Abstract
Phosphatidylinositol (PtdIns) lipids have been identified as key signaling mediators for random cell migration as well as chemoattractant-induced directional migration. However, how the PtdIns lipids are organized spatiotemporally to regulate cellular motility and polarity remains to be clarified. Here, we found that self-organized waves of PtdIns 3,4,5-trisphosphate [PtdIns(3,4,5)P3] are generated spontaneously on the membrane of Dictyostelium cells in the absence of a chemoattractant. Characteristic oscillatory dynamics within the PtdIns lipids signaling system were determined experimentally by observing the phenotypic variability of PtdIns lipid waves in single cells, which exhibited characteristics of a relaxation oscillator. The enzymes phosphatase and tensin homolog (PTEN) and phosphoinositide-3-kinase (PI3K), which are regulators for PtdIns lipid concentrations along the membrane, were essential for wave generation whereas functional actin cytoskeleton was not. Defects in these enzymes inhibited wave generation as well as actin-based polarization and cell migration. On the basis of these experimental results, we developed a reaction-diffusion model that reproduced the characteristic relaxation oscillation dynamics of the PtdIns lipid system, illustrating that a self-organization mechanism provides the basis for the PtdIns lipids signaling system to generate spontaneous spatiotemporal signals for random cell migration and that molecular noise derived from stochastic fluctuations within the signaling components gives rise to the variability of these spontaneous signals.
Keywords: phosphatase and tensin homolog, spontaneous polarization, relaxation oscillation, traveling wave, noise
How the biological functions of living cells arise spontaneously and intrinsically from the dynamics of intracellular reaction networks is a central question in cell biology. Random cell migration under no or uniform external cues, which is a fundamental form of cell motility, is an example of this spontaneity (1–4). To induce motile activity in random directions, internal signals need to be generated without any external cues. Specific intracellular processes are likely responsible for such spontaneous signal generation. Therefore, it is important to understand the internal processes responsible for spontaneous signal generation and their regulatory mechanisms in the absence of external stimuli.
Stochastic fluctuations in molecular reaction networks may be an origin for the internal signals that drive random cell migrations. For example, in the case of paramecium cells, a mechanism for spontaneous signal generation has been proposed in which a hierarchical organization exists ranging from small thermal fluctuations in membrane ion channels to large spike-like fluctuations in membrane potentials that generate signals to stochastically turn their direction of motion in homogeneous environments (1). In bacteria, the noise amplification of signaling reactions through high gain responses has been implicated in the generation of stochastic signals for flagella motors (2, 5). In contrast to these stochasticity-based forms of random cell motility, cells that exhibit crawling movements over surfaces using actin-based motile machinery such as pseudopods and tails need to be organized both spatially and temporally to generate cell polarity and motility (6). Therefore, the spontaneous signal generation in these crawling-type cells should include a spatiotemporal organization mechanism as well as a stochastic mechanism, distinguishing them from swimming-type cells like the aforementioned bacteria. To explain the cytoskeletal organization relating to cellular polarity and motility, a self-organization concept has been suggested (7–12). A spatiotemporal self-organization has been previously described in actin-based motile machinery in which the collective behaviors of cytoskeletal proteins and molecular motors give rise to highly organized supramolecular structures such as actin waves in living cells (7–12). However, how signaling mediators working upstream of actin-based motile machinery are involved in the self-organization process is unknown.
