Abstract
Mouse mammary tumor virus (MMTV) is a complex murine retrovirus that encodes an HIV Rev-like export protein, Rem, from a doubly spliced version of envelope (Env) mRNA. Previously, the N-terminal 98-amino acid sequence of Rem, which is identical to Env signal peptide (SP), and full-length Rem were shown to be functional in a reporter assay that measures a postexport function. Here we show that MMTV-infected cells or cells transfected with rem or env cDNAs express SP, which is the active component in the reporter assay. Uncleaved Rem was partially glycosylated, but mutations in both glycosylation sites within the C terminus prevented Rem function. Mutations that reduced Rem or Env cleavage by signal peptidase greatly reduced SP levels and functional activity in the reporter assay and allowed accumulation of the uncleaved protein. Fluorescence microscopy revealed that GFP-tagged cleavage-site mutants are unstable and lack fluorescence compared with wild-type Rem, suggesting improper folding. Proteasome inhibitors allowed accumulation of uncleaved Rem relative to SP and increased reporter activity, consistent with SP retrotranslocation and proteasome escape before nuclear entry. Expression of a dominant-negative p97 ATPase did not alter levels of unprocessed Rem and SP but decreased reporter activity, suggesting p97-facilitated retrotranslocation of SP. Our results provide an example of a SP that is processed by signal peptidase and retrotranslocated to allow nuclear localization and function.
Keywords: mouse mammary tumor virus, retrotranslocation, retrovirus, signal peptide, ERAD
Mouse mammary tumor virus (MMTV) has provided many insights into cell and cancer biology. MMTV is a complex mouse retrovirus with regulatory features similar to those found in human complex retroviruses, such as HIV and human T-cell leukemia virus (HTLV) (1). These features include the ability to manipulate the immune system and to produce a doubly spliced mRNA encoding a protein (Rem) functionally similar to HIV Rev (1, 2). Rem mRNA is translated in the same reading frame as the MMTV envelope (env) gene but has a deletion of internal sequences encoding the surface (SU) and transmembrane (TM) proteins (1, 2) (Fig. 1A).
Fig. 1.
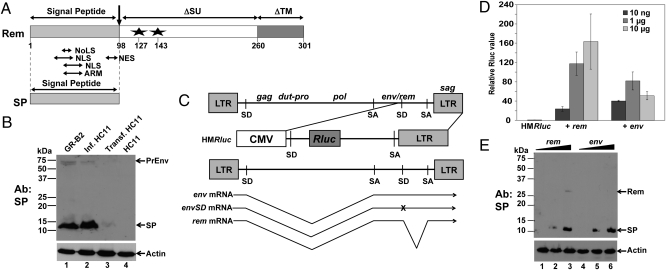
MMTV mRNAs expressing either Rem or Env generate functional SP. (A) Domain structure of Rem and SP. The Rem C terminus contains an in-frame fusion of deleted versions of the Env protein (SU and TM proteins). Stars indicate glycosylation sites. ARM, arginine-rich RNA-binding motif; NES, nuclear export sequence; NoLS, nucleolar localization sequence. (B) MMTV-infected and transfected mammary cells express SP. GR-B2 mammary tumor cell extracts (lane 1) were compared with extracts from HC11 cells infected with MMTV (lane 2) or stably transfected with HYB-MTV (lane 3) by Western blotting with SP-specific antibody. Uninfected HC11 cell extracts are in lane 4. PrEnv, envelope precursor. (C) Structure of the pHMRluc reporter vector relative to the MMTV env and rem mRNAs. The reporter vector is derived from the 3′ end of the C3H-MMTV genome inserted downstream of the CMV promoter. SA, splice acceptor; SD, splice donor; x, mutation of the env splice donor. (D) Dose-dependent activity of the rem and env mRNAs on the pHMRluc vector. HC11 cells were transiently transfected with the indicated amount of expression plasmid. The averages of triplicate assays ± SDs are shown after normalization. Assays using the pHMRluc vector alone (in the absence of Rem or Env) were assigned a relative value of 1. (E) Western blotting of rem- and env-transfected HC11 cells. Western blots of extracts from transfected cells were incubated with SP-specific antibody. SP was not detected in cells transfected with 10 ng of the rem and env expression vectors (lanes 1 and 4).
MMTV Rem is a 301-amino acid protein that regulates expression and export of full-length viral RNA from the nucleus to the cytoplasm using the Crm1 export pathway (1, 2). The Rem protein contains several Rev-like motifs, including a nuclear localization signal (NLS), a nucleolar localization sequence, an arginine-rich RNA-binding motif, and a leucine-rich nuclear export sequence (Fig. 1A). These motifs map to the N-terminal 98 amino acids, which are identical in sequence to the signal peptide (SP) of the MMTV Env protein (1, 2). Similar to HIV Rev or HTLV-1 Rex (3, 4), GFP fusions to Rem or the SP are localized to nucleoli. Both GFPRem and GFP-SP fusions are functional in a reporter assay that measures a postexport function (e.g., translation) (5). Rem-dependent reporter activity requires the NLS as well as the presence of a Rem-responsive element (RmRE) in the reporter construct (1, 6). Together, these data suggest that SP binds to MMTV RNA in the nucleolus for Rem-dependent reporter activity.
