Abstract
More than 150 different point mutations in POLG, the gene encoding the human mitochondrial DNA polymerase γ (pol γ), cause a broad spectrum of childhood and adult onset diseases like Alpers syndrome, Ataxia-Neuropathy syndrome and progressive external ophthalmoplegia. These disease mutations can affect the pol γ enzyme’s properties in numerous ways, thus potentially influencing the severity of the disease. Hence, a detailed characterization of disease mutants will greatly assist researchers and clinicians to develop a clear understanding of the functional defects caused by these mutant enzymes. Experimental approaches for characterizing the wild type (WT) and mutant pol γ enzymes are extensively described in this manuscript. The methods start with construction and purification of the recombinant wild type and mutant forms of pol γ protein, followed by assays to determine its structural integrity and thermal stability. Next, the biochemical characterization of these enzymes is described in detail, which includes measuring the purified enzyme’s catalytic activity, its steady state kinetic parameters and DNA binding activity, and determining the physical and functional interaction of these pol γ proteins with its p55 accessory subunit.
1. Introduction
Of the 16 DNA polymerases in the eukaryotic cell, only pol γ is known to function in animal mitochondria [1–3]. Thus, pol γ is absolutely essential for mitochondrial DNA replication and repair. The holoenzyme of pol γ consists of a catalytic subunit (encoded by POLG at chromosomal locus 15q25) and a dimeric form of its accessory subunit (encoded by POLG2 at chromosomal locus 17q24.1). The catalytic subunit is a 140 kDa enzyme (p140) that contains an N-terminal exonuclease domain connected by a linker region to a C-terminal polymerase domain and has DNA polymerase, 3′→5′ exonuclease and 5′ dRP lyase activities [4]. The accessory subunit is a 55 kDa protein (p55) required for tight DNA binding and processive DNA synthesis [5]. The pol γ holoenzyme functions in conjunction with the mitochondrial DNA helicase (Twinkle) and the mitochondrial SSB to form the minimal replication apparatus [6].
The POLG gene is one of several nuclear genes that is associated with mitochondrial DNA depletion or deletion disorders. To date, more than 150 disease mutations have been identified in the POLG gene and an up-to-date mutation database can be found at http://tools.niehs.nih.gov/polg/, which shows these mutations equally distributed over the length of the protein. Disorders associated with POLG mutations include: 1) Myocerebrohepatopathy spectrum disorder (MCHS), 2) Alpers syndrome, 3) Ataxia neuropathy spectrum disorder (ANS), 4) Myoclonus epilepsy myopathy sensory ataxia (MEMSA), 5) Autosomal recessive progressive external ophthalmoplegia (arPEO), and 6) Autosomal dominant progressive external ophthalmoplegia (adPEO) [7]. Also, alteration of the (CAG)10 repeat in the 2nd exon of POLG has been implicated in male infertility [8].
A summary of previous structure-function studies in pol γ that were performed to address disease mutations has recently been published [9]. Among all disease mutations, A467T mutation is the most common POLG mutation and has been found to be associated with all of the disease symptoms mentioned above. Previous studies have shown that the A467T pol γ possesses only 4% of the wild-type DNA polymerase activity and is compromised for its ability to interact with the p55 accessory subunit [10]. Another mutation, W748S, which has nearly always been found to be in cis with the E1143G mutation, is a frequent cause of ataxia-neuropathy syndrome [11]. The E1143G substitution, a single nucleotide polymorphism (SNP), is found in 4% of European populations. The W748S mutation has intrinsic lower polymerase activity as well as a demonstrated lower affinity for DNA [12]. We have found that the E1143G SNP can modulate the deleterious effect of the W748S mutation [12]. This finding raises the possibility that other SNPs could potentially affect POLG enzymatic activity.
Four adPEO mutations, G923D, R943H, Y955C and A957S that are found in and around motif B in the active site of the polymerase were characterized biochemically [13]. Two of the substitutions (R943H and Y955C), change side chains that interact with the incoming dNTP, and pol γ with either of these substitutions retains less than 1% of the wild-type polymerase activity and displays a severe decrease in processivity. The significant stalling of DNA synthesis and extremely low catalytic activities of both enzymes are the two most likely causes of the severe clinical presentation in R943H and Y955C heterozygotes [13]. The substitution of Tyr955 to cysteine also increases nucleotide misinsertion replication errors 10–100 fold in the absence of exonucleolytic proofreading [14]. Lately, we also characterized six Alpers mutations, four of which are highly conserved and located in the thumb subdomain of the polymerase portion of the enzyme (G848S, T851A, R852C and R853Q [15]. Purified recombinant pol γ proteins containing these point mutations exhibited less than 1% WT enzyme activity levels in addition to reduced DNA binding exhibited by the G848S and R852C enzymes [15]. For the majority of the disease substitutions that have been studied in vitro, the biochemical defects correlate with the severity and age of onset found in patients [9]. Further analysis of disease substitutions as well as structural analysis should aid in the continued understanding of disease mutations in the POLG gene.
