Abstract
Background
Genome-wide association studies have identified multiple independent regions on chromosome 8q24 that are associated with cancers of the prostate, breast, colon, and bladder.
Methods
To investigate their biological basis, we examined the possible association between 164 single nucleotide polymorphism (SNPs) in the 8q24 risk regions, spanning 128,101,433–128,828,043 bp, and serum androgen (testosterone, androstenedione, 3αdiol G, and bioavailable testosterone) and sex hormone-binding globulin levels in 563 healthy, non-Hispanic, Caucasian men (55–74 years old) from a prospective cohort study, the Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial. Age-adjusted linear regression models were used to determine the association between the SNPs in an additive genetic model and log transformed biomarker levels.
Results
Three adjacent SNPs centromeric to prostate cancer risk-region 2 (rs12334903, rs1456310, and rs980171) were associated with testosterone (P<1.1×10−3) and bioavailable testosterone (P<6.3×10−4). Suggestive associations were seen for a cluster of 9 SNPs in prostate cancer risk region 1 and androstenedione (P<0.05).
Conclusions
These preliminary findings require confirmation in larger studies, but raise the intriguing hypothesis that genetic variations in the 8q24 cancer risk regions may correlate with androgen levels.
Impact
These results may provide some clues for the strong link between 8q24 and prostate cancer risk.
Keywords: 8q24, genetic polymorphisms, serum androgens
Introduction
Genome-wide association studies have identified multiple independent regions on chromosome 8q24 that are associated with cancers of the prostate (1–7), breast (8), colon (9–11), and bladder (12). All of these risk regions lie outside of coding regions, with the exception of a psuedogene (POU5F1P1) in prostate cancer risk region 3 (13), and have no known function. The most proximal gene to these risk regions is the well known oncogene MYC at about 30 kb telomeric to the bladder cancer region. However, currently there is no evidence to suggest that 8q24 cancer risk markers are in linkage disequilibrium (LD) with common genetic variants in MYC or its promoters nor is there definitive evidence for an association between 8q24 risk variants and MYC expression (14–16).
To investigate the biological basis for the link between 8q24 risk regions and cancer risk, we examined the correlation between common genetic variants in 8q24 and serum levels of androgen and sex-hormone binding globulin (SHBG) in 563 healthy non-Hispanic Caucasian males within the Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial. Included in the analysis were 164 SNPs encompassing the six identified 8q24 risk regions (spanning chromosome 8:128,101,433–128,828,043 bp) from the NCI Cancer Genetic Markers of Susceptibility (CGEMS) Project and serum androgens (testosterone, 5α-androstane-3α, 17β-diol glucuronide (3αdiol G), and androstenedione), and sex hormone-binding globulin (SHBG).
Methods
Study Subjects
The PLCO Cancer Screening Trial is a randomized trial designed to determine if screening for prostate, lung, colorectal and ovarian cancers leads to mortality reduction for these cancers. Details of the study have been described previously (17, 18). Briefly, the trial includes 155,000 men and women (aged 55 to 74 with no reported history of prostate, lung, colon and ovarian cancer) enrolled between 1992and 2001, and randomized to either the screening or control arm of the trial. Men randomized to the screening arm provided basic risk factor information through the Baseline Questionnaire, and were offered a prostate-specific antigen (PSA) tests and digital rectal exams (DRE) at baseline and annually thereafter for 3 years, followed by 2 years of screening with PSA alone. These men also donated serial blood samples at each of six screening rounds, and were followed by an annual survey for self-reported cancer diagnosis. The study protocol was approved by the institutional review board at each study center and the National Cancer Institute, and participants provided informed consent.
In the current study, we included 563 healthy non-Hispanic Caucasian men with both genotyping (4) and baseline serum androgen (19) data. These men were originally selected as controls for two nested case-control studies of prostate cancer as described previously (4, 19). Briefly, the selection criteria for the controls for these two studies were:1) had been screened for prostate cancer (PSA and/or DRE) at least once before October 1, 2003; 2) had no prior history of prostate cancer before randomization; 3) completed the baseline risk factor questionnaire and returned at least 1 annual study update questionnaire; 4) were non-Hispanic Caucasian; 5) signed an informed consent; 6) provided a blood sample; and 7) selected by incidence-density sampling to be matched to the cases by age of entry in 5-year intervals, time since initial screen (1-year time windows), and year of blood draw at study entry. In total, 1,087 controls were selected for the genotyping study (4) and 889 healthy men were selected for the serum androgen study (19). For this study, only men with both genotyping and serum androgen data were eligible (563men).
