Each cell faces many challenges during its life cycle. One of the most crucial is ensuring that its genetic patrimony is accurately passed on to daughter cells. In bacteria, the most well-dissected DNA segregation mechanisms are those specified by low-copy-number plasmids. Whereas high-copy-number plasmids rely on passive diffusion for their distribution at cell division, low-copy-number plasmids require specialized mechanisms. To this end, they encode a “survival tool kit” enabling them to be delivered to and partitioned into daughter cells.
This tool kit is specified by a dedicated segregation locus, consisting of three essential components: (i) a gene encoding a polymerizing motor protein with an ATP- or GTP-binding motif, (ii) a gene encoding a DNA-binding protein, and (iii) a cis-acting centromere-like site, located either upstream or downstream of the two genes. Segregation loci have been assigned to three main categories (types I–III) on the basis of the genetic organization of the partition cassette and the phylogenetic relationships among the encoded proteins (1–4). Type I systems include a Walker-type ATPase, denoted as ParA: they are the most widespread segregation modules and are encoded by partition cassettes found on low-copy-number plasmids and on many bacterial and archaeal chromosomes (5). Type II modules encode a NTPase, ParM, which is an ancestral homolog of eukaryotic actin (6). Although evolutionarily unrelated, both ParA and ParM proteins are polymerization-based engines that assemble into cytoskeletal structures involved in DNA transport, positioning, and segregation (3, 4). Type III systems were identified more recently: this discovery has opened up unforeseen perspectives on prokaryotic cytoskeletal proteins mediating genome segregation (7–9). In PNAS, Ni et al. shed light on the molecular mechanisms underpinning the function of type III partition cassettes by reporting the structures of TubR and TubZ proteins encoded by the pBtoxis plasmid from Bacillus thuringiensis (10).
Type III segregation modules are found on large plasmids of the Bacillus cereus group of bacteria (7, 9). The partition cassette of plasmid pBtoxis harbors two genes: orf157 encoding an 11.6-kDa specific DNA-binding protein, known as TubR, and orf156 encoding a 54.4-kDa protein, designated as TubZ, which is a distant homolog of both tubulin and FtsZ (7, 8). Both orf157 and orf156 are necessary for plasmid maintenance (11). TubR binds to a cis sequence of four iterons located upstream of the genes (8). TubZ polymerizes in vivo into filaments that exhibit directional growth and translocate within the cell by treadmilling, elongating at one end while retracting at the opposite end (7). TubZ has also been shown to assemble into two-stranded polymers in vitro upon binding GTP (12). The protein is characterized by a strong GTPase activity, and as a consequence, TubZ subunits within the polymers are almost entirely in the GDP-bound form. It was suggested that the growing tip of TubZ filaments may contain a GTP cap that stabilizes the polymer (12).
The report by Ni et al. describes the structure of the TubR protein, the centromere-binding component of the TubRZ system (10). The authors show that TubR forms a highly intertwined dimer. Each monomer harbors five α-helices and three β-ribbons arranged in the sequence β1-α1-α2-α3-α4-β2-β3-α5, which contains a winged helix-turn-helix (HTH) motif structurally similar to that found in bacterial transcriptional repressors of the ArsR family (10). This finding is very intriguing, as to date no DNA segregation protein has been shown to be homologous to the metal-binding transcription factors of the ArsR group. The authors further investigate this parallel by superimposing TubR structure onto that of CzrA from Staphylococcus aureus, the closest structural homolog of the ArsR cluster. The overlay highlights similarities in the winged HTH motif. However, four key differences emerge: (i) TubR forms a dimer that is remarkably distinct from that formed by CzrA; (ii) TubR does not contain the metal-binding motif typical of either the ArsR proteins or any other metal-regulated factor; (iii) the monomer–monomer interface is extremely different in the TubR dimer compared with the CzrA dimer; and (iv) the recognition helices of the HTH domain are solvent exposed in CzrA, whereas they are largely buried in the TubR dimer (10). This last observation raises a unique theme: the recognition helices of the HTH motif mediate dimerization of TubR and in doing so, they remain buried in the monomer–monomer interface, leaving only some N-terminal residues accessible for other interactions. Interestingly, in this case the recognition helices of TubR are involved in protein–protein contacts as opposed to the conventional and well-established role in protein–DNA association.
