Abstract
Objective
Thoracic aortic aneurysms (TAAs) result from dysregulated remodeling of the vascular extracellular matrix (ECM) which may occur as a result of altered resident cellular function. The present study tested the hypothesis that aortic fibroblasts undergo a stable change in cellular phenotype during TAA formation.
Methods
Primary murine aortic fibroblasts were isolated from normal and TAA-induced aortas (4-wks post-induction with 0.5M CaCl2 15 min) by outgrowth method. Normal and TAA cultures were examined using a focused PCR array to determine fibroblast-specific changes in gene expression in the absence and presence of biological stimulation (endothelin-1, phorbol-12-myristate-13-acetate, angiotensin II). The relative expression of 38 genes, normalized to 4 housekeeping genes, was determined and genes displaying a minimum 2-fold increase/decrease or genes with significantly different normalized Ct values were considered to have altered expression.
Results
At steady state TAA fibroblasts revealed elevated expression of several MMPs (Mmp2, Mmp11, Mmp14), collagen genes/elastin (Col1a1, Col1a2, Col3a1, Eln), and other matrix proteins, as well as decreased expression of Mmp3, Timp3, and Ltbp1. Moreover, gene expression profiles in TAA fibroblasts were different than normal fibroblasts after equivalent biological stimuli.
Conclusions
This study demonstrated for the first time that isolated primary aortic fibroblasts from TAA-induced mice possess a unique and stable gene expression profile, and when challenged with biological stimuli, induce a transcriptional response that is different from normal aortic fibroblasts. Together, these data suggest that aortic fibroblasts undergo a stable phenotypic change during TAA development which may drive the enhancement of ECM proteolysis in TAA progression.
Keywords: aneurysm, extracellular matrix, remodeling, phenotype, fibroblast
INTRODUCTION
Thoracic aortic aneurysms (TAAs) develop in response to pathological changes that alter the structure and composition of the aortic extracellular matrix (ECM).(1, 2) These dynamic changes produce an imbalance between matrix degradation and deposition resulting in compromised structural integrity and a propensity to dilate, dissect, or rupture.(2, 3) While the inciting stimuli remain undefined, it has become clear that TAA development is multifactorial and involves both cellular and molecular mechanisms. Current data from clinical TAA specimens and experimental animal models have implicated the matrix metalloproteinases (MMPs) as key mediators of aneurysm formation. While many studies have suggested that inflammatory cells are the major source of MMPs in aneurysm development,(4, 5) changes in MMP abundance can also arise as a result of altered production by the endogenous cellular constituents. For example, LeMaire et al. implicated a role for endogenous cells in mediating elevated MMP expression in ascending aortic aneurysm specimens from patients with bicuspid aortic valves lacking inflammatory infiltrate.(5) Furthermore, work from this laboratory localized MMP-9 promoter activation to fibroblasts/fibroblast-derived cells within the developing murine TAA.(6) With many studies reporting a loss of medial smooth muscle cells (SMC) during TAA development,(7, 8) it is likely that the changes in endogenous cellular constituents play a significant role in mediating TAA formation and progression. In a recent report from this laboratory using a murine model of TAA, aortic dilatation occurred concomitantly with a loss of medial SMCs and the emergence of a subset of fibroblast-derived myofibroblasts.(9)
Accordingly, the present study examined the hypothesis that aortic fibroblasts undergo a phenotypic transformation that results in enhanced degradative capacity and the ability to compensate for the loss of SMCs in the developing aneurysm. The hypothesis was tested through two primary objectives. The first objective examined gene expression differences in primary aortic fibroblasts isolated from either normal mice, or mice 4-weeks following TAA induction surgery. The second objective then determined whether the isolated fibroblasts responded in similar fashion when equivalently challenged with relevant biological stimuli. To accomplish these objectives, a custom quantitative real-time PCR array was constructed and employed to simultaneously determine the relative expression of multiple determinants of ECM degradation and deposition in normal and TAA fibroblasts.
MATERIALS AND METHODS
Experimental design
The present study examined gene expression in primary murine aortic fibroblasts isolated from normal and TAA-induced mice. Four C57BL/6J mice (8–12 weeks old, equal number of males and females) underwent TAA induction surgery with a terminal time-point of 4-wks post-TAA induction. At terminal surgery, the descending thoracic aorta was excised under sterile conditions and processed for fibroblast outgrowth according to established procedures.(10) At the same time, aortas were harvested from four unoperated C57BL/6J age- and gender- matched control mice and processed identically for fibroblast outgrowth. This animal protocol was approved by the Medical University of South Carolina Institutional Care and Use Committee, and all mice were treated and cared for in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publication No. 85-23, revised 1996).
Operative procedure
Murine TAAs were induced as previously described.(11) Briefly, following anesthetic induction, mice were intubated and a surgical plane of anesthesia was maintained using a 2% isoflurane/oxygen mixture. The descending thoracic aorta was exposed through a left-thoracotomy. A sponge soaked in 0.5 M calcium chloride was then placed in direct contact with the periadventitial surface for 15 minutes. The chest was irrigated, closed in layers, and the mice were allowed to recover.
