Abstract
It was recently reported that rs1541160 on chromosome 1q24.2 has a marked effect on survival of amyotrophic lateral sclerosis (ALS) patients by influencing KIFAP3 expression. The cohorts used in that study were collected from ALS specialty clinics. We attempted to replicate these findings in a population-based cohort of 504 Italian ALS patients. None of 140 SNPs genotyped within the KIFAP3 locus (including rs1541160) had an effect on survival (log-rank P value for rs1541160 = 0.47) or on gene expression in that region. These data illustrate the complexities associated with analyzing ALS phenotypes for association.
Keywords: amyotrophic lateral sclerosis, Italian, survival analysis, genome-wide association study, gene expression quantitative trait loci mapping
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterized by rapidly progressive paralysis leading to death from respiratory failure. Population-based epidemiological studies indicate that the median survival of ALS patients is 20–36 mo (1–3). Demographic and clinical features, as well as riluzole and certain symptomatic therapies, are known to modestly modify survival, but the influence of genetics on the rate of disease progression and survival is not known (4). Landers et al. (5) recently reported that the CC genotype of SNP rs1541160 within the kinesin-associated protein 3 (KIFAP3) gene on chromosome 1q24.2 conferred a 14-mo survival advantage on ALS patients. Furthermore, expression data based on 26 occipital lobe brain samples suggested that the CC genotype of this SNP reduced KIFAP3 expression by 41% compared with the TT genotype (5). To confirm the accuracy of this report, we attempted to replicate this finding in a population-based cohort of 504 Italian ALS patients collected using the Piemonte and Valle d'Aosta Registry for ALS (PARALS). We also evaluated the effect of the identified SNP, and all SNPs within the 1-Mb surrounding region, on the expression of KIFAP3 in four brain regions obtained from 144 neurologically normal individuals.
Results
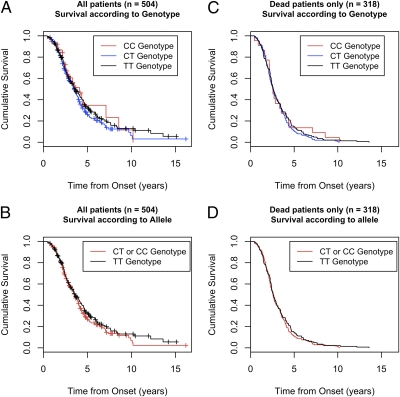
Rs1541160 had no effect on survival in our population-based cohort of 504 Italian ALS cases either under the genotypic model [median survival for CC genotype = 4.09 y; CT genotype = 3.48 y; TT genotype = 3.58 y; log-rank P = 0.48, Peto P = 0.60 (Fig. 1A)] or under the allelic model [log-rank P = 0.33, Peto P = 0.53 (Fig. 1B)]. Similarly, survival analysis limited to the 318 deceased ALS patients failed to demonstrate an effect of rs1541160 on survival either under the genotypic model [log-rank P = 0.55, Peto P = 0.92 (Fig. 1C)] or under the allelic model [log-rank P = 0.42, Peto P = 0.71 (Fig. 1D)]. Linear regression modeling of the deceased-only cohort, incorporating age, gender, site of symptom onset, and population structure (component vectors 1 and 2 from multidimensional scaling) as covariates, was also not significant (β coefficient = 0.024, P = 0.89). Survival analysis limited to the cohort of patients attending a specialized ALS clinic was similarly negative [n = 89 patients (Fig. S1)]. None of the 139 other SNPs within the 1-Mb region surrounding KIFAP3 significantly influenced survival of the Italian ALS cohort (Table S1).
Fig. 1.
Survival from time of symptom onset in a population-based cohort of Italian ALS patients according to rs1541160 status. (A) Kaplan–Meier curves for all 504 patients according to rs1541160 genotype status. (B) Kaplan–Meier curves for all 504 patients according to rs1541160 C allele carrier status. (C) Kaplan–Meier curves restricted to 318 deceased patients according to rs1541160 genotype status. (D) Kaplan–Meier curves for 318 deceased patients according to rs1541160 C allele carrier status. +, censored events.