Transient transfections with an expression construct encoding a C-terminally Myc-tagged Rem allowed detection of three proteins: a glycosylated protein of 38 kDa, which was observed with antibodies to both SP and Myc, an N-terminal SP of 14 kDa, and a C-terminal Myc-tagged glycosylated protein of 32 kDa (7). Both the Myc- and SP-specific epitopes were detected in the nucleoli. These experiments suggested a unique trafficking mechanism in which the SP directs Rem translation to the endoplasmic reticulum (ER) before signal peptidase cleavage and SP retrotranslocation to the cytoplasm (7). Retrotranslocation is a cellular process that is initiated when proteins carrying a signal peptide in the ER lumen or membrane are recognized as misfolded or defective (8). These proteins then are redirected to the cytoplasm using the ATPases associated with diverse cellular activities (AAA ATPase) p97/valosin-containing protein (VCP) and 19S proteasome cap before degradation by the 20S proteasome (ER-associated degradation, ERAD) (9). Many SPs associated with viral and cellular proteins are cleaved by signal peptidase and then processed by the intramembrane SP peptidase before proteasome-mediated degradation (10).
In the current work, we have shown that functional SP is expressed from either rem or env mRNAs and that SP is the dominant Rem-derived peptide found in MMTV-infected cells. Mutation of both Rem glycosylation sites abolished SP function. Further, mutation of consensus signal peptidase cleavage sites in either env or rem prevented SP formation and activity. A dominant-negative (DN) p97 mutant had no effect on Rem cleavage but inhibited SP function. Together, these results suggest that MMTV SP has a trafficking mechanism that requires signal peptidase cleavage of Rem or Env before retrotranslocation and nuclear entry.
Results
SP Is Generated from Either rem or env cDNA.
Previous data indicated that generation of full-length Rem or SP might be dependent on the cell line used for expression (2, 11, 12). Whole-cell extracts from mouse mammary tumor cells (GR-B2) expressing GR-strain MMTV were subjected to Western blotting with antibody prepared against the NLS/RNA-binding domain within the SP. Results showed that only SP, not full-length Rem, was detectable (Fig. 1B, lane 1). Similar results were observed in HC11 cells infected with C3H-MMTV (lane 2). A small amount of SP was detected in HC11 cells stably transfected with an infectious MMTV provirus (13) (lane 3) but not uninfected cells (lane 4), probably because of lower proviral copies. Previous results of rem cDNA transfections into human T lymphoma (Jurkat) cells or HC11 mouse cells yielded two bands of ≈38 and 14 kDa (6), consistent with both full-length Rem and SP (Fig. 1A). As previously reported (7), the full-length product of the doubly spliced rem mRNA is processed to SP.
Jaagsiekte sheep retrovirus (JSRV) appears to express a functional SP from the singly spliced env mRNA rather than a doubly spliced mRNA (14, 15). Therefore, we compared levels of MMTV SP produced from rem and env expression constructs. HC11 mouse mammary cells were transiently transfected with constructs expressing env or rem cDNA as well as the reporter plasmid pHMRluc (Fig. 1C). The pHMRluc construct has the cytomegalovirus (CMV) promoter upstream of the 3′ end of the MMTV genome with a Renilla luciferase gene between the splice donor and acceptor. Because pHMRluc contains the RmRE but lacks the env/rem region encoding SP, luciferase activity is responsive to exogenous Rem expression (1). Transfection of either rem or env expression plasmids showed dose-dependent luciferase activity as compared with empty expression vector at lower DNA levels (Fig. 1D). In agreement with this finding, equal amounts of SP were detected from rem and env cDNA (Fig. 1E). Therefore, like JSRV (14, 15), functional SP is produced from the env mRNA.
Because the singly spliced env mRNA could be spliced further to yield doubly spliced rem mRNA (Fig. 1C), we mutated the second splice donor site in the envelope region to eliminate additional splicing to this site. Transfection of this construct revealed no differences in SP levels or reporter activity in Jurkat human T cells (Fig. S1) or in 293T cells at several different plasmid concentrations. Because SP alone is functional in the luciferase assay (6), these data indicate that both rem and env mRNAs generate functional SP.
Mutation of Glycosylation Sites in the Rem C Terminus Affects SP Function.
