. In contrast, cells defective in cyr1, encoding adenyl cyclase, or in the pka1 gene, encoding the catalytic subunit of PKA, readily initiate sexual development in rich medium [16], [17]. This response is followed by gross changes in gene expression, indicating that several transcription factors are likely to be regulated by PKA either directly or indirectly.
The fission yeast ste11 gene encodes an HMG transcription factor, which is responsible for the expression of many genes required for the initiation of sexual development [18], [19]. Upstream from Ste11, another transcription factor, Rst2, induces the expression of ste11 and is under the regulation of PKA [20]. PKA inhibits Rst2 at two levels: by phosphorylation and by excluding Rst2 from the nucleus [21]. Thus, a decrease in the PKA activity, which naturally results from a lack of environmental nutrients, triggers the activation of Rst2 and, in turn, ste11 expression and sexual differentiation.
In sum, TOR and PKA are two signal transduction pathways that couple nitrogen and carbon source availability, respectively, to regulate diverse cell responses that ultimately drive cell growth and proliferation and inhibit sexual differentiation. In this study, we further investigated the interplay between the TOR and cAMP signaling pathways, which regulate gene expression mainly in relation to sexual differentiation. Our results indicate that the PKA pathway plays a more important role in repressing ste11 than the TOR pathway. Both pathways act in synergy but independently of one another, and converge directly at the level of ste11 expression. This regulatory network probably functions to provide cells with the flexibility necessary to respond to different inputs from two global nutrient sensors.
Materials and Methods
Fission yeast strains and media
All S. pombe strains used in this study are listed in Table S1 in supplementary material. Standard methods were used for growth, transformation and genetic manipulations [22]. The tagged versions of the rst2-gfp, rst2-HA and ste11-gfp genes were generated by a PCR-based method [23]. Except where specifically indicated, all experiments in liquid culture were carried out in Edinburgh Minimal Medium (EMM) containing the required supplements (except when mentioned), starting with a cell density of 2–4×106 cells/ml, corresponding to the mid-exponential phase of growth. Temperature shift experiments were carried out using a shaking water bath at 32°C.
Flow cytometry
Approximately 107 cells were collected by centrifugation, fixed in 70% cold ethanol and processed as described [22]. Flow cytometry analysis (FACS) was performed on a Becton-Dickinson FACScan device, using cells stained with propidium iodide. Cell size measurements were accomplished using the forward light scatter (FSC) data of the FACS.
RNA extraction and Northern blots
Total RNA from cells was isolated by lysis with glass beads in the presence of phenol [22]. 5–10 µg of RNA from each sample was separated on a formaldehyde agarose gel. Northern blot was carried out using Gene ScreenPlus (NEN, Dupont), following manufacturer's instructions. DNA probes were labeled with [32P]dCTPs using the Rediprime II Random Prime Labelling System kit (Amersham).
Protein extraction and Western blots
Protein extracts were obtained using trichloroacetic acid (TCA) extraction, as described previously [24]. For Western blots, 75–100 µg of total protein extract were run on 7,5% SDS-PAGE, transferred to a nitrocellulose filter (Amersham), and probed with mouse anti-HA 12CA5 (Roche Applied Sciences), mouse anti-ste11 (a gift of Dr Olaf Nielsen), mouse anti-GFP (Clontech) and mouse anti-tubulin (a gift of Dr Keith Gull) primary antibodies and, as secondary antibodies, NA 931, anti-mouse IgG, Horseradish Peroxidase (Amersham). Immunoblots were developed using the enhanced chemiluminescence procedure (ECL kit, Amersham). Phos-tag AAL-107 gels (NARD Institute) were used for shifts in the mobility of phosphorylated Rst2 at a final concentration of 100 µM, following the protocol suggested by the manufacturer.
Epifluorescence microscopy of GFP fusion proteins
Epifluorescence microscopy was carried out using a Zeiss Axioplan 2 fluorescence microscope equipped with an Orca-ER camera (Hamamatsu). To visualize nuclei, Hoechst 33342 was used to stain DNA.
Microarrays and data analyses
RNA was purified and cleaned using the RNeasy kit (Qiagen). RNA was analyzed with a 2100 Bioanalyzer (Agilent). The wild-type double-strand cDNA synthesis kit (Affymetrix) was used for the synthesis and purification of cDNA, and a Nanodrop spectrophotometer was used for cDNA quantification. cDNA was labeled and hybridized with a S. pombe Tiling 1.0FR Array, following the Affymetrix protocol. All the probes included in the arrays were aligned according to the S. pombe genome of the Pombe Sanger Institute Project (http://www.sanger.ac.uk/Projects/S_pombe/). To calibrate the sequence-specific probe effect, all data were background-corrected and quantile-normalized. To analyze different levels of transcription, all the normalized data were located in the correct genomic position and were visualized using Artemis (www.sanger.ac.uk/resources/software/artemis/).
