Abstract
The recently elucidated Get proteins are responsible for the targeted delivery of the majority of tail-anchored (TA) proteins to the endoplasmic reticulum. Get4 and Get5 have been identified in the early steps of the pathway mediating TA substrate delivery to the cytoplasmic targeting factor Get3. Here we report a crystal structure of Get4 and an N-terminal fragment of Get5 from Saccharomyces cerevisae. We show Get4 and Get5 (Get4/5) form an intimate complex that exists as a dimer (two copies of Get4/5) mediated by the C-terminus of Get5. We further demonstrate that Get3 specifically binds to a conserved surface on Get4 in a nucleotide dependent manner. This work provides further evidence for a model in which Get4/5 operates upstream of Get3 and mediates the specific delivery of a TA substrate.
Keywords: crystallography, Get pathway, Mdy2, Ubl4a, tail-anchor
Targeted delivery of membrane proteins is a critical process. For a special class of membrane proteins, tail-anchored (TA) proteins, the targeting pathways have only recently begun to be understood. These proteins are a large and diverse class of integral membrane proteins found in all organisms. Examples include SNAREs, apoptosis factors, and protein translocation components. TA proteins are characterized by having a single transmembrane helix (TM) at their extreme C-terminus. Due to this topological constraint, these proteins are not able to follow the signal recognition particle (SRP) dependent cotranslational pathway that typifies most integral membrane proteins. Instead, these proteins must find their correct membrane for insertion posttranslationally (1–4).
The newly characterized Get pathway (Guided Entry of Tail-anchored proteins) is the major targeting pathway for TA proteins in yeast. The first protein identified to specifically recognize a TA protein substrate is the ATPase Get3, which protects TA proteins in the cytoplasm and targets them to the endoplasmic reticulum (5–9). Deletions of this protein lead to mistargeting of TA proteins and growth sensitivity in a variety of conditions (9, 10). Recently, a number of structural studies of Get3 have led to the model where Get3 undergoes a dramatic conformational change upon nucleotide binding shifting from an open to a closed form and generating a TM binding pocket (11–15). Despite the many structures, the precise mechanism of how Get3 binds and releases substrate is not fully understood. At the ER, the Get3 TA protein complex binds two integral membrane proteins, Get1 and Get2, that are thought to act as receptors for the release of the protein substrate (7). Upstream of Get3 are two proteins, Get4 and Get5 that are the subjects of this study.
Get4 [yeast locus Yor164c, human locus C7orf20 and cee in fish (16)] is a highly conserved protein that is estimated to have arisen early in evolution (Fig. S1A) (16). Its high homology, 26% identity from yeast to humans, belies the fact that until recently very little was known about its biological role. It contains no known motifs and has only been annotated based on a series of genome-wide screens. Get4 localizes to the cytoplasm (17) and, although not essential, knockouts in yeast lead to sensitivity in a number of growth conditions (18) whereas disruption of the homologue in Caenorhabditis elegans retards growth (19, 20). Multiple protein interaction studies in yeast have implicated Get4 in binding to Get5 and Get3 and associating with Sgt2 and Hsp90-like proteins (21–25).
Get5/Mdy2 [yeast locus Yol111c, known as GdX/Ubl4a in mammals (26)] is a multidomain protein (Fig. S1B). The N-terminal domain (Get5-N) is found only in fungi where it is conserved. Following that is a ubiquitin-like domain (Get5-Ubl) (27) and a C-terminal domain (Get5-C). Get5 was originally annotated based on a decreased mating phenotype (26, 28). Unlike most yeast proteins that contain a Ubl, Get5 does not interact with polyubiquinated proteins nor does it bind the 26S proteasome (29). A biochemical and genetic study linked Get5 to both Sgt2, a tetratrico peptide repeat (TPR) containing protein (30), and to Ydj1, a J-domain containing Hsp40 homologue that interacts with the cytosolic Hsp70 homologues Ssa1p/Ssa2p (31).
Genomic screens suggest that Get4 and Get5 form a stable complex (referred to here as Get4/5). Using epistatic arrays and biochemistry, Jonikas et al. showed that these two proteins were also involved in the Get pathway (9). These proteins operated upstream of Get3 and, based on a study where Get5 was found bound the ribosome (21), it was suggested that these proteins acted as the ribosome receptor for Get3. Kar2p, a resident ER protein, is secreted in mutants defective in TA targeting. A screen for these mutants confirmed Get4 and Get5 as part of the Get pathway (32).
During the preparation of this manuscript the first structure of yeast Get4 and a fragment of Get5, generated from unintended proteolysis, was published (33). The authors used two-hybrid screens to explore general interactions of Get4/5 implicating Sgt2 and Ydj1 binding to Get5 and Get3 binding to the N-terminal half of Get4. A second structure of Get4 alone from Chaetomium thermophilum has also been published (34).