Recently, we described a summary of assays used to characterize the pol γ catalytic subunit [16]. In this methods review, we have updated those methods and include summaries of the purification schemes required to purify the pol γ catalytic subunit (p140) and the p55 accessory subunit. Furthermore, as an example of how to use these assays to characterize a disease mutation, we include analysis for the R964C mutant pol γ that has been recently described [17].
2. Description of Methods
2.1 Construction of human DNA polymerase γ mutants
QuikChange site-directed mutagenesis kit (Stratagene) was used to introduce mutations in the cDNA encoding the exonuclease deficient (Exo−) pol γ (POLG). The pol γ Exo− background was created by mutating two crucial amino acids in the exonuclease domain (D198A/E200A) to abolish the 3′→5′ exonuclease activity that may interfere with biochemical assays involving nucleic acids [18]. This POLG cDNA without its mitochondrial targeting sequence was cloned into the pVL1393 baculovirus transfer vector and used as the template for the PCR reaction. This pol γ Exo− without any additional mutation will be referred as wild-type (WT) and the pol γ Exo− with a mutation that converts arginine to cysteine at position 964 will be referred as R964C henceforth. The oligonucleotides containing the point mutations (underlined nucleotide) for introducing the R964C mutation in POLG are 5′- CAG CCC TTT GCT GAG TGC TTA CTA ATG CAG-3′ and 5′-CTG CAT TAG TAA GCA CTC AGC AAA GGG CTG-3′. The mutation was confirmed by sequencing the pol γ insert in the baculovirus transfer vector.
2.2 Overexpression and purification of the catalytic subunit of polymerase γ
Highly purified recombinant pol γ catalytic subunit is essential for performing biochemical assays to determine the role of specific disease mutations in the enzyme. Hence, the protein was overexpressed using baculovirus expression in insect cell systems [18]. The baculovirus transfer vector pVL1393 harboring either the WT or R964C mutant Pol γ containing a (His)6-tag at its N-terminus was transfected with the BD Baculogold Linearized Baculovirus DNA (from the BD Baculogold Transfection Kit, BD Bioscience) in Spodoptera frugiperda (Sf9) cells according to the manufacturer’s protocol. The recombinant baculovirus expressing the WT or R964C mutant pol γ was subsequently plaque purified and further amplified using Sf9 insect cells (70–80% confluent) in 36 tissue culture flasks (BD Falcon, 175 cm2). Following incubation of the flasks for 63 h at 27°C for expression of the pol γ protein, the insect cells were centrifuged at 1500 × g and the pellet was frozen and stored at −80°C until further purification.
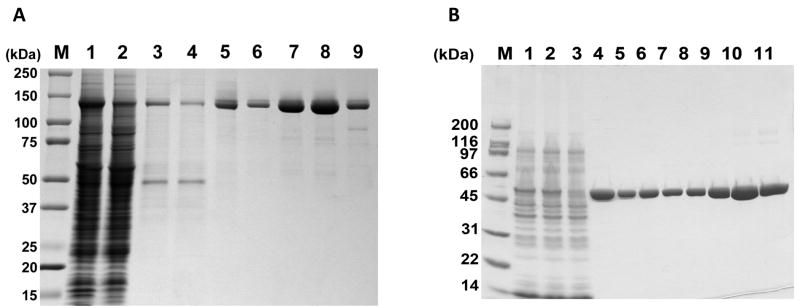
Since pol γ has a functional half-life of less than 3 min at 42°C in the absence of the p55 accessory subunit or DNA, all the steps of enzyme purification were carried out at 4°C and small aliquots were collected after each step and analyzed on a SDS-PAGE gel to monitor the integrity of the protein (Fig 1A). The frozen insect cell pellet from 1 L of infected insect cells was thawed on ice, lysed by resuspending in 5 volumes of lysis buffer (50 mM KPO4 [pH 7.5], 10% glycerol, 400 mM NaCl, 1 mM EDTA, 1 mM 2-mercaptoethanol, 0.1 mM PMSF, 1:1000 dilution of Protease Inhibitor Cocktail (Sigma P8849), 0.5% NP-40) and centrifuged at 20,000 × g for 10 min to remove the cell debris. The supernatant containing the soluble lysate was mixed with 20 ml of phosphocellulose P-11 resin (Whatman) equilibrated in buffer A50 (50 mM KPO4 [pH 7.5], 10% glycerol, 50 mM NaCl, 1 mM EDTA, 1 mM 2-mercaptoethanol, 0.1 mM PMSF) prior to the addition of 10 volumes of Buffer A50 to capture DNA binding proteins like DNA pol γ from the crude extract. The phosphocellulose P-11 resin was subsequently recovered after end-over-end mixing for 45 min by cold vacuum