Results
In S. pombe, the TOR and PKA pathways synergize to prevent exit from the cell cycle
In fission yeast, the TOR and PKA pathways drive cell growth and proliferation and inhibit sexual differentiation. Inactivation of Tor2 in rich medium leads to cell cycle exit, G1 arrest and sexual differentiation, meiosis and sporulation [7], [8], [11], [25]. Similarly, cells defective in the catalytic subunit of PKA (pka1Δ) or adenylate cyclase (cyr1Δ) undergo sexual development, meiosis and sporulation in the absence of nitrogen starvation [26]. In contrast, overexpression of Tor2, or mutants in cgs1 encoding the regulatory subunit of the PKA, reveal a reduction in mating and sporulation in poor medium [7], [15].
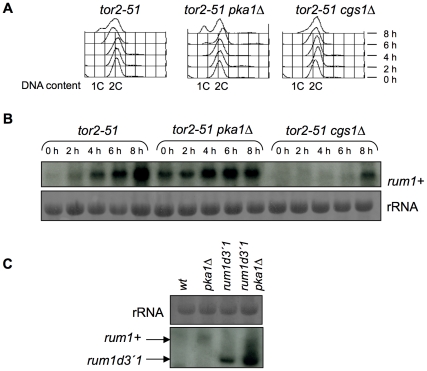
We combined the temperature-sensitive allele of the tor2 gene, tor2-51, with deletions of the catalytic subunit of PKA (pka1Δ) or the regulatory subunit (cgs1Δ) and measured the DNA content by flow cytometry analysis. As shown in Fig. 1A, tor2-51 control cells began to accumulate with 1C DNA content about 8 hours after Tor2 inactivation. The double mutant tor2-51 pka1Δ showed a faster rate of accumulation of cells with 1C DNA content, and G1 cells were observed 4 hours after Tor2 inactivation. Finally, in the tor2-51 cgs1Δ double mutant no G1-arrested cells were observed 8 hours after Tor2 inactivation. These results indicate that the TOR and the PKA pathways cooperate to prevent cell cycle exit and G1 arrest in fission yeast.
Figure 1. The TOR and PKA pathways synergize to prevent cell cycle exit and G1 arrest.
(A) Flow cytometry analysis of tor2-51, tor2-51 pka1Δ and tor2-51 cgs1Δ strains. Cells were grown to mid-exponential phase in minimal medium at 25°C and then shifted to the restrictive temperature of 32°C to inactivate Tor2. Samples were taken at the indicated times after Tor2 inactivation. (B) tor2-51, tor2-51 pka1Δ and tor2-51 cgs1Δ cells were grown to mid-exponential phase at 25°C and shifted to 32°C. Samples were collected at the indicated times for RNA extraction. Northern blot was performed and hybridized with a probe against the rum1+ gene. Ribosomal RNA (rRNA) was stained with methylene blue and used as loading control. (C) Wild-type, pka1Δ, rum1d3′1 and rum1d3′1 pka1Δ cells were collected during mid-exponential phase for RNA extraction. Northern blot was performed and hybridized with a probe against rum1+. rRNA stained with methylene blue was used as loading control.
Consistent with the FACS data, the inactivation of Tor2 led to the accumulation of the G1-specific rum1 transcript [7]. The level of rum1 mRNA was higher in pka1Δ cells and lower in cgs1Δ cells than in the control (Fig. 1B). To determine whether the increase in rum1 mRNA in pka1Δ cells was due to transcription activation or mRNA stabilization, we used a rum1 allele lacking the AU-rich elements in its 3′UTR, called rum1d3′1 [27]. In this mutant, the levels of rum1 mRNA also increased after tor2 inactivation, indicating that PKA activity was repressing rum1 transcription (Fig. 1C).