Here we report an independent structure of Get4 with an N-terminal fragment of Get5 (Get4/5-N) in a unique crystal form. Using the structure as a guide, we show that the full-length Get4/5 complex exists as a dimer (two copies of Get4/5) and identify important functional residues and the binding interface with Get3. Our results further define the structural elements of Get4/5 and provide strong evidence for the current model that has Get4 and Get5 acting as upstream factors of Get3 in the Get targeting pathway.
Results
Purification and structure determination of Get4/5-N.
Full-length Get4/5 was expressed in Escherichia coli and purified using affinity chromatography. We were able to express Get4 alone; however, all of the protein went into inclusion bodies in all tested expression conditions. Further purification of Get4/5 by anion exchange chromatography resulted in two separate peaks. These peaks were stable and were injected onto a size exclusion column where they both ran separately and larger than expected based on molecular weight (see below). Both failed to crystallize.
To address the possibility that disorder might have prevented crystallization, we performed in situ proteolysis by including protease in the crystallization trials. Crystals grew quickly using chymotrypsin and contained nearly full-length Get4 and the N-terminal third of Get5 (Get4/5-N). To improve crystallization, we performed limited proteolysis (Fig. S2A) and then purified Get4/5-N by ion exchange chromatography (Fig. S2 B and C). Initial crystals were hexameric rods with reproducible twists halfway down their length that did not diffract. We found that several additives containing amines generated trigonal crystals (see SI Text, Methods). The final crystals grew using L-proline as an additive and diffracted to 2.8 Å (Fig. S2D).
The structure was solved using seleno-methionine, single wavelength anomalous dispersion (SAD) phasing and 3-fold noncrystallographic symmetry. The final structure contained three almost identical Get4/5-N in the asymmetric unit, main-chain rmsd of approximately 0.6 Å3 (Fig. 1C). The most complete Get4/5-N model contains nearly all of Get4, residues 9-299, and the N-terminus of Get5, residues 3–56 (Fig. 1A). The structure refined to an Rfactor of 18.2% and a Free-Rfactor of 22.4%. Crystallographic statistics are presented in Table S1.
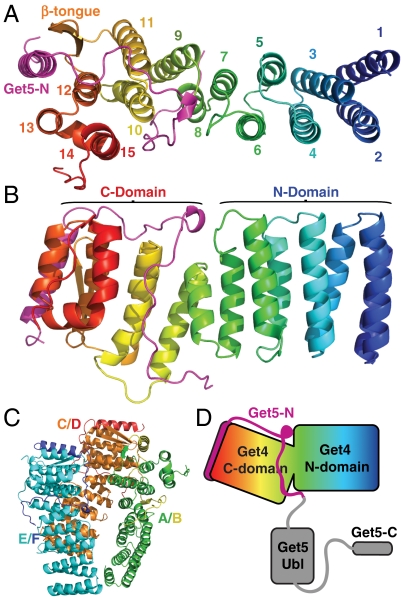
Fig. 1.
The structure of Get4/5-N. (A) Top view of Get4/5-N with Get4 color-ramped from N- (blue) to C-terminus (red) and Get5-N shown in magenta. Secondary structure elements are labeled as in Fig. S1. (B) Side view relative to (A) with N- and C-domains indicated. (C) The asymmetric unit with each chain colored individually and labeled as in the deposited coordinates. (D) A cartoon of the structure based on (B). The missing Ubl- and C-domains of Get5 are shown in gray.
Description of the Structure of the Get4/5-N Complex.
Get4, which has no predicted sequence motifs, is essentially two rectangular blocks formed by right-handed α-helical coils that can be divided into N-terminal and C-terminal domains (N-domain and C-domain) (Fig. 1B). The N-domain consists of the first 7 helices that are similar in length and are reminiscent of the TPR motif (30). Unlike this motif, they do not contain an obvious internal consensus nor do they exhibit any curvature, a common feature of helical repeats.
The C-domain continues with right-handed helical coils; however, the helical length is more diverse. In addition, unlike the short loops of the N-domain, the C-domain loops show more variation in length. The loop between helices α11 and α12 is formed by two β-strands (β-tongue). The helix α13 makes a sharp turn into helix α14 that then bends to form helix α15 generating a U shape. The C-terminus forms an extended peptide that docks against a neighboring molecule.