Serum Androgen and SHBG
Serum androgen and SHBG assays have been previously described (19). Briefly, all assays were performed at the International Agency for Research on Cancer (IARC) on serum drawn at baseline. Total testosterone was measured by direct radioimmunoassay (Immunotech, Marseille, France), androstenedione and 3αdiol G by direct double-antibody radioimmunoassay (Diagnostic Systems Laboratories, Webster, TX), and SHBG by sandwich immunoradiometric assay (CIS-Bio, Gif-sur-Yvette, France); overall coefficients of variations (CVs) from duplicate samples were 14, 11, 14 and 18% for testosterone, 3αdiol G, androstenedione, and SHBG, respectively (19). Total testosterone (nmol/L) and SHBG (nmol/L) were used to calculate bioavailable testosterone using the following equation as described previously (20):
Genotyping
Genotyping was performed under contract by Illumina Corporation (San Diego, CA) as part of the CGEMS Project and coordinated by the NCI Core Genotyping Facility as described previously (4). The assay consists of a fixed panel of 561,494 tag SNPs of which181 SNPs were located in and around the 8q24 cancer risk regions (128,101,433–128,828,043 bp; NCBI Build 36.3). We excluded 19 SNPs with minor allele frequencies (MAFs) less than 5% and/or SNPs that were not in Hardy Weinberg Equilibrium (HWE; p<0.001), leaving 164 SNPs for analysis. These 164 SNPs encompasses the following regions (from centromere to telomere): prostate cancer region 2 (128,101,433–128,279,002 bp; 38 SNPs), prostate cancer region 4 (128,280,411–128,412,048; 36 SNPs), breast cancer region (128,413,806–128,472,696; 17 SNPs), prostate cancer region 3 (128,473,069–128,535,996; 18 SNPs), prostate cancer region 1 (128,541,502–128,614,565; 22 SNPs), bladder cancer region (128,617,860–128,816,653; 31 SNPs), and MYC (128,819,722–128,828,043; 2 SNPs). Quality control evaluations were described in detail in the original CGEMS analysis (4).
Statistical Analysis
We used Stata (version 9.0, Stata Corp) to run all analyses unless stated otherwise. Linear regression analysis was used for determining effect sizes and significance of associations between natural log transformed androgen or SHBG levels and all 164 SNPs. Polytomous regression analysis was also used to determine the associations of the SNPs with androgens or SHBG categorized in quartiles and quintiles. All genotypes were coded as 0, 1, and 2 according to the copy number of the less common allele and were modeled linearly. All regression models were adjusted for age at entry in five-year intervals (55–59, 60–64, 65–69, 70–74); race was not included in the analysis since all study participants were non-Hispanic Caucasian men. P-values for SNP effect were based on two-sided Wald-test statistic. To adjust for multiple comparisons, we used a parametric bootstrap procedure (21) with 20,000 replicates generated under the null for the evaluation of global significance levels accounting for all SNPs tested within each sub-region or for all 164 SNPs tested within the entire8q24 region. For a select number of SNPs identified by linear regression analysis to be most significantly correlated with androgen levels, we used the Kruskall-Wallis non-parametric one-way analysis of variance (ANOVA) to determine if median androgen levels differ by genotype. Pair-wise LD was estimated between SNPs in each of the 8q24 regions based on D′ and r2 statistics calculated in Haplo view (22).
Results
Selected demographic and serologic characteristics of the 563 study participants are shown on Table 1. Over 65% of the men were between the ages of 60 and 70 and the majority (>70%) had at least some college education. In addition, over half of the men were former smokers and 5.4% had a family history of prostate cancer. Furthermore, over half of the men were overweight and about 22% were obese. Means and ranges of serum androgens and SHBG levels for participants of this study were similar to those previously reported in the PLCO (19).