Within Dictyostelium discoideum cells that exhibit random migration using actin-based motile machinery (3, 4, 6), intracellular phosphatidylinositol (PtdIns) lipids have been identified as key signaling mediators for directional cell migration in response to the extracellular chemoattractant cyclic adenosine 3′,5′-monophosphate (cAMP) (13, 14). Membrane-bound PtdIns 3,4,5-trisphosphate [PtdIns(3,4,5)P3] has been described as a key element for the biochemical compass to direct chemotaxis in which the chemoattractant-induced localization of PtdIns(3,4,5)P3 on the membrane serves as a cue for pseudopod formation (13–17). Phosphoinositide-3-kinase (PI3K) and phosphatase and tensin homolog (PTEN), which catalyze PtdIns(3,4,5)P3 production and degradation, respectively, have been revealed to regulate chemotaxis (3, 18–21). In addition to their roles in chemotaxis, PI3K and PTEN have been revealed to regulate random cell motility and polarity in the absence of cAMP (3, 18–21). However, few studies have focused on the intrinsic dynamics of PtdIns lipids in cells with no chemoattractants. To understand how motile signals for random cell migration arise spontaneously from the internal dynamics within intracellular reaction networks, we explored dynamical aspects of the PtdIns lipids signaling system in living Dictyostelium cells in a nonstimulated state. We found that the self-organization of the PtdIns lipids signaling system, which spontaneously generates PtdIns(3,4,5)P3-enriched domains that travel rotationally along the membrane of living cells in an F-actin-independent manner, provides a mechanistic basis for spontaneous cell polarization and migration in the absence of chemoattractants.
Results and Discussion
Self-Organized Traveling Waves in the PtdIns Lipids Signaling System.
To monitor the dynamics of the PtdIns lipids system, we prepared Dictyostelium cells that express the fluorescently labeled pleckstrin-homology domain of Akt/PKB (PHAkt/PKB) and PTEN, where PHAkt/PKB and PTEN were used as indicators for PtdIns(3,4,5)P3 metabolism. PHAkt/PKB can bind to PtdIns(3,4,5)P3 at the membrane, whereas PTEN catalyzes the degradation of PtdIns(3,4,5)P3 and has a binding motif for PtdIns(4,5)P2 (18, 21). PHAkt/PKB was tagged with enhanced green fluorescent protein (PHAkt/PKB-EGFP), whereas PTEN was tagged with HaloTag with the fluorescent dye tetramethylrhodamine (PTEN-TMR). Both proteins were coexpressed in wild-type cells without any obvious defects in growth, chemotaxis, and development. We observed PHAkt/PKB-EGFP and PTEN-TMR in developed cells chemotactically competent to cAMP. Consistent with previous observations (15–17, 19, 20), PHAkt/PKB-EGFP and PTEN-TMR localization exhibited reciprocal polarity in locomoting cells (Fig. 1A and Movie S1). PI3K was also localized at the pseudopod regions (Fig. 1B). Such reciprocal localization of PI3K and PTEN in cells leads to the polarized localization of PtdIns(3,4,5)P3 along the membrane, providing binding sites for PHAkt/PKB.
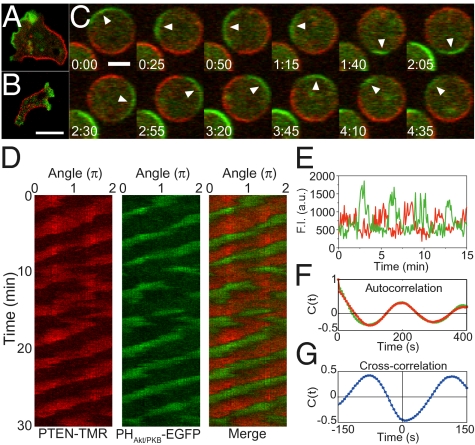
Fig. 1.
Self-organized traveling waves of PHAkt/PKB and PTEN. (A and B) Fluorescent image of Dictyostelium discoideum cells expressing PTEN-TMR (red) and PHAkt/PKB-EGFP (green) (A) and cells expressing PTEN-TMR (red) and PI3K2-EGFP (green) (B). Both PHAkt/PKB-EGFP and PI3K2-EGFP were localized at the pseudopod regions. PTEN-TMR was localized at the lateral and tail regions of migrating cells. (Scale bar, 10 μm.) (C) Time-lapse images of a cell treated with 5 μM latrunculin A. Arrowheads indicate a PHAkt/PKB- enriched domain on the membrane. Time, min:s. (Scale bar, 5 μm.) (D) Kymograph of PTEN-TMR and PHAkt/PKB-EGFP showing traveling waves while maintaining reciprocal localization. (E) Time trajectories of fluorescence intensities of PTEN-TMR (red) and PHAkt/PKB-EGFP (green) on a subdomain of the membrane. (F) Temporal autocorrelation function for the PTEN-TMR (red) and PHAkt/PKB-EGFP (green) shown in D. (G) Temporal cross-correlation function between PTEN-TMR and PHAkt/PKB-EGFP.