TOR and PKA pathways synergize to repress sexual differentiation
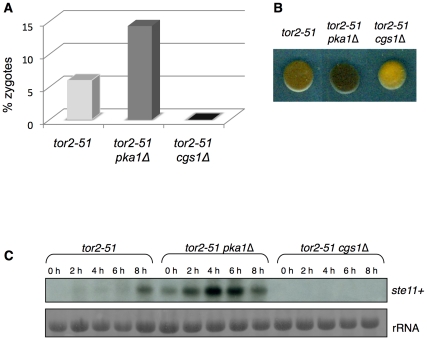
In order to test the relative importance of the TOR and PKA pathways in repressing mating, we analyzed the mating efficiency of homothallic h90 S. pombe cells after Tor2 inactivation in wild-type, pka1Δ and cgs1Δ backgrounds. As shown in Fig. 2A and 2B, mating efficiency was higher in tor2-51 pka1Δ than in tor2.51 and was almost completely abolished in tor2.51 cgs1Δ cells. Therefore, it seems that the TOR and PKA pathways synergize to repress sexual differentiation. In addition, Tor2 inactivation per se was insufficient to promote mating when the PKA activity was up-regulated.
Figure 2. Sexual differentiation in PKA mutants after Tor2 inactivation.
(A and B) h90 tor2-51, h90 tor2-51 pka1Δ and h90 tor2-51 cgs1Δ cells were incubated for 48 hours at 32°C in the presence of nitrogen (YES medium). The percentage of zygotes was determined and represented as a plot (A) and spores were stained with iodine vapour (B). h90 tor2-51 pka1Δ cells showed a higher mating efficiency than h90 tor2-51 cells after Tor2 inactivation. In contrast, h90 tor2-51 cgs1Δ cells were unable to mate after Tor2 inactivation. (C) tor2-51, tor2-51 pka1Δ and tor2-51 cgs1Δ cells were grown to mid-exponential phase in minimal medium, washed several times and incubated in minimal medium lacking nitrogen. Samples were collected at the indicated times to extract RNA and Northern blot was performed and hybridized with a probe against the ste11+ gene. rRNA stained with methylene blue was used as a loading control. In tor2-51 pka1Δ cells, ste11 expression levels increased earlier and to a higher level than in tor2-51. In the tor2-51 cgs1Δ double mutant there was no relevant activation of ste11+ transcription after Tor2 inactivation.
As described previously, inactivation of either Tor2 or mutants in pka1 promoted sexual differentiation by induction of ste11+ mRNA. We analyzed the levels of ste11+ mRNA after Tor2 inactivation at different time-points after the shift of tor2-51 to the restrictive temperature (Fig. 2C). ste11+ mRNA increased 8 hours after Tor2 inactivation in tor2-51, whereas in the tor2-51 pka1Δ ste11+ mRNA levels were already high by 2 hours after the temperature shift. In contrast, in tor2-51 cgs1Δ on which PKA is constitutively active, ste11+ mRNA was not induced even at 8 hours after Tor2 inactivation. This lack of ste11+ mRNA induction is probably the reason of the sterility of tor2-51 cgs1Δ cells.
Lack of PKA activity rescues the sterility of Tor2 overexpression
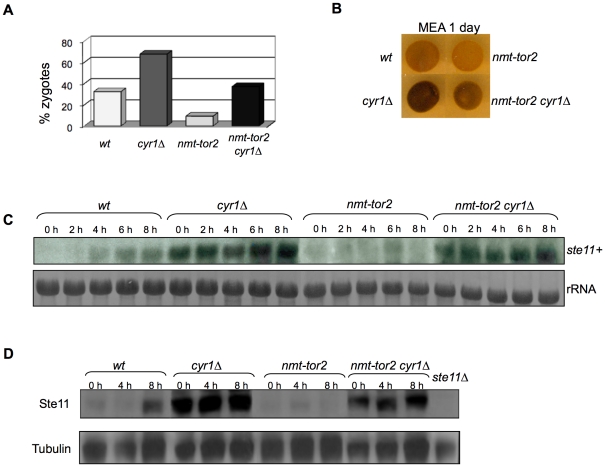
Tor2 overexpression inhibits sexual differentiation and mating [7]. Under nitrogen depletion, cells overexpressing Tor2 show reduced ste11+ mRNA induction and its target gene mei3+ mRNA levels are not detectable, suggesting that Tor2 overexpression is impairing Ste11 function [7]. We therefore overexpressed Tor2 in a cyr1Δ mutant, lacking adenylate cyclase and hence defective in PKA activation, to determine whether the inactivation of PKA would promote sexual differentiation when Tor2 was overexpressed. We measured sexual differentiation by counting the percentage of zygotes and by staining the spores formed after 24 hours under nitrogen starvation with iodine vapors. As shown in Fig. 3A and 3B, inactivation of PKA (cyr1Δ) promoted mating more efficiently than wild-type cells. Tor2 overexpression in the cyr1Δ mutant (nmt-tor2 cyr1Δ) showed a phenotype between that of the wild-type and that of cyr1Δ cells. This result indicates that PKA inactivation is able to promote sexual differentiation even in cells in which Tor2 is overexpressed.