Table 1.
Select characteristics of 563 study participants
| Characteristic | Participants N=563 |
||
|---|---|---|---|
| n | % | ||
| Age at enrollment (years) | |||
| 55–59 | 96 | 16.8 | |
| 60–64 | 192 | 33.7 | |
| 65–69 | 180 | 31.6 | |
| 70–74 | 95 | 16.7 | |
| Education level | |||
| Less than high school | 46 | 8.1 | |
| Completed high school | 99 | 17.4 | |
| Some college or vocational training | 191 | 33.5 | |
| College and above | 227 | 39.8 | |
| Family history of prostate cancer | |||
| No | 524 | 92.1 | |
| Yes | 31 | 5.4 | |
| Cigarette smoking status | |||
| Never | 215 | 37.7 | |
| Former | 293 | 51.4 | |
| Current | 55 | 9.6 | |
| Body mass index (BMI) | |||
| Normal (18.5–24.9) | 145 | 25.6 | |
| Overweight (25–29.9) | 288 | 50.8 | |
| Obese (>30) | 127 | 22.4 | |
| Serum Marker | Mean | SD | Range |
| Total testosterone (nmol/L) | 17.7 | 8.6 | 0.6 – 60.0 |
| Bioavailable testosterone (nmol/L) | 4.9 | 1.9 | 0.2 – 14.5 |
| 4-Androstene-3,17-dione (Androstenedione; ng/dL) | 126.1 | 44.3 | 29.6 – 301.6 |
| 5α-androstane-3α,17β-diol glucuronide (3αdiol G; ng/dL) | 828.6 | 585.6 | 67.6 – 5346.7 |
| Sex hormone-binding globulin (SHBG; nmol/L) | 48.7 | 22.9 | 13.1 – 152.3 |
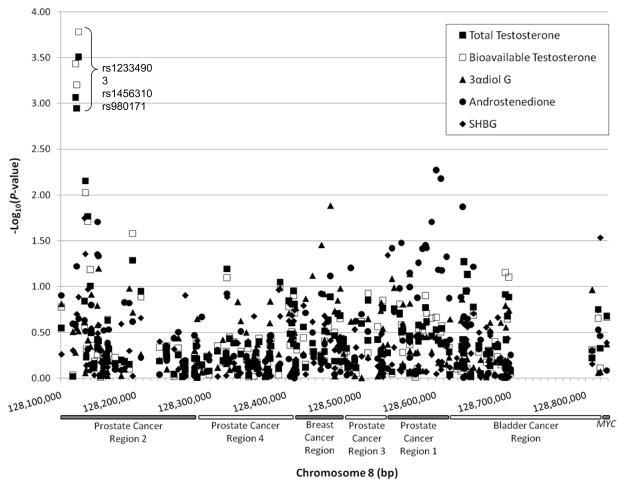
Figure 1 shows the summary graph of -log10 of P-values from linear regression analysis of the associations between all 164 SNPs typed within a 727 kb region of chromosome 8q24 and 4 serum androgen measures and SHBG plotted against the chromosomal locations of each SNP. Of the 164 SNPs analyzed, 24 were associated with serum androgens or SHBG at P<0.05, with 6 reaching P<0.01; a total of 30 tests were significant at P<0.05, with 10 reaching P<0.01. Similar results were found when analyzing androgens and SHBG in quartiles and quintiles or comparing highest to the lowest levels (data not shown). Additional adjustment for hour of blood draw to account for variations in circadian variation serum androgen levels (23, 24) did not change the results (data not shown).
Figure 1.
