Abstract
Background
Micro(mi)RNAs are short RNA sequences, ranging from 16 to 35 nucleotides (miRBase; http://www.mirbase.org). The majority of the identified sequences are 21 or 22 nucleotides in length. Despite the range of sequence lengths for different miRNAs, individual miRNAs were thought to have a specific sequence of a particular length. A recent report describing a longer variant of a previously identified miRNA in Arabidopsis thaliana prompted this investigation for variations in the length of other miRNAs.
Results
In this paper, we demonstrate that a fifth of annotated A. thaliana miRNAs recorded in miRBase V.14 have stable miRNA isoforms that are one or two nucleotides longer than their respective recorded miRNA. Further, we demonstrate that miRNA isoforms are co-expressed and often show differential argonaute complex association. We postulate that these extensions are caused by differential cleavage of the parent precursor miRNA.
Conclusions
Our systematic analysis of A. thaliana miRNAs reveals that miRNA length isoforms are relatively common. This finding not only has implications for miRBase and miRNA annotation, but also extends to miRNA validation experiments and miRNA localization studies. Further, we predict that miRNA isoforms are present in other plant species also.
Background
Micro(mi)RNAs are important for gene regulation [1] and for cell fate decisions during development [2]. Aberrant levels of miRNAs are seen in various disease states [3-6]. miRNAs are transcribed from one strand of their genomic loci into a primary miRNA transcript, which folds into a characteristic bulge with stem-loop conformation [7]. In plants, the primary transcript is cleaved by a Dicer-like (DCL) RNase III enzyme, DCL1, into an approximately 19 bp duplex with a two-nucleotide (nt) overhang at either end [8]. Of the two strands forming the duplex, one strand, designated miRNA*, is typically degraded while the other is incorporated into the argonaute (AGO)-containing effector complex [9,10]. Co-immunoprecipitation experiments demonstrate an enrichment of miRNAs in AGO1, whereas AGO2 shows depletion of miRNAs compared with non-immunoprecipitated samples [11].
The biological significance of sequence length heterogeneity has been recently identified for a mature miRNA in Arabidopsis thaliana, in which ath-MIR168 is processed as miRNAs of 21 and 22 nucleotides in length from its two genomic loci. Vaucheret demonstrated that reducing the amount of 21 nt miRNA greatly reduces homeostasis and leads to developmental defects of the plant, especially in environmentally challenging conditions [12]. In general, it is appreciated that there is variation in the lengths of different miRNAs, as the mature miRNAs listed in miRBase http://www.mirbase.org/ are between 16 and 35 nucleotides in length [13]. In miRBase V.14 there are 209 small RNA sequences identified for in A. thaliana, of which 7%, 79%, 11% and 3% are 20, 21, 22 and 24 nt in length, respectively. The reason and function for this heterogeneity is unclear and we are unaware of any systematic investigation into non-uniform length distributions of individual miRNAs. Each annotated miRNA in miRBase is a single defined sequence, and there are no details on the possibility of variable sequence length. Sequence length variation may have been overlooked previously, as small variations in the sequence length might not have been thought to alter the function of individual miRNAs, as they are directed to their targets by base pairing.
Recent reports show however, that alterations in miRNA length can potentially lead to dramatic effects on miRNA function in organisms such as A. thaliana, in which the identity of the first 5' nucleotide of the miRNA is the major determinant for AGO protein association [11,14]. Sequence-specific AGO association has been characterized for most A. thaliana AGO complexes [11,14,15]. Of these, AGO1 is the major AGO in the pathway of miRNA post-transcriptional gene silencing [16-19], whereas AGO4 functions in repeat-associated silencing of RNA accumulation and in regulating loci- specific DNA methylation [20,21].
To investigate the frequency with which additional nucleotides on the 5' ends of miRNAs are observed, we queried several published A. thaliana small RNA datasets collected by pyrophosphate and Solexa/Iillumina http://www.illumina.com sequencing techniques. The approach of analyzing small RNA sequencing datasets has previously proven successful for the identification of post-transcriptional modifications in small RNAs [22-25]. Using similar methods, we queried all of the annotated miRNAs from A. thaliana (miRBase V.14) for 5' extensions of one to three nucleotides based on nucleotides present in the pre-miRNA hairpin.
The datasets investigated were from three small RNA sequencing studies including a small RNA transcriptome that responds to changing phosphate levels [26], an RNA analysis of the dicer (DCL2/DCL3/DCL4) triple mutant [27], and a study on RNAs that are co-immunoprecipitated with different AGO proteins [11]. In total, these datasets contained 51,907,309 redundant small RNA sequences.