Terminal surgical procedures and fibroblast isolation
Mice were transferred to a laminar flow biosafety cabinet, and euthanized under deep anesthesia by exsanguination induced by right atriotomy. The animals were then systemically perfused with sterile saline until the perfusate was clear and the liver was blanched. The descending thoracic aorta was excised, rinsed in sterile saline, and cut longitudinally. The endothelial cell layer was removed by gently rubbing the luminal surface with a sterile swab. The aorta was then cut into approximately ten 1 × 2 mm pieces and carefully placed onto the surface of a dry tissue culture flask (T-75, Cat#13-680-65; BD Falcon, Fisher Scientific, Pittsburgh, PA). The tissue was allowed to adhere in the absence of medium for 5–10 min, then 10 ml of fibroblast growth medium (Fibroblast Growth Medium, Promocell Cat#C39315, Heidelberg, Germany), containing Fibroblast Growth Supplement, (Promocell Cat#C23010; consisting of 1.0 ng/ml basic fibroblast growth factor, and 0.5 μg/ml insulin at final concentration) and 20% heat-inactivated fetal-calf serum (Invitrogen, Cat# 10082-147, Carlsbad, CA), was carefully added to the flask. The flask was placed in a humidified 5% CO2 incubator and the fibroblasts were allowed to grow out of the individual tissue chunks. Evidence of fibroblast outgrowth was typically observed within 7 days of plating. Of the eight cell lines initiated, four normal aortic fibroblast cell lines and three TAA cell lines reached confluence within 22±1 days after plating. Fibroblasts were identified as spindle-shaped cells during log phase growth, and their identity was confirmed by staining log-phase cells with phalloidin to observe cellular architecture and with cell-type specific markers to verify purity (DDR2, prolyl-4-hydroxylase, and heavy chain myosin). The established cell lines were maintained in culture and split into new flasks when the cell density reached approximately 90% confluence. Fibroblasts in passages 3–6 were used for experimental studies.
Cell stimulation
For steady-state analysis normal and TAA fibroblasts were grown to approximately 80% confluence, then placed in serum-reduced medium (fibroblast growth medium + fibroblast growth supplement) containing 0.1% bovine serum albumin for 24 hr before harvest. The cells were scraped into cold (4°C) phosphate buffered saline, and collected by centrifugation (3000 × g). The cell pellet was resuspended in 300 μl of RNAprotect Cell Reagent (Cat#76526, Qiagen, Inc., Valencia, CA) and stored at 4°C for 24 hr.
For the cellular stimulation studies, normal and TAA fibroblasts were grown to approximately 70% confluence, then placed in serum-reduce medium containing 0.1% bovine serum albumin (as detailed above) for 24 hrs. The following day, the serum-reduced medium was aspirated and replaced with 5 ml of the same medium containing either 1 nM endothelin-1 (ET-1; Cat# E-7764; Sigma Chemical Co, St. Louis, MO), 100 nM angiotensin-II (AngII; Cat# A-9525; Sigma Chemical Co, St. Louis, MO), or 100 nM phorbol-12-myristate-13-acetate (PMA; Cat# P-8139; Sigma Chemical Co, St. Louis, MO)). The cells were then allowed to grow for an additional 24 hrs. At the end of the stimulation period, the cells were processed as detailed above.
Gene expression analysis
Total RNA was isolated using the Qiagen RNeasy Plus Mini Kit (Cat#74134, Qiagen, Inc, Valencia, CA). RNA quality and quantity was analyzed with the Experion Automated Electrophoresis System (Bio-Rad Laboratories, Hercules, CA) using an Experion RNA StdSens Analysis Kit (Cat#700-7103, Bio-Rad, Hercules, CA). One μg of high-quality RNA from each cell line was reverse-transcribed to generate cDNA using an RT2 First Strand Kit (Cat#C-03, SABiosciences, Frederick, MD), and the cDNA was immediately assayed for gene expression by quantitative real-time PCR (QPCR).
In order to easily assess the expression of numerous genes from the established primary cell lines, a custom RT2 Profiler™ PCR Array (Custom Services; SABiosciences, Frederick, MD) was designed to test 42 different genes including 4 housekeeping control genes, in a 96-well plate format. The generated cDNA was diluted into RT2 qPCR Master Mix (Cat#PA-011, containing Hot-Start Taq polymerase and a SYBR Green/Fluorescein mix specific for Bio-Rad QPCR systems; SABiosciences, Frederick, MD) according the manufacturer’s instructions, and was applied to a 96-well PCR Array plate. Quantitative PCR was performed using a MyiQ Single-Color Real-Time PCR Detection System (Bio-Rad, Hercules, CA) with the following cycling parameters: initial denaturation for 10 min at 95°C, was followed by 40 cycles of 15 sec at 95°C, 40 sec at 55°C, and 30 sec at 72°C. A melt curve was established immediately following the conclusion of the cycling program to allow for confirmation of a single QPCR product for each gene-specific primer set (1 min at 95°C, 2 min at 65°C, followed by 60 cycles of 15 sec at 65°C with an increase of 0.5°C per cycle). Negative controls were run on each plate to verify the absence of genomic DNA contamination (no reverse transcription control), and the absence of overall DNA contamination in the PCR system and working environment (no template control).
Data analysis
Cycle threshold (Ct) values were recorded and fold change in steady-state gene expression between TAA fibroblasts and normal fibroblasts was calculated using the ΔΔCt method (see Table 1 for a complete listing of genes examined, including housekeeping genes). Fold expression values greater than 2.0 or less than 0.5 were considered to have a significant change in gene expression. Additionally, any gene that displayed a significant difference in mean ΔCt value (TAA vs. normal by 2-sided, 2-tailed t-test; p<0.05) was likewise considered to have a significant change in expression. Ct values of 35.0 or greater were considered non-cycling and were removed from analysis. Of the genes tested, primers for Mmp8, Mmp12, Mmp7, and Timp4 consistently produced multiple peaks on melt-curve analysis indicating that more than one product was being amplified in those wells. Accordingly, these genes were also removed from analysis. Additionally, Mmp9, Timp1, and Lamb3 did not consistently cycle either due to low target concentration or poor primer design. To measure Mmp9 and Timp1 gene expression, TaqMan primer/probe sets were used with same quantitative PCR conditions indicated above (Mmp9, Cat# Mm00442991_m1; Timp1, Cat# Mm00441818_m1; Applied Biosystems, Foster City, CA). There were two incidents of stimulus-induced increase in Ct values (PMA, Mmp15; and AngII, Junb). In PMA treated cells, each stimulated control cell line yielded a measurable Ct value for Mmp15, while each stimulated TAA cell line produced Ct values of >35.0. In AngII treated cells, Junb expression was suppressed in both normal and TAA fibroblasts following stimulation.