filtration. The resin was further washed two times with 50 ml buffer A50, two times with 50 ml buffer Alow (50 mM KPO4 [pH 7.5], 10% glycerol, 50 mM NaCl) by vacuum filtration and transferred to a Erlenmeyer flask containing 60 ml of buffer Ahigh (300 mM KPO4 [pH 7.5], 10% glycerol, 50 mM NaCl) to elute pol γ. Following 10 min of occasional swirling of the slurry and vacuum filtration, the eluate was adjusted to a final concentration of 250 mM NaCl, 0.005% (v/v) NP-40, and 20 mM imidazole by the addition of concentrated stock solutions. The eluate was then mixed with 2 ml Ni-NTA resin (Qiagen) pre-equilibrated in buffer N (50 mM KPO4 [pH 7.5], 10% glycerol, 500 mM NaCl, 0.005% NP-40), rotated end-over-end for 30 min and the resin was collected by centrifugation at 100 × g for 10 min. After two washes with 10 ml of buffer N, the Ni-NTA resin was applied to Econo-Pac 10 disposable column (Bio-Rad laboratories) and washed further with 10 ml of buffer N. The pol γ protein was then eluted with 5 ml of buffer N containing 250 mM imidazole. To bring down the salt concentration of buffer N for further purification, buffer exchange was performed using a Fast Desalt HR 10/10 column (GE Healthcare) equilibrated in buffer A (50 mM Tris-HCl [pH 7.5], 10% glycerol, 1 mM EDTA, 1 mM 2-mercaptoethanol, 0.005% NP-40) containing 100 mM KCl. The last step involved an ion exchange chromatography in which the protein was applied to a 1.0 ml HR 5/5 Mono Q (GE Healthcare) FPLC column equilibrated in a buffer A containing 100 mM KCl. The column was washed with 3 volumes of column equilibration buffer A containing 100 mM KCl and eluted with a 15 ml linear gradient of KCl (100 to 500 mM) in buffer A. Both the WT and mutant pol γproteins were eluted at ~220 mM KCl, and peak fraction was frozen in small aliquots with liquid nitrogen and stored at −80 °C until further biochemical analysis. Typically, a single protein preparation yields 2–3 mg of purified recombinant human pol γ.
Figure 1.
Purification of recombinant pol γ subunits. (A) Catalytic subunit of pol γ enzyme was purified and samples from various steps were analyzed on a 4–20% gradient SDS-PAGE gel. Lane 1, whole cell extract; lane 2, soluble lysate; lane 3, phosphocellulose P-11 flow-through; lane 4, Ni-NTA column flow-through; lane 5, Fast Desalt HR 10/10 column eluate; lanes 6–9, peak fractions eluted from Mono Q column. M, Molecular weight markers. (B) Purification of human p55 from BL21(DE3) E. coli. Samples were analyzed by 4–20% SDS-PAGE and stained with Coomassie Brilliant Blue. M, SDS-PAGE Molecular weight markers and the positions of molecular weight standards in kDa are indicated; Lane 1, E. coli whole cell extract generated by French Press; Lane 2, soluble E. coli lysate following 30,000 × g centrifugation; Lane 3, unbound proteins following batch binding of soluble lysate to Ni-NTA agarose; Lane 4, Ni-NTA agarose eluate pool; Lanes 5–11, MonoS fractions 24–30. Fractions 29 and 30 were pooled and subsequently used for biochemical analyses.
2.3 Overproduction and purification of the p55 accessory subunit of pol γ
The full-length human p55 without its mitochondrial targeting sequence was originally cloned, expressed and purified from E. coli as an N-terminal (His)6-tagged protein in a pQE vector [5]. To improve expression and quantity of protein, several rare codons in the cDNA were modified and the (His)6-tag moved to the C-terminus as described [19]. E. coli BL21(DE3) that had been transformed with a plasmid expressing the C-terminally tagged p55 was grown at 37 °C in 1.5 L of LB media containing 100 μg/ml ampicillin to an OD595 of 1.0, then cultures were chilled to 30 °C and induced for 17 h with 1 mM isopropyl thiogalactoside. Cells were harvested by centrifugation, washed once with 10 mM Tris [pH 7.5], 50 mM NaCl and 1mM EDTA, frozen with liquid nitrogen, and stored at −80 °C.
