Abstract
Knowledge of the orientation of molecules within biological structures is crucial to understanding the mechanisms of cell function. We present a method to image simultaneously the positions and fluorescence anisotropies of large numbers of single molecules with nanometer lateral resolution within a sample. Based on a simple modification of fluorescence photoactivation localization microscopy (FPALM), polarization (P)-FPALM does not compromise speed or sensitivity. We show results for mouse fibroblasts expressing Dendra2-actin or Dendra2-hemagglutinin.
Light microscopy allows noninvasive imaging of multiple species in biological specimens with single-molecule sensitivity, but diffraction normally limits the resolution to ~150–250 nm. As many biological processes occur on smaller length scales, techniques that can image below the diffraction limit and yield single-molecule information are becoming increasingly important.
Recently developed methods can break the diffraction barrier by stimulated emission depletion1 or by localization of large numbers of single molecules, and achieve effective resolution in the 10–40 nm range2–4. In localization-based methods, small subsets of photoactivatable fluorescent molecules are stochastically activated in the sample by illumination with an activation laser. Photoactivated molecules are illuminated by a second laser, imaged and then deactivated, either actively or by spontaneous photobleaching. The process is repeated until data have been acquired on a sufficiently large number of molecules or all possible molecules. Image analysis is then used to measure the position of each molecule and determine its intensity.
Localization-based methods can now image living cells5,6, three-dimensional specimens7,8 and multiple species. These methods, however, do not provide information about the orientation and rotational freedom of individual molecules, which can be used to test the degree of interaction between molecules in biological systems. Furthermore, understanding organization and functionality of molecular machines often requires determination of the orientation of molecules within cellular structures and relative to one another. Previous imaging of single molecule anisotropies has relied on near-field methods9, shape analysis of molecular images obtained by diffraction-limited techniques10,11 or other methods of imaging relatively sparse distributions of molecules. We set out to augment the capabilities of localization-based microscopy to obtain high-density maps of single-molecule positions and anisotropies.
We present a method for imaging single-molecule polarization anisotropy (a measure of the orientation of the transition dipole moment of a fluorescent molecule) in biological specimens with resolution below the diffraction limit. Our method is based on fluorescence photoactivation localization microscopy (FPALM)2 with a modified detection path, and we termed it polarization-FPALM(P-FPALM). The addition of a polarizing beam splitter into the detection path allows simultaneous, spatially separate imaging of the emission polarized parallel and perpendicular to a particular axis within the sample. The two detection paths are adjusted to have the same total length from the microscope tube lens. Analysis of the relative intensities of molecules in the two images yields the anisotropy of each localized molecule. Others have implemented a similar approach to study the rotational mobility of individual fluorescent molecules during single-particle tracking experiments12.
For imaging, we placed the sample on the stage of an inverted microscope with a ×60, 1.2 numerical aperture (NA) water-immersion objective and illuminated it using two lasers: 405 nm activation and 488 nm readout for the photoactivatable green fluorescent protein (PA-GFP)13 or 405 nm activation and 556 nm readout for the photoswitchable protein Dendra2 (ref. 14), which can be photoactivated from a green-fluorescent form to a red-fluorescent form. We focused the lasers in the objective back-aperture to cause a large area of the sample to be illuminated with an approximately Gaussian profile with linear polarization along the x, x and y directions for the 405, 488 and 556 nm beams, respectively (Supplementary Figs. 1 and 2 online). Fluorescence detected by the same objective is filtered by the dichroic mirror and interference filters (Supplementary Table 1 online), focused by the tube lens to form an intermediate image, which is magnified by a telescope consisting of +60 mm and +200 mm achromatic lenses, to result in an overall magnification of ~192 and an effective pixel size in object space of 83.3 nm. The magnified image was detected with an electron-multiplying charge-coupled device (EMCCD) camera at 10–32 frames per second for ~20–600 s (Supplementary Table 2 online). The use of a water-immersion lens is an advantage because it minimizes aberrations when imaging a sample that is also in water. Illumination using a relatively unfocused Gaussian beam is also advantageous because it reduces the tipping of the polarization toward the z axis, which results from a high-NA diffraction-limited focus. Because single molecules are being localized using two detection channels (three channels are required to determine the full orientation in three dimensions), anisotropies measured for molecules that are oriented with a component out of the x-y plane will only be approximate, owing to tipping of the polarization by the high-NA objective15. As a result, the anisotropies we acquired could not be interpreted directly as an angle relative to the laser polarization axis, but calculations accounting for the effects of polarization tipping (Supplementary Figs. 3 and 4 online) allowed specification of the range of orientations the molecule could have (Supplementary Fig. 5 online), within experimental error, when close to the center of the field. Activation and illumination pulse protocols are described in Supplementary Methods and Supplementary Table 2 online.