Table 1.
List of genes assayed in normal and TAA fibroblasts by QPCR.
| Target | Gene Symbol | UniGene | RefSeq | Description | |
|---|---|---|---|---|---|
| MMPs/TIMPs | 1 | Mmp2 | Mm.29564 | NM_008610 | Matrix metallopeptidase 2 |
| 2 | Mmp3 | Mm.4993 | NM_010809 | Matrix metallopeptidase 3 | |
| 3 | Mmp7 | Mm.4825 | NM_010810 | Matrix metallopeptidase 7 | |
| 4 | Mmp8 | Mm.16415 | NM_008611 | Matrix metallopeptidase 8 | |
| 5 | Mmp9 | Mm.4406 | NM_013599 | Matrix metallopeptidase 9 | |
| 6 | Mmp11 | Mm.4561 | NM_008606 | Matrix metallopeptidase 11 | |
| 7 | Mmp12 | Mm.2055 | NM_008605 | Matrix metallopeptidase 12 | |
| 8 | Mmp13 | Mm.5022 | NM_008607 | Matrix metallopeptidase 13 | |
| 9 | Mmp14 | Mm.280175 | NM_008608 | Matrix metallopeptidase 14, (MT1-MMP) | |
| 10 | Mmp15 | Mm.217116 | NM_008609 | Matrix metallopeptidase 15, (MT2-MMP) | |
| 11 | Timp1 | Mm.8245 | NM_011593 | Tissue inhibitor of metalloproteinase 1 | |
| 12 | Timp2 | Mm.206505 | NM_011594 | Tissue inhibitor of metalloproteinase 2 | |
| 13 | Timp3 | Mm.4871 | NM_011595 | Tissue inhibitor of metalloproteinase 3 | |
| 14 | Timp4 | Mm.255607 | NM_080639 | Tissue inhibitor of metalloproteinase 4 | |
| ECM Proteins | 15 | Col1a1 | Mm.277735 | NM_007742 | Procollagen, type I, alpha 1 |
| 16 | Col1a2 | Mm.277792 | NM_007743 | Procollagen, type I, alpha 2 | |
| 17 | Col3a1 | Mm.249555 | NM_009930 | Procollagen, type III, alpha 1 | |
| 18 | Col4a1 | Mm.738 | NM_009931 | Procollagen, type IV, alpha 1 | |
| 19 | Col6a1 | Mm.2509 | NM_009933 | Procollagen, type VI, alpha 1 | |
| 20 | Eln | Mm.275320 | NM_007925 | Elastin | |
| 21 | Ltbp1 | Mm.269747 | NM_019919 | Latent transforming growth factor-beta binding protein 1 | |
| 22 | Ltbp2 | Mm.3900 | NM_013589 | Latent transforming growth factor beta binding protein 2 | |
| 23 | Fbn1 | Mm.271644 | NM_007993 | Fibrillin 1 | |
| 24 | Lamb1-1 | Mm.172674 | NM_008482 | Laminin B1 subunit 1 | |
| 25 | Lamb2 | Mm.425599 | NM_008483 | Laminin, beta 2 | |
| 26 | Lamb3 | Mm.435441 | NM_008484 | Laminin, beta 3 | |
| 27 | Fn1 | Mm.193099 | NM_010233 | Fibronectin 1 | |
| 28 | Spp1 | Mm.288474 | NM_009263 | Secreted phosphoprotein 1, (Osteopontin) | |
| 29 | Thbs1 | Mm.4159 | NM_011580 | Thrombospondin 1 | |
| 30 | Sparc | Mm.291442 | NM_009242 | Secreted acidic cysteine rich glycoprotein, (SPARC) | |
| 31 | Ager | Mm.3383 | NM_007425 | Advanced glycosylation end product receptor, (RAGE) | |
| Transcription Factors | 32 | Sp1 | Mm.4618 | NM_013672 | Trans-acting transcription factor 1, (Sp1) |
| 33 | Sp3 | Mm.124328 | NM_011450 | Trans-acting transcription factor 3, (Sp3) | |
| 34 | Jun | Mm.275071 | NM_010591 | Jun oncogene, (Jun) | |
| 35 | Junb | Mm.1167 | NM_008416 | Jun-B oncogene, (Jun b) | |
| 36 | Fos | Mm.246513 | NM_010234 | FBJ osteosarcoma oncogene, (Fos) | |
| 37 | Fosb | Mm.248335 | NM_008036 | FBJ osteosarcoma oncogene B, (Fos b) | |
| 38 | Etv4 | Mm.5025 | NM_008815 | Ets variant gene 4 (E1A enhancer binding protein), (PEA-3) | |
| House-keeping Genes | 39 | Gusb | Mm.3317 | NM_010368 | Glucuronidase, beta |
| 40 | Hprt1 | Mm.299381 | NM_013556 | Hypoxanthine guanine phosphoribosyl transferase 1 | |
| 41 | Hspb1 | Mm.13849 | NM_013560 | Heat shock protein 1, (Hsp27) | |
| 42 | Gapdh | Mm.343110 | NM_008084 | Glyceraldehyde-3-phosphate dehydrogenase |
Data compiled using Rebhan et al.(27)
Gene expression in each stimulated cell line was determined in a similar manner. To more easily represent the expression differences between normal and TAA fibroblasts, genes were clustered into similar gene families, and total gene expression was depicted as an area profile on a radar plot. Each axis displays the sum of the relative fold change in gene expression for a given gene cluster in the TAA fibroblasts as compared to the control fibroblasts. The genes included in each cluster were: 1) MMP/TIMP genes (Mmp2, Mmp3, Mmp9, Mmp11, Mmp13, Mmp14, Mmp15, Timp1, Timp2, Timp3); 2) Collagen/Elastic Architecture genes (Col1a1, Col1a2, Col3a1, Col4a1, Col6a1, Eln, Ltbp1, Ltbp2, Fbn1); and 3) Other ECM genes (Lamb1-1, Lamb2, Lamb3, Fn1, Spp1, Thbs1, Sparc, Ager).