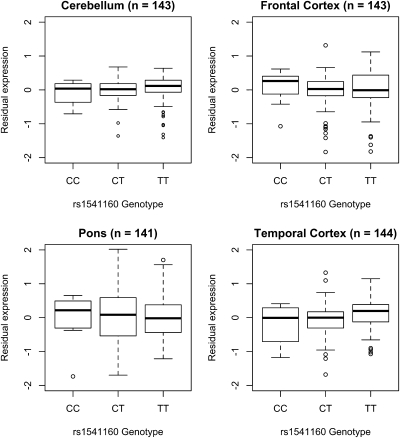
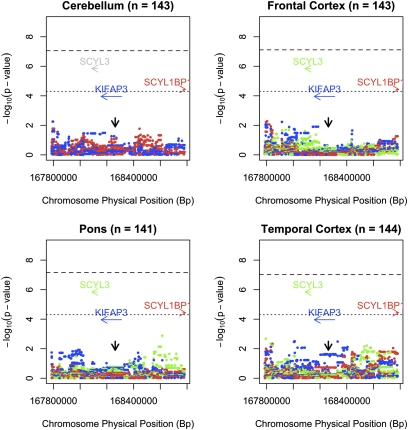
Expression quantitative trait loci (eQTL) mapping of the chromosome 1q24 region demonstrated that rs1541160 had no effect on KIFAP3 expression within the frontal cortex, temporal cortex, pons, or cerebellum of ∼144 neurologically normal brains [r2 in cerebellum = 0.01, P = 0.232; r2 in frontal cortex = 7.0 × 10−4, P = 0.76; r2 in pons = 2.1 × 10−5, P = 0.96; r2 in temporal cortex = 0.035, P = 0.03 (Fig. 2)]. Furthermore, none of the other SNPs within the locus influenced KIFAP3 expression or expression of any nearby gene (Fig. 3). To confirm that this discrepancy with the work of Landers et al. (5) was not due to methodological differences, we quantified expression of KIFAP3 in our 143 frontal cortex samples using the same Taqman gene expression assays used in their work. These experiments confirmed the lack of effect of rs1541160 on KIFAP3 expression levels [hs00183973, r2 = 0.003, P = 0.86; hs00946074, r2 = 0.018, P = 0.38 (Fig. S2)]. We also attempted to replicate the semiquantitative Western blot analysis of KIFAP3 published by Landers et al. (5) using frontal cortex samples from neurologically normal individuals who were homozygous for either the minor or major allele of rs1541160. These experiments used the same Santa Cruz Biotechnology KIFAP3 antibody as in the original work, and a second KIFAP3 antibody from BD Biosciences. Neither antibody demonstrated a significant effect of SNP genotype on protein level [Santa Cruz Biotechnology antibody sc-55598, P = 0.88; BD Biosciences antibody 610637, P = 0.57 (Fig. S3)].
Fig. 2.
Boxplots illustrating the dose relationship between allele load at rs1541160 and expression of KIFAP3 in human cerebellar tissue samples (n = 143), frontal cortex samples (n = 143), pons samples (n = 141), and temporal cortex samples (n = 144). Binary logarithm of residual expression is shown on the y axis, and rs1541160 genotypes are listed along the x axis. Notably, rs1541160 has no effect on KIFAP3 expression in any of the tested tissue types.
Fig. 3.
Expression quantitative trait loci (eQTL) across the KIFAP3 locus on chromosome 1q24 measured in human cerebellar tissue samples (n = 143), frontal cortex samples (n = 143), pons samples (n = 141), and temporal cortex samples (n = 144). In this analysis the allelic load at each of the 1,004 polymorphisms across the locus is tested for association with transcript levels of each gene within the locus. The results of the analysis are shown as log transformed P values (based on Cochran–Armitage test for trend) that are color-coded to match the transcript of interest. Notably, genotypes across this locus are not associated with KIFAP3 (blue), SCYL3 (green), or SCYL1BP1 (red) expression levels. Only transcripts expressed in each tissue are displayed. SCYL3 expression was not detected in cerebellar tissue. The dashed horizontal line represents the threshold for genome-wide significance (8.85 × 10−8), and the dotted horizontal line represents the threshold for significance correcting for 1,004 SNPs within the locus [0.05/(1,004 × 4 tissues) = 1.25 × 10−5]. The vertical black arrow indicates the position of rs1541160.