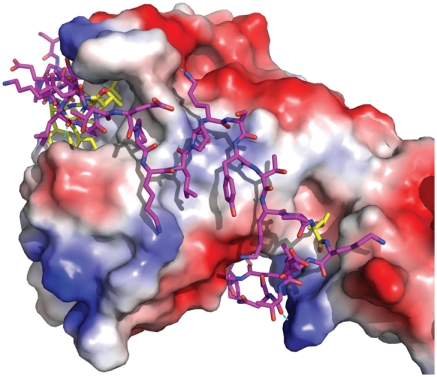
Get5-N forms an extended peptide that wraps tightly around the C-domain of Get4 (Fig. 2). It begins with a helix that docks in a groove formed by helices α12, α13, and the β-tongue of Get4 (Fig. 1A). Outside of the conserved hydrophobic interactions of the helix, the rest of Get5-N forms relatively few specific interactions to Get4 (Fig. 2). The helix is followed by an extended, highly ordered loop that follows a groove contacting the loops formed by α12/α13, α10/α11, and α8/α9 in Get4 via backbone contacts. Comparing the structure to sequence conservation, the lengths of these Get4 loops are highly conserved implying that this platform is important in higher eukaryotes as well. The rest of Get5-N follows a groove formed by α8, α10, and α15 and then finally contacting the loop between α7/α8 of Get4. α8 is book-ended by Get5-N and the short length of this helix appears to be conserved.
Fig. 2.
Binding of Get5-N. Get4 is shown as an accessible surface colored from positive (blue) to negative (red) Coulombic charge in an orientation similar to Fig. 1A. Get5-N is shown as sticks in magenta with residues that are conserved making specific contacts in yellow.
Comparison to Other Get4 and Get5-N Structures.
When the three molecules in our asymmetric unit are aligned based on their N-domain there is a clear twist in Get4 at α7/α8 that results in a relative bend in the C-domain. The greatest difference is between the A molecule and the C molecule with a relative rotation of about 5° and a maximal shift of about 4 Å (Fig. S3A). In the Get4/5-N structure by Chang et al., there is very little difference between the four molecules in their asymmetric unit [Protein Data Bank (PDB) ID code 2 wpv] (Fig. S3B) (33). When aligned individually relative to our structure, there are no major differences in the overall fold of the N-domain (Fig. S3B) or the C-domain (Fig. S3C); however, there is a larger twist between the two domains that results in a 10° rotation and a 6 Å shift (Fig. S3B). Part of this rotation is taken up by a shift in α10 (Fig. S3C). Other differences are extensions of most of the termini in the structure reported here. The structure of Get4 reported by Bozkurt et al. shows the conservation of the overall fold of Get4 and appears to be in a conformation similar to our A molecule; however, there are some significant distortions, presumably the result of the missing Get5-N (PDB ID code 3lpz) (Fig. S3D) (34).
Surface Features of Get4.
As noted, based on sequence the overall fold of Get4 appears to be conserved across eukaryotes excluding the β-tongue (Fig. S1A). The internal fold accounts for the majority of the highly conserved residues. Only two surfaces at the ends of the molecule have a high level of conservation (Fig. S4A). The C-domain conserved surface contributes to the binding of Get5-N despite the lack of Get5-N in higher eukaryotes. The largest conserved surface is the N-domain face (Fig. S4A). The overall surface of Get4/5-N is acidic with a single basic patch directly correlating to the conserved face of the Get4 N-domain (Fig. S4B).
Dimerization.
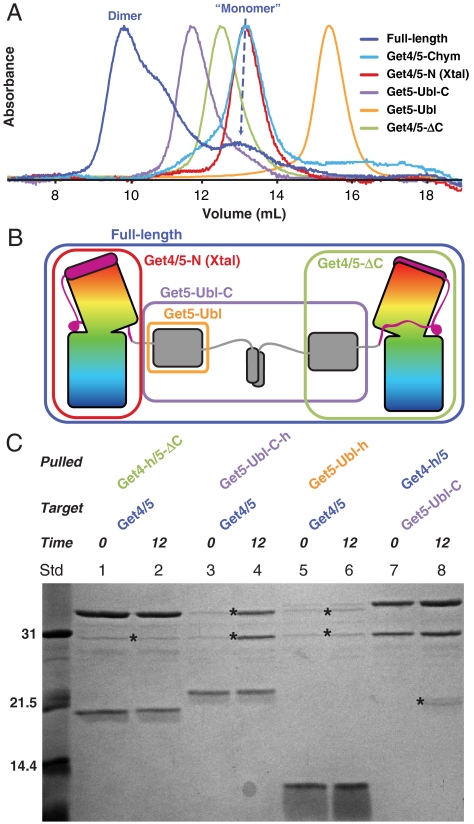
As stated above, Get4/5 eluted as two peaks by ion exchange chromatography. Each peak ran anomalously large on a size exclusion column in the expected size range of 4–8 copies relative to typical globular proteins (Fig. 3A, blue trace). We confirmed that the lower molecular weight peak corresponded to a single copy of Get4/5 and the higher molecular weight peak to two copies of Get4/5 by multiangle light scattering (MALS) (Fig. S5A). We refer to these two forms as the “monomer” and dimer in the rest of the text. We noted that the dimer peak was stable while the monomer peak partially converted to the dimer peak over time. These results suggest that the proteins have exaggerated hydrodynamic properties relative to what would be predicted based on molecular weight.
