Abstract
Neuroticism is a moderately heritable personality trait considered to be a risk factor for developing major depression, anxiety disorders and dementia. We performed a genome-wide association study in 2,235 participants drawn from a population-based study of neuroticism, making this the largest association study for neuroticism to date. Neuroticism was measured by the Eysenck Personality Questionnaire. After Quality Control, we analysed 430,000 autosomal SNPs together with an additional 1.2 million SNPs imputed with high quality from the Hap Map CEU samples. We found a very small effect of population stratification, corrected using one principal component, and some cryptic kinship that required no correction. NKAIN2 showed suggestive evidence of association with neuroticism as a main effect (p<10−6) and GPC6 showed suggestive evidence for interaction with age (p≈10−7). We found support for one previously-reported association (PDE4D), but failed to replicate other recent reports. These results suggest common SNP variation does not strongly influence neuroticism. Our study was powered to detect almost all SNPs explaining at least 2% of heritability, and so our results effectively exclude the existence of loci having a major effect on neuroticism.
Introduction
Neuroticism is a personality trait that has been associated with several psychiatric disorders, and is considered one of the risk factors for the development of depression, anxiety disorders [1], [2], [3], [4] and dementia [5]. When measured by the Eysenk Personality Questionnaire (EPQ) [6], neuroticism has been shown to be relatively stable, with a stability of 0.62 over 20 years [7], although there is some evidence of a moderate decline in older ages [8], [9]. Neuroticism is consistently higher in females than males [10], [11], but it is unclear whether this is a direct effect of gender or a consequence of different environments experienced by males and females [8], [12]–[14]. The heritability of neuroticism has been estimated in the range 0.30 to 0.50, based on twin studies [7], [15]–[17] that also show a genetic covariance of neuroticism with anxiety and depression [10], [13], [18]–[22].
Significant or suggestive linkage has been reported within regions on chromosomes 1p, 1q, 4q, 6p, 7p, 11p, 11q, 12q, 13q and 15q [23]–[26]. Shifman [27] reported a genome-wide association study for EPQ-N in 2,054 extreme-scoring individuals from a large cohort (patient registers of UK general practitioners) using eight DNA pools. No significant SNPs were identified under linkage peaks derived from sib pairs in the same cohort [14], [23]. They reported an association at SNP rs702543, in the phosphodiesterase 4D gene (PDE4D) on chromosome 5q, but even after individual genotyping of a further 1534 individuals from the same cohort, the combined p-value was not genome-wide significant (p = 2×10−6). Further analysis of rs702543 in a combined sample from three populations (Australian twins, Virginia Adult Twin registry, Netherlands twin families), using a measure of neuroticism that is similar to EPQ-N, showed a non-significant trend for an excess of the risk allele (A) in high neuroticism-score individuals. A family-based analysis revealed overtransmission of the A allele in high-score individuals from the Australian sample (p = 0.04), but not in the Netherlands sample. Thus, the association of rs702543 with neuroticism is notable but requires confirmation. In a genome-wide study of US-based and German samples, van den Oord [28] found association between neuroticism and four SNPs in the chr 14 gene MAMDC1 (combined p<10−6). To date, the most recent association study [29] failed to replicate an association between the candidate gene NPY and stress response and emotion (reported by Zhou [30], using instruments other than the EPQ).
We conducted the largest genome-wide association study to date, with 2,235 individuals from a population-based collection, for whom EPQ-N score and 430K SNP genotypes were available. We also analysed an additional 1.2 M well-imputed SNPs.
Results
The sample consists of individuals who previously participated in the ‘CoLaus’ study [31] a community survey of 6 738 randomly-selected residents of the city of Lausanne (Switzerland). Of the 3 400 Caucasians who accepted the psychiatric evaluation (55% participation rate), 2 239 individuals also completed and returned the EPQ questionnaire (66% return rate), and had genotype data available.
The mean age of the study individuals was 51.9 (SD 8.85; range 36–70), and 55.4% were female. These compare with mean age 51.2 (SD 8.82; range 36–70) and 53.0% female among all 3,307 PsyCoLaus individuals, indicating that responders to the questionnaire are slightly older and more likely to be female than non-responders.