The p55 protein was purified to homogeneity as described [5], with the modifications listed in [19]. A frozen cell pellet from 1.5 L of induced cells was thawed on wet ice and resuspended in 80 ml of a lysis buffer containing 50 mM Tris-HCl [pH 7.5], 300 mM NaCl, 25 mM imidazole, 1 % Triton X-100, 3 mg/ml lysozyme, and 1:1000 dilution of Protease Inhibitor Cocktail (Sigma P8849). The cell suspension was passed through a 40 ml French Pressure Cell (Amicon) at 18,000 psi, and the resulting whole cell lysate was sedimented at 30,000 × g for 15 min at 4°C. The cleared supernatant was mixed for 30 min at 4°C with 2 ml of Ni-NTA agarose slurry (Qiagen) pre-equilibrated in the same buffer. The resin was batch washed two times with a 50 ml solution composed of 50 mM Tris-HCl [pH 7.5], 500 mM NaCl, 25 mM imidazole and 1% Triton X-100, followed by pouring the resin into a disposable column, and further washed in the column with the same buffer except the Triton X-100 was replaced with 0.01% NP-40. Protein was then eluted from the Ni-NTA agarose by 50 mM Tris-HCl [pH 9.0], 250 mM imidazole, followed by passage of the eluate through a Fast Desalt HR10/10 column (GE Healthcare) equilibrated in Buffer A (25 mM KPO4 (pH 7.0), 10% glycerol, 1 mM EDTA, 1 mM 2-mercaptoethanol) also containing 0.005% NP-40 and 100 mM NaCl. This highly purified protein was applied to a 1.0 ml Mono S HR 5/5 FPLC column (GE Healthcare) equilibrated in the same buffer. The column was washed with 4 ml equilibration buffer and developed with a 20 ml linear gradient of NaCl (0.1–1 M) in Buffer A. Peak fractions containing homogenous p55 were frozen with liquid nitrogen in small aliquots and stored at −80°C. A Coomassie gel depicting the steps in this purification is shown in Fig 1B.
2.4 Evaluation of protein stability by circular dichroism analysis
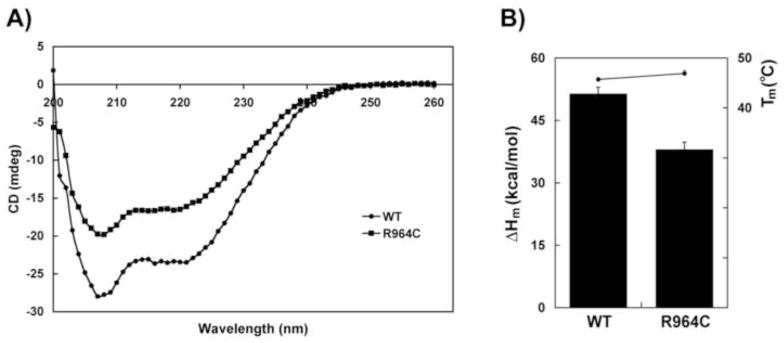
Prior to in vitro biochemical analysis the structural integrity of both purified WT and R964C mutant enzymes have to be confirmed. The effects of various point mutations on the secondary structure of pol γ were assessed by circular dichroism (CD) spectroscopy. This technique would measure the relative amounts of α-helices, β-sheets and random coils in any protein, in addition to measuring its intrinsic stability. The CD studies were performed with a Jasco 810 Spectropolarimeter equipped with a Peltier thermal controller (Jasco, Inc., MD), following the method described by Chan et al., [12] with WT and R964C mutant pol γ. Briefly, 1 ml of purified recombinant pol γ protein diluted to 20 ug/ml with 10 mM potassium phosphate buffer [pH 7.5], 5% glycerol, 200 mM NaCl, 0.1 mM EDTA and 0.1 mM 2-mercaptoethanol was placed in the cuvette and the CD spectra was measured from 260 to 200 nm at 4°C with constant stirring. Pol γ prefers a high concentration of glycerol and NaCl for storage and since these reagents were not in optimal concentrations for the CD spectra analysis as it will produce higher backgrounds at shorter wavelengths (<200 nm), only the amount of α-helix was measured. In addition, since pol γ is highly α-helical, the thermal stability of the α-helices in pol γ was determined by measuring the ellipticity at 220 nm by increasing the temperature from 26°C to 60°C by 1°C/min. Using a graphical program with sigmoidal curve fitting, the equation was derived for each thermal denaturation curve, where the melting temperature (Tm) is the point of inflection and the change in enthalpy for protein folding (ΔHm) is the slope of the curve for each pol γ protein. The CD spectroscopy and thermal stability of the WT and R964C pol γ enzymes are shown in Fig 2.
Figure 2.
Secondary structure of WT and R964C pol γ proteins and their stability. (A) CD spectroscopy of WT (circles) and R964C (squares) proteins. (B) Thermal stability of WT and R964C mutant proteins. The enthalpy change during protein folding (ΔHm), measured at 220 nm is represented by bars with standard deviations. The melting temperature (Tm) is represented by circles. The results are an average of three independent experiments.
