Abstract
The vitamin D receptor (VDR) mediates the actions of 1,25-dihydroxyvitamin D3 (1,25(OH)2D3) in target cells and tissues by orchestrating the expression of gene networks responsible for vitamin D-induced phenotypes. The molecular mechanisms of these regulatory systems have been studied for decades under the principle that transcriptional regulation occurs near the transcriptional start site of the gene. However, this now appears to be an outdated view of transcriptional control. In this study, we examined the genome-wide chromatin immunoprecipitation on microarray (ChIP-chip) across pre-osteoblastic cells for VDR, retinoid X receptor (RXR), RNA polymerase II, and histone H4 acetylation (H4ac). We uncovered potential regulatory mechanisms for genes important to osteoblast biology as well as skeletal formation under the control of 1,25(OH)2D3. We found that VDR, along with RXR and H4ac, binds to distal regions 43% of the time; and within gene introns and exons 44%, leaving only 13% of activation at traditional promoter regions. Here, we briefly summarize our findings for all the VDR/RXR cis-acting transcriptional elements (VDR/RXR cistrome) in pre-osteoblastic cells, MC3T3-E1, provide a few examples of this dynamic control by VDR and 1,25(OH)2D3, and demonstrate that distal transcriptional control contributes to the majority of vitamin D3-mediated transcription.
Keywords: VDR; RXR; ChIP-chip; genome-wide; RNA Polymerase II; Cyp24a1; Spp1; Opn; Rarβ; Prkca; Cistrome; 1,25(OH)2D3; vitamin D3; epigenetics; histone acetylation; distal enhancers
1. Introduction
The active metabolite of vitamin D3, 1,25 dihydroxyvitamin D3 (1,25(OH)2D3), plays essential roles throughout the body in maintenance of calcium and phosphate homeostasis [1]. These effects are most dramatic in the intestine, kidney and bone [1]. 1,25(OH)2D3, along with the parathyroid hormone, play critical roles in the proper mineralization of bone through its genomic actions in osteoclasts and osteoblasts [1]. The biological effects of 1,25(OH)2D3 are manifested by regulation of gene expression through its binding to and activation of the vitamin D receptor (VDR), a member of the steroid hormone receptor family [2]. After activation by 1,25(OH)2D3, VDR binds to DNA with its heterodimeric partner the retinoid X receptor (RXR) to specific hexameric DNA sequence response elements (VDREs) that activate transcription of nearby genes [3, 4]. These VDREs share a common sequence similarity and the ideal can be described as AGGTCAxxxAGGTCA [5, 6]; however, VDREs can be quite diverse in nature [7, 8]. It is the diversity of sequence of these 15 base pairs that makes predictions of active VDREs difficult by in vitro and in silico derived methodology alone. Methods such as multimerized elements reporter assays of in silico derived VDREs remove the regulatory element from its natural environment and dechromatinize the surrounding sequence making the results largely artificial. These methods, however are a valuable validation through mutagenesis once a legitimate element is discovered. Only an in vivo binding assay such as that obtained by using the chromatin immunoprecipitation assay (ChIP) combined with in silico analysis can yield truly biologically active VDREs [9-11]. Through the advancement of the hybridization of ChIP DNA to microarrays (ChIP-chip) or the massively parallel sequencing (ChIP-seq) of ChIP DNA, all VDR/RXR cis-acting elements throughout the genome (VDR/RXR cistrome) can be discovered similar to current studies with the estrogen receptor [12].
In this investigation, we define the VDR/RXR cistrome for pre-osteoblastic cells (MC3T3-E1) upon activation with 1,25(OH)2D3. We analyzed the genome for the ability of VDR, RXR, RNA Polymerase II (RNA Pol II) and histone H4 acetylation (H4ac) to be recruited to specific loci and genes. We were also able to successfully pair these results with gene expression analysis to mate DNA binding to those genes that are differentially expressed (DE). We will summarize these results and offer a few examples of new regulatory sites for genes important in skeletal biology.