RESULTS
Relative gene expression analysis at steady-state
Primary aortic fibroblasts from normal and TAA-induced mice were analyzed to determine the relative expression of 38 genes and 4 housekeeping genes (Table 1) under steady-state culture conditions, using a single custom QPCR array plate per cell line. The TAA mice displayed a relative increase in Mmp2, Mmp11, and Mmp14 expression, and a relative decrease of Mmp3, Mmp9, and Timp3 expression (Table 2, Steady-State). Furthermore, the steady-state expression of several collagen genes and elastin was also elevated (Col1a1, Col1a2, Col1a3, Col4a1, and Eln), while the expression of Ltbp-1 was decreased (Table 2). Additionally, several other ECM proteins displayed elevated expression (Lamb2, Fn1, Spp1, and Sparc) (Table 2). Lastly, the relative expression of several transcription factors known to be involved in regulating matrix turnover was assessed. The TAA fibroblasts demonstrated increased steady-state expression of Fos and Fosb (Table 2).
Table 2.
Relative fold gene expression in TAA fibroblasts as compared to normal fibroblasts at steady-state and following stimulation with various growth factors at the indicated concentrations; ND = not detected.
| Fold Expression (TAA Fibroblasts vs. Normal Fibroblasts) | |||||
|---|---|---|---|---|---|
| Gene Symbol | Steady-State | ET-1 (1 nM) | PMA (100 nM) | AngII (100 nM) | |
| MMPs/TIMPs | Mmp2 | 6.54 (p = 0.0055) | 4.46 (p = 0.0239) | 3.65 (p = 0.0472) | 5.94 (p = 0.0171) |
| Mmp3 | 0.41 (p = 0.3622) | 0.26 (p = 0.2470) | 0.06 (p = 0.0137) | 0.22 (p = 0.2382) | |
| Mmp9 | 0.09 (p = 0.0488) | 0.04 (p = 0.0012) | 0.21 (p = 0.1109) | 0.09 (p = 0.1828) | |
| Mmp11 | 9.78 (p = 0.0027) | 2.78 (p = 0.2959) | 2.03 (p = 0.1184) | 7.26 (p = 0.0278) | |
| Mmp13 | 1.11 (p = 0.8461) | 0.97 (p = 0.9514) | 0.11 (p = 0.0280) | 0.42 (p = 0.7376) | |
| Mmp14 | 2.01 (p = 0.1014) | 2.13 (p = 0.0008) | 1.78 (p = 0.1450) | 1.87 (p = 0.1579) | |
| Mmp15 | 1.17 (p = 0.8590) | 0.66 (p = 0.3911) | ND | 0.23 (p = 0.0902) | |
| Timp1 | 1.41 (p = 0.2415) | 0.99 (p = 0.9766) | 1.53 (p = 0.1271) | 1.39 (p = 0.4637) | |
| Timp2 | 1.11 (p = 0.5787) | 0.95 (p = 0.8977) | 0.95 (p = 0.8377) | 1.27 (p = 0.3828) | |
| Timp3 | 0.33 (p = 0.1490) | 0.49 (p = 0.3012) | 0.43 (p = 0.0799) | 0.30 (p = 0.0896) | |
| Collagen/Elastic Architecture | Col1a1 | 2.52 (p = 0.0583) | 1.83 (p = 0.0793) | 2.99 (p = 0.0015) | 1.84 (p = 0.2291) |
| Col1a2 | 3.45 (p = 0.0082) | 2.90 (p = 0.0007) | 3.61 (p = 0.0047) | 3.46 (p = 0.0049) | |
| Col3a1 | 4.83 (p = 0.0083) | 3.51 (p = 0.0102) | 4.52 (p = 0.0120) | 4.05 (p = 0.0109) | |
| Col4a1 | 2.82 (p = 0.0039) | 2.36 (p = 0.0284) | 2.22 (p = 0.3715) | 1.94 (p = 0.4345) | |
| Col6a1 | 1.67 (p = 0.2485) | 1.47 (p = 0.4708) | 1.52 (p = 0.5061) | 1.41 (p = 0.4827) | |
| Eln | 2.30 (p = 0.2558) | 2.32 (p = 0.2918) | 0.79 (p = 0.7256) | 2.24 (p = 0.4751) | |
| Ltbp-1 | 0.09 (p = 0.0564) | 0.21 (p = 0.0323) | 0.04 (p = 0.0033) | 0.19 (p = 0.0955) | |
| Ltbp-2 | 1.71 (p = 0.2016) | 1.39 (p = 0.4174) | 1.44 (p = 0.3357) | 1.63 (p = 0.3735) | |