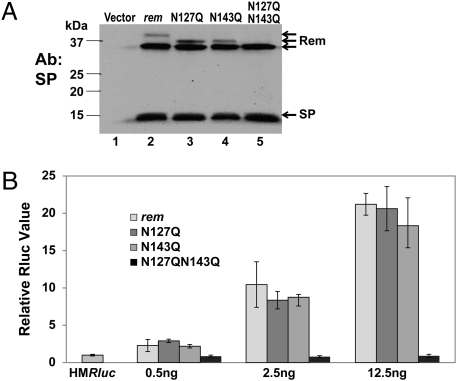
Rem appears to be glycosylated on the C terminus (7), a region shared with SU protein (1, 2). To determine if glycosylation affects Rem cleavage or processing, we mutated one or both consensus glycosylation sites (Fig. 1A). The wild-type rem and mutant constructs were transiently cotransfected into 293T cells with pHMRluc and, after 48 h, extracts were tested for glycosylation. Under these conditions, Rem was partially glycosylated, as indicated by the slower mobility band compared with unglycosylated Rem (Fig. 2A, lane 2). Mutation of asparagine at position 127 or 143 (N127Q or N143Q, respectively) resulted in a decreased mass (ca. 2.5 kDa) for glycosylated Rem. Furthermore, mutations at both positions eliminated detectable Rem glycosylation (compare lanes 2 and 5). No effect was observed on SP cleavage. To test the effect of these mutations on Rem function, transfections were repeated using lower amounts of the expression vectors to obtain dose-dependent reporter activity. Wild-type rem expression resulted in up to a 20-fold increase in luciferase levels, and mutation of individual glycosylation sites gave similar increases in activity (Fig. 2B). However, the double mutation (N127Q/N143Q) eliminated detectable Rem-mediated elevation of reporter activity. Thus, at least one glycosylation site in Rem is essential for function at lower protein levels.
Fig. 2.
Glycosylation of the Rem C terminus is required for Rem function but not for cleavage. (A) Western blotting of 293T cells transfected with 250 ng of pHMRluc, 250 ng of pGL3, and 5.5 μg of wild-type rem or glycosylation-site mutants as indicated. The three forms of Rem detected with SP-specific antibody (upper arrows) include Rem glycosylated at positions 127 and 143 (lane 2; 38-kDa band), at amino acid 143 or 127 only (lanes 3 and 4), or at neither position (lane 5). (B) Rem function requires at least one functional glycosylation site. Transient transfections of 293T cells were performed using the indicated amount of wild-type rem or glycosylation-site mutant constructs. Luciferase assays are reported as in Fig. 1D.
Mutation of the Rem Signal Peptidase Cleavage Site Blocks Rem Processing and Function.
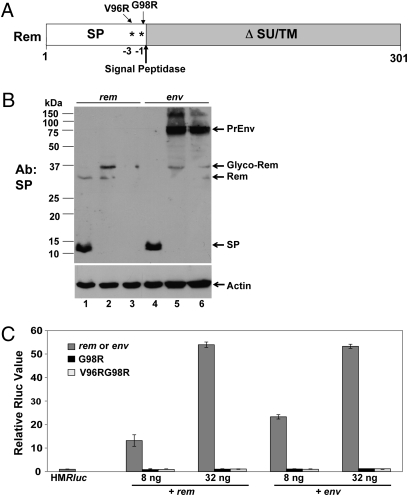
Previous results from in vitro translation suggested that mutation of the consensus site for signal peptidase cleavage prevents Rem processing (7). To determine if alteration of the signal peptidase cleavage site blocks Rem cleavage and activity in whole cells, we prepared rem expression constructs with mutations at position −1 (G98R) or both positions −1 and −3 (V96RG98R) relative to the predicted cleavage site (Fig. 3A). The mutant and wild-type constructs were transiently transfected into HC11 cells before Western blotting with SP-specific antibody. As expected, the wild-type rem construct allowed SP expression (Fig. 3B, lane 1), whereas each of the mutants (lanes 2 and 3) showed lower expression than the wild-type proteins. More strikingly, no cleavage product was detected with the G98R mutant. Instead, bands of ca. 37 and 33 kDa were observed, consistent with uncleaved glycosylated and unglycosylated Rem, respectively (Fig. 3B, lane 2). The rem construct expressing the double mutation was not detectable at this DNA concentration (lane 3), but lack of cleavage was documented with higher amounts of transfected DNA (Fig. 4C). In addition, no cleavage product was detected using either the single G98R or double V96RG98R mutation in the env expression construct, although both the glycosylated and unglycosylated Env precursors were present (Fig. 3B, lanes 5 and 6). Therefore, signal peptidase cleavage-site mutations in both rem and env genes prevented SP generation.
Fig. 3.
Signal peptidase cleavage is required for Rem function. (A) Rem diagram showing signal peptidase cleavage-site mutations. (B) Western blotting with SP-specific antibody reveals that cleavage-site mutations in either the rem or env expression vectors prevent SP generation. Constructs were transiently transfected in HC11 cells. Wild-type, G98R, or V96RG98R proteins are in lanes 1 and 4, lanes 2 and 5, and lanes 3 and 6, respectively. In the upper panel, the positions of the Env precursor (PrEnv), glycosylated Rem, unglycosylated Rem, and SP are shown. Rem detected in lanes 5 and 6 results from additional splicing of env mRNA. The lower panel shows Western blotting of the same extracts using actin-specific antibody. (C) Mutation of the signal peptidase cleavage site in either the rem or env expression construct prevents SP function. Transient transfections in HC11 cells were performed with concentrations of each expression plasmid as indicated. Luciferase assays are reported as in Fig. 1D.
Fig. 4.