Discussion
Our data clearly show that SNP rs1541160 does not modify survival within our cohort of 504 Italian ALS cases. This result is surprising because Landers et al. (5) reported that the survival advantage associated with this SNP on chromosome 1q24 was large enough (14.9 mo representing a 44.5% increase in median survival of patients) to be consistently observed among the different, similar sized cohorts included in their analysis. One possible explanation for this discrepancy may arise from the manner in which the different cohorts were ascertained; the Italian cohort was collected in a population-based manner (6), whereas the cohorts in the original work were mostly drawn from patients attending ALS clinics (i.e., clinic series) (5). Survival statistics based on such clinic cohorts are known to consistently underestimate ALS mortality by up to one-third because of referral bias (4, 7). Longer living patients are more likely to attend an ALS specialty clinic, which in turn means they are more likely to access the disease-modifying medication riluzole and to have access to symptomatic therapies that further prolong their survival (8, 9). In contrast, the population-based cohorts are more reflective of the true survival pattern within the general ALS population. Another possible explanation for our observations is that the original association of KIFAP3 with ALS survival was a false-positive finding arising by chance from the several hundred thousand tests performed as part of any GWAS (10).
Population-specific genetic variability within the chromosome 1 region is unlikely to account for the discrepancy between our data and the original Landers et al. report (5). It is unlikely that rs1541160 would influence survival within the four European-ancestry populations reported in the original work and have no effect within a similarly sized Italian cohort. Furthermore, the minor allele frequency of rs1541160 in the Italian population (C allele frequency = 30.8%) was almost identical to those reported for the European and US populations in the original work (28.6–30.2%, respectively) and observed within the CEPH HapMap population (26.7%, release 23, 60 founders). It is also possible that the specific SNP that influences survival within the chromosome 1 locus varies from population to population. However, our data effectively exclude this scenario because none of the 1,004 SNPs (140 genotyped SNPs plus 864 imputed SNPs) within the 1 Mb surrounding the KIFAP3 locus had a significant effect on survival within the Italian cohort.
Our eQTL mapping data and Taqman gene expression assays failed to demonstrate an effect of rs1541160 or any SNP within the locus on KIFAP3 expression within ∼144 samples from four distinct brain regions. This observation agrees with three previously published eQTL datasets, including a study involving 427 liver samples (11), a previous study from our laboratory that analyzed 279 additional frontal cortex samples (12), and an analysis of data derived from 210 lymphoblast cell lines (Fig. S4) (13). In contrast, Landers et al. (5) found that the CC genotype of rs1541160 decreased expression of KIFAP3 by 41% based on 26 brain samples. Population differences cannot account for these contrasting results, as all 144 brains used to generate our expression data were non-Hispanic Europeans drawn from a similarly representative US population sample as used in the work of Landers et al. (5). Instead, the larger number of samples included in our expression analysis resulted in increased power to detect true quantitative trait loci, and to exclude false-positive associations.
In conclusion, our data did not replicate the reported effect of rs1541160 on ALS survival or on KIFAP3 expression in a population-based Italian sample. To date, none of the reported associations for ALS have been successfully replicated, suggesting that ALS may be a more genetically heterogeneous disease than previously recognized, and that results of genome-wide studies should be interpreted with caution. This may be especially true in the analysis of phenotype aspects of ALS (e.g., survival, age at onset, rate of progression) based on patients attending specialty ALS clinics, because the demographics of such cohorts may be influenced by referral bias.
Material and Methods
Samples.
Included in the study were 504 cases diagnosed with probable or definite ALS according to the El Escorial criteria (14). These cases were ascertained through PARALS, an ongoing population-based epidemiological study of ALS based in two regions of northwestern Italy (population = 4,332,842 in 2001) (6). The cohort consisted of 270 (53.6%) men and 234 (46.4%) women. 367 (72.8%) described limb-onset symptoms, whereas the remaining 137 (27.2%) presented with bulbar-onset disease. Mean age at onset was 61.5 y (SD = 11.1, range = 20.5–87.3), and 318 (63.1%) patients were deceased or had undergone tracheostomy at the time of last follow-up. Of the entire cohort, 449 (89.1%) were taking riluzole medication; of deceased patients, 283 (89.0%) were taking riluzole. Median survival of the entire cohort (n = 504) was 3.54 y (95% CI, 3.33–3.95 y), whereas median survival of the deceased-only cohort (n = 318) was 2.58 y (95% CI, 2.42–2.8 y). Median survival and riluzole use by rs1541160 genotype status is listed in Table S2. Written informed consent for genetic analysis was obtained from each individual, and appropriate institutional review board approval was obtained concerning human subjects.