2.5 Estimation of the catalytic activity of DNA pol γ
DNA polymerase γ can synthesize DNA on a variety of primed DNA substrates including activated calf thymus DNA, singly primed M13 ssDNA and homopolymeric DNA. However, the most sensitive substrate for determining pol γ activity is the homopolymeric primer template substrate like poly(dA)-oligo(dT)12-18 (GE Healthcare). Polymerase activity was determined in reaction mixtures (50 μl) containing 25 mM HEPES-KOH [pH 8.0], 25 mM NaCl, 0.05 mg/ml acetylated BSA, 2 mM 2-mercaptoethanol, 1 mM MgCl2, 0.02 mg/ml poly(dA)-oligo(dT)12-18, 0.05 mM dTTP, 5 nM [α–32P] dTTP (NEN Radiochemicals, Perkin Elmer) and 10–100 ng of purified WT or R964C pol γ enzyme. Reactions were stopped after incubation at 37 °C for 10 min by transferring to 4°C and adding 1 ml of stop solution (500 mM NaOH, 100 mM Sodium pyrophosphate, 0.1 mg/ml sonicated calf thymus DNA, 0.5 mg/ml BSA). DNA was precipitated for 5 min after adding 1ml of 20% TCA and the mixture was filtered through Whatman GF/C filters, washed with 1 N HCl, rinsed with 100% ethanol and dried. The TCA-insoluble radioactivity was determined by liquid scintillation counting. The specific enzymatic activity of both WT and R964C mutant enzymes are represented as units/ng of the pol γ protein, where 1 unit is defined as the number of pmol of dTTP incorporated per ng of the pol γ protein per hour at 37°C. For steady-state kinetic values, the same assay was performed with fixed concentration of pol γ (10 ng) in the presence and absence of the p55 accessory subunit using poly(dA)-oligo(dT)12-18 as substrate, with varying concentrations of dTTP (0, 0.5, 1, 2, 4, 8, 16, 32 and 64 μM). Table 1 shows the activity and steady-state kinetic parameters (Km and kcat) for the WT and R964C mutant pol γ.
Table 1.
Specific activities, steady-state kinetic parameters and DNA binding affinities of WT and R964C pol γ enzymes were determined as described in Methods. The average values of three independent experiments are shown with errors expressed as standard deviations. ND, not determined.
| Protein | Activity (units/ng) | Steady state kinetics | DNA binding Kd (nM) | ||
|---|---|---|---|---|---|
| Km (mM dTTP) | kcat (s−1) | kcat/Km | |||
| WT | 207 ± 4 | 12 ± 1.2 | 13 ± 1 | 1.1 ± 0.1 | 31 ± 1 |
| WT + p55 | ND | 4.4 ± 0.4 | 4.8 ± 0.3 | 1.1 ± 0.04 | ND |
| R964C | 33 ± 1 | 8.5 ± 0.7 | 2.1 ± 0.2 | 0.25 ± 0.01 | 75 ± 4 |
| R964C + p55 | ND | 8.6 ± 2.4 | 1.5 ± 0.01 | 0.18 ± 0.04 | ND |
2.6 Analysis of DNA binding ability of human pol γ
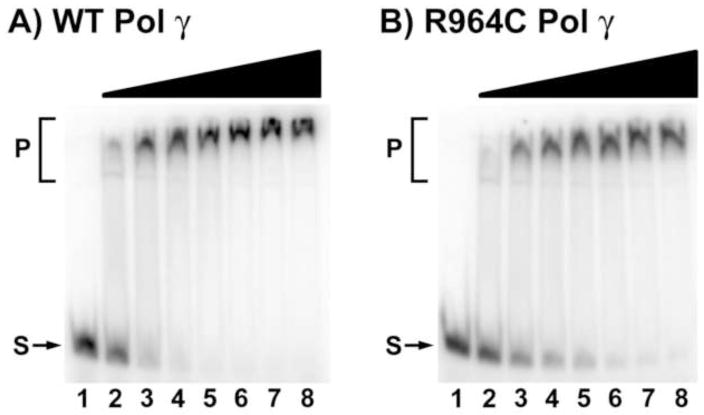
WT pol γ has been shown to tightly bind to the 3′ end of primer template substrates. This property of the enzyme has been greatly exploited in delineating the biochemical deficiencies of mutant pol γ proteins that cause various diseases in humans in many studies. For example, A467T, W748S, G848S and R852C point mutations in pol γ have been previously shown to have a compromised DNA binding ability that affects its overall functional efficiency [10, 12, 15]. The dissociation constant (Kd) for DNA binding of the WT and R964C mutant form of pol γ were determined by electrophoretic mobility shift assay (EMSA). Briefly, double-stranded primer-template molecules were constructed by hybridizing a 5′-32P- end-labeled 38-mer (5′-TTA TCG CAC CTA CGT TCA ATA TTA CAG GCG AAC ATA CT-3′) to a 1.2-fold molar excess of an unlabeled, complementary 34-mer (5′-GTA TGT TCG CCT GTA ATA TTG AAC GTA GGT GCG A-3′). Reaction mixtures (20 μl) were assembled at room temperature and contained 10 mM Tris-HCl [pH 8.0], 0.2 mg/ml acetylated BSA, 2 mM dithiothreitol, 50 nM of primer-template, and 0, 25, 50, 75, 100, 150, 200 or 250 nM of WT or R964C mutant pol γ protein. After 5 min incubation at room temperature, 5 μl of 5X loading buffer (10 mM Tris-HCl [pH 8.0], 50% glycerol, 0.1% bromophenol blue) was added and aliquots of the reaction mixture were subjected to electrophoresis for 1 h at 180 V at 4 °C through an 8% TBE polyacrylamide gel (Invitrogen) in 0.5X TBE (45 mM Tris, 45 mM boric acid, 1 mM EDTA) buffer. The gels were dried and exposed on a phosphor storage screen. Radioactivity was imaged on a Typhoon 9400 phosphorimager (GE Healthcare), and the intensity of the radiolabeled bands were quantified with NIH Image software. Representative gels for WT and R964C mutant are shown in Fig 3 and the Kd values for DNA binding are shown in Table 1.