Figure 3. Sexual differentiation in PKA mutants overexpressing Tor2.
(A and B) Cells of the opposite mating types of wild-type, cyr1Δ, nmt-tor2 and nmt-tor2 cyr1Δ were grown to mid-exponential phase, spotted onto malt extract plates and incubated for 24 hours at 25°C. (A) Percentage of zygotes, (B) Iodine staining. Tor2 overexpression impaired sexual differentiation. This phenotype was rescued in a cyr1Δ background. (C and D) Wild-type, cyr1Δ, nmt-tor2 and nmt-tor2 cyr1Δ cells were grown to mid-exponential phase in minimal medium at 25°C, washed several times and incubated in minimal medium lacking nitrogen. Cells were collected at the indicated times and RNA and protein were extracted. (C) Northern blot hybridized with a probe against ste11+ gene. (D) Western blot using anti-Ste11 antibody. Cells overexpressing Tor2 showed lower ste11+ mRNA and protein levels than the wild-type. ste11 mRNA and protein levels are clearly increased in cells lacking Cyr1 (nmt-tor2 cyr1Δ). rRNA and tubulin were used as loading controls.
The Ste11 transcription factor activates the expression of many genes that are essential for mating in fission yeast, including itself [20], [28]. We analyzed the induction of ste11+ mRNA in cells overexpressing Tor2 after nitrogen starvation (Fig. 3C). No clear induction of ste11+ mRNA was observed in nmt-tor2 cells. However, ste11+ mRNA was induced to an even higher level than in the wild-type in nmt-tor2 cyr1Δ cells. We also observed high levels of Ste11 protein when PKA was inactive (cyr1Δ), regardless of the Tor2 expression levels (Fig. 3D). Thus, the PKA pathway seems to control sexual differentiation independently of the Tor2 pathway, playing a more important role in repressing ste11+ transcription.
PKA controls Rst2 nuclear localization under nitrogen deprivation and after Tor2 inactivation
Rst2 is a transcription factor that is required to activate ste11+ transcription [20]. PKA negatively regulates Rst2 in rich medium by promoting its exclusion from the nucleus [21], thus repressing ste11+ expression.
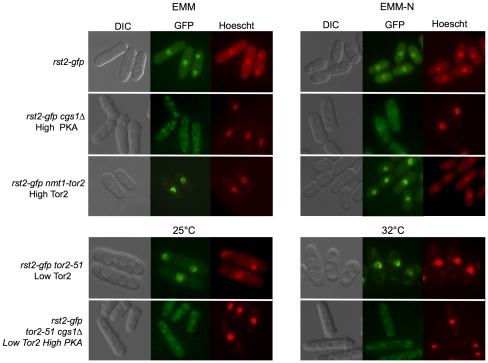
Rst2 protein levels did not change when cells were grown in medium withouth nitrogen or after Tor2 inactivation (Figure S1A, supplementary material). We analyzed the subcellular localization of Rst2 in mutants with high PKA (cgs1Δ) or high TOR activity (nmt1-tor2). In mutants with high PKA activity Rst2 was always cytoplasmic, regardless of the growth conditions (Fig. 4). This pattern of Rst2 localization was maintained in cells with low or high Tor2 activity, indicating that Tor2 does not regulate Rst2 subcellular localization.
Figure 4. Rst2 subcellular localization is not regulated by Tor2.
rst2-gfp, rst2-gfp cgs1Δ, rst2-gfp nmt-tor2 cells were grown to mid-exponential phase in minimal medium and shifted to minimal medium lacking nitrogen for 4 hours. In all cases, Rst2-GFP was located in the nucleus, except in the cgs1Δ mutant. rst2-gfp tor2-51 and rst2-gfp tor2-51 cgs1Δ cells were grown to mid-exponential phase in minimal medium at 25°C and then shifted to 32°C to inactivate Tor2. Nuclei were stained with Hoescht.
The PKA and TOR pathways regulate Ste11 localization
Ste11 shuttles between the nucleus and the cytoplasm. In growing cells, Ste11 is present at low levels and is pancellular. Mating pheromones and nutrient starvation trigger nuclear accumulation and increase the expression of Ste11 and its target genes [29].