Results
MiRNAs with SNE
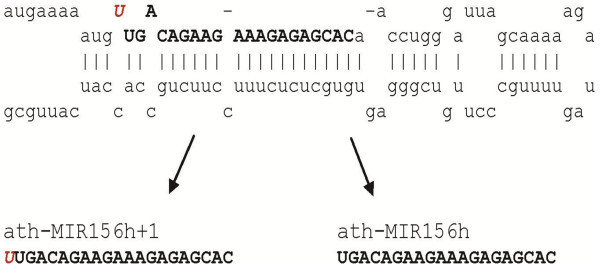
For our in silico northern blot analysis, we queried each dataset mentioned above with each miRNA sequence for A. thaliana listed in miRBase V.14. In addition to the recorded mature miRNA sequence, we extended each mature miRNA at the 5' by 1, 2 and three nucleotide(s) according to the hairpin sequence of the miRNA. To our surprise, numerous miRNAs encompass a 5' single nucleotide extension (SNE) compared with the recorded mature miRNA; for example, ath-MIR156h. The SNE of ath-MIR156h is an additional 5' uridine/uracil (U) that is not reported in the annotated mature miRNA sequence [13,28,29], but is present in the parental pre-miRNA hairpin (Figure 1); the extended form of ath-MIR156h is henceforth referred to as ath-MIR156h+1. Both ath-MIR156h and ath-MIR156h+1 were present in small RNA samples, independent of genetic background, environmental effects, tissue types and sequencing technologies (see Table 1). A consistent cloning ratio of 7:3 (ath-MIR156h+1:ath-MIR156h) was observed, despite large variations in total abundance in different genetic backgrounds and tissues. One exception to the 7:3 ratio was found in small RNA cloning data originating from the plant root, in which the two miRNAs were found in a 1:1 ratio (Table 1). The high frequency of occurrence of the ath-MIR156h+1 sequence and the reproducibility between datasets suggests a biological role for this long variant of ath-MIR156h.
Figure 1.
Two mature micro(mi)RNA sequences derived from the ath-MIR156h pre-miRNA hairpin. The primary hairpin ath-MIR156h gives rise to two mature isoforms of ath-MIR156h, which differ by one nucleotide on the 5' terminus.
Table 1.
ath-MIR156h exists in two isoforms across seven datasets
| ath-MIR156h+1, %, (n) | ath-MIR156h, %, (n) | Reference | |
|---|---|---|---|
| Col-0 | 71.7 (2,445) | 28.2 (962) | [27] |
| SUr2a | 69.9 (25,789) | 30.0 (11,102) | [27] |
| S234a | 70.2 (24,472) | 29.7 (10,802) | [27] |
| Shoot +Pi | 75.9 (328) | 24.1 (104) | [26] |
| Shoot -Pi | 74.1 (413) | 25.9 (144) | [26] |
| Root -Pi | 54.5 (54) | 45.5 (45) | [26] |
| Root +Pi | 50.4 (70) | 49.6 (71) | [26] |
Ath-MIR156h+1 has a single extra nucleotide on the 5' terminus compared with ath-MIR156h. Both isoforms co-exist across various genetic backgrounds and under various environmental conditions. WT (Col-0), ΔRDR2 (Sur2a) and DCL2/DCL3/DCL4-triple (S234a) mutant Arabidopsis thaliana [27], root and shoot small RNA transcriptomes under varying phosphate levels GEO: GSE17741 [26].
The distribution of ath-MIR156h+1 in various AGO complexes differs from that of the parental ath-MIR156h miRNA. AGO association was analyzed by determining the frequencies with which ath-MIR156h and ath-MIR156h+1 were identified in previously published datasets of miRNAs co-purified with AGO1, AGO2, AGO4 and AGO5 [11]. The current model for A. thaliana miRNAs predicts that ath-MIR156h should be mostly present in AGO1-RISC complexes, as the miRNA possesses a 5' U nucleotide. As predicted, over half of ath-MIR156h miRNAs reside in AGO1 complexes (54%), whereas the remainder are split into AGO5 (31%) and AGO4 (15%) effector complexes. No association with AGO2 was found. However, in addition to a 10-fold increased frequency of detection, ath-MIR156h+1 was detected almost exclusively in AGO5 complexes (84.1%), with few sequences detected in AGO1 (8%) and AGO4 (7%) datasets (Figure 2). This shift in association with AGOs was not initially predicted, as ath-MIR156h+1 still has a 5' U, nonetheless a shift in the frequency of AGO association was observed.