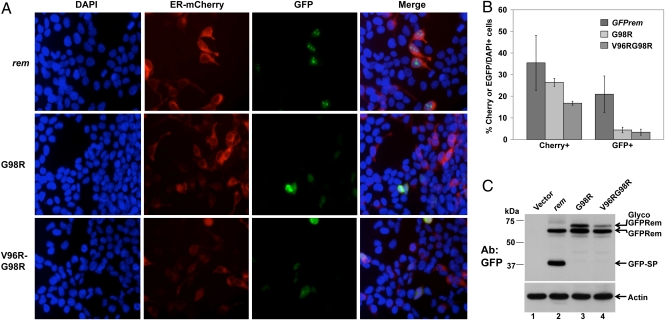
GFP-tagged rem constructs defective for signal peptidase cleavage lack fluorescence. (A) Single and double cleavage-site mutants lack fluorescence in 293T cells. Cells were transiently transfected with 0.25 μg of wild-type rem or 6 μg of mutant constructs together with 0.2 μg of the expression construct ER-mCherry. Cells were stained with DAPI and visualized by fluorescence microscopy. (B) Quantitation of fluorescence by wild-type EGFP-tagged Rem compared with cleavage-site mutants. The percentage of mCherry+ and GFP+ cells compared with the number of DAPI+ cells in each of three fields was determined. The averages and SDs are given. The percentage of mCherry+ cells was statistically lower for the double mutant than for the wild-type rem or G98R constructs. The percentage of GFP+ cells was statistically lower for the mutants than for wild-type transfectants. (C) Western blotting of GFP-tagged Rem and cleavage-site mutants. Cells were transfected as described in A (ca. 20-fold higher levels of mutant DNA than in the wild type); extracts were used for Western blotting with GFP- or actin-specific antibodies.
To determine if the cleavage-site mutations eliminate functional Rem activity, we cotransfected wild-type rem, env, or mutant expression constructs into HC11 cells with the pHMRluc vector (1). As expected, both wild-type rem and env constructs showed a dose-dependent induction of luciferase expression, which requires the presence of the RmRE (5). In contrast, no Rem-dependent activity was detectable using either single or double mutants from the rem or env constructs, even at elevated DNA concentrations (Fig. 3C). These experiments demonstrate that SP cleavage is required for activity in the pHMRluc assay. Further, because Rem activity requires the NLS (1), our results suggest that SP is processed in the ER before nuclear localization.
Previous data indicate that SP is localized primarily in nucleoli (1). To determine the localization of the Rem cleavage mutants, GFP-tagged Rem or the single G98R or double V96RG98R expression constructs were cotransfected with the ER-mCherry construct into 293T cells before fluorescence microscopy. The ER-mCherry expression plasmid was used to identify the ER (16), whereas DAPI was used to stain DNA within nuclei (1). As expected, GFP-tagged Rem protein was easily visible within the nucleoli after transfection of 250 ng of wild-type rem construct (Fig. 4A; also see Fig. S2 for a separate transfection). Under these conditions, ≈35% of the cells expressed ER-mCherry, whereas ca. 20% of cells showed expression of GFPRem (Fig. 4B). Although the G98R mutant had no effect on the transfection efficiency, the double mutant showed significantly lower mCherry expression. No GFP fluorescence was detectable with either of the mutants using the same amount of DNA for transfection as the wild-type rem construct. However, ≈20-fold higher levels of the mutant expression vectors gave similar levels of wild-type and mutant proteins by Western blotting (Fig. 4C). At the higher mutant protein levels, many dead cells were noted, and a low level of diffuse fluorescence was observed. Quantitation showed a statistically significant decrease in GFP-positive mutant-transfected cells compared with those transfected with the wild-type rem construct (Fig. 4B and Fig. S3). Confocal microscopy was performed on wild-type and mutant-transfected cells (Fig. S4). Although wild-type Rem showed distinct nucleolar localization with low levels of transfected DNA, localization of G98R and V96RG98R mutants required both higher levels of DNA and artificial intensification of the signal. In rare cells with detectable red and green fluorescence, the G98R mutant was localized in the cytoplasm, but ER-mCherry was observed in both the nucleus and cytoplasm. Rare cells positive for both GFP-V96RG98R and ER-mCherry showed red fluorescence only in the cytoplasm and GFP signal throughout the cells. Together, these experiments suggest that the cleavage-site mutants were degraded more rapidly than wild-type Rem, perhaps because of improper folding and ERAD. Overexpression of these mutants may lead to a general disruption of ER trafficking.
Proteasome Inhibitors and P97/VCP DN Proteins Affect Rem Processing and Activity.
The requirement for Rem processing by signal peptidase and unfolding of cleavage-site mutants is consistent with SP retrotranslocation from the ER to the cytoplasm before nuclear localization (7). ERAD is a process that involves recognition of incorrectly folded proteins and retrotranslocation through the ER membrane using ATPase activity from p97 and/or the proteasome followed by degradation (9). To determine if inhibition of the proteasome affects Rem processing or SP activity, vectors expressing GFPRem or a known retrotranslocation substrate (the α1-antitrypsin mutant NHKQQQ) (16) were transfected into HC11 mouse mammary cells. Cells were treated with the proteasome inhibitor lactacystin before Western blotting of cell extracts. Both NHKQQQ and a larger form accumulated in cell extracts in the presence of lactacystin (Fig. S5). Cell extracts from rem-transfected cells showed a similar accumulation of Rem, but the amount of full-length Rem compared with SP increased significantly in the presence of the inhibitor (Fig. S5, lanes 1 and 2). Using the proteasome inhibitor MG132, Western blotting with the GFP-specific antibody again showed that proteasome inhibition gave increased levels of Rem relative to SP (Fig. S6). Similar results were obtained in several different experiments using either GFP-tagged or untagged Rem and Western blotting with GFP or SP-specific antibody. Although the amount of SP increased in the presence of MG132 after transfection of the env expression plasmid, no increase in Env precursor was detected. These results suggested that the proteasome degrades Rem preferentially as compared with SP.