This polarized localization of both PHAkt/PKB-EGFP and PTEN-TMR was observed even in the absence of external cAMP and cellular motile activities (Fig. 1C). To exclude any spatiotemporal effects from actin-based shape changes or motility on the dynamics of the PtdIns lipids system, cells were treated with 5 μM of the actin polymerization inhibitor latrunculin A, leading to a spherical shape lacking both motile and protrusive activities. The cells were also observed in the presence of 4 mM caffeine, which inhibits the self-production of cAMP catalyzed by intracellular adenylyl cyclase (SI Discussion and Fig. S1) (22). Thus, the observed dynamics are thought to be independent of actin-based motility and extracellular chemoattractant-evoked events. As shown in Fig. 1C, both PHAkt/PKB-EGFP- and PTEN-TMR-enriched domains were not stationary. Rather, they traveled along the membrane while maintaining their reciprocal polarity (Movie S2). We performed kymograph analysis in which fluorescence intensities of PHAkt/PKB-EGFP and PTEN-TMR along the cell periphery were plotted with time to show the wave propagation clearly (Fig. 1D) (SI Materials and Methods). These localized domains exhibited continuous propagation in one direction at an almost constant speed along the membrane for >30 min. Fluorescence intensities at a given position of the membrane exhibited a periodic transition between PHAkt/PKB- and PTEN-enriched states (Fig. 1E). The autocorrelation functions of the two time series showed an oscillatory nature with a period of ~200 s (Fig. 1F). Their cross-correlation function exhibited anticorrelation, showing a clear reciprocal relationship between PHAkt/PKB-EGFP and PTEN-TMR membrane levels (Fig. 1G).
A variety of spatiotemporal patterns in PHAkt/PKB-EGFP and PTEN-TMR localization were observed in individual cells (Fig. 2). Cells exhibited not only continuous propagation of localized domains, but also the generation, separation, and fusion of domains and reversal in their propagation directions. The spatiotemporal auto- and cross-correlation functions of these patterns revealed several modes of ordered patterns such as traveling waves (Fig. 2 A and C) and spatiotemporal oscillations (standing waves) (Fig. 2 B and D). However, even in these cases, the reciprocal localization of both PHAkt/PKB-EGFP and PTEN-TMR was maintained. For these observations (n = 95 cells), >92% of the cells exhibited the ordered patterns whereas the remainder exhibited a membrane uniformly covered with PTEN-TMR lacking a distinctive PHAkt/PKB-EGFP domain (Movie S3). Although the expression levels of both proteins vary among individual cells even when a cloned cell line was used, cells expressing the proteins at an extremely low level still exhibited the ordered patterns (Movie S3). In fact, wild-type cells expressing either PHAkt/PKB-EGFP or PTEN-TMR showed similar spatiotemporal dynamics, indicating that the exogenous expression of both proteins was not essential for these patterns to occur.
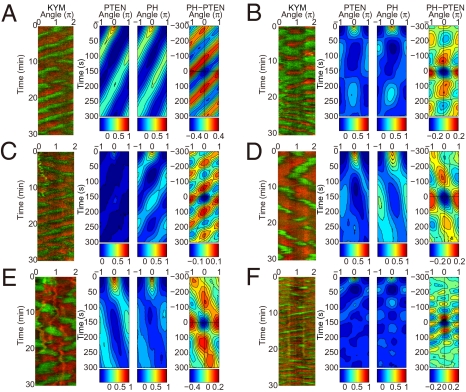
Fig. 2.