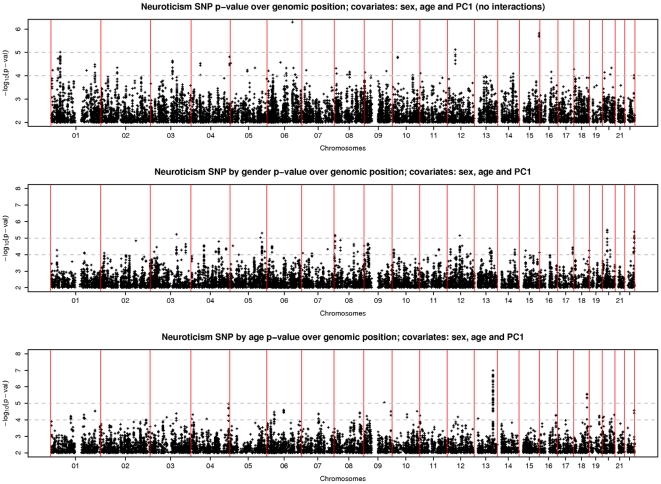
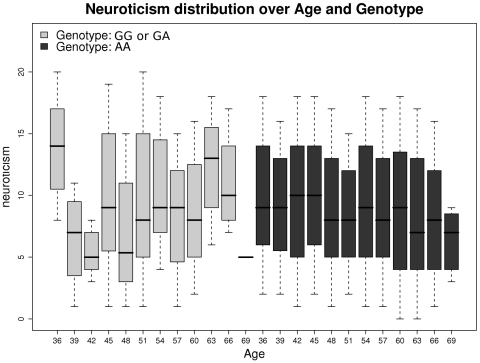
A QQ plot of the p-values for the genotyped SNPs (supplementary material File S1) suggested negligible inflation due to population structure or cryptic relatedness (genomic control λ = 1.014), but also little evidence of p-values below those expected under the null hypothesis. Results from the main-effects analyses are shown in Figure 1a, and Table 1. Both genotyped and imputed SNPs within the gene ARRDC4, at the distal end of 15q, almost reached p = 10−6. We identified several imputed SNPs with p<10−6 on 6q, within the gene NKAIN2, which has not previously been reported in linkage or association studies, although the strongest association at a genotyped SNP (rs11154221) in this gene has p≈10−4. No SNP showed a notable interaction with sex (Figure 1b). SNP by age interaction analyses revealed multiple associations at the distal end of 13q within gene GPC6 (Figure 1c, Table 1). The strongest signal (p≈10−7) was at a genotyped SNP, rs9561329; EPQ score tends to increase with age for carriers of the minor allele, and decreases for non-carriers (Figure 2). Conditional regression analyses revealed that the signal at each of the SNPs reported in Table 1 effectively disappeared when the most strongly-associated SNP at that locus was included as a predictor in the regression, suggesting at most one causal variant underlying each signal.
Figure 1. P-values over genomic location.
-log10(p-value) for association with EPQ-N, against genomic location for 1.7 M genotyped and well-imputed SNPs. Top: main SNP effect; middle: sex by SNP interaction; bottom: age by SNP interaction.
Table 1. Most significant main effect and interaction results for association with neuroticism (EPQ-N).
| SNP main effect, p<2.5×10−6 | |||||||
| chr | position | snp | -log10(p-val) | Beta (SE) | gene | Alleles (MAF) | genotyped? |
| 6 | q21 | rs12204812 | 6.43 | −0.91 (0.18) | NKAIN2 | A/C (0.33) | |
| 6 | q21 | rs9491140 | 6.47 | −0.91 (0.18) | NKAIN2 | C/T (0.33) | |
| 6 | q21 | rs9491142 | 6.45 | −0.91 (0.18) | NKAIN2 | A/G (0.33) | |
| 6 | q21 | rs12192208 | 6.29 | −0.90 (0.18) | NKAIN2 | A/C (0.33) | |
| 15 | q26.3 | rs1043372 | 5.68 | −0.82 (0.17) | ARRDC4 | C/T (0.38) | Y |
| 15 | q26.3 | rs1043374 | 5.75 | −0.83 (0.17) | ARRDC4 | C/A (0.38) | Y |
| 15 | q26.3 | rs4965121 | 5.83 | −0.84 (0.17) | ARRDC4 | C/G (0.38) | |
Figure 2. EPQ-N distribution by 3-year age groups according to minor allele carrier status at rs9561329.
SNP rs702543 in gene PDE4D, reported by Shifman [27] to be associated with neuroticism, was not present among our 1.7 M genotyped or imputed SNPs. However, we obtained p = 0.006 for association at the imputed SNP rs296410 that showed the highest R2 with rs702543 in the Hap Map CEU samples (R2 = 0.51); in our study, the major allele (T, frequency 0.52) at rs296410 shows a positive linear association with EPQ-N score. We could not replicate the association found by van den Oord [28] for the gene MAMDC1 on 14q. Three of the four SNPs they reported had a perfect proxy in our SNP set, but our lowest p-value in this gene was p = 0.43. We tested 3 genotyped SNPs in the gene NPY studied by Zhou30, including a SNP, rs16126, in high LD (R2≈0.9) with two of their reported SNPs, but we found p>0.4 at all three SNPs. Finally, we found no significant enrichment of a GO functional category among SNPs with p<10−4.