To determine whether proteasome inhibitors also affect SP function, different amounts of the env or rem expression vectors were cotransfected with the reporter vector pHMRluc into HC11 cells. Both vectors showed a dose-dependent increase in luciferase activity as well as statistically significant increases (ca. 2- to 3-fold) of GFP-SP in the presence of MG132 as compared with vehicle-treated cells (Fig. S7), suggesting that the proteasome partially controls SP levels through Rem degradation.
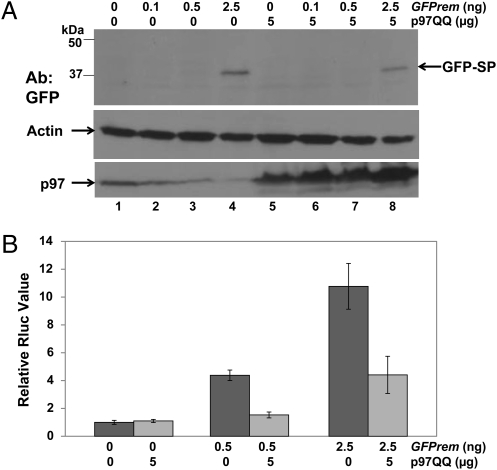
Retrotranslocation from the ER requires energy to extract proteins from the membrane. The AAA ATPase p97/VCP may provide this requirement (17). Therefore, we determined whether a DN p97 protein carrying mutations in both ATPase domains (p97QQ) (18) could interfere with SP processing or activity. Rem-expression constructs were cotransfected with p97QQ, and extracts were tested by Western blotting with GFP-specific antibody. Results indicated that extracts with or without expression of the p97 mutant protein had equivalent amounts of GFP-SP (Fig. 5A Top). Additional Western blots confirmed that p97 was present at higher levels after DN protein expression (Bottom) relative to actin as a control for protein loading (Middle). Control transfections with a mutant α1-antitrypsin revealed that DN p97 was functional at this concentration (Fig. S8). These data indicate that Rem processing to SP is not affected by the p97 ATPase.
Fig. 5.
A DN ATPase p97/VCP decreases SP activity but not cleavage. (A) Western blotting of 293T cells transfected with different amounts of GFPrem construct in the absence or presence of a DN p97 (p97QQ). Extracts were subjected to Western blotting with antibodies specific for GFP (Top), actin (Middle), or p97 (Bottom). (B) DN p97QQ inhibits Rem function. Two different amounts of GFPrem were transfected with or without the p97QQ expression construct. Luciferase activity is reported as in Fig. 1D.
If p97 is required for SP extraction from the membrane, the DN protein might be predicted to have no effect on Rem processing but to prevent SP trafficking to the nucleus for RNA binding. Thus, similar cotransfection experiments with p97QQ were performed in the presence of the pHMRluc reporter plasmid at two different levels of the rem expression vector (Fig. 5B). The results indicated that p97QQ inhibited Rem function, despite similar levels of SP in the presence and absence of the DN protein. These experiments support a role for p97/VCP in SP retrotranslocation from the ER membrane to the cytoplasm.
Discussion
In this article, we showed that functional SP is generated by both rem and env mRNAs (Fig. 1E). Previous results have shown that Rem activity in the pHMRluc assay measures a postexport function that requires both a nuclear localization signal and a binding site (RmRE) localized at the env–U3 border of MMTV RNA (1, 6). Both the NLS and the RNA-binding motif are localized within SP. Mutation of the second splice donor site in the env mRNA, which should prevent additional splicing to give rem mRNA, did not affect generation of SP or activity in the pHMRluc assay (Fig. S1). Therefore, our results strongly argue that functional SP is generated from both Env and Rem proteins.