Various ordered patterns of PHAkt/PKB and PTEN. (A–F) Gallery of kymographs of PHAkt/PKB-EGFP and PTEN-TMR (Left, KYM). Each kymograph corresponds to different single cells. Developed cells (A–D) and vegetative cells in the absence (E) or the presence (F) of 4 mM caffeine are shown. Each cell shows different patterns despite the same experimental conditions. The corresponding spatiotemporal autocorrelation of PTEN-TMR (PTEN) and PHAkt/PKB-EGFP (PH) and the spatiotemporal cross-correlation between PHAkt/PKB-EGFP and PTEN-TMR (Right, PH-PTEN), plotted as a function of time and angle differences, are shown. Intensities are explained below each kymograph.
Because Dictyostelium cells have been seen to exhibit random migration in their vegetative stages similar to developmental stages by the PI3K/PTEN pathway (3), it follows that if the PtdIns(3,4,5)P3/PTEN waves are associated with random motility, then the vegetative cells should exhibit similar waves. As shown in Fig. 2, vegetative cells treated with latrunculin A exhibited ordered patterns even in the absence of 4 mM caffeine (Fig. 2 E and F). In locomoting cells without latrunculin A, PHAkt/PKB and PTEN were localized with a reciprocal polarity (Fig. S1C). These observations in vegetative cells are almost the same as those in developed cells, suggesting that the self-organizing properties of the PtdIns lipids signaling system in vegetative cells are shared at least in part with those in developed cells. Furthermore, the fact that vegetative cells do not use autocrine signaling through self-production of cAMP suggests that developmental stage-specific components such as cAMP receptors and adenylyl cyclase are not essential for the wave generation.
Involvement of PI3K and PTEN in the PtdIns Lipids Wave Generation As Well As Random Cell Migration.
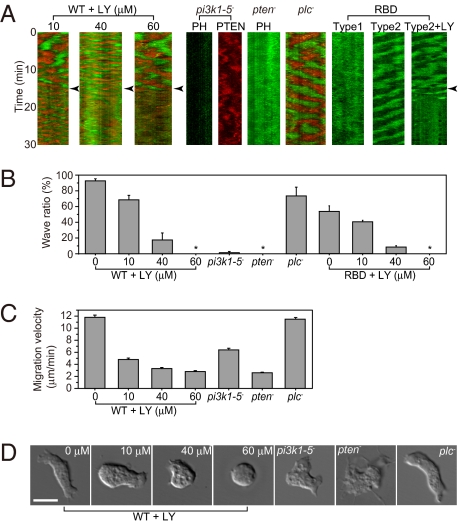
We studied the roles of three key enzymes, PI3K, PTEN, and phospholipase C (PLC), on both the self-organized dynamics of the PtdIns lipids system and random cell migration under no external chemoattractant. When cells were treated with the PI3K inhibitor LY294002, the oscillation periods of their waves modulated in ~30% and 80% of the cell populations at 10 μM and 40 μM LY294002, respectively, but disappeared in most cells at 60 μM LY294002 (Fig. 3 A and B). Because PtdIns(3,4,5)P3 production is inhibited partially at 10 μM LY294002 but completely at 60 μM (23), the results suggest PI3K activity is associated with PtdIns(3,4,5)P3/PTEN wave generation. Because LY294002 has been known to significantly inhibit kinases other than PI3K at higher concentrations, we examined the pi3k1-5-null cells lacking all PI3K genes (24). In these cells, PHAkt/PKB-EGFP localized uniformly in the cytosol but not on the membrane (Fig. 3 A and B). Furthermore, PTEN-TMR localized on the membrane in a polarized manner, but these membrane localizations showed no continuous traveling waves (Fig. 3 A and B and Movie S4). Spatiotemporal correlation analysis of a kymograph revealed continuous PTEN-TMR wave generation in wild-type cells but not in the pi3k1-5-null cells (SI Discussion and Fig. S2). These results indicate that PI3K is required for the generation of PtdIns(3,4,5)P3/PTEN waves. In pten-null cells, enhanced accumulation of PHAkt/PKB-EGFP was observed uniformly over the entire cell surface with no traveling waves (Fig. 3 A and B). This result is probably because of an abundance of PtdIns(3,4,5)P3 on the membrane due to a loss of PTEN activity. In contrast to PI3K and PTEN, PLC was not essential for the generation of PtdIns(3,4,5)P3/PTEN waves, as PtdIns(3,4,5)P3/PTEN waves in plc-null cells were indistinguishable from those in wild-type cells (Fig. 3 A and B). These results demonstrate that the enzymes PI3K and PTEN are involved in the self-organized traveling waves of the PtdIns lipids system.