A separate analysis was carried out on the X chromosome, including the pseudo-autosomal region. Genotypes were coded as 0, 1, and 2 for females, and 0 and 2 for males, and sex was included as a covariate in the linear regression model. No significant associations were found.
Discussion
We present a comprehensive analysis of a population-based sample for genome-wide association with neuroticism. The evidence from 19 cryptic first-degree relative pairs in our study suggests a heritability for EPQ-N of at most 0.22, lower than the previously-reported range of 0.3 to 0.5. We identified SNPs in NKAIN2 showing evidence for an additive main effect, but short of genome-wide significance. This gene has been linked with neurological phenotypes [41], though little functional or molecular information is available. Several SNPs in gene GPC6 on 13q gave a signal for an interaction with age. GPC6 is known to be involved in the regulation of the Wnt pathway [42] that plays a role in intracellular signalling both during neural development [43] and also in the mature nervous system with possible implication in synaptic modulation and plasticity. There is some evidence that abnormalities of signal transduction may be implicated in the pathophysiology of psychiatric disorders, including mood disorders and schizophrenia [44], [45]. Several studies also suggest that Wnt-pathway abnormalities may play a role in neurodegenerative process and Alzheimer disease [46], [47]. Neuroticism, alone or in combination with other personality traits or lifestyle, has been shown to be predictive for Alzheimer [5], [48].
It is difficult to speculate why our novel, population-based heritability estimate is so much lower than those from previous family- and studies. Common environment may partly explain a familial effect that may have been misattributed to genetics. Like all studies, ours has a number of limitations. The EPQ-N was assessed from self-administered questionnaires posted back to the researchers. Only 66% of the eligible subjects sent back the questionnaire, which may have generated a response bias, although we have shown that there are only minor differences in sex and age between responders and non-responders. Our study has a cross-sectional design, and individuals suffering a neurological condition such as dementia may be less likely to complete and return the EPQ. However, all participants in the PsyCoLaus had a psychiatric assessment and completed a Mini Mental Stage Examination [49]. If a locus has pleiotropic effects, affecting both neuroticism and dementia in old age, this could generate an age-dependent response-bias towards protective alleles, leading to a genotype by age interaction such as we observed at GPC6.
Our study was powered to detect almost all SNPs explaining at least 2% of phenotypic variation, and so our results effectively exclude the hypothesis that there are major loci affecting neuroticism in the Lausanne population. The lack of strong genetic association with EPQ-N score might imply that neuroticism is controlled by many loci each of relatively small effect, as is the case for many other complex phenotypes. There may also be important environmental interactions with risk factors not available in our study. If confirmed by additional studies, the suggestive age by genotype interaction that we report raises interesting methodological and biological questions.
Materials and Methods
Sample and assessment
The CoLaus was designed to assess cardiovascular disease risk factors and to collect plasma samples for the study of genetic variants and biomarkers. The random sampling procedure was based on a list of Lausanne inhabitants aged 35–75 years (n = 56 694 in 2003), obtained from the population register of the city. From the original study, all 35 to 66 year-old participants were invited to participate in the psychiatric arm of the study; for more details on the rationale, design and methods of the study see Preisig [32]. All subjects who were sufficiently fluent in French or English and agreed to participate were included into the PsyCoLaus sub-study and underwent the psychiatric assessment between 2004 and 2008. The present analysis was restricted to Caucasians (92% of the sample). The study was approved by the Local Ethics Committee and was supported in part by GlaxoSmithKline. All participants were duly informed about this sponsorship, and consented for the use of biological samples and data by GlaxoSmithKline and its subsidiaries.
All the participants were interviewed using the Diagnostic Interview for Genetic Studies [33] (DIGS), in order to formulate DSM-IV and subthreshold diagnoses. Additional assessment included: medical records, family history [Family History-Research Diagnostic Criteria (FH-RDC) developed by Andreasen [34]], several self-report questionnaires including EPQ and STAI, and life events interview.
The EPQ is a 90-item self-report personality inventory [6] that has been widely used for personality assessment in psychiatry and other medical fields. Factor analysis of both English and French versions revealed four dimensions that have been labelled Extraversion, Neuroticism, Psychoticism, and Social desirability (Lie scale). Reliabilities of the resulting scales, and correlations between factors, were found to be similar between English and French versions. In particular, using three different French samples, Eysenck [6] reported Cronbach alpha coefficients of 0.78 to 0.87 for neuroticism.