Fig. 3.
Dimerization by Get5-C. (A) Size exclusion chromatograms from various constructs of Get4/5 after affinity purification. “Chym” represents Get4/5 after a 15 minute chymotrypsin digest. The absorbances are normalized to the highest recorded value. The dimer and monomer peaks for full-length Get4/5 are indicated. (B) Cartoon of the Get4/5 dimerization by Get5-C. Constructs used in A and C are indicated by colored boxes. (C) Domain swapping by dimerization. His-tagged proteins, indicated by h, with and without Get5-C are mixed with untagged protein containing Get5-C for 0 and 12 h.
Analyzing the sequence features of Get5, there are two probable flexible loops connecting the Ubl to the N- and C-domains that could account for the larger radius. Addition of chymotrypsin to either monomer or dimer immediately cleaved the loop between Get5-N and the Get5-Ubl-C fragment; however, the loop connecting to the C-domain was resistant to cleavage (Fig. S2A). When this proteolyzed mix was run on the size exclusion column the peak shifted dramatically to a much smaller size with a small shoulder that eluted earlier (Fig. 3A, cyan trace). The two dominant proteolysis fragments, Get4/5-N and Get5-Ubl-C, could be purified by ion exchange chromatography (Fig. S2 B and C). When these fragments were run on the size exclusion column Get4/5-N (Fig. 3A, red trace) corresponded to the bulk of the proteolysis peak and Get5-Ubl-C was the leading shoulder (purple trace). These peaks were clearly resolved; therefore, Get4/5-N does not form a stable complex with Get5-Ubl-C. The estimated extinction coefficient is much lower for Get5-Ubl-C and the smaller leading shoulder in the protease fragment reflects this (cyan trace). Get5-Ubl-C (15.9 kDa), a dimer by MALS, ran anomalously large on the sizing column even ahead of the larger Get4/5-N (40 kDa) (Fig. S2A). This implies a model that involves dimerization of Get4/5 by the Get5-Ubl-C fragment (Fig. 3B).
The mass of the Get4/5 monomer peak by MALS was approximately 8 kDa larger than that predicted for Get4/5 and contained a small peptide in the range of 7 kDa (Fig. S5). We tested the peptide by mass spectrometry and N-terminal sequencing and determined that it corresponded to an internal start site at the single methionine in our Get5-Ubl-C construct; i.e. it was an expressed Get5-C-domain. The extra C-domain in the monomer prevented dimer formation with full-length Get4/5 leading to the conclusion that this domain was involved in dimerization. To further explore this we expressed and purified the additional constructs of Get5-Ubl-C, Get4/5-ΔC and Get5-Ubl (Fig. 3B). The constructs without C-domains both ran as monomers as predicted (green and orange traces, respectively). Get5-Ubl-C behaved similarly to the cleaved Get5-Ubl-C fragment and we again detected a fraction of the protein bound to a Get5-C peptide similar to the full-length monomer (Fig. S5B). Get5-C, including its disordered loop, is found in all eukaryotes and in yeast the sequence of its C-terminal helical region is consistent with a possible coiled-coil, as predicted by COILS (35).
We hypothesized that two different constructs of Get4/5 may exchange their C-domains to form mixed Get4/5 dimers. To test this, various constructs of Get4/5 (Fig. S6A) were mixed with full-length Get4/5 and the products were analyzed after a 12 h incubation (Fig. 3C). An affinity tagged Get4/5-ΔC was unable to capture a full-length Get4/5 after 12 h (lane 2 compared to lane 1); however, Get5-Ubl-C was able to capture Get4/5 (lane 4) and Get4/5 could capture Get5-Ubl-C (lane 8). Get5-Ubl alone was unable to capture Get4/5 (lane 6). These results verify that the C-domain was responsible for Get4/5 dimerization and rule out a role for the Ubl-domain.
A coiled-coil prediction for the Get5 C-terminal region was found in higher eukaryotes but was not seen in all fungi. An example is Aspergillus fumigatus Get4/5 and we wanted to see if Get4/5 formed a dimer in this species. By MALS, the purified full-length AfGet4/5 corresponded to a stable dimer (Fig. S5A). We never detected a monomer fraction for this species nor did we ever see the equivalent of the Get5-C fragment (Fig. S5B). To confirm dimerization by the C-domain, we generated AfGet4/5 constructs with the C-domain removed (Af4/5-ΔC) and the Get5 Ubl-C-domain alone (Af5-Ubl-C). All three Af constructs behaved the same as their Sc equivalents on a size exclusion column (Fig. S7 A and B). Additionally, the Af constructs were able to exchange based on the presence of the C-domain (Fig. S7C, lanes 1-10). Not surprisingly, the Sc5-Ubl-C was not able to swap with the Af5-Ubl-C confirming that dimerization is conserved independent of sequence (Fig. S7C, lanes 13 & 14).