Fig. 3.
Involvement of PI3K and PTEN in signal generation for cell polarization and migration. (A) PHAkt/PKB-EGFP and/or PTEN-TMR dynamics in cells in the presence of latrunculin A and an inhibited PtdIns lipids system. At the time points indicated by the arrowheads, wild-type cells were treated with LY294002 (+LY). pi3k1-5-null (pi3k1-5−), pten-null (pten−), and plc-null (plc−) cells were also examined. GFP-RBD was observed in wild-type cells. (B) Summary of the self-organized wave inhibition. From left to right: wild-type cells (95 cells, n = 16) at 10 μM (136 cells, n = 4), 40 μM (119 cells, n = 3), and 60 μM (78 cells, n = 8) LY294002; pi3k1-5-null cells (60 cells, n = 5), pten-null cells (55 cells, n = 6), and plc-null cells (82 cells, n = 4); and RBD wave (121 cells, n = 10) at 10 μM (84 cells, n = 6), 40 μM (47 cells, n = 3), and 60 μM (51 cells, n = 3) LY294002. Data are mean ± SEM; n, number of independent experiments; *, no waves. (C) Migration velocity of the cells: wild type (n = 157), at 10 μM (n = 150), 40 μM (n = 174), and 60 μM (n = 137) LY294002; and pi3k1-5-null (n = 103), pten-null (n = 109), and plc-null (n = 104). Data are mean ± SEM; n, number of cells. (D) Cell shapes observed in the absence of latrunculin A. (Scale bar, 10 μm.)
For PI3K activation, Ras has been revealed to be essential in the random migration of Dictyostelium cells, which functions as a component in the Ras-PI3K-PtdIns(3,4,5)P3 positive feedback loop in an F-actin-dependent manner (3). Even when using latrunculin A to inhibit actin polymerization, PHAkt/PKB still shows localization along the membrane under our experimental conditions, which is conceivable as a result of PtdIns(3,4,5)P3 accumulation produced by activated PI3K. To see how PI3K is activated for the PtdIns(3,4,5)P3/PTEN wave generation, we examined Ras activation by using the GFP-labeled Ras-binding domain of Raf1 (GFP-RBD), which acts as an indicator for activated Ras (25). We observed two types of GFP-RBD dynamics in wild-type cells (Fig. 3 A and B). In type 1, GFP-RBD was localized uniformly over the entire cell surface with no obvious traveling waves, showing uniform activation of Ras along the membrane. In type 2, GFP-RBD exhibited traveling waves with an oscillation period similar to those of PHAkt/PKB-EGFP and PTEN-TMR, showing localized activation of Ras. The PI3K inhibitor LY294002 suppressed GFP-RBD waves in a dose-dependent manner, indicating that RBD waves are somehow coupled to PtdIns(3,4,5)P3/PTEN waves (Fig. 3 A and B). However, because the number of cell populations categorized as type 2 and exhibiting RBD waves (54%) was significantly less than that exhibiting PtdIns(3,4,5)P3/PTEN waves (92%), we conclude that RBD waves are not prerequisite for PtdIns(3,4,5)P3/PTEN waves and that uniform activation of Ras and PI3K along the membrane is sufficient for the generation of PtdIns(3,4,5)P3/PTEN waves. Even if the Ras-PI3K-PtdIns(3,4,5)P3 feedback loop somehow functions in latrunculin A-treated cells, the effects of the feedback loop on the generation of both PtdIns(3,4,5)P3/PTEN and RBD waves would only be partial, which can thus explain why the generation of RBD waves was observed only in some cells. Uniform activation of Ras and PI3K along the membrane was probably due to the presence of caffeine because GFP-RBD was not observed on the membrane in the absence of caffeine (3).