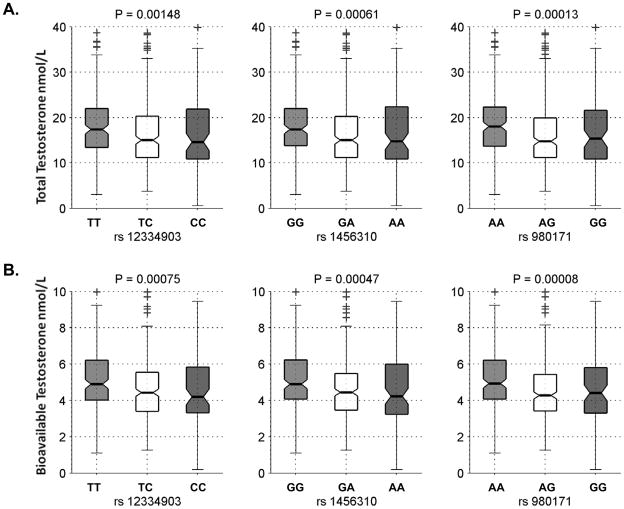
The strongest associations seen are for 3 adjacent SNPs at the centromeric end of prostate cancer risk region 2 (rs12334903, rs1456310, and rs980171) with total and bioavailable testosterone (P ≤ 1.13×10−3 and 6.28×10−4, respectively; Table 2 and Supplemental Table 1). These three SNPs are in relatively strong LD with each other (D′ >0.88, r2>0.69) have similar minor allele frequencies (MAFs; 39–43%), and similar effects on testosterone (each copy of the risk allele is associated with an 8–10% decrease in mean testosterone levels). All three SNPs remained significantly associated with testosterone after adjusting for multiple comparison of 38 SNPs tested within region 2 (P<0.03); after adjusting for all 164 SNPs tested, rs980171 remained associated with total and bioavailable testosterone (P=0.039 and 0.023, respectively) and rs12334903 with bioavailable testosterone (P=0.049). The Kruskal-Wallis non-parametric ANOVA also show that median total and bioavailable testosterone levels differed significantly among groups defined by the genotypes of the 3 SNPs (nominal P<1.48×10−3 and 7.5×10−4, respectively; Figure 2). Also of note is the association of a cluster of 9 SNPs (128,541,502–128,607,399 bp) in prostate cancer region 1 with androstenedione (nominal P<0.05; Supplemental Table 1), with five of the 9 SNPs (rs1447295, rs4242382, rs4242384, rs7017300, and rs11988857) in relatively strong LD (D′ >0.89, r2>0.71); these SNPs did not remain significant after adjusting for multiple testing of 22 SNPs in region 1 (P<0.34).
Table 2.
Associations between three most significant SNPs in 8q24 cancer risk regions and serum testosterone measures in 563 male subjects
| Serum Androgen | SNP (major/minor alleles) | Location (bp) | MAF1 | β2 | SE2 | P2 | P3 | P4 |
|---|---|---|---|---|---|---|---|---|
| Total testosterone | rs12334903(C) | 128,119,695 | 0.40 | −0.10 | 0.03 | 8.69×10−4 | 0.023 | 0.100 |
| rs1456310(A) | 128,121,615 | 0.43 | −0.10 | 0.03 | 1.13×10−3 | 0.030 | 0.127 | |
| rs980171(G) | 128,123,704 | 0.39 | −0.11 | 0.03 | 3.14×10−4 | 9.60×10−3 | 0.039 | |
| Bioavailable testosterone | rs12334903(C) | 128,119,695 | 0.40 | −0.09 | 0.03 | 3.73×10−4 | 0.012 | 0.049 |
| rs1456310(A) | 128,121,615 | 0.43 | −0.08 | 0.02 | 6.28×10−4 | 0.019 | 0.076 | |
| rs980171(G) | 128,123,704 | 0.39 | −0.09 | 0.03 | 1.66×10−4 | 4.95×10−3 | 0.023 |
MAF = minor allele frequency
Based on age-adjusted linear regression analysis of log-transformed androgen and SHBG measurements using SNPs in an additive genetic model
Adjusted for multiple comparisons using a parametric bootstrap procedure with 20,000 replicates generated under the null for the evaluation of significance levels accounting for all 38 SNPs tested within the prostate cancer region 2
Adjusted for multiple comparisons using a parametric bootstrap procedure with 20,000 replicates generated under the null for the evaluation of significance levels accounting for all 164 SNPs tested in 8q24 region
Figure 2.