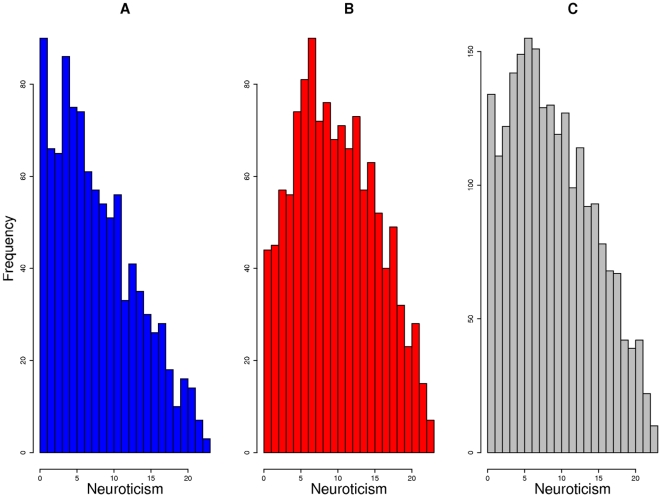
Neuroticism scores range between 0 and 24, with higher scores indicating more severe neuroticism. See Figure 3 for the distribution of EPQ-N in our study. The overall mean score was 9.33 (SD 5.61), but there was a highly-significant sex difference (males: mean 8.17, median 7, SD 5.50; females: mean 10.26, median 10, SD 5.53; Mann-Whitney test p<10−16). There was no correlation between age and EPQ-N (r = −0.02, p = 0.26).
Figure 3. Distribution of EPQ-N score across the subjects.
A: Males. B: Females. C: Both sexes.
Genotyping and quality controls
Venous blood samples were taken at the time of the participation in the CoLaus study. Nuclear DNA was extracted from whole blood. Participants consented for the genetic data to be used for the study of cardiovascular risk factors and psychiatric disorders. The samples were genotyped on the Affymetrix GeneChip Human Mapping 500K Array Set according to the Affymetrix published protocol. Genotypes were determined by the Bayesian Robust Linear Model with Mahalanobis Distance algorithm (BRLMM). Details of the genotyping procedures are given by Li [35]. A QC procedure was carried out on the sample to remove SNPs with poor amplification and individuals with an excess of missing genotypes. First, a total of 60,545 SNPs with >5% missing genotypes were removed. Next, 4 individuals with missingness >5% among the remaining SNPs were excluded. This left 2,235 individuals genotyped at 430,193 autosomal SNPs. An additional 9,828 SNPs were available on the X chromosome, but only in 2,168 individuals. Overall we flagged 1,893 autosomal SNPs showing significant deviation from HWE (p<10−4). None of the SNPs highlighted here were among these 1,893.
Statistical analysis
Assuming an additive genetic model and a Gaussian-distributed phenotype, 2 235 individuals provide 95% power to detect a genotyped SNP with heritability h2 = 0.02 at significance level 5×10−7, and 39% power for h2 = 0.01. Thus we expect to detect almost all SNPs with h2>0.02 and a large proportion of those with h2>0.01.
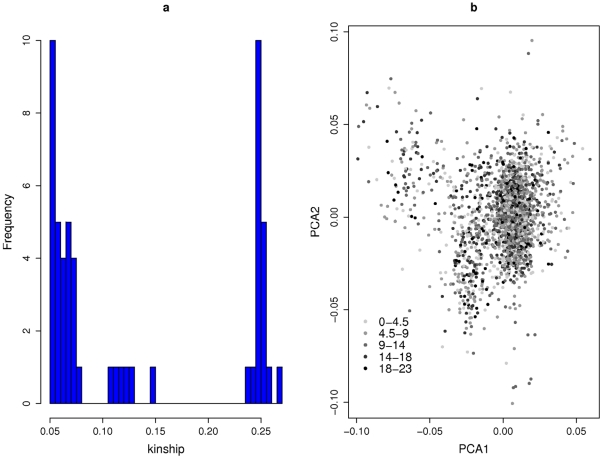
In order to investigate any possible population structure or cryptic relatedness among our study subjects, we selected a subset of genotyped SNPs to produce a kinship matrix and perform Principal Component Analysis (PCA). The SNPs were chosen using PLINK [36] to have minor allele fraction >5% and pairwise R2≤0.5, in a window size of 50 SNPs sliding 5 SNPs at a time. This process chose 75,046 SNPs for kinship and PCA analyses. For these analyses, missing genotypes were replaced with the mean genotype score for that SNP. The kinship matrix is K = XXT/2n, where n is the number of SNPs and X is a matrix with rows corresponding to individuals, while each column contains the genotype score for a SNP (i.e. the number of copies of minor allele, 0, 1 or 2), standardised to mean zero and unit variance. The (i, j) element of K is the excess allele sharing for alleles drawn from individuals i and j, beyond that expected for unrelated individuals given the allele fraction estimates [37]. The eigenvectors of K are the principal components, used to diagnose and correct for population structure. The first principal component (PC1) showed some dispersion of individuals away from the main cluster (Figure 4(b)), which may correspond to admixture. This was the only one of the first five PCs that was significantly associated with neuroticism (p<10−4), explaining 0.5% of the phenotypic variance. From K we also identified 54 pairs of individuals with kinship estimate >0.05, which corresponds approximately to at least a cousin relationship in an outbred population (Figure 4(a)). Of these 54 kin pairs, 19 had kinships around 0.25, consistent with a first-degree relationship (sibling, or parent/offspring).