2. Materials and Methods
2.1 Reagents
1,25(OH)2D3 was obtained from Tetrionics, Inc. (Madison, WI). Antibodies to VDR (C-20) and RXR (DN-197) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Anti-tetra-acetyl H4 antibody (06-866) was acquired from Upstate (Charlottesville, VA). Anti-RNA polymerase II antibody (8WG16) was obtained from Covance (Emeryville, CA). All quantitative real-time PCR (qPCR) reagents (Power SYBR green) were obtained from ABI (Foster City, CA) or Fast Start SYBR Green Master Mix (w/ rox) from Roche (Indianapolis, IN). All qPCR was conducted on the RealPlex 2.0 from Eppendorf AG (Hamburg, Germany). DNA microarrays for ChIP and gene expression were obtained from Roche NimbleGen (Indianapolis, IN, Madison, WI). Primers were obtained from IDT (Coralville, IA).
2.2 Cell Culture
MC3T3-E1 cells were obtained from ATCC (Manassas, VA). MC3T3-E1 cells were α-MEM supplemented with 10% fetal bovine serum (FBS) from Hyclone (Logan, UT) [13].
2.3 ChIP-chip analysis (ChIP coupled to DNA microarray)
ChIP assays were performed as previously described [8, 14, 15]. ChIP-chip methodology was performed as previously described (Zella, et. al. Dec. 2009, Mol Endocrinology, accepted). One notable exception, Roche NimbleGen’s whole genome tiling arrays for mouse (mm8) in the HD2 format (high-density 2.1 million probe arrays, 24μg of each sample (Cy5 and Cy3) were used for hybridization. Data were extracted using the NimbleScan (version 2.5) software (Roche NimbleGen) and normalized using lowess normalization in R. The log2-ratios of test versus experimental data were calculated for each point and peaks were called using CMARRT algorithms [16]. Data shown are representative of two or more ChIP-chip analyses performed for each experimental design. All data were visualized using the Generic Genome Browser, Gbrowse (www.gmod.org/wiki/Gbrowse) [17].
2.4 RNA isolation and Gene Expression Analysis
MC3T3-E1 cells were grown to confluency and treated with ethanol vehicle (Veh) or 100nM 1,25(OH)2D3 for 24 hours prior to RNA isolation. RNA was isolated using the TRI-Reagent protocol (MRC, Cincinnati, OH) and double stranded cDNA (dscDNA) was prepared by the double stranded cDNA synthesis kit (Invitrogen, Carlsbad, CA). DscDNA was then labeled as described above in ChIP-chip using only Cy3 labeled dye. Labeled samples were hybridized to mouse (mm8) 385k microarrays (Roche NimbleGen). All samples were completed in triplicate. Samples were processed and differentially expressed genes (DE) were determined with the moderated t-statistic [18] using the limma package in R and Arraystar v3.0 (DNAstar, Madison, WI) [19, 20]. We used a confidence interval of 99% and only analyzed genes with a 2 fold or greater expression. qPCR was performed using primers specific to the DE genes for validation (data not shown).
3. Results and Discussion
3.1 VDR/RXR cistrome is defined by transcription factor binding as well as markers of transcriptional activation
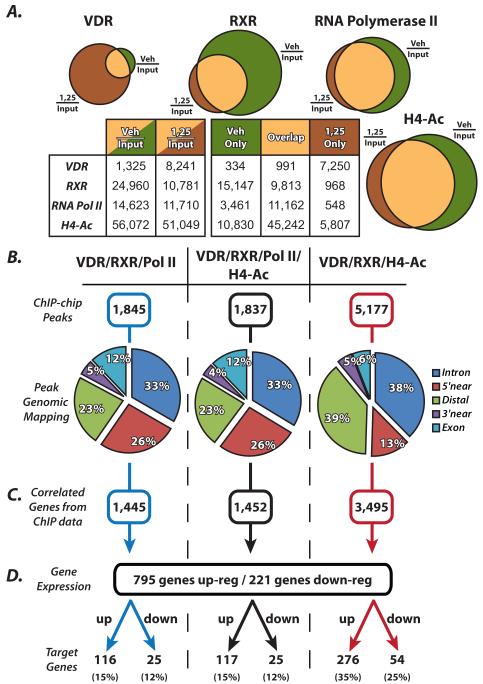
We established the VDR/RXR cistrome through successive rounds of ChIP-chip using antibodies directed at VDR, RXR, RNA polymerase II (RNA Pol II) and histone H4 acetylation (H4ac). MC3T3-E1 cells were grown to confluency and treated with 100nM 1,25(OH)2D3 or ethanol vehicle for 3 hours prior to ChIP assay. The output DNA was amplified, labeled and hybridized as indicated in Materials and Methods. Statistically significant peaks were called from the data using the CMARRT algorithm and the peaks were tabulated in Figure 1. Data are represented by Veh/Input, which is the vehicle treated cells over the assay Input cohybridization and will be referred to as “basal” binding or ligand independent binding (LIB); or 1,25/Input, which is the 1,25(OH)2D3 treated cells over the same Input sample and will be referred to as “activated” binding or ligand dependent binding (LDB). Figure 1A shows a graphical venn diagram representation of the data listed in the table below in Figure 1A. VDR was found to bind the genome in the basal LIB condition (Veh/Input) at 1,325 sites; this number then increases to 8,241 in the presence of activated binding, LDB (1,25/Input). By parsing those data out further, 7,250 sites upregulated were de novo VDR binding with 1,25(OH)2D3 treatment (1,25 only), where VDR was not bound previously. These data are similar to those that have been collected for the estrogen receptor [12, 21, 22]. There is significant LIB of VDR to the genome basally, which was unexpected, however all of the 1,325 sites of basal VDR binding are also coordinated with RXR in the LIB condition. An opposite trend was observed with RXR, which binds basally to the DNA at many more sites than when activated by 1,25(OH)2D3. RXR is a heterodimer partner for many transcription factors and is involved in many cellular processes so this result was expected [4]. It was interesting to find that many VDR bound sites in “1,25 only” condition, were pre-marked by the presence of RXR at those site. In fact, only 968 RXR sites arose de novo with the 7,250 sites for VDR, therefore greater than 6,000 sites were prebound by RXR before VDR arrived. There were similar trends for RNA pol II and H4ac as they are important to processes independent of 1,25(OH)2D3 activation.
Figure 1.
Summary of genome-wide ChIP-chip and gene expression studies reveal the VDR/RXR cistrome. A, Schematic venn diagram representations of the data displayed in the table below summarizing the number of peaks found with vehicle treatment (Veh/Input) and with 1,25(OH)2D3 treatment (1,25/Input) as well as Vehicle only, 1,25 only, and an overlap of regions from ChIP-chip data for VDR, RXR, RNA polymerase II (RNA Pol II) and histone H4 acetylation (H4ac). MC3T3-E1 cells treated for 3 hours prior to ChIP with vehicle or 100nM 1,25(OH)2D3. B, Overlapping peaks were tabulated for VDR/RXR/RNA Pol II (left column), VDR/RXR/RNA Pol II/H4ac (center column) and VDR/RXR/H4ac (right column). Peaks were mapped to their surrounding genes and categorized into intragenic (Intron or Exon), 5′near (within 5kb upstream of the 5′ end of gene), 3′near (within 5kb downstream of 3′ end of gene) or Distal (any region not within the gene or within 5kb of the gene at either end). C, the peaks were mapped to their closest surrounding gene and listed as correlated genes from ChIP-chip data. D, gene expression analysis was performed on MC3T3-E1 cells for 24 hours with either vehicle or 100nM 1,25(OH)2D3. These genes were then cross referenced with the genes that were associated with the ChIP-chip peaks and displayed as either up- or down-regulated genes.
For the current study, overlapping peak locations were compared for three distinct groups. In Figure 1B, peak locations were compared for LDB VDR (1,25/Input) in combination with RXR and RNA Pol II (left column), VDR, RXR and H4ac (right column) as well as all 4 antibodies together (middle column). Sites were then tabulated for the above combination and listed in Figure 1B next to ChIP-chip peaks. We found that there were far fewer binding sites associated with RNA Pol II compared to that of H4ac. This is due to the genomic distribution of these sites as diagramed in Figure 1B. ChIP-chip peaks were mapped to their surrounding gene locations and were placed into the following categories: “Intron” and “Exon” refer to the intragenic regions, “5′ near” and “3′ near” refer to the first 5kb upstream of the gene TSS and 5kb downstream of the 3′ end of the gene, and finally, “Distal” refers to an region not within the gene and greater than 5kb up or down stream of the gene. Distal regions are often referred to as “enhancers”, however, we have not yet proven transactivation capabilities for most of these elements and therefore will refer to them as “distal”. Binding sites that contain RNA Pol II activity are largely found within genes or at gene promoters. We find a smaller percentage, 23%, present at distal locations and 26% present at the promoters. Those peaks that contain RNA pol II binding are focused at the promoters of genes and are all accompanied by H4ac, as can be seen in Figure 1A. There are only 8 peaks in the genome that contain RNA Pol II that do not contain H4ac. Binding sites were found more in the distal region 39% and less at the promoter of genes (13%) when only VDR/RXR/H4ac were studied. This grouping is a better representation of all activated sites whether they are at the promoter or at distal sites, since H4ac associates with both. Interestingly however, several distal regions do contain some RNA Pol II binding as well. From these mapping data, we are also able to list all the genes that are potentially affected by these peaks of binding activity. For example, in the right column, 5,177 peaks of ChIP-chip data correspond to 3,495 genes, shown in Figure 1C.