For analysis, the images T and R (Fig. 1a and Supplementary Fig. 1), corresponding to the fluorescence transmitted and reflected by the polarizing beamsplitter, respectively, are first correlated with each other using images of fluorescent beads. Image R is shifted, rotated, and stretched linearly in the x and y directions (conserving the total number of detected photons), to produce the best normalized cross-correlation with image T. The transformation parameters measured from the bead images are then used to transform all subsequent images.
Figure 1.
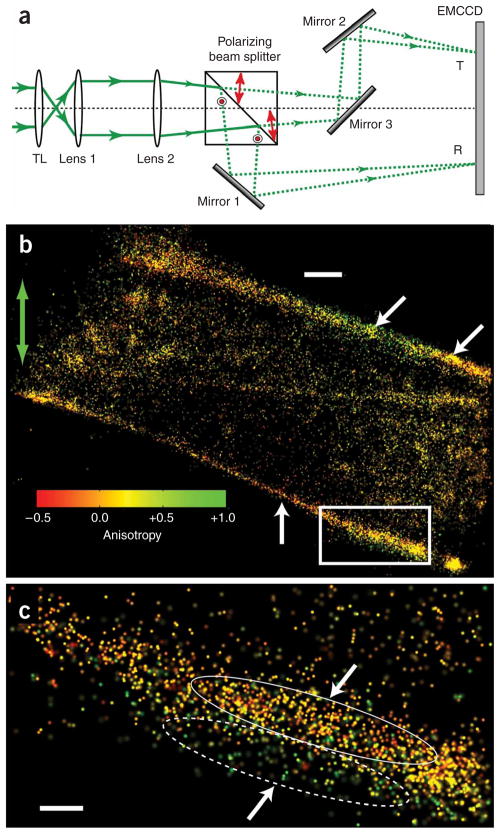
P-FPALM detection path and P-FPALM imaging of a fixed fibroblast expressing Dendra2-actin. (a) The intermediate image formed by tube lens (TL) is expanded by lenses 1 and 2 before entering the polarizing beam splitter. The reflected beam (R; lower path, light polarized in the x direction at the sample) is directed to the EMCCD by mirror 1 while the transmitted beam (T; upper path, light polarized in the y direction at the sample) is directed to the same EMCCD by mirrors 2 and 3. Mirrors 2 and 3 are adjusted so that the total length of both paths is equal. Red arrows and circles show the polarization of the light in the T and R paths, respectively. (b) P-FPALM image of Dendra2-actin expressed in a fixed fibroblast (21,525 molecules). Green arrow indicates the direction of polarization of the readout beam. (c) Magnification of the boxed region in b. Gradients in single-molecule anisotropy (2,015 molecules) are marked by arrows and ellipses. White arrows in b and c point to regions within the cell with consistently negative or consistently positive anisotropy values. Compare for example the two regions marked by arrows on the upper right of b. Compare also the lower fiber bundle in b labeled with the arrow, and the regions outlined by ellipses in c. Color bar indicates anisotropy scale for b and c. Scale bars, 1 μm (b) and 250 nm (c).