Figure 4. Kinship plots.
Plot (a): histogram of kinship coefficients >0.05; Plot (b): Principal component scores obtained from the kinship matrix. Each point represents an individual with EPQ-N score represented by grey scale as indicated on plot.
To analyse all the study subjects without having to remove one of each kin pair, we considered implementing a mixed-regression association analysis in which phenotypic correlation was proportional to K [38], [39], while age and sex were fitted as fixed effects. The motivation for this model is that any polygenic component of disease causation should be shared among individuals according to their kinships. However, in fitting the mixed model the maximum likelihood estimate of the component of variance attributable to kinship was zero. In fact, among the 19 pairs of apparent first-degree relatives, the correlation in neuroticism was −0.36, which leads to an upper 97.5% confidence limit for the heritability of neuroticism of 0.22. As a consequence of this negative correlation in our study, there is no inflation of test statistics if we ignore kinship and analyse all the study subjects using standard linear regression. After adjusting for age and sex, the EPQ-N score was close normal (Gaussian) and no transformation was necessary.
The genotypes of 2 149 individuals were imputed at over 2 million SNPs using the CEU HapMap samples and the IMPUTE program [40]. We retained for analysis the 1 705 237 genotyped and imputed SNPs with, on average over individuals, probability ≥0.9 assigned to their most likely genotype. These were analysed using a standard linear model implemented in R, with SNP coded as the expected minor allele count. In addition to PC1, age and sex were included as covariates in each model, and the interactions of these terms with the SNP were also tested. The age term used for the linear model was the age at the time of psychological assessment, not that at recruitment.
Finally, we used a Gene Ontology (GO) tree machine (Vanderbilt University) to explore the possible enrichment of any pre-defined functional category among the genes that included a SNP with p<10−4.
Supporting Information
LogQQ plot of the p-values of the SNP main effect.
(5.49 MB TIF)
Acknowledgments
The authors would like to express their gratitude to the Lausanne inhabitants who volunteered to participate in the PsyCoLaus study. We would also like to thank all the CoLaus investigators, and the PsyColaus investigators who have contributed to the recruitment, Marie-Louise Matthey, Christiane Shalofsky, Georgette Jungo, Stéphane Rothen, Lajos Oros and Gioia Roccia, as well as many GSK employees who contributed to the execution of this study including Alun McCarthy, Paul Matthews, and especially Emiliangelo Ratti for supporting this work within GSK Drug Discovery. Special thanks also go to Allen Roses and Lefkos Middleton. Thanks to William Astle for providing code and assistance for the mixed-model analysis.
Footnotes
Competing Interests: F. Tozzi, V. Mooser, D. Waterworth and N. W. Galwey are GlaxoSmithKline (GSK) full-time employees. P. Muglia was a GlaxoSmithKline full-time employee at the time when this study was performed. M. Johnson was a GSK consultant at the time when the project was initiated. F. Calboli was supported by a GSK grant to Imperial College London. The PsyCoLaus study was supported by grants from the Swiss National Science Foundation (#3200B0-105993, #3200B0-118308, 33CSCO-122661) and from GlaxoSmithKline (Psychiatry Center of Excellence for Drug Discovery and Genetics Division, Drug Discovery - Verona, R&D). GSK's role does not alter the authors' adherence to the PLoS ONE policies on sharing data.