If the so-called recognition helices are not available to make contacts with DNA, how does TubR bind to the centromere? The Schumacher team has identified a large basic patch on the surface of TubR: this region includes residues Arg-74, Arg-77, and Lys-79 located in the wing (the loop between β2 and β3) adjacent to the HTH and Lys-43 in the α3 helix preceding the recognition helix, α4, in the HTH signature (10). A mutational analysis confirmed that these positively charged residues are crucial for DNA binding, indicating that this patch represents the bona fide DNA-binding domain of TubR. In addition, a model illustrating the mechanistic interaction of TubR with DNA was constructed by docking a DNA duplex onto the basic patch of TubR. The model reveals that the wings (one from each monomer) interact with consecutive minor grooves of the DNA, whereas the N termini of the recognition helices are inserted into a single major groove (10). The classical architecture of HTH-DNA complexes involves the introduction of the recognition helices into successive major grooves of the DNA. Therefore, the strategy adopted by TubR to recognize DNA with its winged HTH domain is a unique finding that has major implications not only in the field of genome segregation, but also in that of mechanisms of protein–DNA interaction.
Once a centromere-binding protein has found and bound its cargo, either plasmid or chromosome, it associates with the partner motor protein (2, 3). Ni et al. show that TubZ binds the TubR–plasmid complex by using its C terminus. To understand the role and behavior of this distant relative of tubulin and FtsZ, the authors solve the structure of pBtoxis TubZ (1–428) in its apo- and GTPγS-bound forms. The structures reveal that TubZ is monomeric and confirm that TubZ is a member of the tubulin/FtsZ family of cytoskeletal proteins (10). Although substantially similar, the structures of tubulin, FtsZ, and TubZ display significant differences in their N and C termini, which reflect the divergent function-dependent specialization and the diverse interactions of these proteins with various binding partners. The N terminus of TubZ comprises a helix, H0, which is also present in the N-terminal region of Methanococcus jannaschii FtsZ. However, whereas H0 is rather flexible and involved in monomer–monomer association during protofilament assembly in M. jannaschii FtsZ, the H0 helix of TubZ is tightly tethered to its C terminus and exhibits no changes in conformation following polymerization. Further, the extreme C-terminal tips of tubulin, FtsZ, and TubZ are structurally very different. Again, divergent structures have evolved for diverse tasks, as the FtsZ C terminus mediates the binding of FtsA and ZipA, which anchor the FtsZ ring to the membrane (13); the C-terminal domain of tubulin interacts with multiple proteins, including microtubule-associated proteins (MAPs), which modulate microtubule dynamics (14); and the C terminus of TubZ associates with TubR (10). Another interesting finding that emerges from the data of Ni et al. concerns the assembly of TubZ into protofilaments. The authors suggest that, upon GTP binding, TubZ forms protofilaments, in which longitudinal monomer–monomer contacts are quite different from those in FtsZ and tubulin polymers. They also suggest that TubZ does not assemble into higher-order structures like microtubules.
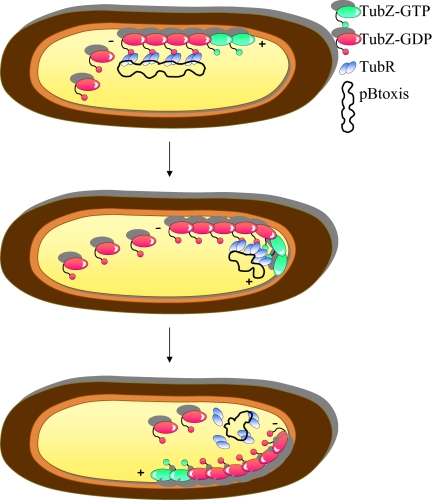
How does the TubR–TubZ machine drive segregation of the pBtoxis plasmid at cell division? The study by Ni et al. provides key insights into this mechanism. Quadruplet TubR dimers bind to their attachment site on the pBtoxis plasmid and hold this cargo (Fig. 1). Apo-TubZ monomers bind GTP and assemble into a linear protofilament that can be described as a moving cable car. The TubR-pBtoxis complex hops on the TubZ polymer via the interaction of TubR with the surface-exposed, flexible C-terminal tip of TubZ subunits. GTP hydrolysis within the TubZ polymer generates treadmilling, elongation at the plus end and retraction at the minus end, resulting in the translocation of the plasmid to the cell pole. How does pBtoxis hop off the TubZ cable car once the cell pole is reached? Ni et al. propose that the bending of the TubZ filament at the cell pole exerts strain on the bound cargo, causing the detachment of TubR-pBtoxis.
Fig. 1.