In Vitro Interaction of Get4/5 and Get3.
Previous studies have demonstrated that Get4/5 can form a complex with Get3; however, these studies did not address the specifics of the interaction nor did they address the role of nucleotide in binding (9, 36). We further explored this interaction using purified components. Initially, an affinity-his-tagged Get4/5 was used to test the binding of Get3 (Fig. 4A). Using Ni-affinity beads, very little Get3 could be captured by Get4/5 when mixed in the absence of nucleotide (lane 2). The addition of ADP (lane 3) or ATP (lane 4) dramatically increased the amount of bound Get3. The structures of the apo form of Get3 were always in an open form (11, 12, 14); however, structures of Get3 adopt both open and closed forms in the presence of nucleotide (11–13). The fact that binding is enhanced by nucleotide would imply that nucleotide has shifted the equilibrium from the open to the closed form, and it is the closed form that is recognized by Get4/5. Get3 was able to capture Get4/5 (lane 6) but this was not enhanced by nucleotide (lanes 7 and 8). Our Get3 has an N-terminal affinity-tag and binding this to beads may also shift the equilibrium to the closed form. Get3 captured only Get4/5-N from our proteolyzed pool and not Get5-Ubl-C (lane 9); therefore, Get3 binds specifically to Get4/5-N.
Fig. 4.
Binding of Get4/5 to Get3. (A) Tagged wild-type Get4/5 and Get3 are incubated for 2 h at room temperature then bound to Ni-beads. “D” represents incubation in the presence of 2 mM ADP-Mg2+, and “T” in the presence of 2 mM ATP-Mg2+. (B) His-tagged Get4/5 and Get3 mutants are incubated in the presence of 2 mM ADP-Mg2+ as in (A). The YYE/AAA Get4 mutant lacks a TEV cleavage site and is slightly smaller than the other tagged mutants. (C) As in (B), using his-tagged Get3. (D) View from the N-terminal face of Get4. Mutated residues are displayed as spheres with carbons and labels colored based on wild-type level (cyan), weak (yellow), or weakest (red) interactions with Get3. (E) The closed state of Get3 (PDB ID code 2WOJ) with mutated residues displayed and colored as in (D).
Based on these results, we searched for mutants that would affect Get4/5 binding to Get3. The most conserved surface of Get4 is on the N-terminus where there is a patch of highly conserved positive residues (Fig. S4). To evaluate potential effects of these charged residues in the Get3 interaction, we mutated pairs of positive charges to aspartates to fully disrupt possible interfaces (K12D/K15D, R19D/K23D, H33D/R37D, and R42D/R45D). In addition, to rule out the general effects of charge swapping, we generated another pair of mutants of unconserved residues (K65D/K67D) as a control. Finally, we mutated the highly conserved Tyr 29, Tyr 30, and Glu 31 to alanines (YYE/AAA) (16). All of these mutants were purified and behaved similar to wild type on a size exclusion column (Fig. S6C).
The purified mutants were mixed with Get3 and captured using Ni-affinity beads. All of the conserved mutants showed a significant loss in the ability to capture Get3 (Fig. 4B, lanes 1–6). Two of the mutants, H33D/R37D (lane 3) and YYE/AAA (lane 7), captured Get3 at a markedly reduced level. As expected, Get5-Ubl-C was unable to capture Get3 (lane 8). By reversing the experiment we saw the same pattern of mutant Get4/5 capture by Get3 (Fig. 4C, lanes 2–7). These results center the binding of Get3 to the positive N-terminal face of Get4 (Fig. 4D and Fig. S8 A and B).
In our previous study of Get3, we had mutated a number of conserved Get3 surface residues that were unable to rescue a Δget3 knockout (11). These residues could not be explained by contacts in the Get3 dimer in either the open or closed state and many localized to a negative surface (Fig. S8 C–E). We tested several for Get4 binding by alanine mutation (Y250, E253, E258, D265, and K297). All of the mutants expressed well and behaved similar to wild type on the size exclusion column (Fig. S6D). When wild-type Get4/5 was used to capture the Get3 mutants only two (Y250A and E253A) showed a significant decrease in binding (Fig. 4B, lanes 9–13 compared to 1). These two mutants are near the interface of the Get3 dimer and presumably destabilize the closed state (Fig. 4E). All of the Get3 mutants were able to capture Get4/5 (Fig. 4C, lanes 8–12). The bead bound Get3 is less sensitive to nucleotide; therefore, the closed form may be favored despite the mutants.