We calculated the anisotropy (r) from the ratio of fluorescence emitted by the molecule and detected with polarization parallel (I||) and perpendicular (I⊥) to the laser, respectively16:
| (1) |
The I|| and I⊥ were background subtracted, and corrected for bleed-through and relative detection efficiency (see Supplementary Methods for details on experimental methods and comparison with simulations). As a control, we determined mean anisotropy values for rhodamine B in both low- and high-viscosity solutions; these values agreed within uncertainty (±0.025 in anisotropy value) with results published previously17, demonstrating the accuracy of the method on a sample with known anisotropy values (Supplementary Fig. 6a online). Anisotropies measured for caged-fluorescein suspended in agarose gel also agreed with theoretical calculations (Supplementary Fig. 7 online).
PA-GFP molecules imaged on glass showed little spatial dependence of the anisotropy (Supplementary Fig. 8 online). In contrast, molecules localized in cells transfected with Dendra2-actin had elongated filamentous structures with clear patterns in the distribution and anisotropy values of molecules (Fig. 1b,c and Supplementary Fig. 9 online). We expected actin fiber bundle density to affect the measured anisotropy by limiting or permitting certain probe orientations.
The effective resolution (Supplementary Methods, equation S3) of ~26 nm calculated for the structure shown in Figure 1c was limited by the localization precision (~7 nm median value; Supplementary Fig. 10 online), but more so by the density of localized molecules (~25 nm median nearest neighbor distance). We improved the localization precision compared to our previous FPALM work by reducing background by prebleaching imaging buffers with UV light, using a lower-noise camera and increasing the excitation intensity. Also, consistent with other reports in the field, we report the one-sigma uncertainty in positions using the s.d. of the point spread function (Supplementary Methods, equation S1), rather than the 1/e2 radius of the point spread function we used previously5. The reported localization precision values had been calculated from measured numbers of photons, and do not take into account other sources of uncertainty, such as drift and detector heterogeneity. Under the criterion for effective resolution given in reference 6, where twice the nearest neighbor distance had been used as a measure of the Nyquist limit, the effective resolution for the data in Figure 1c would be ~50 nm.
Molecules localized in the extended fiber bundles had obvious gradients in their anisotropy; some regions contained mostly molecules emitting parallel to the direction of the excitation (Fig. 1b,c). In another region within the same structure, the majority of molecules emitted fluorescence polarized perpendicular to the excitation direction (Fig. 1b). We observed the opposite trend in anisotropy close to the edge of the cell (Fig. 1b,c). In contrast, the interior of the cell had fewer visible fiber bundles but instead had actin-rich regions >1 μm wide without extended linear structures (Supplementary Fig. 9b), resembling actin imaged by PALM in lamellipodia3. Confocal images of cells labeled with both Dendra2-actin and Alexa-680–phalloidin confirmed co-localization of Dendra2-actin with actin structures (Supplementary Fig. 11 online).
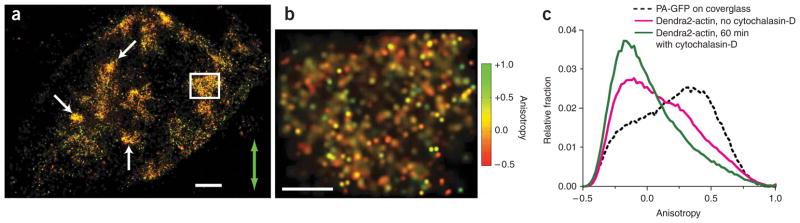
To test whether these trends in anisotropy correspond to filamentous actin structures, we treated cells with 1 μM cytochalasin-D for 60 min to disrupt the actin cytoskeleton before fixing and imaging them. The cell structure changed drastically (Figs. 2a,b and Supplementary Figs. 12 and 13 online): the cells rounded up, the fiber bundles were no longer visible, and the remaining structures showed very little trend in the anisotropy. Among all treated cells that we imaged, none showed distinct filamentous structures or anisotropy patterns like the ones observed in untreated cells. Within the ~1 μm-sized globular clusters that were visible, we observed a mixture of molecules emitting parallel and perpendicular to the excitation (Fig. 2b). Overall histograms of anisotropy values for all treated and all untreated cells showed substantial differences resulting from cytochalasin-D treatment (Fig. 2c and Supplementary Fig. 14 online). Note that all structures visible below ~250 nm would be unresolved in a conventional fluorescence microscope.