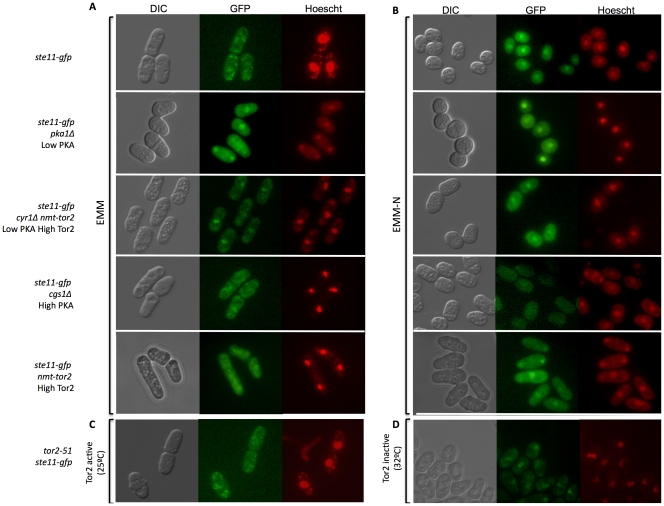
To determine the effects of Tor2 and PKA activities on Ste11 localization, we tagged the chromosome copy of Ste11 with GFP in the wild-type, in cells lacking PKA (pka1Δ), in cells expressing a constitutively active version of PKA (cgs1Δ), in cells overexpressing Tor2 (nmt-tor2), and in the tor2-51 mutant. Exponentially growing wild-type cells showed a pancellular localization of Ste11-GFP (Fig. 5A). In cells lacking PKA, Ste11-GFP accumulated in the nucleus, even when Tor2 was overexpressed (Fig. 5A). Therefore, PKA activity inhibits Ste11 nuclear accumulation, regardless of Tor2 activity.
Figure 5. Ste11-GFP subcellular localization is regulated by PKA and TOR pathways.
(A) Ste11-GFP was observed with fluorescence microscopy in wild-type (ste11-gfp), ste11-gfp pka1Δ, ste11-gfp cyr1Δ nmt-tor2, ste11-gfp cgs1Δ and ste11-gfp nmt-tor2 cells grown in minimal medium at 32°C, and (B) after nitrogen starvation for 4 hours at 25°C. DNA was stained with Hoescht. (C and D) Ste11-GFP was also observed in tor2-51 ste11-gfp cells grown to mid-exponential phase at 25°C (C) and then shifted to the 32°C for 6 hours to inactivate Tor2 (D).
Under nitrogen deprivation, Ste11-GFP accumulated in the nucleus (Fig. 5B). In cells where PKA is constitutively active (cgs1Δ), Ste11 levels were very low and no nuclear accumulation was observed, even under nitrogen deprivation. When the PKA pathway was inactive (pka1Δ), Ste11 was located in the nucleus in rich medium. After Tor2 inactivation, Ste11 also accumulated in the nucleus, even in rich medium (active PKA) (Fig. 5C). When Ste11 is located in nucleus, Ste11 levels increase in the cells (Fig. S1B, supplementary material). Accordingly, both the PKA and the Tor2 pathways inhibit sexual differentiation by preventing Ste11 nuclear accumulation.
Ste11-dependent gene expression is controlled by the TOR and PKA pathways
The above observations suggested that the TOR and PKA pathways were inhibiting Ste11 activity by excluding it from nucleus. Ste11 is a transcription factor that induces the expression of ste11+ itself, the mating type genes, and the master regulator of meiosis, Mei2 [20], [28]. To confirm that the PKA and TOR pathways were indeed inhibiting Ste11 activity, we performed microarray analyses in cells growing in rich medium after Tor2 inactivation. Under these conditions, ste11 and many genes involved in mating and meiosis are induced, mimicking the nitrogen starvation response [9]. In addition, we analyzed the gene expression profile in cells overexpressing Tor2 after nitrogen starvation. Table 1 lists the genes that were induced owing to the loss of Tor2 and not induced in cells overexpressing Tor2 after nitrogen starvation. Most of these genes are Ste11 targets [9]. We therefore conclude that Tor2 inhibits mating and meiosis by repressing the Ste11 transcription factor.