Our data also demonstrate that SP generation and functional activity in the pHMRluc assay require the consensus site for cleavage by signal peptidase, which resides in the ER (19). The necessity for signal peptidase cleavage for SP generation is consistent with retrotranslocation from the ER to the cytosol before nuclear trafficking. Previous data show that SP generation is not inhibited by SP peptidase (7), an intramembrane protease that can cleave SPs (20). Our experiments also indicate that SP is retrotranslocated following translation of either Rem or Env at the ER membrane. First, mutation of both glycosylation sites in the Rem C-terminus allows SP generation at expression levels detectable by Western blotting but inhibits SP activity at low levels of transfected DNA (Fig. 2). Inappropriate glycosylation is known to target proteins for retrotranslocation and degradation by cytosolic proteasomes (21), suggesting that unglycosylated Rem may be targeted to the proteasome. Second, mutations in the signal peptidase cleavage site in either rem or env expression constructs prevent SP generation as well as functional activity (Fig. 3). The Rem cleavage-site mutants tagged with EGFP also are unstable and show little fluorescence even after overexpression (Fig. 4), suggesting that the GFP portion of the fusion is unfolded and may be degraded. Third, proteasome inhibitors increased the ratio of Rem relative to SP (Figs. S5 and S6). Because uncleaved Rem is not observed in infected cells (Fig. 1B), unglycosylated Rem accumulation in the presence of proteasome inhibitors is consistent with its selective targeting for ER-associated degradation. Fourth, a DN p97, which abolishes ATPase activity known to participate in protein extraction from the ER for proteasome degradation (17, 22), did not alter processing of Rem to SP (Fig. 5A). However, the DN p97 repressed Rem functional activity, suggesting that p97 is necessary for efficient extraction of SP from the ER membrane but not for cleavage by signal peptidase. Therefore, these experiments are consistent with a completely unique trafficking mechanism for Rem or Env-generated SP involving synthesis on ER-associated polysomes and insertion into the translocon. After cleavage by signal peptidase, SP, which appears to escape polyubiquitination, is extracted from the membrane into the cytoplasm where the NLS is recognized for nuclear import. Trafficking to the nucleolus allows SP binding to all MMTV mRNAs via the RmRE before nuclear export.
Because functional SP appears to be generated from both env and rem mRNAs, why would MMTV make a doubly spliced rem mRNA? The obvious explanation is that env and rem mRNAs generate different C-terminal products after cleavage by signal peptidase. Previous data indicate that a glycosylated 33-kDa product was generated after transfection of a C-terminally Myc-tagged rem construct. These experiments indicated that at least a portion of the Rem C-terminus (Rem-CT) is glycosylated, folded, and probably secreted from MMTV-infected cells (7). Based on results with MG132, we speculate that during normal viral infections, unglycosylated Rem is degraded rapidly by the proteasome, whereas a smaller portion is glycosylated in the ER, cleaved, and folded to allow escape from ERAD. Cleavage-site mutation alters the ratio of glycosylated to unglycosylated Rem (Fig. 4C), suggesting that rapid signal peptidase cleavage may decrease the amount of glycosylated Rem-CT. Rem may generate SP to increase expression of all viral mRNAs as well as Rem-CT, which may have an immune modulatory function early in infection. On the other hand, Env precursor generates larger amounts of Env, which is cleaved to SU and TM, as well as SP later in the infectious cycle. Higher quantities of SP probably are needed in virus-producing cells for RNA export and expression.
The trafficking pattern of MMTV SP appears to be unique. Previous experiments suggest that human endogenous retrovirus K (HERV-K) and JSRV, which also are betaretroviruses, make functional SPs either through env mRNA (JSRV) (14, 15) or through singly spliced env and/or doubly spliced rec mRNAs (HERV-K) (23). The trafficking pattern of these other betaretrovirus SPs has not been described, although, like MMTV SP, these SPs appear to be localized primarily to the nucleolus (24). Other viruses (e.g., hepatitis C virus) make capsid proteins that localize to the cytoplasm after precursor cleavage by the ER intramembrane protease, SP peptidase (25). Therefore, the processing of MMTV and betaretrovirus SPs appears to be fundamentally different from other positive-stranded RNA viruses. Because defects in protein folding and ERAD are associated with many human diseases (26), further studies of betaretrovirus SP trafficking will continue to illuminate many important aspects of cell biology and pathogenic processes.
Materials and Methods
Cell Lines and Transfections.
Growth of the HEK cells (293T), human T lymphoma cells (Jurkat), GR-B2 mouse mammary tumor cells, and HC11 normal mouse mammary cell lines has been described previously (6, 27). HC11 cells were infected by coculture with Jurkat cells producing the cloned provirus HYB-MTV (13). The 293T cells were transfected by the calcium phosphate method using 6 μg of total DNA for 5 × 105 cells. Jurkat or HC11 cells (1 × 107) were transfected by electroporation with 20 μg of total DNA (260 V, 1,050 μF, 4-mm gap cuvettes for Jurkat; 140 V, 1,750 μF, 2-mm gap cuvettes for HC11) in a BTX ECM600 instrument. Each transfection was performed in triplicate using the same amount of total DNA for every experiment, and all experiments (except for lactacystin) were performed at least twice with similar results. MG132 or lactacystin (Boston Biochem) was dissolved in DMSO and added to a concentration of 10 μM ≈24 h posttransfection. After a 12-h treatment, cells were harvested, and protein extracts were prepared as described (6) for luciferase assays or Western blots.
Constructs and Reporter Assays.