Figure 2.
Effect of cytochalasin-D on structure and anisotropy of Dendra2-actin. (a) P-FPALM image of a fixed fibroblast after a 60-min treatment with 1 μM cytochalasin-D (32,553 molecules). Green arrow indicates the direction of polarization of the readout beam. White arrows indicate globular clusters of Dendra2-actin. (b) Magnification of the boxed region in a (1,878 molecules) highlighting a mixture of molecules emitting parallel and perpendicular to the excitation. Color bar indicates anisotropy scale in a and b. Scale bars, 1 μm (a) and 250 nm (b). (c) Cumulative distribution of single-molecule anisotropies for PA-GFP on coverglass (n = 10; 108,399 total molecules), and in fixed fibroblasts expressing Dendra2-actin without cytochalasin-D treatment (n = 30; 496,844 total molecules) and after a 60-min treatment with 1 μM cytochalasin-D (n = 5; 187,457 total molecules).
When interpreting anisotropy values, probe rotational mobility is an important consideration. Even in fixed samples, fluorescent probes not attached to cell structures by multiple fixative crosslinks may be capable of limited motion. Because the rotational time constant for fluorescent proteins in cells18 is typically on the nanosecond timescale, the emission from a given orientation of the probe will be sampled thousands of times during a single frame. Hence, the measured anisotropy will reflect the range of orientations accessible to the probe. For fixed samples, fewer orientations will be accessible, and the anisotropy values will be substantially different from the values observed for freely diffusing molecules in solution16.
To evaluate P-FPALM using a different biological application we obtained images of hemagglutinin in fixed fibroblasts. This yielded clustered distributions of hemagglutinin consistent with previous FPALM imaging7 and also provided orientational information for individual hemagglutinin molecules within clusters. The anisotropy of PA-GFP–tagged hemagglutinin showed an example of a cluster of molecules with similar anisotropy values, positioned near the edge of the cell, approximately ~1 × 2 μm in size (Supplementary Fig. 15 online). The surrounding clusters of molecules had larger values of anisotropy. P-FPALM imaging of Dendra2-tagged hemagglutinin (Supplementary Figs. 16 and 17 online) also showed hemagglutinin clustering. In some cases, we observed differences in anisotropy values from cluster to cluster. Whereas anisotropy measurements of some tagged molecules could be tag-dependent, we neither confirmed nor excluded this possibility. These spatially dependent differences in anisotropy could be useful for understanding the formation of clusters of hemagglutinin in membranes, which are used by the influenza virus to gain entry into host cells and to assemble the components to build a new virus that will eventually bud from the host cell.
The ability to image the anisotropy with resolution below the diffraction limit presents several captivating opportunities, most importantly the ability to image short-range order and to quantify the degree of preferential orientation of molecules. As long as the limitations of the method are taken into account, we can use the anisotropy to estimate the degree of alignment (but not the precise angle) between the transition-dipole moment of the emitting molecule (the fluorescent probe) and a particular coordinate axis16. Interactions between membrane domains and the cytoskeleton, such as those found in focal adhesions6, are expected to result in preferential orientation of molecules, but the size of those structures is generally well below the diffraction limit19. The improved resolution in P-FPALM will allow quantification of order of proteins and lipids in membrane domains at length scales inaccessible to standard methods.
The ability to image both nanoscale structure and quantify relative molecular orientation can address biological questions where improved resolution is crucial but insufficient by itself. In principle, P-FPALM will be compatible with live-cell FPALM, with PALM and STORM using widefield excitation, and with multicolor imaging. It requires only the addition of a polarizing beam splitter, three mirrors and two lenses to the original FPALM geometry. Detection efficiency is not substantially reduced.
In future live-cell applications, the anisotropy could be used to distinguish between molecules that are bound and unbound: for example, a ligand that binds a membrane receptor will not sample all orientations and will in some cases show greater anisotropy than an unbound copy of the same molecule. This kind of approach will be useful for studies of protein-protein interactions, polymerization, depolymerization, growth and collapse of intracellular structures, and lateral organization in membranes. As high excitation intensities are potentially damaging to cells, users of P-FPALM will certainly need to make appropriate control experiments to check for any effects of the high intensity illumination on cell viability.