In Vivo Effects of Get4 and Get5 Mutants.
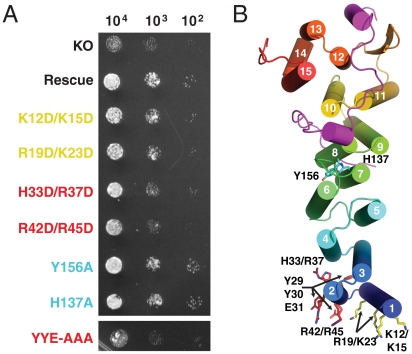
To assess the mutants in vivo, we utilized the fact that deletion of either Get4 or Get5 showed growth phenotypes under stress conditions and tested for rescue by our constructs (10). We rescued the Δget4 sensitivity to temperature and copper sulfate by expression of wild-type Get4 using the native promoter on a plasmid (Fig. 5A). With this construct, we generated all of the mutants used in the binding experiment. All of these mutants were unable to completely rescue the phenotype. The poorest rescue was by H33/R37, R42/R45, and YYE/AAA, which are on the highly conserved helix α2 (Fig. 5B). We generated two mutants at the interface of the Get4 N- and C-domain (H137A and Y156A) that showed no phenotype (Fig. 5).
Fig. 5.
Get4 rescue. (A) Spot plate growth assays of YEp-352 derived rescue plasmids under control of genomic promoters in the BY4741 Get4::KanMX background. Plates consisted of Sc-Ura supplemented with 2 mM CuSO4 and were incubated at 37 °C. The panel is generated from a single plate. “KO” represents transformations with empty YEp-352 vector and “Rescue” represents wild-type Get4. Rescue mutants labels are colored based on no (cyan), moderate (yellow), and strong (red) phenotypes. (B) Get4/5-N with helices shown as cylinders. Sets of mutated residues are shown as sticks with carbons colored based according to phenotype.
The structure of the Ubl domain from the human homologue of Get5, Ubl4a/GDX, has been solved by NMR and deposited to the PDB (ID code 2dzi). Using this as a template we generated a homology model of Get5-Ubl using SWISS-MODEL and our alignment (Fig. S9A) (37). In ubiquitin isoleucine 44 is highly conserved and always involved in protein binding interactions (38). In Get5 homologues, this residue is highly conserved as a leucine or methionine (L120) (Fig. S1B). Based on the homology model, we noted that L120 was part of a conserved interface that included a number of positively charged residues including the conserved K124, which corresponds to the commonly conjugated K48 in ubiquitin (Fig. S9 A–C). Get5 deletion mutants are viable in rich media but show phenotypes under stress conditions (10). Mutation of these residues to alanines (L120A/K124A) was not able to fully rescue the Δget5 at 37 °C (Fig. S9D) despite the protein being stable in solution (Fig. S6C).
Discussion
Get4 and Get5 are two highly conserved proteins whose functions have only recently begun to be understood. The growing consensus is a direct role for these proteins operating upstream of Get3 in the TA protein targeting pathway. Here we have presented a structure of the yeast Get4/5-N complex along with a model for the structural elements of Get5-Ubl-C. We have shown that the purified Get4/5 complex dimerizes mediated by the Get5 C-domain. The Get4 N-terminal face forms part of the recognition interface with Get3, apparently preferring to bind to the closed form, and this surface is important in vivo.
Get4 and the N-domain of Get5 form a stable and intimate complex. The fact that Get5-N appears to be important in yeast is somewhat surprising considering the absence of this domain in Get5 homologues in higher eukaryotes. One might speculate that in higher eukaryotes another protein can perform a similar interaction with Get4. An attractive option would be another protein that could bridge between Ubl4a and the mammalian Get4 homologue. This theoretical protein would contain a Get5-N-like region for binding to Get4, a dimerization domain for binding to Ubl4a and possibly a second Ubl. This complex would retain many of the features of Get4/5.
Get3 is a soluble protein that transiently interacts with Get4. In the current model, Get4/5 facilitates binding of TA proteins to Get3. The preference for a closed state of Get3 fits nicely into the model where Get4/5 act as mediators to the ribosome and bind Get3 in a state competent for TA protein binding. The dimerization of Get4/5 adds the additional possibility that the 2-fold symmetrical Get3 dimer presents a binding site for each of the Get4 binding sites in the Get4/5 dimer. A mechanism such as this would lead to cooperative binding with a much higher affinity for Get3 in the correct state.