Figure 2.
AGO association of ath-MIR156h isoforms. Mature miRNA ath-MIR156h resides mostly in the argonaute (AGO)1 effector complex, whereas the majority of ath-MIR156h+1 resides in the AGO5 effector complex. The cloning ratios are given in percentage and absolute numbers. Statistical analysis on the AGO association frequencies using the Freeman-Halton extension of the Fisher exact probability test [33] revealed a highly significant result (P = 2.5 × 10-5) for the observed difference in AGO association. Data originated from the AGO association datasets [11].
MiRNAs with a double nucleotide extension
In our first example, we demonstrated that both ath-MIR156h and ath-MIR156h+1 coexist within the plant at constant ratios, with each miRNA isoforms showing preferential AGO association. A second class of miRNAs identified possess two additional 5' nucleotides. An example of this class is ath-MIR775, which exists as both ath-MIR775 and ath-MIR775+2; the latter has two additional 5' U nucleotides, with both of these nucleotides present in the pre-miRNA hairpin. The parental miRNA and ath-MIR775+2 were found at comparable frequencies in all the datasets (1858 and 1587 occurrences, respectively, in the AGO association database). Conversely, there was a negligible occurrence of the +1 miRNA. There are two possible explanations for this exclusive occurrence of ath-MIR775 and ath-MIR775+2. Cleavage events generating the mature miRNAs might generate the two variable length miRNAs forms (0 and +2) exclusively. Alternatively, all three lengths (0, +1 and +2) might be generated, but with only the 0 and +2 forms being stabilized and protected from degradation.
Analysis of the AGO associations of ath-MIR775 and ath-MIR755+2 revealed a difference in the identity of preferential AGO association. In more than 95% of results, the ath-MIR775 sequence was found to be associated with AGO1, whereas the ath-MIR775+2 variant was associated with AGO5 in nearly 70% of cases (Table 2).
Table 2.
Cloning frequencies of ath-MIR775 sequences in the argonaute (AGO) association datasets
| AGO1 | AGO2 | AGO4 | AGO5 | |
|---|---|---|---|---|
| ath-MIR775 | 1788 | 4 | 22 | 44 |
| ath-MIR775+1 | 38 | 0 | 7 | 81 |
| ath-MIR775+2 | 409 | 0 | 86 | 1092 |
Not all miRNAs are heterogeneously processed
Heterogeneity in mature miRNA lengths is not the rule, as many do not exhibit detectable amounts of variable length processing. Examples include ath-MIR168b which was observed 86,634 times in the four different AGO association datasets, whereas the ath-MIR168b+1 sequence was observed only 34 times. This observed frequency is within the 3% insertion/deletion error rate of pyrophosphate sequencing [30]. To date, there have been no detailed analyses by Illumina or Solexa sequencing of the frequency of insertion and deletion errors. The presence of both types of miRNAs (variable and homogeneous lengths) suggests that the variable lengths of some miRNAs are not simply the result of 'ragged end' processing of all miRNAs, but are a specific process for a subset of miRNAs.
Overall frequency of variable length miRNAs
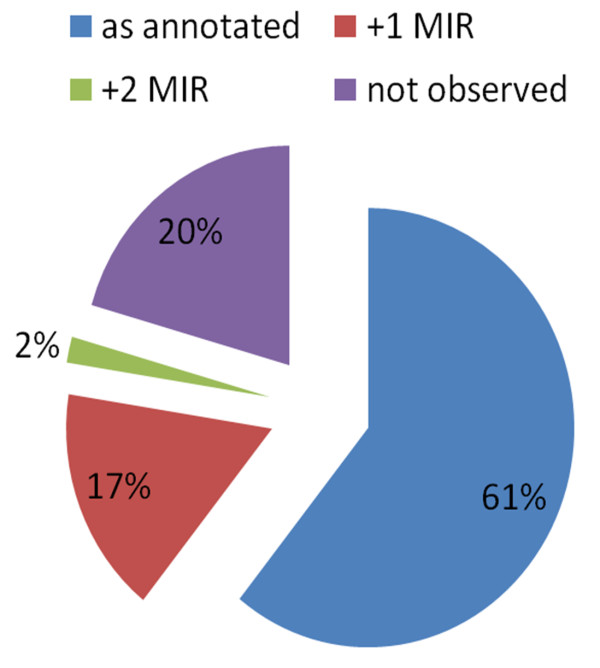
In addition to the two examples outlined above, we systematically queried the entire ath-MIR dataset from miRBase V.14 in an in silico northern blot analysis. The presence and frequency of each miRNA sequence, including the +1, +2 and +3 extended miRNA forms, was queried against the database (Figure 3; see Additional File 1). The sequence had to be present in a dataset at least six times to be counted. Of the 209 annotated miRNAs in miRBase, 166 were found in this analysis. Of the observed miRNA sequences, 35 were found to have a single nucleotide addition, and four were observed had two nucleotides added. In total, nearly 20% of the annotated miRNAs had additional 5' nucleotides. These 5' extensions were not simply misannotated miRNAs, as isoforms of various lengths co-existed. In addition to identifying the miRNAs, we examined miRNAs exhibiting length isoforms for changes in AGO association (see Additional File 2).