Longer acquisitions may allow higher molecular densities to be observed in well-immobilized samples. Extension of the technique to three-dimensional imaging would be both possible and useful, considering that structures such as actin will span many focal planes. Hence, three-dimensional imaging would capture larger numbers of total molecules in different focal planes and allow extended structures to be visualized even more comprehensively. P-FPALM takes advantage of the information encoded in the polarization of light to show the first nanoscale images of the orientations of molecules within biological structures.
Supplementary Material
Acknowledgments
We thank C. Fang-Yen, P. Blank, J. Bewersdorf, J. Zimmerberg and M. Mason for useful discussions, G. Patterson (US National Institute of Child Health and Human Development) for providing the construct encoding the PA-GFP protein, J. Shim, J. Rochira and E. Allgeyer for laboratory assistance, A. McGinn, T. Tripp and P. Byard for professional services. This work was supported by grants K25-65459 from the US National Institute of Allergy and Infectious Diseases, CHE-0722759 from the National Science Foundation, start up funds from the University of Maine (S.T.H.), and by grants GM070358 and GM073913 from the National Institute of General Medical Sciences (V.V.V.).
Footnotes
AUTHOR CONTRIBUTIONS
T.J.G. and M.S.G. conceived the method, performed and analyzed experiments, wrote and edited the manuscript. M.V.G. performed experiments and edited the manuscript. V.V.V. and S.-R.Y. created genetic constructs and edited the manuscript. J.A.G. assisted with experiments and analysis, and edited the manuscript. S.T.H. conceived the method, performed and analyzed experiments, wrote and edited the manuscript.
COMPETING INTERESTS STATEMENT
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturemethods/.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
Note: Supplementary information is available on the Nature Methods website.
References
- 1.Hell SW, Wichmann J. Opt Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 2.Hess ST, Girirajan TP, Mason MD. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Betzig E, et al. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 4.Rust MJ, Bates M, Zhuang X. Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hess ST, et al. Proc Natl Acad Sci USA. 2007;104:17370–17375. doi: 10.1073/pnas.0708066104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shroff H, Galbraith CG, Galbraith JA, Betzig E. Nat Methods. 2008;5:417–423. doi: 10.1038/nmeth.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang B, Wang W, Bates M, Zhuang X. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Juette MF, et al. Nat Methods. 2008;5:527–529. doi: 10.1038/nmeth.1211. [DOI] [PubMed] [Google Scholar]
- 9.Betzig E, Chichester RJ. Science. 1993;262:1422–1425. doi: 10.1126/science.262.5138.1422. [DOI] [PubMed] [Google Scholar]
- 10.Bartko AP, Dickson RM. J Phys Chem B. 1999;103:11237–11241. [Google Scholar]
- 11.Bohmer M, Enderlein J. J Opt Soc Am B. 2003;20:554–559. [Google Scholar]
- 12.Harms GS, Cognet L, Lommerse PHM, Blab GA, Schmidt T. Biophys J. 2001;80:2396–2408. doi: 10.1016/S0006-3495(01)76209-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patterson GH, Lippincott-Schwartz J. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 14.Gurskaya NG, et al. Nat Biotechnol. 2006;24:461–465. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- 15.Fourkas JT. Opt Lett. 2001;26:211–213. doi: 10.1364/ol.26.000211. [DOI] [PubMed] [Google Scholar]
- 16.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3. Springer Science; New York: 2006. [Google Scholar]
- 17.Weber G. J Opt Soc Am. 1956;46:962–970. [Google Scholar]
- 18.Hess ST, Sheets ED, Wagenknecht-Wiesner A, Heikal AA. Biophys J. 2003;85:2566–2580. doi: 10.1016/s0006-3495(03)74679-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobson K, Mouritsen OG, Anderson RG. Nat Cell Biol. 2007;9:7–14. doi: 10.1038/ncb0107-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.