| Fbn1 | 1.83 (p = 0.2042) | 2.13 (p = 0.1269) | 2.33 (p = 0.1862) | 2.55 (p = 0.1643) | |
| Other ECM | Lamb1-1 | 1.66 (p = 0.1437) | 1.61 (p = 0.3753) | 0.99 (p = 0.9830) | 2.02 (p = 0.1812) |
| Lamb2 | 21.96 (p = 0.0378) | 1.36 (p = 0.7058) | 0.05 (p = 0.0194) | 0.56 (p = 0.3979) | |
| Fn1 | 2.23 (p = 0.1582) | 1.96 (p = 0.1730) | 1.56 (p = 0.3862) | 1.83 (p = 0.4078) | |
| Spp1 | 2.43 (p = 0.2790) | 3.15 (p = 0.3129) | 0.17 (p = 0.6975) | 1.15 (p = 0.9537) | |
| Thbs1 | 0.88 (p = 0.5526) | 0.59 (p = 0.0240) | 1.39 (p = 0.2732) | 0.70 (p = 0.3291) | |
| Sparc | 2.10 (p = 0.1208) | 2.25 (p = 0.0624) | 1.40 (p = 0.5110) | 0.90 (p = 0.8262) | |
| Ager | 0.72 (p = 0.4922) | 0.69 (p = 0.5648) | 0.73 (p = 0.5835) | 0.90 (p = 0.8769) | |
| Transcription Factors | Sp1 | 1.68 (p = 0.1245) | 1.21 (p = 0.5112) | 1.22 (p = 0.5883) | 0.97 (p = 0.8670) |
| Sp3 | 0.98 (p = 0.9197) | 1.13 (p = 0.7552) | 1.03 (p = 0.9300) | 0.94 (p = 0.8385) | |
| Jun | 1.49 (p = 0.6169) | 3.71 (p = 0.0106) | 2.63 (p = 0.0460) | 2.28 (p = 0.0174) | |
| Junb | 0.82 (p = 0.7838) | 1.28 (p = 0.7831) | 1.46 (p = 0.4151) | ND | |
| Fos | 2.14 (p = 0.7734) | 1.71 (p = 0.4049) | 0.46 (p = 0.4656) | 0.97 (p = 0.9632) | |
| Fosb | 2.41 (p = 0.3959) | 3.40 (p = 0.2602) | 5.21 (p = 0.1182) | 1.67 (p = 0.6711) | |
| Etv4 | 0.91 (p = 0.7819) | 1.14 (p = 0.7663) | 0.44 (p = 0.2232) | 0.82 (p = 0.6757) | |
Relative gene expression analysis following stimulation with ET-1, PMA, and AngII
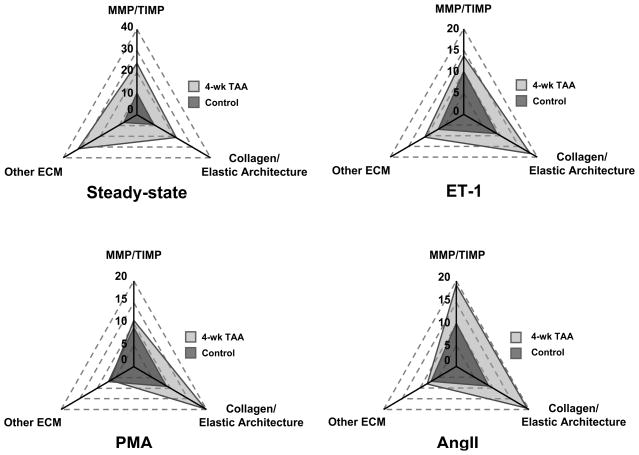
In order to determine whether normal and TAA fibroblasts respond in similar fashion to equivalent biological stimuli (ET-1, PMA, AngII), both sets of fibroblasts were stimulated for 24 hrs, and the relative changes in gene expression were examined. In almost all cases, the TAA fibroblasts responded more robustly than the normal fibroblasts to biological stimulus (Table 2, ET-1, PMA, AngII). In order to more easily demonstrate the relative changes in gene expression, the results in Table 2 were further analyzed by cluster analysis in the following functional groups; MMP/TIMP, Collagen/Elastic Architecture, and Other ECM. Figure 1 reveals the relative fold expression results for normal and TAA fibroblasts showing that the gene expression profiles for the TAA fibroblasts were different from normal fibroblasts at steady-state and following exposure to equivalent biological stimuli.
Figure 1. Analysis of gene expression following cellular stimulation.
Relative gene expression in normal and TAA fibroblast cell lines was analyzed in the absence (steady-state) or presence of biological stimuli. Cells were treated with either 1 nM ET-1, 100 nM AngII, or 100 nM PMA for 24 hrs, and then analyzed by QPCR. Relative fold expression results were clustered in functional groups (MMP/TIMP, Collagen/Elastin, Other ECM), and total gene expression was depicted as an area profile on a radar plot. Each axis displays the sum of the relative fold change in gene expression for a given gene cluster in the TAA fibroblasts as compared to the control fibroblasts.