Funding: The PsyCoLaus study was supported by grants from the Swiss National Science Foundation (#3200B0-105993, #3200B0-118308, 33CSCO-122661) and from GlaxoSmithKline (Psychiatry Center of Excellence for Drug Discovery and Genetics Division, Drug Discovery - Verona, R&D). F. Calboli was supported by a GSK grant to Imperial College London. GlaxoSmithKline had a role in study design, data analysis, decision to publish, and preparation of the manuscript: F. Tozzi, V. Mooser, D. Waterworth and N. W. Galwey are GlaxoSmithKline full-time employees. P. Muglia was a GlaxoSmithKline full-time employee at the time when this study was performed. M. Johnson was a GSK consultant at the time when the project was initiated.
References
- 1.Christensen MV, Kessing LV. Do personality traits predict first onset in depressive and bipolar disorder? Nord J Psychiatry. 2006;60:79–88. doi: 10.1080/08039480600600300. [DOI] [PubMed] [Google Scholar]
- 2.Hirschfeld RM, Klerman GL, Lavori P, Keller MB, Griffith P, et al. Premorbid personality assessments of first onset of major depression. Arch Gen Psychiatry. 1989;46:345–350. doi: 10.1001/archpsyc.1989.01810040051008. [DOI] [PubMed] [Google Scholar]
- 3.Ising M, Lauer CJ, Holsboer F, Modell S. The Munich vulnerability study on affective disorders: premorbid psychometric profile of affected individuals. Acta Psychiatr Scand. 2004;109:332–338. doi: 10.1111/j.1600-0447.2004.00309.x. [DOI] [PubMed] [Google Scholar]
- 4.Kendler KS, Kuhn J, Prescott CA. The interrelationship of neuroticism, sex, and stressful life events in the prediction of episodes of major depression. Am J Psychiatry. 2004;161:631–636. doi: 10.1176/appi.ajp.161.4.631. [DOI] [PubMed] [Google Scholar]
- 5.Wang HX, Karp A, Herlitz A, Crowe M, Kareholt I, et al. Personality and lifestyle in relation to dementia incidence. Neurology. 2009;72:253–259. doi: 10.1212/01.wnl.0000339485.39246.87. [DOI] [PubMed] [Google Scholar]
- 6.Eysenck HJ, Eysenck SBG. Manual for Eysenck Personality Questionnaire (adult and junior) Digits: San Diego 1975.
- 7.Birley AJ, Gillespie NA, Heath AC, Sullivan PF, Boomsma DI, et al. Heritability and nineteen-year stability of long and short EPQ-R Neuroticism scales. Personality and Individual Differences. 2006;40:737–747. [Google Scholar]
- 8.Lake RI, Eaves LJ, Maes HH, Heath AC, Martin NG. Further evidence against the environmental transmission of individual differences in neuroticism from a collaborative study of 45850 twins and relatives on two continents. Behav Genet. 2000;30:223–233. doi: 10.1023/a:1001918408984. [DOI] [PubMed] [Google Scholar]
- 9.Viken RJ, Rose RJ, Kaprio J, Koskenvuo M. A developmental genetic analysis of adult personality: extraversion and neuroticism from 18 to 59 years of age. J Pers Soc Psychol. 1994;66:722–730. doi: 10.1037//0022-3514.66.4.722. [DOI] [PubMed] [Google Scholar]
- 10.Jardine R, Martin NG, Henderson AS. Genetic covariation between neuroticism and the symptoms of anxiety and depression. Genet Epidemiol. 1984;1:89–107. doi: 10.1002/gepi.1370010202. [DOI] [PubMed] [Google Scholar]
- 11.Jorm AF. Sex differences in neuroticism: a quantitative synthesis of published research. Aust NZ J Psychiatry. 1987;21:501–506. doi: 10.3109/00048678709158917. [DOI] [PubMed] [Google Scholar]
- 12.Eaves LJ, Heath AC, Neale MC, Hewitt JK, Martin NG. Sex differences and non-additivity in the effects of genes on personality. Twin Res. 1998;1:131–137. doi: 10.1375/136905298320566267. [DOI] [PubMed] [Google Scholar]
- 13.Fanous A, Gardner CO, Prescott CA, Cancro R, Kendler KS. Neuroticism, major depression and gender: a population-based twin study. Psych Med. 2002;32:719–728. doi: 10.1017/s003329170200541x. [DOI] [PubMed] [Google Scholar]
- 14.Martin N, Goodwin G, Fairburn C, Wilson R, Allison D, et al. A population-based study of personality in 34000 sib-pairs. Twin Res. 2000;3:310–315. [PubMed] [Google Scholar]
- 15.Bouchard TJ, Jr, Loehlin JC. Genes evolution and personality. Behav Genet. 2001;31:243–273. doi: 10.1023/a:1012294324713. [DOI] [PubMed] [Google Scholar]