Figure 3.
Frequency of extra 5' nucleotides. Several small Arabidopsis thaliana RNA datasets were analyzed for the occurrence of miRNAs with extra 5' nucleotides. From these datasets, 61% of the A. thaliana miRNAs listed in miRBase had no significant occurrence of extra nucleotides, 20% of the annotated miRNAs were not observed in the datasets at all, and 19% of miRNA had a significant proportion of either one (+1 MIR) or two (+2 MIR) nucleotides.
In addition to our presented in silico data, previous work using a genetic approach also suggests co-existence of miRNA and miRNA+1 and the importance of their co-expression. A recent report described and confirmed the occurrence of a long (22 nt) form of ath-miR168 [12]. In addition, experiments by Vaucheret and data from other studies also reveals evidence of long miRNA variants; for example, careful examination of previously published miRNA northern blots found the presence of double bands for some miRNAs, such as for ath-miR169, ath-miR156 and ath-miR172 [31].
Conclusions
We have presented evidence arising from several small RNA sequencing experiments that supports the co-existence of mature miRNAs and their 5' extended forms in A. thaliana. Our results expand the previous genetic evidence of variability in miRNA sequence length [12] by revealing that nearly a fifth of miRNAs identified in A. thaliana have additional nucleotide(s) on their 5' ends. These 5' extended miRNAs are not simply misannotated, as both longer and shorter forms of the miRNAs co-exist. Additionally, we provide evidence that the 5' end variations can result in changes in the type of AGOs with which these miRNA isoforms preferentially associate. Differences in AGO associations suggest alterations in the biological functioning of the different observed forms of these miRNAs. These variable length miRNAs could essentially be considered miRNA isoforms and should be included in any annotation of miRNAs.
Methods
A Perl script mapped each mature miRNA to their respective hairpin, recorded the hairpin sequence, then appended one, two or three nucleotide(s) to the 5' of the mature miRNA. For miRNA, the Perl script recorded five sequences: hairpin, mature miRNA, +1 miRNA, +2 miRNA and +3 miRNA. All * sequences were ignored and not used for analysis. Scripts are available online under GPLV.2 http://www.bioinformatics.org/ebbie. The output file in FASTA format was used for an in silico northern blot, which probed all computer-generated small RNA sequences in various datasets (GEO:GSE17741 [26], GEO:GSE5343 [32] and ath-sbs [27]) using a modified Ebbie-(mis)match-AGO v1 [22] script. To determine AGO complex affiliation, the computer-generated small RNAs were similarly compared against the AGO1, AGO2, AGO4 and AGO5 small RNA datasets [11] using Ebbie-(mis)match-AGO v2. Computation was performed on an IBM system (Model x3850; IBM Computers, Markham, ON, Canada).
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
HAE designed experiments and analyzed data. AF wrote the Perl scripts. HAE and RPF wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Validation of computer-generated miRNA isoforms. An in silico northern blot was performed of all computer-generated miRNA isoforms by scanning seven different datasets for the expression of each in silico miRNA isoform.
Validated miRNA isoforms and their AGO association. All validated miRNA isoforms from Additional file 1 were scanned across various argonaute complexes.
Contributor Information
H Alexander Ebhardt, Email: ebhardt@ualberta.ca.
Amber Fedynak, Email: afedynak@gmail.com.
Richard P Fahlman, Email: rfahlman@ualberta.ca.
Acknowledgements
HAE is a Natural Sciences and Engineering Research Council of Canada (NSERC) Post-Doctoral Fellow. This project has been made possible through a grant from the Alberta Health Services and the Alberta Cancer Foundation to RPF.