Figure 3.
DNA binding affinity of WT and R964C pol γ proteins. Representative gels showing electrophoretic mobility shift assays performed using (A) WT and (B) R964C mutant pol γ enzymes as described in Methods to estimate the Kd (DNA) values. Lanes 1–8 contained 50 nM substrate; Lanes 2–8 had 0, 25, 50, 75, 100, 150, 200 or 250 nM of WT (A) or R964C mutant (B) pol γ protein. S, substrate, P, protein-DNA complex.
2.7 Examination of physical interaction of the catalytic subunit of pol γ with its accessory subunit
Studies have shown that interaction of the accessory subunit (p55) with the pol γ catalytic subunit enhances the processivity and DNA binding properties of the catalytic subunit, and the most common POLG mutation, the A467T substitution, has been demonstrated to impair this association [10]. Hence, examining the physical association of p55 with the mutant pol γ enzyme is crucial in characterizing these mutants. There are numerous ways by which protein-protein interactions could be determined. However, the two routine assays used in the laboratory are the co-immunoprecipitation and N-ethylmaleimide (NEM) inhibition assays.
2.7.1 Immunoprecipitation assay
Rabbit polyclonal antibodies raised against recombinant human p55 accessory subunit [5] were immobilized on Protein G Sepharose beads (GE Healthcare), pre-equilibrated in phosphate buffered saline NP-40 (PBSN)–BSA buffer consisting of 150 mM KPO4 [pH 7.5], 150 mM NaCl, 0.1% NP-40, and 0.1 mg/ml BSA. The reaction was carried out at 4 °C for 6 h followed by three washes with PBSN buffer before storing the beads in the same buffer. Prepared Protein G Sepharose beads suspension (20 μl) mixed with purified p55 accessory subunit (3 μg) and either with pol γ WT or R964C mutant protein (3 μg) in 400 μl with PBSN-BSA buffer was rotated end-over-end for 45 min at 4 °C. The beads were collected by centrifugation at 5,000 rpm for 2 min at 4°C, were washed twice with PBSN-BSA and once with PBSN lacking BSA and resuspended in 25 μl 2X lithium dodecyl sulfate (LDS) loading buffer (4X LDS loading buffer from Invitrogen, made 2X with PBSN lacking BSA). The samples were heated for 5 min at 95°C before analysis on 4–12% NuPAGE Bis-Tris polyacrylamide gels (Invitrogen). After electrophoresis, the proteins were electrotransferred to an Immobilon-P PVDF (polyvinylidene fluoride) membrane (Millipore). The membrane was then washed in TNT buffer (50 mM Tris-HCl [pH 7.5], 500 mM NaCl, and 0.1% Triton X-100) for 15 min and was blocked with 5% dried milk in TN buffer (50 mM Tris-HCl [pH 7.5] and 500 mM NaCl) at room temperature. The blot was incubated with 0.2 mg/ml anti-Penta-His monoclonal antibody (Qiagen) in TN containing 0.1 mg/ml BSA for 2 h, was washed three times with TN for 10 min, and incubated in a 1/3,000 dilution of goat anti-mouse alkaline phosphatase–conjugated secondary antibody (Bio-Rad) for 1 h. After three 10-min washes in TNT and three 10-min washes in TN, bands were visualized with Western Blue reagent (Promega). An example of this assay in the analysis of several Alper’s disease mutations in the pol γ catalytic subunit was described previously [15].