The wild-type rem expression construct was modified with a leucine (the consensus residue at position 71) rather than proline (formerly known as “P71L”) (6) and inserted into the EGFPC1 vector (Clontech) to create GFPrem. An untagged rem expression construct was obtained by deletion of EGFP sequences. The MMTV env construct (Q61) (28) in the pcDNA3.1 vector was derived from C3H-MMTV–infected cells (kindly provided by Susan Ross, University of Pennsylvania, Philadelphia, PA). Both constructs were used for PCR-based site-directed mutagenesis as previously described (29). A plasmid expressing the α1-antitrypsin mutant NHKQQQ tagged on the C terminus with EGFP and the reporter plasmid for ER localization, ER-mCherry, were generously provided by N. Hosokawa (Kyoto University, Kyoto, Japan) (16). The DN p97/VCP clone (p97QQ) was kindly provided by Yihong Ye (NIH, Bethesda, MD) (18). The reporter assays included pHMRluc (1) and a second reporter plasmid (pGL3-Control) that lacks the RmRE and expresses firefly luciferase from the SV40 promoter (Promega). Reporter levels were assessed per 100 μg of protein using the Dual Luciferase Assay Kit (Promega). Firefly luciferase levels were used to normalize Renilla luciferase activity for general effects on transfection efficiency and expression as previously described (6). The average of triplicate assays ± SDs containing expression constructs is reported relative to transfections containing only reporter vectors (assigned a relative value of 1).
Western Blotting and Antibodies.
Western blots were performed as previously described (1). Commercially obtained antibodies were used as follows: GFP (Clontech), p97 (Cell Signaling Technology), actin (Calbiochem), and HRP-conjugated donkey anti-rabbit or goat anti-mouse Ig (Jackson ImmunoResearch). Antibody to SP was generated in rabbits by Cocalico Biologicals following immunization with a peptide (QRPHLALRRKRRREC) (Sigma-Genosys) spanning the RNA-binding domain/NLS; the peptide was conjugated to keyhole limpet hemocyanin (KLH). In some cases, antibody was preadsorbed using KLH-agarose or by incubation with Western blots of extracts from uninfected cells. Results from the Western Lightning plus ECL chemiluminescence kit (Perkin-Elmer) were quantitated using AlphaImager software (Cell Biosciences).
Fluorescence Microscopy.
Transfected cells were fixed in 4% paraformaldehyde for 15 min, permeabilized with 0.05% Triton X-100 for 30 min, stained with 200 nM DAPI (Invitrogen), and examined under an Olympus IX70 fluorescence microscope. For each expression construct, ca. 200 cells stained with DAPI in each field were examined for expression of EGFP or mCherry; the averages of at least three fields then were plotted with SDs. The ratio of mCherry- to DAPI-positive cells was considered a measure of transfection efficiency.
Statistics.
Two-tailed Student t tests were used to compare results of triplicate transfections between mutant and wild-type constructs. A value of P ≤ 0.05 was considered to be significant.
Supplementary Material
Acknowledgments
We thank Rachel Lay for help with experiments. Jon Huibregtse, Tom Hope, and Bill Skach provided useful suggestions. This work was supported by National Institutes of Health Grant R01 CA116813.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004303107/-/DCSupplemental.
References
- 1.Mertz JA, Simper MS, Lozano MM, Payne SM, Dudley JP. Mouse mammary tumor virus encodes a self-regulatory RNA export protein and is a complex retrovirus. J Virol. 2005;79:14737–14747. doi: 10.1128/JVI.79.23.14737-14747.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Indik S, Günzburg WH, Salmons B, Rouault F. A novel, mouse mammary tumor virus encoded protein with Rev-like properties. Virology. 2005;337:1–6. doi: 10.1016/j.virol.2005.03.040. [DOI] [PubMed] [Google Scholar]
- 3.Stauber R, Gaitanaris GA, Pavlakis GN. Analysis of trafficking of Rev and transdominant Rev proteins in living cells using green fluorescent protein fusions: Transdominant Rev blocks the export of Rev from the nucleus to the cytoplasm. Virology. 1995;213:439–449. doi: 10.1006/viro.1995.0016. [DOI] [PubMed] [Google Scholar]
- 4.Heger P, Rosorius O, Hauber J, Stauber RH. Titration of cellular export factors, but not heteromultimerization, is the molecular mechanism of trans-dominant HTLV-1 rex mutants. Oncogene. 1999;18:4080–4090. doi: 10.1038/sj.onc.1202762. [DOI] [PubMed] [Google Scholar]
- 5.Mertz JA, Lozano MM, Dudley JP. Rev and Rex proteins of human complex retroviruses function with the MMTV Rem-responsive element. Retrovirology. 2009;6:10–22. doi: 10.1186/1742-4690-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mertz JA, Chadee AB, Byun H, Russell R, Dudley JP. Mapping of the functional boundaries and secondary structure of the mouse mammary tumor virus Rem-responsive element. J Biol Chem. 2009;284:25642–25652. doi: 10.1074/jbc.M109.012476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dultz E, et al. The signal peptide of the mouse mammary tumor virus Rem protein is released from the endoplasmic reticulum membrane and accumulates in nucleoli. J Biol Chem. 2008;283:9966–9976. doi: 10.1074/jbc.M705712200. [DOI] [PubMed] [Google Scholar]