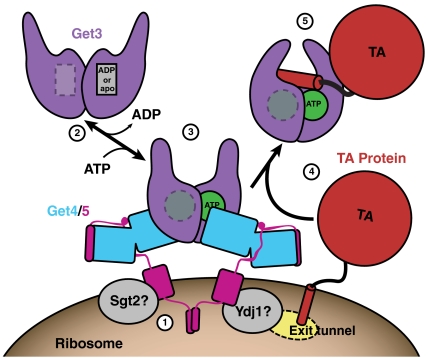
The results presented here allow us to clarify the role of Get4 and Get5 as intermediaries in TA targeting (Fig. 6). Get4/5 are able to recognize the nucleotide state of Get3 and localize the closed form of Get3 to the ribosome dependent on an emerging TA substrate. Binding of the TA protein to Get3 leads to a conformational change that releases the Get3/TA complex from Get4/5 and the ribosome. Sgt2 and cellular chaperones either facilitate this transfer of substrate or act as parts of an alternate pathway.
Fig. 6.
A model for the role of Get4/5. (1) Dimeric Get4/5 presumably binds the ribosome near the exit tunnel. Sgt2 and Ydj1 associate with the Get5-Ubl-C-domains. (2) Get3 in the open state is free in the cytoplasm. (3) Get3, in a closed state, is recruited to the Get4/5 complex upon ATP binding. (4) Tail-anchored proteins emerge from the ribosome and become associated with the Get3/Get4/5 complex. (5) The soluble Get3/TA protein complex is released to the cytoplasm for targeting to the ER.
The steps in the Get targeting pathway continue to become clear but there are many outstanding questions that remain. The yeast Get4/5 appears to interact with the ribosome via Get5 (21); however, it remains to be demonstrated that this is a direct interaction. The role of nucleotide hydrolysis in the targeting pathway is not clear and may be involved in fidelity of substrate selection at the ribosome or in release of the TA protein at the ER. Finally, although the precise role of dimerization remains to be elucidated, the fact that Get4/5 forms dimers is a provocative result in light of the symmetry of the Get3 dimer.
Methods
Detailed descriptions of experiments are provided in SI Text, Methods. Briefly, Saccharomyces cerevisiae Get4 and Get5 genes were synthesized, expressed in E. coli, and purified using Ni-affinity, anion exchange, and size exclusion chromatography. The Get4/5-N fragment could be crystallized after a limited proteolysis with chymotrypsin and the structure was determined using SAD. The oligomerization states of various constructs were assayed using size exclusion chromatography. For the in vitro capture experiments, Get3 and Get4/5 were incubated at room temperature, bound to Ni-NTA agarose and washed rapidly. Yeast rescue mutants were cloned into YEp-352 and introduced to BY4741 Get4::KanMX strains and Get5::KanMX strains. Growth defects were observed by plating on copper sulfate containing media and incubating at 37 °C.
Supplementary Material
Acknowledgments.
We thank J. Howard, A. Müller, and A. Palazzo for discussion and critical comments on the manuscript, and T. Walton for help with MALS. We thank Gordon and Betty Moore for support of the Molecular Observatory at Caltech. All data collection was performed at beamline 9-2 and 11-1 at SSRL. Operations at Stanford Synchrotron Radiation Laboratory are supported by the US Department of Energy and the National Institutes of Health. W.M.C. is supported by the Searle Scholar program and a Burroughs–Wellcome Fund Career Award for the Biological Sciences.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006036107/-/DCSupplemental.
Data deposition: The atomic coordinates and structure factors for the refined crystal structure have been submitted to the Research Collaboratory for Structural Bioinformatics Protein Data Bank with accession number 3LKU.