DISCUSSION
Previous clinical and experimental aneurysm studies have demonstrated significant changes in aortic structure and composition in the developing TAA. These changes include disruption of the medial elastic lamellae,(12, 13) alterations in collagen deposition,(12–14) changes in cellular content characterized by the loss of smooth muscle cells,(7, 8) and alterations in aortic transcriptional profiles.(6, 15, 16) Additionally, previous studies from this laboratory have identified fibroblasts/fibroblast-derived cells as a potential source for enhanced MMP transcriptional activity during TAA development in mice.(6, 9) Taken together, this led us to hypothesize that aortic fibroblasts are an important cellular component within the aortic wall, which undergo a stable phenotypic change during TAA development, allowing them to adapt to the rapidly degrading conditions and mediate the vascular remodeling process. To test this hypothesis, primary thoracic aortic fibroblasts were isolated from normal and TAA-induced mice, and transcriptional profiles of key determinants of matrix degradation and deposition were assessed in the absence and presence of relevant biological stimuli. The unique outcomes of this study are three-fold. First, the present study demonstrated significant transcriptional changes in TAA fibroblasts as compared to normal aortic fibroblasts. These results are consistent with expectations of enhanced degradation and remodeling of the vascular ECM during TAA development which may ensue, in part, due to altered fibroblast function. Second, when isolated fibroblasts were challenged by treatment with relevant biological stimuli (ET-1, AngII, PMA), differential transcriptional responses were observed between normal and TAA fibroblasts, suggesting that intracellular signaling pathways in the TAA fibroblasts may be altered, and thus may respond differently to equivalent stimuli. Last, the observed gene expression differences between normal and TAA fibroblasts were evident in multiple cell lines following the several cell passages required to establish the in vitro cultures. Using an established fibroblast outgrowth procedure,(10) multiple primary cell lines from both normal and TAA-induced animals were established in vitro. The highly reproducible transcriptional differences between the TAA and normal fibroblasts are therefore not a byproduct of selection during in vitro establishment or as a direct result of cell passaging, but due to a stable alteration in transcriptional activity induced during the process of aneurysm development in vivo. Taken together, the results of this study support the hypothesis that aortic fibroblasts undergo a stable phenotypic transformation during TAA development, and that these changes in gene expression may alter normal fibroblast function and contribute to TAA formation and progression.
Steady-state changes gene expression
To assess transcriptional differences between normal and TAA fibroblasts, quantitative real-time PCR (QPCR) was performed using a custom designed PCR array to assess the gene expression of several critical determinants of matrix degradation and deposition, known to be involved in TAA development. Previous studies from this laboratory have examined aortic tissue from normal and TAA mice, and have demonstrated TAA-dependent changes in gene transcription and protein abundance of MMPs and TGF-β pathway components.(6, 15) Accordingly, it was hypothesized that if the aortic fibroblast plays a dominant role in TAA development, isolated aortic fibroblasts from TAA tissue would likewise display an altered transcriptional profile as compared to normal aortic fibroblasts. Indeed, our results were consistent with that hypothesis. The TAA fibroblasts displayed a distinct pattern of gene expression reflecting enhanced expression of several MMPs (Mmp2, Mmp11, and Mmp14), matrix proteins (Col1a1, Col1a2, Col3a1, Col4a1, Eln, Lamb2, Fn1, Spp1, and Sparc), and transcription factors (Fos and Fosb), along with the decreased expression of Mmp3, Mmp9, Timp3, and Ltbp1. The observed elevated expression of MMP genes was highly consistent with previous studies from this laboratory demonstrating a direct role for increased MMP production during TAA development in this model.(6, 17) Thus, the stable elevation of MMP transcription in TAA fibroblasts argues that this differentiated cell-type plays a critical role in degrading the ECM during TAA formation and progression.
While many studies analyzing clinical TAA specimens have demonstrated elevated collagenase activity (MMP-dependent),(1, 5, 18, 19) few have examined aortic collagen content or expression directly.(14, 20) The present study demonstrates a robust elevation of collagen gene expression in the TAA fibroblasts. While counterintuitive to the enhanced ECM degradation that takes place during TAA development, others have described elevated collagen expression and regional deposition in TAA tissue. For example, Iliopoulos and coworkers have suggested that collagen content, while diminished in the aortic media, is elevated in the adventitial region of the aorta, resulting in no net gain or loss of total collagen content.(13) Similarly, Della Corte et al. have demonstrated regional differences in collagen content in the greater versus lesser curvature of the ascending aorta from aneurysmal patients with bicuspid aortic valves.(12) Collectively, these studies suggest that the enhancement of collagen expression may coincide with TAA development and may result in the regional deposition of newly formed collagen fibers. In a recent report from this laboratory, medial and adventitial collagen content was measured over the 16-wk time-course of TAA-development in this model.(9) No net change was observed in either the medial or adventitial compartment. While this may suggest a lack of collagen degradation, given the present results demonstrating elevated MMP and collagen gene expression in the isolated TAA fibroblasts, it is more likely that collagen degradation and deposition are balanced in regard to the total content. This is further supported by the observation that Sparc (secreted protein acidic and rich in cysteine) expression is elevated in the TAA fibroblasts, suggesting that these cells possess a synthetic phenotype, primed to deposit newly processed collagen fibers. The organizational structure of the collagen matrix on the other hand, may be significantly remodeled during TAA development, as has previously been demonstrated in the post-infarct myocardium.(21) Of interest, the present study also identified decreased expression of Ltbp-1 (latent transforming growth factor-β binding protein-1); an ECM structural protein that serves to bind and sequester the small latent complex of transforming growth factor-beta (TGF-β). TGF-β is best known for its ability to induce collagen gene expression, but may also play a role in exacerbating vascular remodeling during TAA development.(22) Accordingly, it follows that a decrease in Ltbp-1 expression may lead to an increase in the extracellular levels of TGF-β, and enhancement of collagen gene expression. Moreover, elevated TGF-β signaling has also been associated with the induction of MMP expression and abundance.(23, 24)
To further demonstrate the stability of the observed phenotypic change between isolated normal and TAA fibroblasts, the expression levels of several transcription factors implicated in regulating MMP-dependent transcription were assessed. Interestingly, Fos and Fosb, family members and components of the AP-1 transcription factor, were both stably induced and may play a direct role in the observed change in transcriptional profile of the TAA fibroblasts.