- 8.Johnson AE, Haigh NG. The ER translocon and retrotranslocation: Is the shift into reverse manual or automatic? Cell. 2000;102:709–712. doi: 10.1016/s0092-8674(00)00059-3. [DOI] [PubMed] [Google Scholar]
- 9.Nakatsukasa K, Brodsky JL. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic. 2008;9:861–870. doi: 10.1111/j.1600-0854.2008.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolfe MS. Intramembrane-cleaving proteases. J Biol Chem. 2009;284:13969–13973. doi: 10.1074/jbc.R800039200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoch-Marchaim H, Hasson T, Rorman E, Cohen S, Hochman J. Nucleolar localization of mouse mammary tumor virus proteins in T-cell lymphomas. Virology. 1998;242:246–254. doi: 10.1006/viro.1997.8997. [DOI] [PubMed] [Google Scholar]
- 12.Hoch-Marchaim H, et al. The leader peptide of MMTV Env precursor localizes to the nucleoli in MMTV-derived T cell lymphomas and interacts with nucleolar protein B23. Virology. 2003;313:22–32. doi: 10.1016/s0042-6822(03)00236-8. [DOI] [PubMed] [Google Scholar]
- 13.Shackleford GM, Varmus HE. Construction of a clonable, infectious, and tumorigenic mouse mammary tumor virus provirus and a derivative genetic vector. Proc Natl Acad Sci USA. 1988;85:9655–9659. doi: 10.1073/pnas.85.24.9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caporale M, et al. The signal peptide of a simple retrovirus envelope functions as a posttranscriptional regulator of viral gene expression. J Virol. 2009;83:4591–4604. doi: 10.1128/JVI.01833-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofacre A, Nitta T, Fan H. Jaagsiekte sheep retrovirus encodes a regulatory factor, Rej, required for synthesis of Gag protein. J Virol. 2009;83:12483–12498. doi: 10.1128/JVI.01747-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosokawa N, et al. Human XTP3-B forms an endoplasmic reticulum quality control scaffold with the HRD1-SEL1L ubiquitin ligase complex and BiP. J Biol Chem. 2008;283:20914–20924. doi: 10.1074/jbc.M709336200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halawani D, Latterich M. p97: The cell's molecular purgatory? Mol Cell. 2006;22:713–717. doi: 10.1016/j.molcel.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- 19.Paetzel M, Karla A, Strynadka NC, Dalbey RE. Signal peptidases. Chem Rev. 2002;102:4549–4580. doi: 10.1021/cr010166y. [DOI] [PubMed] [Google Scholar]
- 20.Golde TE, Wolfe MS, Greenbaum DC. Signal peptide peptidases: A family of intramembrane-cleaving proteases that cleave type 2 transmembrane proteins. Semin Cell Dev Biol. 2009;20:225–230. doi: 10.1016/j.semcdb.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirayama H, Seino J, Kitajima T, Jigami Y, Suzuki T. Free oligosaccharides to monitor glycoprotein endoplasmic reticulum-associated degradation in Saccharomyces cerevisiae. J Biol Chem. 2010;285:12390–12404. doi: 10.1074/jbc.M109.082081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlson EJ, Pitonzo D, Skach WR. p97 functions as an auxiliary factor to facilitate TM domain extraction during CFTR ER-associated degradation. EMBO J. 2006;25:4557–4566. doi: 10.1038/sj.emboj.7601307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magin C, Hesse J, Löwer J, Löwer R. Corf, the Rev/Rex homologue of HTDV/HERV-K, encodes an arginine-rich nuclear localization signal that exerts a trans-dominant phenotype when mutated. Virology. 2000;274:11–16. doi: 10.1006/viro.2000.0438. [DOI] [PubMed] [Google Scholar]
- 24.Cullen BR. Using retroviruses to study the nuclear export of mRNA. Results Probl Cell Differ. 2002;35:151–168. doi: 10.1007/978-3-540-44603-3_8. [DOI] [PubMed] [Google Scholar]
- 25.Targett-Adams P, Hope G, Boulant S, McLauchlan J. Maturation of hepatitis C virus core protein by signal peptide peptidase is required for virus production. J Biol Chem. 2008;283:16850–16859. doi: 10.1074/jbc.M802273200. [DOI] [PubMed] [Google Scholar]
- 26.Vij N. AAA ATPase p97/VCP: Cellular functions, disease and therapeutic potential. J Cell Mol Med. 2008;12(6A):2511–2518. doi: 10.1111/j.1582-4934.2008.00462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ringold GM, Yamamoto KR, Tomkins GM, Bishop M, Varmus HE. Dexamethasone-mediated induction of mouse mammary tumor virus RNA: A system for studying glucocorticoid action. Cell. 1975;6:299–305. doi: 10.1016/0092-8674(75)90181-6. [DOI] [PubMed] [Google Scholar]
- 28.Katz E, et al. MMTV Env encodes an ITAM responsible for transformation of mammary epithelial cells in three-dimensional culture. J Exp Med. 2005;201:431–439. doi: 10.1084/jem.20041471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bramblett D, et al. A redundant nuclear protein binding site contributes to negative regulation of the mouse mammary tumor virus long terminal repeat. J Virol. 1995;69:7868–7876. doi: 10.1128/jvi.69.12.7868-7876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.