Discussion
In this study of 563 healthy men, we found several SNPs in the 8q24 cancer risk regions associated with circulating levels of androgens. Specifically, SNPs centromeric to prostate cancer region 2 were associated with testosterone measures while those in prostate cancer region 1 were suggestively associated with androstenedione. Since many of the SNPs within each region are in strong LD with each other, the effect of the SNPs within each region on serum androgens may reflect their link with the same causal variant. These findings suggest a potential relationship between the 8q24 cancer risk regions and serum androgens, which in turn may provide some biological clues into the strong link between 8q24 variants and cancer risk. Future studies with larger sample size are needed to confirm these findings.
It is noteworthy that 8q24 SNPs identified in this study to be associated with androgens are also associated with or are in LD with SNPs that are associated with prostate cancer risk (2–6, 25–27), suggesting that serum testosterone may be related to the link between 8q24 variants and prostate cancer. Although the exact mechanism is unclear, data from a recent study showed that the 8q24 cancer risk regions harbor several androgen-responsive transcriptional enhancers, one of which contained a SNP, rs11986220, in prostate cancer region 1 that facilitated stronger androgen responsiveness in vitro (28). This particular SNP was not genotyped in our study, but is in moderate to strong LD with several SNPs identified in the study to be associated with androstenedione (D′ =1 and r2>0.54 based on the International HapMap Project data (29)). Interactive effects of 8q24variants and serum androgens on prostate cancer risk warrants further investigation, in particular in studies with large samples size.
Our study has several strengths. First, we had high-quality genotyping data as suggested by high concordance and high completion rates, thereby minimizing misclassification of genotyping (4). Second, we had high-quality serum assays of biomarkers, with low intra-and inter-assay variation (coefficients of variation <18%) (19), which minimizes misclassification of the outcome. And third, effects of disease status (prostate cancer) is minimized by including only participants of the screening arm of the PLCO who have not been diagnosed with prostate cancer at the time of subject selection.
Limitations of the study should also be noted. Our findings are only borderline significant after adjusting for the tests on 164 SNPs with 5 biomarker measures. However, the 164 SNPs tested were over 5 distinct and independent regions, as defined by LD structure, and we conservatively accounted for these in our assessment of the statistical analyses. Given the relatively small sample size of our study and the existing strong evidence on the relevance of 8q24 region to various cancers, our findings require confirmation in studies with larger sample sizes to better understand the relationship between 8q24 and androgens. Future studies are also needed to determine the relationship between 8q24 and androgens in other ethnic groups.
In summary, our study showed that variations in the 8q24 cancer risk regions may be associated with serum androgen levels, which raises the intriguing hypothesis that serum androgens may be related to the reported association between the 8q24 risk regions and cancer, especially that of the prostate. Future studies with larger sample sizes and in other ethnic populations are needed to confirm these findings and enable generalization to other populations. In addition, future investigations into the mechanisms underlying the link between the 8q24 cancer risk variants, androgen levels and prostate carcinogenesis are warranted.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics and contracts from the Division of Cancer Prevention, National Cancer Institute, National Institutes of Health, Department of Health and Human Services. The authors thank Drs. Christine Berg and Philip Prorok, Division of Cancer Prevention, National Cancer Institute; the Screening Center investigators and staff of the Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial; Mr. Tom Riley and staff, Information Management Services, Inc.; Ms. Barbara O’Brien and staff, West at, Inc.; Drs. Bill Kopp and Dr. Wen Shao, and staff (Science Applications International Corporation-Frederick); and Ms. Josiane Bouzac, Ms. Priscilia Amouyal and Mr. David Achaintre (IARC). Most importantly, we acknowledge the study participants for their contributions to making this study possible.