We then performed traditional gene expression analysis on the mouse genome using Roche Nimblegen gene expression arrays. MC3T3-E1 cells were treated with vehicle or 100nM 1,25(OH)2D3 for 24 hours. RNA was collected, processed, reverse transcribed to double stranded cDNA and hybridized to arrays as described in Materials and Methods. We found through our statistical analysis that there were 795 genes up-regulated and 221 genes down-regulated greater than 2-fold (99% CI). Figure 1D combines these gene expression analyses with the ChIP-chip analysis. Each subset of ChIP-chip genes were further separated into up- and down-regulated gene groups. We find that the most interesting target genes reside in this final grouping at the end of Figure 1D. Again, taking the right column, we see that 3,495 ChIP-chip genes are focused down to 276 genes up-regulated and 54 genes down-regulated from the gene expression analysis. These analyses allowed focused groups for further study. These gene groups were also extensively studied through gene ontology (GO) for correlation to cellular processes and interactions throughout the genome (data not shown). A few of the largest GO terms associated with these groups of genes was that of Skeletal Formation and Biology (GO:0001501) as well as Differentiation (GO:0030154).
3.2 Gene regulation occurs at the promoter as well as distal sites within intergenic regions
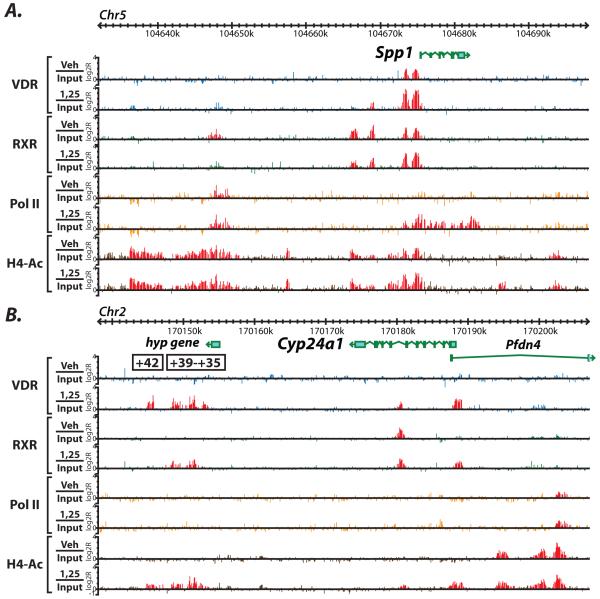
From this GO term search, we selected a few genes to display as an example of the power of genome-wide analyses as demonstrated in Figure 2 and Figure 3. Figure 2 shows the actual ChIP-chip raw data for VDR, RXR, RNA Pol II and H4ac in MC3T3-E1 cells and is focused on two genes that show activities at the promoter as well as in distal regions. These are part of the data that comprised the analysis in Figure 1. Statistically significant peaks are highlighted in red. In Figure 2A, we examined the genomic region around the gene Spp1, also known as Opn or Osteopontin precursor. It is known that there are a set of regulatory elements directly upstream of the Spp1 promoter region as shown by two distinct red peaks in the VDR Veh/Input and 1,25/Input tracks [9]; highlighting an example of ligand independent binding (LIB) of VDR that is further increased by 1,25(OH)2D3 stimulation. This is also accompanied by RNA Pol II increases as well as H4ac in the same region. As can be seen in Figure 2A, there are peaks of RXR, RNA Pol II and H4ac upstream of Spp1 that are unknown regulatory regions. We believe that these regions may control Spp1 activity and are under further investigation.
Figure 2.