2.7.2 NEM inhibition assay
NEM inhibits the catalytic activity of pol γ by irreversibly and covalently modifying the sulfhydryl groups suggesting that solvent accessible cysteine residues are crucial for the enzyme activity. However, since cysteine residues are not located in the active site of the enzyme, NEM could inhibit pol γ activity by interfering with the formation of a ternary complex between the enzyme, the primer-template, and the incoming dNTP substrate. Previous studies from our laboratory have shown that the p55 accessory subunit protects the catalytic subunit from NEM inhibition by over 100-fold [5]. This property of p55 has been exploited for elucidating the physical interaction between these proteins in many studies including the analysis of the G451E disease mutation in POLG2 [19]. With the G451E-p55, NEM sensitivity was used, in part, to determine the loss of binding of this p55 mutant protein to the p140 catalytic subunit [19]. The standard polymerase assays with poly (dA)-oligo (dT)12-18 (GE Healthcare) substrates, described in section 2.5 were performed with varying concentrations of NEM (typically, 0–0.5 mM) for both WT and R964C pol γ enzymes. Physical association between the catalytic and accessory subunits could be determined by the level of inhibition caused by NEM in the presence and absence of p55 accessory subunit. The results from this experiment revealed that similar to the WT enzyme, the R964C mutant was also protected from NEM inhibition in the presence of p55 accessory subunit indicating no defect in protein-protein interactions (data not shown).
2.8 Determination of functional interaction of pol γ catalytic subunit with p55 accessory subunit
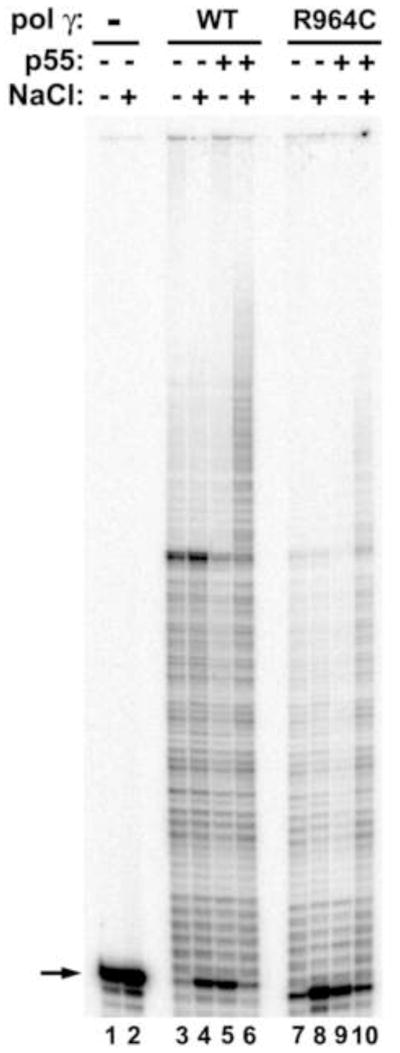
To determine whether the physical interaction of pol γ (WT and mutants) with the p55 accessory subunit translates into functional interaction, in vitro primer extension assays can be carried out using pol γ enzymes in the presence or absence of p55. The processivity of the catalytic subunit is stimulated as much as 50-fold upon interaction with the accessory subunit in the presence of salt [5]. The primer extension ability of pol γ can be determined by monitoring the extension of a 5′-32P- end-labeled 35-mer oligonucleotide (5′-CCA GTG CCA AGC TTG CAT GCC TGC AGG TCG ACT CT-3′) primer hybridized to M13mp18 DNA. Briefly, the 10 μl reaction contained 25 mM HEPES-KOH [pH 7.6], 5 mM 2-mercaptoethanol, 5 mM MgCl2, 0.05 mg/ml heat-treated BSA, 0 or 75 mM NaCl, 0.025 mM each dNTP, 2 nM of the labeled oligonucleotide, 5 nM of purified WT or R964C pol γ enzyme in the presence or absence of 10 nM of the p55 accessory subunit. Following incubation at 37 °C for 20 min, reactions were terminated with 10 μl formamide stop solution and products were analyzed by electrophoresis on a 12% denaturing polyacrylamide gel. The reaction products were visualized by phosphorimaging with a Typhoon 9400 PhosphorImager (Molecular Dynamics). This assay can reveal functional interaction of the p55 accessory subunit with the WT and mutant pol γ catalytic subunit. Figure 4 shows the functional interaction between WT and R964C mutant pol γ with its p55 accessory subunit.
Figure 4.
Functional interaction of WT and R964C pol γ enzymes with the p55 accessory subunit. Primer extension assays were performed using WT and R964C pol γ enzymes in the presence and absence of p55 accessory subunit on singly primed M13 DNA as described in Methods. Reactions contained 2 nM substrate (all lanes), 5 nM pol γ enzymes (lanes 3–6, WT; lane 7–10, R964C), 10 nM p55 accessory subunit (lanes 5, 6, 9 and 10). Activity was measured at 0 mM NaCl (odd numbered lanes) and at 75 mM NaCl (even numbered lanes). Lanes 1 and 2 had no enzyme. The arrow indicates the position of the unextended 35-mer primer.