Stimulated changes in gene expression
In an effort to further define the phenotypic change of the TAA fibroblasts, gene expression profiles were also determined following treatment with relevant biological stimuli (ET-1,(25) AngII,(26) PMA). When comparing relative gene expression between treated normal and TAA fibroblasts within gene family clusters, clear differences in overall transcriptional profiles were observed following treatment with each agent. While it was anticipated that the MMP genes in the TAA fibroblasts would be robustly induced in response to some stimuli (e.g. ET-1, PMA), the blunted response observed may be a direct result of altered downstream signaling pathways. Given that these genes were already highly expressed in the TAA fibroblast, this may further suggest that the intracellular signaling pathways have become uncoupled from receptor activation in these cells. These results further substantiate the hypothesis that the TAA fibroblasts have undergone a stable phenotypic change, and may suggest that the alterations in transcriptional response are a direct result of reconfigured intracellular signaling pathways in the TAA fibroblasts.
Limitations
Because the present study examines gene expression profiles in cultured primary aortic cell lines several limitations must be noted. First, care must be taken in extrapolating changes in gene expression with altered protein levels. While increased or decreased gene expression often equates to coordinate changes protein translation, transcriptional and translation regulation must be considered. Second, while this murine model of TAA has been well described and recapitulates many of the hallmarks of human aneurysmal disease, some aspects such as atherosclerosis and intraluminal thrombosis, are not replicated at the time-points studied. Accordingly, care should be taken in the extrapolation of these results to human TAAs. Lastly, while a significant effect of gender on aortic dilatation has not previously been demonstrated in this model system, gender dependent differences in aortic gene expression cannot be ruled out. Additional studies will be required to ascertain whether a significant interaction between gender and gene expression exist.
Conclusions
Despite limitations, the present study demonstrated for the first time that isolated primary aortic fibroblasts from TAA mice possess a stable and unique phenotype defined by altered gene expression profiles at steady-state and in response to biological stimuli. This distinctive cellular population may emerge as the major cellular mediator of vascular remodeling during TAA development and may drive the enhancement of ECM proteolysis in TAA progression.
Acknowledgments
This work was supported by the NIH/NHLBI R01 HL075488 (to J.S.I.), and R01 HL81692 (to F.G.S.) and by the Department of Veterans Affairs through a Career Development Award to J.A.J. (BLRD-CDA-2) and a Merit Award to F.G.S..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barbour JR, Spinale FG, Ikonomidis JS. Proteinase systems and thoracic aortic aneurysm progression. J Surg Res. 2007 May 15;139(2):292–307. doi: 10.1016/j.jss.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 2.Dapunt OE, Galla JD, Sadeghi AM, Lansman SL, Mezrow CK, de Asla RA, et al. The natural history of thoracic aortic aneurysms. J Thorac Cardiovasc Surg. 1994 May;107(5):1323–32. discussion 32–3. [PubMed] [Google Scholar]
- 3.Davies RR, Gallo A, Coady MA, Tellides G, Botta DM, Burke B, et al. Novel measurement of relative aortic size predicts rupture of thoracic aortic aneurysms. Ann Thorac Surg. 2006 Jan;81(1):169–77. doi: 10.1016/j.athoracsur.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 4.Freestone T, Turner RJ, Coady A, Higman DJ, Greenhalgh RM, Powell JT. Inflammation and matrix metalloproteinases in the enlarging abdominal aortic aneurysm. Arterioscler Thromb Vasc Biol. 1995 Aug;15(8):1145–51. doi: 10.1161/01.atv.15.8.1145. [DOI] [PubMed] [Google Scholar]
- 5.LeMaire SA, Wang X, Wilks JA, Carter SA, Wen S, Won T, et al. Matrix metalloproteinases in ascending aortic aneurysms: bicuspid versus trileaflet aortic valves. J Surg Res. 2005 Jan;123(1):40–8. doi: 10.1016/j.jss.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 6.Jones JA, Barbour JR, Lowry AS, Bouges S, Beck C, McClister DM, Jr, et al. Spatiotemporal expression and localization of matrix metalloproteinase-9 in a murine model of thoracic aortic aneurysm. J Vasc Surg. 2006 Dec;44(6):1314–21. doi: 10.1016/j.jvs.2006.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Della Corte A, Quarto C, Bancone C, Castaldo C, Di Meglio F, Nurzynska D, et al. Spatiotemporal patterns of smooth muscle cell changes in ascending aortic dilatation with bicuspid and tricuspid aortic valve stenosis: focus on cell-matrix signaling. J Thorac Cardiovasc Surg. 2008 Jan;135(1):8–18. e1–2. doi: 10.1016/j.jtcvs.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Ihling C, Szombathy T, Nampoothiri K, Haendeler J, Beyersdorf F, Uhl M, et al. Cystic medial degeneration of the aorta is associated with p53 accumulation, Bax upregulation, apoptotic cell death, and cell proliferation. Heart. 1999 Sep;82(3):286–93. doi: 10.1136/hrt.82.3.