References
- Carthew RW. Gene regulation by microRNAs. Curr Opin Genet Dev. 2006;16:203–208. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAS and their regulatory roles in plants. Ann Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Hagen JW, Lai EC. microRNA control of cell-cell signaling during development and disease. Cell Cycle. 2008;7:2327–2332. doi: 10.4161/cc.6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AE. Functional aspects of animal microRNAs. Cell Mol Life Sci. 2008;65:545–562. doi: 10.1007/s00018-007-7355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Lu Y, Wang Z. Control of cardiac excitability by microRNAs. Cardiovasc Res. 2008;79:571–580. doi: 10.1093/cvr/cvn181. [DOI] [PubMed] [Google Scholar]
- Ebhardt HA, Thi EP, Wang MB, Unrau PJ. Extensive 3' modification of plant small RNAs is modulated by helper component-proteinase expression. Proc Natl Acad Sci USA. 2005;102:13398–13403. doi: 10.1073/pnas.0506597102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, Matzke M, Ruvkun G, Tuschl T. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ, Griffiths-Jones S, Jacobsen SE, Mallory AC, Martienssen RA, Poethig RS, Qi Y, Vaucheret H, Voinnet O, Watanabe Y, Weigel D, Zhu JK. Criteria for annotation of plant microRNAs. Plant Cell. 2008;20:3186–3190. doi: 10.1105/tpc.108.064311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, Chen S, Hannon GJ, Qi Y. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5' terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H. AGO1 homeostasis involves differential production of 21-nt and 22-nt miR168 species by MIR168a and MIR168b. PLoS One. 2009;4:e6442. doi: 10.1371/journal.pone.0006442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–8. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- Havecker ER, Wallbridge LM, Hardcastle TJ, Bush MS, Kelly KA, Dunn RM, Schwach F, Doonan JH, Baulcombe DC. The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell. 2010;22:321–334. doi: 10.1105/tpc.109.072199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crete P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Denli AM, Hannon GJ. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell. 2005;19:421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel JB, Godon C, Mourrain P, Beclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- Qi Y, He X, Wang XJ, Kohany O, Jurka J, Hannon GJ. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- Ebhardt HA, Tsang HH, Dai DC, Liu Y, Bostan B, Fahlman RP. Meta-analysis of small RNA-sequencing errors reveals ubiquitous post-transcriptional RNA modifications. Nucleic Acids Res. 2009;37:2461–2470. doi: 10.1093/nar/gkp093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJ, Taft RJ, Hashimoto T, Kanamori-Katayama M, Kawaji H, Kawano M, Kishima M, Lassmann T, Faulkner GJ, Mattick JS, Daub CO, Carninci P, Kawai J, Suzuki H, Hayashizaki Y. Cross-mapping and the identification of editing sites in mature microRNAs in high-throughput sequencing libraries. Genome Res. 2010;20:257–264. doi: 10.1101/gr.095273.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida K, Jin H, Zhu JK. Bioinformatics analysis suggests base modifications of tRNAs and miRNAs in Arabidopsis thaliana. BMC Genomics. 2009;10:155. doi: 10.1186/1471-2164-10-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantano L, Estivill X, Marti E. SeqBuster, a bioinformatic tool for the processing and analysis of small RNAs datasets, reveals ubiquitous miRNA modifications in human embryonic cells. Nucleic Acids Res. 2010;38(5):e34. doi: 10.1093/nar/gkp1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY, Li WH, Chiou TJ. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009;151:2120–2132. doi: 10.1104/pp.109.147280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- sbs database. http://mpss.udel.edu/at_sbs/
- Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Mallory AC, Bartel DP. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol Cell. 2006;22:129–136. doi: 10.1016/j.molcel.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Kulkarni K, Souret FF, MuthuValliappan R, Tej SS, Poethig RS, Henderson IR, Jacobsen SE, Wang W, Green PJ, Meyers BC. MicroRNAs and other sm all RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 2006;16:1276–1288. doi: 10.1101/gr.5530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman GH, Halton JH. Note on exact treatment of contingency goodness of fit and other problems of significance. Biometrika. 1951. pp. 141–149. [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Validation of computer-generated miRNA isoforms. An in silico northern blot was performed of all computer-generated miRNA isoforms by scanning seven different datasets for the expression of each in silico miRNA isoform.
Validated miRNA isoforms and their AGO association. All validated miRNA isoforms from Additional file 1 were scanned across various argonaute complexes.