ChIP-chip data for genes important for skeletal biology reveal novel enhancers at the promoter as well as distal to the TSS. A, the genomic location for the Spp1 (Opn) gene are shown for chromosome 5 with genomic base pairs given in kilobases (k). ChIP-chip data are listed for each antibody in the basal (Veh/Input) or activated (1,25/Input) state and are displayed as log 2 ratios (log2R). Antibodies used are VDR (blue), RXR (green), RNA Pol II (orange), H4ac (brown). Statistically significant peaks were called and are highlighted in red as described in Materials and Methods. B, the same analysis is shown for the Cyp24a1 gene. Putative regulatory regions discovered are highlighted with their position relative to the transcriptional start site of the gene.
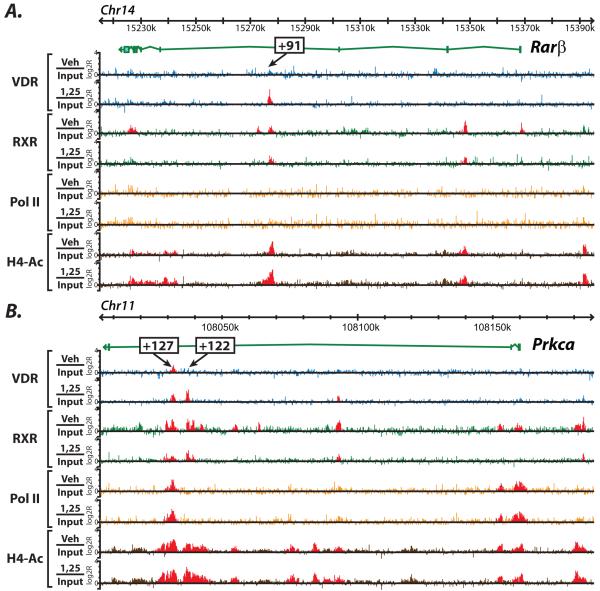
Figure 3.
Transcriptional control can occur within intragenic regions at introns and exons. A, the genomic location for the Rarβ gene are shown for chromosome 5 with genomic base pairs given in kilobases (k). ChIP-chip data are listed for each antibody in the basal (Veh/Input) or activated (1,25/Input) state and are displayed as log 2 ratios (log2R). Antibodies used are VDR (blue), RXR (green), RNA Pol II (orange), H4ac (brown). Statistically significant peaks were called and are highlighted in red as described in Materials and Methods. B, the same analysis is shown for the Prcka (Protein Kinase Cα) gene. Putative regulatory regions discovered are highlighted with their position relative to the transcriptional start site of the gene.
We were also able to describe distal regulation of the most classic vitamin D3-responsive gene, Cyp24a1. For nearly two decades, Cyp24a1 has been believed to be transactivated solely by two proximal VDREs located at −165bp and −265bp upstream of the TSS [23, 24]. As can be seen in Figure 2B, VDR is not only present at the promoter region of the gene when stimulated by 1,25(OH)2D3, but also far downstream +35 to +41kb from the TSS, highlighting the ligand dependent binding (LDB). It was also found that these enhancers recruit RXR as well as H4ac. RNA Pol II was not significantly bound to any of these regions given its low inducible expression in MC3T3-E1 cells (3 fold). Through successive rounds of traditional reporter analysis as well as a large (~200kb) BAC clone reporter for the Cyp24a1 gene which keeps all the elements in their natural chromatin environment, we found that these regions worked synergistically with the promoter elements in full transactivation of the Cyp24a1 gene (data not shown, publication in preparation). We have also defined putative VDREs in these regions through pairing our techniques with in silico analysis. As can be seen in Figure 2B, there is also a peak of VDR activity within the gene itself that is premarked by the presence of RXR. This site is also under investigation for its contributing activities. These techniques were also successfully used to investigate the autoregulation of the Vdr gene itself [25]. We found that the human CYP24A1 gene is under an even more complex regulation than the mouse Cyp24a1 gene (publication in preparation).