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones JA, Beck C, Barbour JR, Zavadzkas JA, Mukherjee R, Spinale FG, et al. Alterations in Aortic Cellular Constituents during Thoracic Aortic Aneurysm Development: Myofibroblast-Mediated Vascular Remodeling. Am J Pathol. 2009 Sep 3;175(4):1746–56. doi: 10.2353/ajpath.2009.081141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindsey ML, Goshorn DK, Squires CE, Escobar GP, Hendrick JW, Mingoia JT, et al. Age-dependent changes in myocardial matrix metalloproteinase/tissue inhibitor of metalloproteinase profiles and fibroblast function. Cardiovasc Res. 2005 May 1;66(2):410–9. doi: 10.1016/j.cardiores.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 11.Ikonomidis JS, Gibson WC, Gardner J, Sweterlitsch S, Thompson RP, Mukherjee R, et al. A murine model of thoracic aortic aneurysms. J Surg Res. 2003 Nov;115(1):157–63. doi: 10.1016/s0022-4804(03)00193-8. [DOI] [PubMed] [Google Scholar]
- 12.Della Corte A, De Santo LS, Montagnani S, Quarto C, Romano G, Amarelli C, et al. Spatial patterns of matrix protein expression in dilated ascending aorta with aortic regurgitation: congenital bicuspid valve versus Marfan’s syndrome. J Heart Valve Dis. 2006 Jan;15(1):20–7. discussion 7. [PubMed] [Google Scholar]
- 13.Iliopoulos DC, Kritharis EP, Giagini AT, Papadodima SA, Sokolis DP. Ascending thoracic aortic aneurysms are associated with compositional remodeling and vessel stiffening but not weakening in age-matched subjects. J Thorac Cardiovasc Surg. 2009 Jan;137(1):101–9. doi: 10.1016/j.jtcvs.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 14.Maleszewski JJ, Miller DV, Lu J, Dietz HC, Halushka MK. Histopathologic findings in ascending aortas from individuals with Loeys-Dietz syndrome (LDS) Am J Surg Pathol. 2009 Feb;33(2):194–201. doi: 10.1097/PAS.0b013e31817f3661. [DOI] [PubMed] [Google Scholar]
- 15.Jones JA, Barbour JR, Stroud RE, Bouges S, Stephens SL, Spinale FG, et al. Altered Transforming Growth Factor-Beta Signaling in a Murine Model of Thoracic Aortic Aneurysm. J Vasc Res. 2008 Apr 23;45(6):457–68. doi: 10.1159/000127437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taketani T, Imai Y, Morota T, Maemura K, Morita H, Hayashi D, et al. Altered patterns of gene expression specific to thoracic aortic aneurysms: microarray analysis of surgically resected specimens. Int Heart J. 2005 Mar;46(2):265–77. doi: 10.1536/ihj.46.265. [DOI] [PubMed] [Google Scholar]
- 17.Barbour JR, Stroud RE, Lowry AS, Clark LL, Leone AM, Jones JA, et al. Temporal disparity in the induction of matrix metalloproteinases and tissue inhibitors of metalloproteinases after thoracic aortic aneurysm formation. J Thorac Cardiovasc Surg. 2006 Oct;132(4):788–95. doi: 10.1016/j.jtcvs.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 18.Ikonomidis JS, Jones JA, Barbour JR, Stroud RE, Clark LL, Kaplan BS, et al. Expression of matrix metalloproteinases and endogenous inhibitors within ascending aortic aneurysms of patients with Marfan syndrome. Circulation. 2006 Jul 4;114(1 Suppl):I365–70. doi: 10.1161/CIRCULATIONAHA.105.000810. [DOI] [PubMed] [Google Scholar]
- 19.Ikonomidis JS, Jones JA, Barbour JR, Stroud RE, Clark LL, Kaplan BS, et al. Expression of matrix metalloproteinases and endogenous inhibitors within ascending aortic aneurysms of patients with bicuspid or tricuspid aortic valves. J Thorac Cardiovasc Surg. 2007 Apr;133(4):1028–36. doi: 10.1016/j.jtcvs.2006.10.083. [DOI] [PubMed] [Google Scholar]
- 20.de Figueiredo Borges L, Jaldin RG, Dias RR, Stolf NA, Michel JB, Gutierrez PS. Collagen is reduced and disrupted in human aneurysms and dissections of ascending aorta. Human pathology. 2008 Mar;39(3):437–43. doi: 10.1016/j.humpath.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Spinale FG. Myocardial matrix remodeling and the matrix metalloproteinases: influence on cardiac form and function. Physiol Rev. 2007 Oct;87(4):1285–342. doi: 10.1152/physrev.00012.2007. [DOI] [PubMed] [Google Scholar]
- 22.Jones JA, Spinale FG, Ikonomidis JS. Transforming growth factor-beta signaling in thoracic aortic aneurysm development: a paradox in pathogenesis. J Vasc Res. 2009;46(2):119–37. doi: 10.1159/000151766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim ES, Kim MS, Moon A. TGF-beta-induced upregulation of MMP-2 and MMP-9 depends on p38 MAPK, but not ERK signaling in MCF10A human breast epithelial cells. Int J Oncol. 2004 Nov;25(5):1375–82. [PubMed] [Google Scholar]
- 24.Safina A, Vandette E, Bakin AV. ALK5 promotes tumor angiogenesis by upregulating matrix metalloproteinase-9 in tumor cells. Oncogene. 2007 Apr 12;26(17):2407–22. doi: 10.1038/sj.onc.1210046. [DOI] [PubMed] [Google Scholar]
- 25.Wagner A, Domanovits H, Holzer M, Kofler J, Roggla M, Mullner M, et al. Plasma endothelin in patients with acute aortic disease. Resuscitation. 2002 Apr;53(1):71–6. doi: 10.1016/s0300-9572(01)00502-0. [DOI] [PubMed] [Google Scholar]
- 26.Daugherty A, Rateri DL, Cassis LA. Role of the renin-angiotensin system in the development of abdominal aortic aneurysms in animals and humans. Ann N Y Acad Sci. 2006 Nov;1085:82–91. doi: 10.1196/annals.1383.035. [DOI] [PubMed] [Google Scholar]
- 27.Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D. GeneCards: integrating information about genes, proteins and diseases. Trends Genet. 1997 Apr;13(4):163. doi: 10.1016/s0168-9525(97)01103-7. [DOI] [PubMed] [Google Scholar]