- 16.Floderus-Myrhed B, Pedersen N, Rasmuson I. Assessment of heritability for personality based on a short-form of the Eysenck Personality Inventory: a study of 12898 twin pairs. Behav Genet. 1980;10:153–162. doi: 10.1007/BF01066265. [DOI] [PubMed] [Google Scholar]
- 17.Jang KL, Livesley WJ, Vernon PA. Heritability of the big five personality dimensions and their facets: a twin study. J Pers. 1996;64:577–591. doi: 10.1111/j.1467-6494.1996.tb00522.x. [DOI] [PubMed] [Google Scholar]
- 18.Hettema JM, Prescott CA, Kendler KS. Genetic and environmental sources of covariation between generalized anxiety disorder and neuroticism. Am J Psychiatry. 2004;161:1581–1587. doi: 10.1176/appi.ajp.161.9.1581. [DOI] [PubMed] [Google Scholar]
- 19.Hettema JM, Neale MC, Myers JM, Prescott CA, Kendler KS. A population-based twin study of the relationship between neuroticism and internalizing disorders. Am J Psychiatry. 2006;163:857–864. doi: 10.1176/ajp.2006.163.5.857. [DOI] [PubMed] [Google Scholar]
- 20.Huezo-Diaz P, Tandon K, Aitchison KJ. The genetics of depression and related traits. Curr Psychiatry Rep. 2005;7:117–124. doi: 10.1007/s11920-005-0008-5. [DOI] [PubMed] [Google Scholar]
- 21.Kendler KS, Neale MC, Kessler RC, Heath AC, Eaves LJ. Major depression and generalized anxiety disorder Same genes (partly) different environments? Arch Gen Psychiatry. 1992;49:716–722. doi: 10.1001/archpsyc.1992.01820090044008. [DOI] [PubMed] [Google Scholar]
- 22.Kendler KS, Neale MC, Kessler RC, Heath AC, Eaves LJ. A longitudinal twin study of personality and major depression in women. Arch Gen Psychiatry. 1993;50:853–862. doi: 10.1001/archpsyc.1993.01820230023002. [DOI] [PubMed] [Google Scholar]
- 23.Fullerton J, Cubin M, Tiwari H, Wang C, Bomhra A, et al. Linkage analysis of extremely discordant and concordant sibling pairs identifies quantitative-trait loci that influence variation in the human personality trait neuroticism. Am J Hum Genet. 2003;72:879–890. doi: 10.1086/374178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo PH, Neale MC, Riley BP, Patterson DG, Walsh D, et al. A genome-wide linkage analysis for the personality trait neuroticism in the Irish affected sib-pair study of alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2007;144:463–468. doi: 10.1002/ajmg.b.30478. [DOI] [PubMed] [Google Scholar]
- 25.Nash MW, Huezo-Diaz P, Williamson RJ, Sterne A, Purcell S, et al. Genome-wide linkage analysis of a composite index of neuroticism and mood-related scales in extreme selected sibships. Hum Mol Genet. 2004;13:2173–2182. doi: 10.1093/hmg/ddh239. [DOI] [PubMed] [Google Scholar]
- 26.Neale BM, Sullivan PF, Kendler KS. A genome scan of neuroticism in nicotine dependent smokers. Am J Med Genet B Neuropsychiatr Genet. 2005;132:65–69. doi: 10.1002/ajmg.b.30095. [DOI] [PubMed] [Google Scholar]
- 27.Shifman S, Bhomra A, Smiley S, Wray NR, James MR, et al. A whole genome association study of neuroticism using DNA pooling. Mol Psychiatry. 2008;13:302–312. doi: 10.1038/sj.mp.4002048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van den Oord EJ, Kuo PH, Hartmann AM, Webb BT, Möller HJ, et al. Genomewide association analysis followed by a replication study implicates a novel candidate gene for neuroticism. Arch Gen Psychiatry. 2008;65:1062–71. doi: 10.1001/archpsyc.65.9.1062. [DOI] [PubMed] [Google Scholar]