3.3 Downstream elements within introns and exons are found to contribute to overall gene activation
Our previous studies demonstrate the control of VDR to regulate its own gene Vdr through several intronic regions far downstream of the TSS [25]. Similar intronic gene regulation occurs throughout the genome. This can be seen in Figure 3A and 3B, where the genes Rarβ and Prkca (Protein Kinase C α) are also examined. These genes were upregulated by 1,25(OH)2D3 and were included as genes important for skeletal formation as defined by the GO term grouping. There is little known about the mechanism of regulation by 1,25(OH)2D3 of these genes to date. Utilizing our genome-wide ChIP-chip studies, however, we were able to uncover potential regulatory elements far downstream of the transcriptional start site (TSS). In the absence of the ChIP-chip tiling approach, these regulatory regions would have been difficult to discover given this distance from the TSS. In Figure 3A, we see a region at +91kb from the TSS in Rarβ that binds VDR in the activated state. This region also shows RXR binding in the activated as well as basal states. There does not appear to be RNA Pol II at this putative enhancer, there is however, strong H4ac in this region basally with a small increase after hormone treatment. In Prkca, Figure 3B, we find a set of potential elements at +127 and +122kb from the TSS. At +127, VDR is basally bound (LIB) and then increases upon hormone stimulation, which is different than the regulation at the +122 site. It is interesting to note that although these two potential enhancers are only 5kb away from each other, they display different binding profiles of RNA Pol II and H4ac. In fact, RNA Pol II is strongly present at the +127 site and completely absent at the +122 site. For all peaks discovered in Figure 2 and 3, an in silico analysis has identified putative VDREs for each of the locations under the peaks.
We have demonstrated that ChIP-chip at the genome-wide level can provide significant understanding of the VDR/RXR cistrome in MC3T3-E1 pre-osteoblastic cells. These data will be key in understanding the molecular mechanisms for all genes that are regulated by 1,25(OH)2D3 in these cells and provide a platform to study these differential activities and chromatin conformations in alternative cell types. Although, not covered in this study, the genes that are not regulated by 1,25(OH)2D3, but do bind VDR and RXR and accumulate RNA Pol II or H4ac are a very interesting set of regulatory elements that are very poorly understood. We believe studies such as these hold the key to unlocking the broader mechanisms in chromatin control during 1,25(OH)2D3 stimulation and better our knowledge of molecular mechanisms that will lead to improved therapeutic strategies.
4. Acknowledgements
We would like to thank members of the Pike Lab for their helpful discussions and review of this manuscript. We would also like to thank William Pulec for Gbrowse and linux server administration.
Footnotes
The authors have nothing to declare or disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
5. References
- [1].Jones G, Strugnell SA, DeLuca HF. Current understanding of the molecular actions of vitamin D. Physiol Rev. 1998;78(4):1193–1231. doi: 10.1152/physrev.1998.78.4.1193. [DOI] [PubMed] [Google Scholar]
- [2].Jurutka PW, Whitfield GK, Hsieh JC, Thompson PD, Haussler CA, Haussler MR. Molecular nature of the vitamin D receptor and its role in regulation of gene expression. Rev Endocr Metab Disord. 2001;2(2):203–216. doi: 10.1023/a:1010062929140. [DOI] [PubMed] [Google Scholar]
- [3].Malloy P, Pike J, Feldman D. The vitamin D receptor and the syndrome of hereditary 1,25-dihydroxyvitamin D-resistant rickets. Endocr Rev. 1999;20(2):156–188. doi: 10.1210/edrv.20.2.0359. [DOI] [PubMed] [Google Scholar]
- [4].Mangelsdorf D, Evans R. The RXR heterodimers and orphan receptors. Cell. 1995;83(6):841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- [5].Malloy PJ, Pike JW, Feldman D. The vitamin D receptor and the syndrome of hereditary 1,25-dihydroxyvitamin D-resistant rickets. Endocr Rev. 1999;20(2):156–188. doi: 10.1210/edrv.20.2.0359. [DOI] [PubMed] [Google Scholar]
- [6].Ozono K, Liao J, Kerner S, Scott R, Pike J. The vitamin D-responsive element in the human osteocalcin gene. Association with a nuclear proto-oncogene enhancer. J Biol Chem. 1990;265(35):21881–21888. [PubMed] [Google Scholar]