Genotyping.
Two hundred sixty-eight samples were genotyped on Infinium HumanHap550 beadchips (Illumina) (15), and 236 samples were genotyped on Infinium HumanHap610-Quad beadchips (Illumina) according to the manufacturer's specifications; 535,468 SNPs were common across both platforms. These included rs1541160 and 140 other SNPs extending from rs10800456 (167.7 Mb) to rs1928716 (168.7 Mb, genome build 36.3) on chromosome 1q24.2.
Statistical Analysis.
Kaplan–Meier survival modeling (16) and differences in survival were measured by the logrank sum test (all cases weighted equally) and the Peto & Peto Generalized Wilcoxon test (earlier events weighted more heavily) using the survival package within R statistical software (version 2.9.0). Calculations were performed using the date of onset as day 0, and the primary endpoint was tracheostomy-free survival. Date of last follow up of patients was first August 2009. Multivariate linear regression models examining minor allele dosage associations with survival duration were adjusted for age at onset, gender, site of onset, and the first two components vectors generated from multidimensional scaling. Identical covariates were used in subsequent iterations of the time-dependent models. A cohort of 504 patients has 99.99% potential power to detect a 41% difference in survival assuming a 70% 5-y mortality rate in controls and a minor allele frequency of 0.308 (see Fig. S5 for power curves).
Expression Analysis.
As part of a larger project in our laboratory, frozen tissue samples of the frontal cortex, temporal cortex, pons, and cerebellum were obtained from 144 neurologically normal, American subjects of European ancestry. One hundred- to 200-mg aliquots of frozen tissue were sub-dissected from each of the samples and used for genotyping and expression assays. Genotyping was performed using Infinium HumanHap550 beadchips (Illumina) followed by imputation to ∼1.6 million SNPs after data cleaning. Profiling of 22,000 mRNA transcripts was performed using HumanRef-8 Expression beadchips (Illumina). A regression analysis was performed on the expression intensities generated for mRNA. Gender, age at symptom onset, postmortem interval, tissue source, and hybridization batch were included as covariates. Residuals from the regression analysis for each probe were then used as the quantitative trait for that probe in genome-wide association analysis looking for eQTLs, performed using the assoc function within PLINK, which correlates allele dosage with change in the trait (17, 18).
Taqman Expression Assays of KIFAP3.
Total RNA was isolated from human frontal cortex samples using TRIzol (Invitrogen). RT-PCR was performed from 0.5 μg of RNA using oligo-dT primers according to the manufacturer's instructions (SuperscriptIII; Invitrogen). After generating cDNA, quantitative RT-PCR was performed with four replicates per sample using Taqman gene expression primers for KIFAP3 (Applied Biosystems; hs00183973 for human KIFAP3, hs00946074 for human KIFAP3, and 4326317E for human GAPDH).
Semiquantitative Western Blot Analysis of KIFAP3.
Brain lysates were prepared by adding 500 mL of RIPA buffer with protease inhibitor (Protease Inhibitor Mixture Tablets; Roche Applied Science) to ≈50 mg of tissue followed by dounce homogenization. The lysate was then sonicated by twenty 1-s pulses and kept on ice for 30 min. After centrifugation (5,000 × g for 10 min at 4 °C), the supernatant was stored in aliquots at −80 °C. The concentration of each sample was determined by a BCA assay. Twenty micrograms of lysate was separated by SDS/PAGE and blotted onto PVDF membrane. The membranes were hybridized with a monoclonal antibody direct against KIFAP3 (sc-55598; Santa Cruz Biotechnology) at a 1:100 dilution. The BD Biosciences KIFAP3 antibody (#610637) was used at 1:500. Rabbit polyclonal antibody directed against β-Actin (A1978; Sigma-Aldrich) was used at a 1:2,000 dilution. Detection was performed using the ECL Plus Western Blotting Detection System (GE Healthcare) in conjunction with a Storm 860 Imaging System (Molecular Dynamics). Quantification of band intensity was performed using ImageQuant software (GE Healthcare).