References
- 1.Amundadottir LT, Sulem P, Gudmundsson J, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–8. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 2.Gudmundsson J, Sulem P, Manolescu A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–7. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 3.Haiman CA, Patterson N, Freedman ML, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–44. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeager M, Orr N, Hayes RB, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–9. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 5.Thomas G, Jacobs KB, Yeager M, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–5. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 6.Eeles RA, Kote-Jarai Z, Giles GG, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–21. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 7.Yeager M, Chatterjee N, Ciampa J, et al. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat Genet. 2009;41:1055–7. doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haiman CA, Le Marchand L, Yamamato J, et al. A common genetic risk factor for colorectal and prostate cancer. Nat Genet. 2007;39:954–6. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomlinson I, Webb E, Carvajal-Carmona L, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–8. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 11.Zanke BW, Greenwood CM, Rangrej J, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–94. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 12.Kiemeney LA, Thorlacius S, Sulem P, et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat Genet. 2008;40:1307–12. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panagopoulos I, Moller E, Collin A, Mertens F. The POU5F1P1 pseudogene encodes a putative protein similar to POU5F1 isoform 1. Oncol Rep. 2008;20:1029–33. [PubMed] [Google Scholar]
- 14.Sole X, Hernandez P, de Heredia M, et al. Genetic and genomic analysis modeling of germline c-MYC overexpression and cancer susceptibility. BMC Genomics. 2008;9:12. doi: 10.1186/1471-2164-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pomerantz MM, Beckwith CA, Regan MM, et al. Evaluation of the 8q24 Prostate Cancer Risk Locus and MYC Expression. Cancer Res. 2009;69:5568–74. doi: 10.1158/0008-5472.CAN-09-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuupanen S, Turunen M, Lehtonen R, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41:885–90. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 17.Prorok PC, Andriole GL, Bresalier RS, et al. Design of the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Control Clin Trials. 2000;21:273–309S. doi: 10.1016/s0197-2456(00)00098-2. [DOI] [PubMed] [Google Scholar]
- 18.Hayes RB, Reding D, Kopp W, et al. Etiologic and early marker studies in the prostate, lung, colorectal and ovarian (PLCO) cancer screening trial. Control Clin Trials. 2000;21:349S–55S. doi: 10.1016/s0197-2456(00)00101-x. [DOI] [PubMed] [Google Scholar]
- 19.Weiss JM, Huang WY, Rinaldi S, et al. Endogenous sex hormones and the risk of prostate cancer: A prospective study. Int J Cancer. 2008;122:2345–50. doi: 10.1002/ijc.23326. [DOI] [PubMed] [Google Scholar]
- 20.Morris PD, Malkin CJ, Channer KS, Jones TH. A mathematical comparison of techniques to predict biologically available testosterone in a cohort of 1072 men. Eur J Endocrinol. 2004;151:241–9. doi: 10.1530/eje.0.1510241. [DOI] [PubMed] [Google Scholar]
- 21.Efron B, Tibshirani RJ. An introduction to the bootstrap. Chapman & Hall/CRC; 1993. [Google Scholar]
- 22.Barrett JC, Fry B, Maller J, Daly MJ. Haplo view: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 23.Lejeune-Lenain C, Van Cauter E, Desir D, Beyloos M, Franckson JR. Control of circadian and episodic variations of adrenal androgens secretion in man. J Endocrinol Invest. 1987;10:267–76. doi: 10.1007/BF03348129. [DOI] [PubMed] [Google Scholar]
- 24.Plymate SR, Tenover JS, Bremner WJ. Circadian variation in testosterone, sex hormone-binding globulin, and calculated non-sex hormone-binding globulin bound testosterone in healthy young and elderly men. J Androl. 1989;10:366–71. doi: 10.1002/j.1939-4640.1989.tb00120.x. [DOI] [PubMed] [Google Scholar]
- 25.Zheng SL, Sun J, Cheng Y, et al. Association between two unlinked loci at 8q24 and prostate cancer risk among European Americans. J Natl Cancer Inst. 2007;99:1525–33. doi: 10.1093/jnci/djm169. [DOI] [PubMed] [Google Scholar]
- 26.Salinas CA, Kwon E, Carlson CS, et al. Multiple independent genetic variants in the 8q24 region are associated with prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17:1203–13. doi: 10.1158/1055-9965.EPI-07-2811. [DOI] [PubMed] [Google Scholar]
- 27.Al Olama AA, Kote-Jarai Z, Giles GG, et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat Genet. 2009;41:1058–60. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 28.Jia L, Landan G, Pomerantz M, et al. Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS Genet. 2009;5:e1000597. doi: 10.1371/journal.pgen.1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PIW. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–9. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.