Table 1. Mating and meiosis genes induced by loss of Tor2 and not induced after nitrogen starvation in cells overexpressing Tor2.
| Systematic name | Gene name | Biological Process (GeneDB) |
| SPAC13G7.13c | msa1 | negative regulation of conjugation with cellular fusion |
| SPBC29B5.02c | isp4 | conjugation with cellular fusion |
| SPAC1039.09 | isp5 | conjugation with cellular fusion |
| SPAC513.03 | mfm2 | regulation of conjugation with cellular fusion by signal transduction |
| SPAC25B8.13c | isp7 | induction of conjugation upon nitrogen starvation |
| SPBC1711.02 | matmc_1 | conjugation with cellular fusion |
| SPBC23G7.09 | matmc_2 | conjugation with cellular fusion |
| SPAC11E3.06 | map1 | regulation of transcription, mating-type specific |
| SPAC4A8.04 | isp6 | conjugation with cellular fusion |
| SPAC1565.04c | ste4 | regulation of conjugation with cellular fusion by signal transduction |
| SPBC32C12.02 | ste11 | conjugation with cellular fusion |
| SPBPJ4664.03 | mfm3 | regulation of conjugation with cellular fusion by signal transduction |
| SPAC11H11.04 | mam2 | pheromone-dependent signal transduction involved in conjugation with cellular fusion |
| SPAPB8E5.05 | mfm1 | conjugation with cellular fusion |
| SPBPJ4664.03 | mfm3 | regulation of conjugation with cellular fusion by signal transduction |
| SPBC25B2.02c | mam1 | peptide pheromone export |
| SPBC24C6.06 | gpa1 | conjugation with cellular fusion |
| SPAC23E2.03c | ste7 | conjugation with cellular fusion |
| SPAC27D7.03c | mei2 | positive regulation of meiosis |
| SPAC22F3.12c | rgs1 | negative regulation of signal transduction involved in conjugation with cellular fusion |
As we have shown above, inactivation of the PKA pathway was able to rescue the sterility of Tor2 overexpression (Fig. 3A and 3B). We performed microarray analyses of wild-type cells and of Tor2-overexpressing cells with and without PKA activity after nitrogen starvation. As shown in Table S2 (supplementary material), inactivation of the PKA pathway increased the transcription levels of most Ste11 target genes, even in cells overexpressing Tor2. We thus surmise that the PKA pathway plays a more critical role than Tor2 in repressing Ste11-dependent transcription.
Discussion
Control of cell growth and differentiation is a complex process governed by nutrient availability and growth factors that are sensed and transmitted by specialized signal transduction pathways. In fission yeast, the TOR and the PKA pathways activate cell growth and inactivate sexual differentiation. Nitrogen starvation triggers cell cycle arrest in G1 and induces the expression of the Ste11 transcription factor that is required for sexual differentiation. Here we show that inactivation of the TOR and PKA pathways synergize to promote the nitrogen starvation response and the activation of ste11 transcription.
In fission yeast, inactivation of Tor2 promotes sexual differentiation, even in rich medium, by increasing ste11 mRNA levels [7]. We analyzed the mating efficiency of cells after Tor2 inactivation in the wild-type and in cells lacking PKA and observed that when PKA was inactive, cells mated more efficiently than wild-type cells. This could be explained because ste11 mRNA levels are higher in the pka1Δ tor2-51 double mutant than in any of the single mutants or in the wild-type. On the other hand, when the PKA pathway was constitutively active, cells were completely sterile because there was no induction of ste11 mRNA. Tor2 overexpression also impairs mating in cells under nitrogen starvation because there is no induction of ste11 mRNA [7]. However, this phenotype was rescued when the PKA pathway was inactivated and in this background, cells showed higher levels of ste11 mRNA and protein.
We also analyzed whether there was any cross-talk between the TOR and the PKA signaling pathways. The Rst2 transcription factor is negatively regulated by PKA and induces transcription of ste11 [20], [21]. Rst2 is located in the nucleus but constitutively active PKA promotes its exclusion from the nucleus [20], [21]. We analyzed Rst2 localization after Tor2 inactivation and under nitrogen starvation when Tor2 was overexpressed. In all cases, Rst2 was normally localized, indicating that Tor2 does not control Rst2 localization. We also analyzed Rst2 phosphorylation and we observed that Rst2 was not hyperphosphorylated after Tor2 inactivation (data not shown). Therefore, it may be concluded that Tor2 does not control Ste11 transcription by repressing Rst2.