Supplementary Material
Acknowledgments
This work was supported by Ministero della Salute, Ricerca Sanitaria Finalizzata 2007 (to A. Chiò, G.R., and G.M.), Fondazione Vialli e Mauro for ALS, Torino (to A. Chiò and G.M.), and Regione Piemonte, Progetti Finalizzati (to G.R.), and the Intramural Research Program of the National Institutes of Health, National Institute on Aging (Z01-AG000949-02), National Institute of Neurological Disorders and Stroke, the Packard Center for ALS Research at Hopkins (to B.J.T.), and the ALS Association (to B.J.T.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The data have been deposited in the dbGaP database, phs000101.v2.p1.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0914079107/-/DCSupplemental.
References
- 1.Chancellor AM, et al. The prognosis of adult-onset motor neuron disease: A prospective study based on the Scottish Motor Neuron Disease Register. J Neurol. 1993;240:339–346. doi: 10.1007/BF00839964. [DOI] [PubMed] [Google Scholar]
- 2.del Aguila MA, Longstreth WT, Jr, McGuire V, Koepsell TD, van Belle G. Prognosis in amyotrophic lateral sclerosis: A population-based study. Neurology. 2003;60:813–819. doi: 10.1212/01.wnl.0000049472.47709.3b. [DOI] [PubMed] [Google Scholar]
- 3.Millul A, et al. Survival of patients with amyotrophic lateral sclerosis in a population-based registry. Neuroepidemiology. 2005;25:114–119. doi: 10.1159/000086353. [DOI] [PubMed] [Google Scholar]
- 4.Chiò A, et al. Prognostic factors in ALS: A critical review. Amyotroph Lateral Scler. 2008;10:1–14. doi: 10.3109/17482960802566824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landers JE, et al. Reduced expression of the Kinesin-Associated Protein 3 (KIFAP3) gene increases survival in sporadic amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2009;106:9004–9009. doi: 10.1073/pnas.0812937106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiò A, et al. PARALS. Epidemiology of ALS in Italy: A 10-year prospective population-based study. Neurology. 2009;72:725–731. doi: 10.1212/01.wnl.0000343008.26874.d1. [DOI] [PubMed] [Google Scholar]
- 7.Lee JR, Annegers JF, Appel SH. Prognosis of amyotrophic lateral sclerosis and the effect of referral selection. J Neurol Sci. 1995;132:207–215. doi: 10.1016/0022-510x(95)00154-t. [DOI] [PubMed] [Google Scholar]
- 8.Chiò A, Bottacchi E, Buffa C, Mutani R, Mora G PARALS. Positive effects of tertiary centres for amyotrophic lateral sclerosis on outcome and use of hospital facilities. J Neurol Neurosurg Psychiatry. 2006;77:948–950. doi: 10.1136/jnnp.2005.083402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Traynor BJ, Alexander M, Corr B, Frost E, Hardiman O. Effect of a multidisciplinary amyotrophic lateral sclerosis (ALS) clinic on ALS survival: A population based study, 1996-2000. J Neurol Neurosurg Psychiatry. 2003;74:1258–1261. doi: 10.1136/jnnp.74.9.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao R, Boehnke M. Quantifying and correcting for the winner's curse in genetic association studies. Genet Epidemiol. 2009;33:453–462. doi: 10.1002/gepi.20398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schadt EE, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myers AJ, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 13.Veyrieras JB, et al. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brooks BR. El Escorial World Federation of Neurology criteria for the diagnosis of amyotrophic lateral sclerosis. Subcommittee on Motor Neuron Diseases/Amyotrophic Lateral Sclerosis of the World Federation of Neurology Research Group on Neuromuscular Diseases and the El Escorial “Clinical limits of amyotrophic lateral sclerosis” workshop contributors. J Neurol Sci. 1994;124(Suppl):96–107. doi: 10.1016/0022-510x(94)90191-0. [DOI] [PubMed] [Google Scholar]
- 15.Chiò A, et al. A two-stage genome-wide association study of sporadic amyotrophic lateral sclerosis. Hum Mol Genet. 2009;18:1524–1532. doi: 10.1093/hmg/ddp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaplan E, Meier P. Non-parametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 17.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibbs JR, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PloS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.