- [7].Pike J, Meyer M, Watanuki M, Kim S, Zella L, Fretz J, Yamazaki M, Shevde N. Perspectives on mechanisms of gene regulation by 1,25-dihydroxyvitamin D3 and its receptor. J Steroid Biochem Mol Biol. 2007;103(3-5):389–395. doi: 10.1016/j.jsbmb.2006.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Meyer M, Watanuki M, Kim S, Shevde N, Pike J. The human transient receptor potential vanilloid type 6 distal promoter contains multiple vitamin D receptor binding sites that mediate activation by 1,25-dihydroxyvitamin D3 in intestinal cells. Mol Endocrinol. 2006;20(6):1447–1461. doi: 10.1210/me.2006-0031. [DOI] [PubMed] [Google Scholar]
- [9].Kim S, Shevde NK, Pike JW. 1,25-Dihydroxyvitamin D3 stimulates cyclic vitamin D receptor/retinoid X receptor DNA-binding, co-activator recruitment, and histone acetylation in intact osteoblasts. J Bone Miner Res. 2005;20(2):305–317. doi: 10.1359/JBMR.041112. [DOI] [PubMed] [Google Scholar]
- [10].Wells J, Farnham PJ. Characterizing transcription factor binding sites using formaldehyde crosslinking and immunoprecipitation. Methods. 2002;26(1):48–56. doi: 10.1016/S1046-2023(02)00007-5. [DOI] [PubMed] [Google Scholar]
- [11].Shang Y, Myers M, Brown M. Formation of the androgen receptor transcription complex. Mol Cell. 2002;9(3):601–610. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- [12].Lupien M, Eeckhoute J, Meyer C, Krum S, Rhodes D, Liu X, Brown M. Coactivator function defines the active estrogen receptor alpha cistrome. Mol Cell Biol. 2009;29(12):3413–3423. doi: 10.1128/MCB.00020-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zella L, Shevde N, Hollis B, Cooke N, Pike J. Vitamin D-binding protein influences total circulating levels of 1,25-dihydroxyvitamin D3 but does not directly modulate the bioactive levels of the hormone in vivo. Endocrinology. 2008;149(7):3656–3667. doi: 10.1210/en.2008-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kim S, Yamazaki M, Zella L, Meyer M, Fretz J, Shevde N, Pike J. Multiple enhancer regions located at significant distances upstream of the transcriptional start site mediate RANKL gene expression in response to 1,25-dihydroxyvitamin D3. J Steroid Biochem Mol Biol. 2007;103(3-5):430–434. doi: 10.1016/j.jsbmb.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Meyer M, Zella L, Nerenz R, Pike J. Characterizing early events associated with the activation of target genes by 1,25-dihydroxyvitamin D3 in mouse kidney and intestine in vivo. J Biol Chem. 2007;282(31):22344–22352. doi: 10.1074/jbc.M703475200. [DOI] [PubMed] [Google Scholar]
- [16].Kuan P, Chun H, Keleş S. CMARRT: a tool for the analysis of ChIP-chip data from tiling arrays by incorporating the correlation structure. Pac Symp Biocomput. 2008:515–526. [PMC free article] [PubMed] [Google Scholar]
- [17].Stein L, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich J, Harris T, Arva A, Lewis S. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12(10):1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Smyth G. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- [19].Bolstad B, Irizarry R, Astrand M, Speed T. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- [20].Irizarry R, Hobbs B, Collin F, Beazer-Barclay Y, Antonellis K, Scherf U, Speed T. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- [21].Welboren W, Stunnenberg H, Sweep F, Span P. Identifying estrogen receptor target genes. Mol Oncol. 2007;1(2):138–143. doi: 10.1016/j.molonc.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Welboren W, van Driel M, Janssen-Megens E, van Heeringen S, Sweep F, Span P, Stunnenberg H. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28(10):1418–1428. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zierold C, Darwish H, DeLuca H. Two vitamin D response elements function in the rat 1,25-dihydroxyvitamin D 24-hydroxylase promoter. J Biol Chem. 1995;270(4):1675–1678. doi: 10.1074/jbc.270.4.1675. [DOI] [PubMed] [Google Scholar]
- [24].Zierold C, Mings J, DeLuca H. Regulation of 25-hydroxyvitamin D3-24-hydroxylase mRNA by 1,25-dihydroxyvitamin D3 and parathyroid hormone. J Cell Biochem. 2003;88(2):234–237. doi: 10.1002/jcb.10341. [DOI] [PubMed] [Google Scholar]
- [25].Zella L, Kim S, Shevde N, Pike J. Enhancers located in the vitamin D receptor gene mediate transcriptional autoregulation by 1,25-dihydroxyvitamin D3. J Steroid Biochem Mol Biol. 2007;103(3-5):435–439. doi: 10.1016/j.jsbmb.2006.12.019. [DOI] [PubMed] [Google Scholar]