References
- 1.Kutay U, Hartmann E, Rapoport TA. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 1993;3(3):72–75. doi: 10.1016/0962-8924(93)90066-a. [DOI] [PubMed] [Google Scholar]
- 2.Kutay U, Ahnert-Hilger G, Hartmann E, Wiedenmann B, Rapoport TA. Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 1995;14(2):217–223. doi: 10.1002/j.1460-2075.1995.tb06994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borgese N, Brambillasca S, Colombo S. How tails guide tail-anchored proteins to their destinations. Curr Opin Cell Biol. 2007;19(4):368–375. doi: 10.1016/j.ceb.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 4.Rabu C, Schmid V, Schwappach B, High S. Biogenesis of tail-anchored proteins: The beginning for the end? J Cell Sci. 2009;122(Pt 20):3605–3612. doi: 10.1242/jcs.041210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuldiner M, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123(3):507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 6.Stefanovic S, Hegde RS. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128(6):1147–1159. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 7.Schuldiner M, et al. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Favaloro V, Spasic M, Schwappach B, Dobberstein B. Distinct targeting pathways for the membrane insertion of tail-anchored (TA) proteins. J Cell Sci. 2008;121(11):1832–1840. doi: 10.1242/jcs.020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jonikas MC, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323(5922):1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hillenmeyer ME, et al. The chemical genomic portrait of yeast: Uncovering a phenotype for all genes. Science. 2008;320(5874):362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suloway CJ, Chartron JW, Zaslaver M, Clemons WM., Jr Model for eukaryotic tail-anchored protein binding based on the structure of Get3. Proc Natl Acad Sci USA. 2009;106(35):14849–14854. doi: 10.1073/pnas.0907522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mateja A, et al. The structural basis of tail-anchored membrane protein recognition by Get3. Nature. 2009;461(7262):361–366. doi: 10.1038/nature08319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bozkurt G, et al. Structural insights into tail-anchored protein binding and membrane insertion by Get3. Proc Natl Acad Sci USA. 2009;106(50):21131–21136. doi: 10.1073/pnas.0910223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu J, Li J, Qian X, Denic V, Sha B. The crystal structures of yeast get3 suggest a mechanism for tail-anchored protein membrane insertion. PLoS ONE. 2009;4(11):e8061. doi: 10.1371/journal.pone.0008061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamagata A, et al. Structural insight into the membrane insertion of tail-anchored proteins by Get3. Genes Cells. 2010;15(1):29–41. doi: 10.1111/j.1365-2443.2009.01362.x. [DOI] [PubMed] [Google Scholar]
- 16.Fernandes JMO, Macqueen DJ, Lee H-T, Johnston IA. Genomic, evolutionary, and expression analyses of cee, an ancient gene involved in normal growth and development. Genomics. 2008;91(4):315–325. doi: 10.1016/j.ygeno.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 17.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425(6959):686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 18.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418(6896):387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 19.Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421(6920):231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 20.Simmer F, et al. Genome-wide RNAi of C elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1(1):E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fleischer TC, Weaver CM, McAfee KJ, Jennings JL, Link AJ. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 2006;20(10):1294–1307. doi: 10.1101/gad.1422006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costanzo M, et al. The genetic landscape of a cell. Science. 2010;327(5964):425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440(7084):637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 24.Ito T, et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98(8):4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McClellan AJ, et al. Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell. 2007;131(1):121–135. doi: 10.1016/j.cell.2007.07.036. [DOI] [PubMed] [Google Scholar]
- 26.Hu Z, Potthoff B, Hollenberg CP, Ramezani-Rad M. Mdy2, a ubiquitin-like (UBL)-domain protein, is required for efficient mating in Saccharomyces cerevisiae. J Cell Sci. 2006;119(Pt 2):326–338. doi: 10.1242/jcs.02754. [DOI] [PubMed] [Google Scholar]
- 27.Toniolo D, Persico M, Alcalay M. A “housekeeping” gene on the X chromosome encodes a protein similar to ubiquitin. Proc Natl Acad Sci USA. 1988;85(3):851–855. doi: 10.1073/pnas.85.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iwanejko L, Smith KN, Loeillet S, Nicolas A, Fabre F. Disruption and functional analysis of six ORFs on chromosome XV: YOL117w, YOL115w (TRF4), YOL114c, YOL112w (MSB4), YOL111c, and YOL072w. Yeast. 1999;15(14):1529–1539. doi: 10.1002/(SICI)1097-0061(199910)15:14<1529::AID-YEA457>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 29.Saeki Y, Saitoh A, Toh-e A, Yokosawa H. Ubiquitin-like proteins and Rpn10 play cooperative roles in ubiquitin-dependent proteolysis. Biochem Biophys Res Commun. 2002;293(3):986–992. doi: 10.1016/S0006-291X(02)00340-6. [DOI] [PubMed] [Google Scholar]
- 30.D’Andrea LD, Regan L. TPR proteins: The versatile helix. Trends Biochem Sci. 2003;28(12):655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 31.Liou ST, Cheng MY, Wang C. SGT2 and MDY2 interact with molecular chaperone YDJ1 in Saccharomyces cerevisiae. Cell Stress Chaperon. 2007;12(1):59–70. doi: 10.1379/CSC-220R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Čopič A, et al. Genomewide analysis reveals novel pathways affecting endoplasmic reticulum homeostasis, protein modification, and quality control. Genetics. 2009;182(3):757–769. doi: 10.1534/genetics.109.101105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang Y-W, et al. Crystal structure of Get4/Get5 complex and its interactions with Sgt2, Get3, and Ydj1. J Biol Chem. 2010:1–19. doi: 10.1074/jbc.M109.087098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bozkurt G, et al. The structure of Get4 reveals an alpha-solenoid fold adapted for multiple interactions in tail-anchored protein biogenesis. FEBS Lett. 2010 doi: 10.1016/j.febslet.2010.02.070. [DOI] [PubMed] [Google Scholar]
- 35.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252(5009):1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 36.Yu H, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322(5898):104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 38.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6(8):610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.