- 29.Cotton CH, Flint J, Campbell TG. Is there an association between NPY and neuroticism? Nature. 2009;458:E6. doi: 10.1038/nature07927. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Z, Zhu G, Hariri AR, Enoch MA, Scott D, et al. Genetic variation in human NPY expression affects stress response and emotion. Nature. 2008;452:997–1001. doi: 10.1038/nature06858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Firmann M, Mayor V, Vidal PM, Bochud M, Pécoud A, et al. The CoLaus study: a population-based study to investigate the epidemiology and genetic determinants of cardiovascular risk factors and metabolic syndrome. BMC Cardiovasc Disord. 2008;8:6. doi: 10.1186/1471-2261-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Preisig M, Waeber G, Vollenweider P, Bovet P, Rothen S, et al. The PsyCoLaus study: methodology and characteristics of the sample of a population-based survey on psychiatric disorders and their association with genetic and cardiovascular risk factors. BMC Psychiatry. 2009;9:9. doi: 10.1186/1471-244X-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nurnberger JI, Jr, Bleharm MC, Kaufmann CA, York-Cooler C, Simpson SG, et al. Diagnostic interview for genetic studies Rationale unique features and training NIMH Genetics Initiative. Arch Gen Psychiatry. 1994;51:849–59. doi: 10.1001/archpsyc.1994.03950110009002. [DOI] [PubMed] [Google Scholar]
- 34.Andreasen NC, Endicott J, Spitzer RL, Winokur G. The family history method using diagnostic criteria Reliability and validity. Arch Gen Psychiatry. 1977;34:1229–35. doi: 10.1001/archpsyc.1977.01770220111013. [DOI] [PubMed] [Google Scholar]
- 35.Li M, Reilly C. Assessing the quality of hybridized RNA in Affymetrix GeneChips using linear regression. J Biomol Tech. 2008;19:122–8. [PMC free article] [PubMed] [Google Scholar]
- 36.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Astle W, Balding DJ. Population structure and cryptic relatedness in genetic association studies, Statist Sci. 2009;24(4):451–471. [Google Scholar]
- 38.Yu JM, Pressoir G, Briggs WH, Bi IV, Yamasaki M, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 2006;38:203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- 39.Chen WM, Abecasis GR. Family-based association tests for genomewide association scans. Am J Hum Genet. 2007;81:913–926. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies via imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 41.Bocciardi R, Giorda R, Marigo V, Zordan P, Montanaro D, et al. Molecular characterization of a t(2;6) balanced translocation that is associated with a complex phenotype and leads to truncation of the TCBA1 gene. Hum Mutat. 2005;26:426–36. doi: 10.1002/humu.20235. [DOI] [PubMed] [Google Scholar]
- 42.Filmus J, Capurro M, Rast J. Genome Biol. 2008;9:224. doi: 10.1186/gb-2008-9-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grove EA, Tole S, Limon J, Yip LW, Ragsdale CW. The hem of the embryonic cerebral cortex is defined by the expression of multiple Wnt genes and is compromised in Gli3-deficient mice. Development. 1998;125:2315–2325. doi: 10.1242/dev.125.12.2315. [DOI] [PubMed] [Google Scholar]
- 44.Beasley C, Cotter D, Khan N, Pollard C, Sheppard P, et al. Glycogen synthase kinase-3 beta immunoreactivity is reduced in the prefrontal cortex in schizophrenia. Neuroscience Letters. 2001;302:117–120. doi: 10.1016/s0304-3940(01)01688-3. [DOI] [PubMed] [Google Scholar]
- 45.Mathew SJ, Manji HK, Charney DS. Novel drugs and therapeutic targets for severe mood disorders. Neuropsychopharmacology. 2008;33:2080–2092. doi: 10.1038/sj.npp.1301652. [DOI] [PubMed] [Google Scholar]
- 46.Inestrosa NC, Varela-Nallar L, Grabowski CP, Colombres M. Synaptotoxicity in Alzheimer's disease: The Wnt signaling pathway as a molecular target. IUBMB Life. 2007;59:316–321. doi: 10.1080/15216540701242490. [DOI] [PubMed] [Google Scholar]
- 47.De Ferrari GV, Moon RT. The ups and downs of Wnt signaling in prevalent neurological disorders. Oncogene. 2006;25:7545–7553. doi: 10.1038/sj.onc.1210064. [DOI] [PubMed] [Google Scholar]
- 48.Wilson RS, Evans DA, Bienias JL, de Leon CFM, Schneider JA, et al. Proneness to psychological distress is associated with risk of Alzheimer's disease. Neurology. 2003;61:1479–1485. doi: 10.1212/01.wnl.0000096167.56734.59. [DOI] [PubMed] [Google Scholar]
- 49.Folstein MF, Folstein SE, MacHugh PR. Mini Mental State: a practical method for grading the cognitive state of patients for the clinician. J Psych Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
LogQQ plot of the p-values of the SNP main effect.
(5.49 MB TIF)