Model for TubRZ-mediated segregation of pBtoxis. Snapshots of three successive stages of the process are shown. The TubR-pBtoxis complex associates with the treadmilling TubZ polymer via the interaction of TubR with the flexible C terminus of TubZ subunits. TubR bound to its cargo is handed over to successive TubZ subunits of the polymer in the (+) elongating direction. The bending of the TubZ filament at the cell pole exerts strain on the bound cargo, causing detachment of TubR-pBtoxis.
Although the picture of TubRZ-mediated DNA segregation is taking shape, many tesserae of the mosaic are still missing. How does the TubZ filament find the TubR-pBtoxis complex? Is this complex “waiting” to hop on at specific stops along the route of the TubZ cable car? Is the TubZ polymer somehow tethered to the cell membrane or is it a “floating” filament? How are the newly replicated plasmids delivered to opposite cell halves? Is TubR involved in TubZ polymer remodelling dynamics? All these questions, and more, represent future challenges that will widen the horizons of the molecular mechanisms of genome segregation and of the dynamics of prokaryotic cytoskeletal structures.
Acknowledgments
Work in my laboratory is supported by the Biotechnology and Biological Sciences Research Council (Grant BB/F012004/1) and the Medical Research Council (Grant G0801162).
Footnotes
The author declares no conflict of interest.
See companion article on page 11763 in issue 26 of volume 107.
References
- 1.Ebersbach G, Gerdes K. Plasmid segregation mechanisms. Annu Rev Genet. 2005;39:453–479. doi: 10.1146/annurev.genet.38.072902.091252. [DOI] [PubMed] [Google Scholar]
- 2.Hayes F, Barillà D. The bacterial segrosome: A dynamic nucleoprotein machine for DNA trafficking and segregation. Nat Rev Microbiol. 2006;4:133–143. doi: 10.1038/nrmicro1342. [DOI] [PubMed] [Google Scholar]
- 3.Schumacher MA. Structural biology of plasmid partition: Uncovering the molecular mechanisms of DNA segregation. Biochem J. 2008;412:1–18. doi: 10.1042/BJ20080359. [DOI] [PubMed] [Google Scholar]
- 4.Hayes F, Barillà D. In: Bacterial Chromatin. Dame RT, Dorman CJ, editors. The Netherlands: Springer, Dordrecht; 2010. pp. 49–70. [Google Scholar]
- 5.Thanbichler M, Shapiro L. Chromosome organization and segregation in bacteria. J Struct Biol. 2006;156:292–303. doi: 10.1016/j.jsb.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Bork P, Sander C, Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci USA. 1992;89:7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larsen RA, et al. Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis. Genes Dev. 2007;21:1340–1352. doi: 10.1101/gad.1546107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang M, Bideshi DK, Park H-W, Federici BA. Iteron-binding ORF157 and FtsZ-like ORF156 proteins encoded by pBtoxis play a role in its replication in Bacillus thuringiensis subsp. israelensis. J Bacteriol. 2007;189:8053–8058. doi: 10.1128/JB.00908-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anand SP, Akhtar P, Tinsley E, Watkins SC, Khan SA. GTP-dependent polymerization of the tubulin-like RepX replication protein encoded by the pXO1 plasmid of Bacillus anthracis. Mol Microbiol. 2008;67:881–890. doi: 10.1111/j.1365-2958.2007.06100.x. [DOI] [PubMed] [Google Scholar]
- 10.Ni L, Xu W, Kumaraswami M, Schumacher MA. Plasmid protein TubR uses a distinct mode of HTH DNA-binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition. Proc Natl Acad Sci USA. 2010;107:11763–11768. doi: 10.1073/pnas.1003817107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang M, Bideshi DK, Park H-W, Federici BA. Minireplicon from pBtoxis of Bacillus thuringiensis subsp. israelensis. Appl Environ Microbiol. 2006;72:6948–6954. doi: 10.1128/AEM.00976-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Erickson HP. In vitro assembly studies of FtsZ/tubulin-like proteins (TubZ) from Bacillus plasmids: Evidence for a capping mechanism. J Biol Chem. 2008;283:8102–8109. doi: 10.1074/jbc.M709163200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Margolin W. FtsZ and the division of prokaryotic cells and organelles. Nat Rev Mol Cell Biol. 2005;6:862–871. doi: 10.1038/nrm1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Downing KH. Structural basis for the interaction of tubulin with proteins and drugs that affect microtubule dynamics. Annu Rev Cell Dev Biol. 2000;16:89–111. doi: 10.1146/annurev.cellbio.16.1.89. [DOI] [PubMed] [Google Scholar]