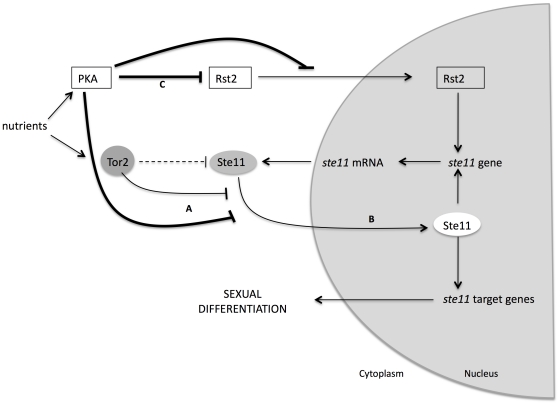
Another possibility is that the PKA and TOR signaling pathways cross-talk directly at the level of the Ste11 transcription factor. Ste11 was present at very low level and had pancellular localization in rich medium. Mating pheromones and nitrogen starvation trigger Ste11 nuclear accumulation and increase its own expression [29]. We observed that PKA inhibited Ste11 nuclear accumulation. When PKA signaling was inactive (pka1Δ), Ste11-GFP accumulated in the nucleus, even in rich medium. The same pattern was observed when Tor2 was inactivated, indicating that in both cases there was no nutritional starvation requirement to promote Ste11-GFP localization in nucleus. Therefore, both nutritional signaling pathways cross-talk at the level of Ste11 by regulating Ste11 localization (Fig. 6). In the case of PKA, there was another level of control: the inhibition of Ste11 expression through the Rst2 transcription factor (Fig. 6). Tor2 physically interacts with Ste11 when Tor2 is active under good nutritional conditions [7]. After Tor2 inactivation, Ste11 accumulates in the nucleus. If PKA is constitutively active, the level of Ste11 is so low (because Rst2 is inactive) that Tor2 inactivation is not sufficient to allow the accumulation Ste11 in the nucleus and the cells are unable to mate. The present study provides evidence that the subcellular distribution of Ste11 is a regulatory mechanism in which TOR and PKA pathways converge to regulate sexual differentiation in response to the nutritional environment.
Figure 6. The PKA and TOR pathways regulate Ste11 subcellular localization.
(A) Under good nutritional conditions, the PKA and TOR pathways inhibit Ste11 nuclear accumulation. (B) When the cells are starved for nitrogen Ste11 accumulates in the nucleus, activates the transcription of its target genes, and promotes sexual differentiation. (C) When the PKA pathway is constitutively active it represses Rst2 activity, and there is no Ste11-dependent gene expression. In this situation, sexual differentiation is repressed even if Tor2 is inactivated.
Supporting Information
(3.69 MB TIF)
(0.06 MB DOC)
(0.07 MB DOC)
Acknowledgments
We thank Drs Keith Gull and Olaf Nielsen for the anti-tubulin and anti-ste11 antibodies, Alberto de Luis for the microarray analysis, and Nicholas Skinner for corrections to the manuscript. N.V. is supported by a postdoctoral grant from the Carlos III Institute, Ministerio de Sanidad. Our group is supported by grants BFU2008-01808, Consolider CSD2007-00015 and Junta de Castilla y León Grupo de Excelencia GR 265.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: N.V. is supported by a postdoctoral grant from the Carlos III Institute, Ministerio de Sanidad. Our group is supported by grants from la Junta de Castilla y Leon (Grupo de Excelencia grant GR265) and the Spanish Ministry of Science and Innovation (BFU2008-01808 and Consolider Ingenio CSD2007-00015). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Curr Biol. 2005;15:702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- 2.Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem. 2005;280:18717–18727. doi: 10.1074/jbc.M414499200. [DOI] [PubMed] [Google Scholar]
- 3.Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 4.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 5.Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 6.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 7.Alvarez B, Moreno S. Fission yeast Tor2 promotes cell growth and represses cell differentiation. J Cell Sci. 2006;119:4475–4485. doi: 10.1242/jcs.03241. [DOI] [PubMed] [Google Scholar]
- 8.Uritani M, Hidaka H, Hotta Y, Ueno M, Ushimaru T, et al. Fission yeast Tor2 links nitrogen signals to cell proliferation and acts downstream of the Rheb GTPase. Genes Cells. 2006;11:1367–1379. doi: 10.1111/j.1365-2443.2006.01025.x. [DOI] [PubMed] [Google Scholar]
- 9.Matsuo T, Otsubo Y, Urano J, Tamanoi F, Yamamoto M. Loss of the TOR kinase Tor2 mimics nitrogen starvation and activates the sexual development pathway in fission yeast. Mol Cell Biol. 2007;27:3154–3164. doi: 10.1128/MCB.01039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weisman R, Roitburg I, Schonbrun M, Harari R, Kupiec M. Opposite effects of tor1 and tor2 on nitrogen starvation responses in fission yeast. Genetics. 2007;175:1153–1162. doi: 10.1534/genetics.106.064170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayashi T, Hatanaka M, Nagao K, Nakaseko Y, Kanoh J, et al. Rapamycin sensitivity of the Schizosaccharomyces pombe tor2 mutant and organization of two highly phosphorylated TOR complexes by specific and common subunits. Genes Cells. 2007;12:1357–1370. doi: 10.1111/j.1365-2443.2007.01141.x. [DOI] [PubMed] [Google Scholar]