Since the primer extension assays do not contain a trap DNA that can bind the polymerase once it has extended the end-labeled primer-template substrate and dissociated, the polymerase will have multiple opportunities to bind and extend the substrate. This configuration does not reveal the true processivity of the enzyme. However, the true processivity of the WT and mutant proteins can be studied under identical conditions, albeit in the presence of sonicated, heat denatured calf thymus DNA to act as a DNA trap. In this assay, the DNA trap should only be added to the reaction after the enzyme has bound to the end-labeled primer-template substrate. The trap DNA should be in excess (> 500 fold) to the substrate in order to prevent the polymerase from re-binding the substrate following the first dissociation event.
Acknowledgments
The authors would like to thank Drs. Deepti Dwivedi and Rajendra Prasad for critical reading of this manuscript. This work was supported by the NIEHS Intramural Research Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bebenek K, Kunkel TA. Adv Protein Chem. 2004;69:137–65. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- 2.Ropp PA, Copeland WC. Genomics. 1996;36:449–58. doi: 10.1006/geno.1996.0490. [DOI] [PubMed] [Google Scholar]
- 3.Sweasy JB, Lauper JM, Eckert KA. Radiat Res. 2006;166:693–714. doi: 10.1667/RR0706.1. [DOI] [PubMed] [Google Scholar]
- 4.Graziewicz MA, Longley MJ, Copeland WC. Chemical Reviews. 2006;106:383–405. doi: 10.1021/cr040463d. [DOI] [PubMed] [Google Scholar]
- 5.Lim SE, Longley MJ, Copeland WC. J Biol Chem. 1999;274:38197–203. doi: 10.1074/jbc.274.53.38197. [DOI] [PubMed] [Google Scholar]
- 6.Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. Embo J. 2004;23:2423–9. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong LJ, Naviaux RK, Brunetti-Pierri N, Zhang Q, Schmitt ES, Truong C, Milone M, Cohen BH, Wical B, Ganesh J, Basinger AA, Burton BK, Swoboda K, Gilbert DL, Vanderver A, Saneto RP, Maranda B, Arnold G, Abdenur JE, Waters PJ, Copeland WC. Hum Mutat. 2008;29:E150–E72. doi: 10.1002/humu.20824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rovio AT, Marchington DR, Donat S, Schuppe HC, Abel J, Fritsche E, Elliott DJ, Laippala P, Ahola AL, McNay D, Harrison RF, Hughes B, Barrett T, Bailey DM, Mehmet D, Jequier AM, Hargreave TB, Kao SH, Cummins JM, Barton DE, Cooke HJ, Wei YH, Wichmann L, Poulton J, Jacobs HT. Nat Genet. 2001;29:261–2. doi: 10.1038/ng759. [DOI] [PubMed] [Google Scholar]
- 9.Chan SS, Copeland WC. Biochim Biophys Acta. 2009;1787:312–19. doi: 10.1016/j.bbabio.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan SSL, Longley MJ, Copeland WC. J Biol Chem. 2005;280:31341–46. doi: 10.1074/jbc.M506762200. [DOI] [PubMed] [Google Scholar]
- 11.Hakonen AH, Heiskanen S, Juvonen V, Lappalainen I, Luoma PT, Rantamaki M, Goethem GV, Lofgren A, Hackman P, Paetau A, Kaakkola S, Majamaa K, Varilo T, Udd B, Kaariainen H, Bindoff LA, Suomalainen A. Am J Hum Genet. 2005;77:430–41. doi: 10.1086/444548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan SSL, Longley MJ, Copeland WC. Hum Mol Genet. 2006;15:3473–83. doi: 10.1093/hmg/ddl424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graziewicz MA, Longley MJ, Bienstock RJ, Zeviani M, Copeland WC. Nat Struct Mol Biol. 2004;11:770–76. doi: 10.1038/nsmb805. [DOI] [PubMed] [Google Scholar]
- 14.Ponamarev MV, Longley MJ, Nguyen D, Kunkel TA, Copeland WC. J Biol Chem. 2002;277:15225–8. doi: 10.1074/jbc.C200100200. [DOI] [PubMed] [Google Scholar]
- 15.Kasiviswanathan R, Longley MJ, Chan SS, Copeland WC. J Biol Chem. 2009;284:19501–10. doi: 10.1074/jbc.M109.011940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan SS, Copeland WC. Methods Mol Biol. 2009;554:59–72. doi: 10.1007/978-1-59745-521-3_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bailey CM, Kasiviswanathan R, Copeland WC, Anderson KS. Antimicrob Agents Chemother. 2009;53:2610–2. doi: 10.1128/AAC.01659-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Longley MJ, Ropp PA, Lim SE, Copeland WC. Biochemistry. 1998;37:10529–39. doi: 10.1021/bi980772w. [DOI] [PubMed] [Google Scholar]
- 19.Longley MJ, Clark S, Yu Wai Man C, Hudson G, Durham SE, Taylor RW, Nightingale S, Turnbull DM, Copeland WC, Chinnery PF. Am J Hum Genet. 2006;78:1026–34. doi: 10.1086/504303. [DOI] [PMC free article] [PubMed] [Google Scholar]