- 12.Shabb JB. Physiological substrates of cAMP-dependent protein kinase. Chem Rev. 2001;101:2381–2411. doi: 10.1021/cr000236l. [DOI] [PubMed] [Google Scholar]
- 13.Smith FL, Lohmann AB, Dewey WL. Involvement of phospholipid signal transduction pathways in morphine tolerance in mice. Br J Pharmacol. 1999;128:220–226. doi: 10.1038/sj.bjp.0702771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor SS, Kim C, Cheng CY, Brown SH, Wu J, et al. Signaling through cAMP and cAMP-dependent protein kinase: diverse strategies for drug design. Biochim Biophys Acta. 2008;1784:16–26. doi: 10.1016/j.bbapap.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeVoti J, Seydoux G, Beach D, McLeod M. Interaction between ran1+ protein kinase and cAMP dependent protein kinase as negative regulators of fission yeast meiosis. Embo J. 1991;10:3759–3768. doi: 10.1002/j.1460-2075.1991.tb04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamawaki-Kataoka Y, Tamaoki T, Choe HR, Tanaka H, Kataoka T. Adenylate cyclases in yeast: a comparison of the genes from Schizosaccharomyces pombe and Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1989;86:5693–5697. doi: 10.1073/pnas.86.15.5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawamukai M, Ferguson K, Wigler M, Young D. Genetic and biochemical analysis of the adenylyl cyclase of Schizosaccharomyces pombe. Cell Regul. 1991;2:155–164. doi: 10.1091/mbc.2.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugimoto A, Iino Y, Maeda T, Watanabe Y, Yamamoto M. Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev. 1991;5:1990–1999. doi: 10.1101/gad.5.11.1990. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto M. Regulation of meiosis in fission yeast. Cell Struct Funct. 1996;21:431–436. doi: 10.1247/csf.21.431. [DOI] [PubMed] [Google Scholar]
- 20.Kunitomo H, Higuchi T, Iino Y, Yamamoto M. A zinc-finger protein, Rst2p, regulates transcription of the fission yeast ste11(+) gene, which encodes a pivotal transcription factor for sexual development. Mol Biol Cell. 2000;11:3205–3217. doi: 10.1091/mbc.11.9.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higuchi T, Watanabe Y, Yamamoto M. Protein kinase A regulates sexual development and gluconeogenesis through phosphorylation of the Zn finger transcriptional activator Rst2p in fission yeast. Mol Cell Biol. 2002;22:1–11. doi: 10.1128/MCB.22.1.1-11.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 23.Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 24.Foiani M, Marini F, Gamba D, Lucchini G, Plevani P. The B subunit of the DNA polymerase alpha-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol Cell Biol. 1994;14:923–933. doi: 10.1128/mcb.14.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urano J, Sato T, Matsuo T, Otsubo Y, Yamamoto M, et al. Point mutations in TOR confer Rheb-independent growth in fission yeast and nutrient-independent mammalian TOR signaling in mammalian cells. Proc Natl Acad Sci U S A. 2007;104:3514–3519. doi: 10.1073/pnas.0608510104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maeda T, Watanabe Y, Kunitomo H, Yamamoto M. Cloning of the pka1 gene encoding the catalytic subunit of the cAMP-dependent protein kinase in Schizosaccharomyces pombe. J Biol Chem. 1994;269:9632–9637. [PubMed] [Google Scholar]
- 27.Daga RR, Bolanos P, Moreno S. Regulated mRNA stability of the Cdk inhibitor Rum1 links nutrient status to cell cycle progression. Curr Biol. 2003;13:2015–2024. doi: 10.1016/j.cub.2003.10.061. [DOI] [PubMed] [Google Scholar]
- 28.Mata J, Bahler J. Global roles of Ste11p, cell type, and pheromone in the control of gene expression during early sexual differentiation in fission yeast. Proc Natl Acad Sci U S A. 2006;103:15517–15522. doi: 10.1073/pnas.0603403103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin J, Kang W, Leung B, McLeod M. Ste11p, a high-mobility-group box DNA-binding protein, undergoes pheromone- and nutrient-regulated nuclear-cytoplasmic shuttling. Mol Cell Biol. 2003;23:3253–3264. doi: 10.1128/MCB.23.9.3253-3264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(3.69 MB TIF)
(0.06 MB DOC)
(0.07 MB DOC)