We then examined cell shape and motility. In the absence of latrunculin A, which means no direct inhibition of the actin cytoskeleton, cells treated with LY294002 exhibited defects in motility in a dose-dependent manner (Fig. 3C). At 10 μM LY294002, cells were still able to move but the movements were inefficient (Fig. 3 C and D). When treated with 60 μM LY294002, most cells exhibited no protrusive activities, resulting in round, immobile cells (Fig. 3 C and D). Thus, prevention of PtdIns(3,4,5)P3/PTEN waves by LY294002 treatment largely correlated with the suppression of cell motility irrespective of whether LY294002 has off-target effects on kinases other than PI3K. Genetic disruption of PI3K resulted in the reduction of cell migration speed, clearly showing that PI3K is involved in random cell migration, consistent with previous reports (24). Because the defects observed in pi3k-1–5-null cells are less severe than those observed in wild-type cells following 60 μM LY294002 treatment, it can be concluded that some PI3K-independent cell migration is sensitive to LY294002 treatment at higher concentrations. Furthermore, pten-null cells formed abnormal pseudopods along the entire cell surface and moved less effectively (Fig. 3 C and D). This result is consistent with reports showing that defects in PtdIns(3,4,5)P3 metabolism mediate abnormal signaling for pseudopod formation (13–17). On the other hand, plc-null cells exhibited random migration with velocities and morphologies similar to wild-type cells (Fig. 3 C and D). These results suggest that the PI3K/PTEN-dependent self-organizing properties of the PtdIns lipids system are associated in part with cell migration in the absence of an external chemoattractant. In contrast, the PI3K-independent cell migration seen in pi3k-1–5-null cells suggests some other mechanism(s) for cell mobility absent of PtdIns(3,4,5)P3/PTEN waves. Although PLC has been suggested to modulate PtdIns(4,5)P2 levels during chemotactic responses (26), it is not essential for PtdIns(3,4,5)P3/PTEN wave generation and random cell migration in the absence of chemoattractants.
Reconstructed Spatiotemporal Dynamics of the PtdIns Lipids Signaling System.
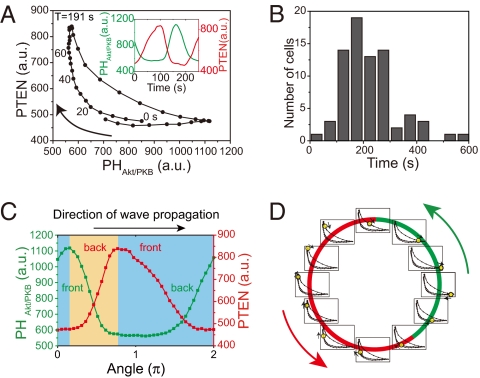
To reconstruct the dynamics of the PtdIns lipids system, we quantitatively extracted the dominant dynamics from the temporal changes in the fluorescence intensities of [PHAkt/PKB] and [PTEN] (SI Materials and Methods and Fig. S3). The reconstructed dynamics shown in [PHAkt/PKB]-[PTEN] coordinates exhibited a reciprocal relation with a characteristic crescent shape and temporal clockwise rotation (Fig. 4A). Note that the temporal progression of the oscillation was not constant along the crescent-shaped trace. The PtdIns lipids system can adopt either a stable PHAkt/PKB-enriched (PTEN-depleted) or a PTEN-enriched (PHAkt/PKB-depleted) state in a local region on the membrane. However, the transition between these two states is fast. This type of oscillation is known as relaxation oscillation (27). Despite the different kymograph patterns (Fig. 2), the characteristic crescent-shaped dynamics were commonly observed in all cells analyzed (n = 75 cells) (Fig. S4 A–L). The period of oscillation was 217.6 ± 99.9 s (Fig. 4B; mean ± SD, n = 75 cells). Such oscillatory dynamics were also observed in cells that partially recovered their actin cytoskeleton using modest treatment of latrunculin A (1–2.5 μM), resulting in similar periods of oscillations (166.5 ± 48.1 s; mean ± SD, n = 11 cells) (Fig. S5 and Movie S5). This partial recovery of the actin cytoskeleton caused spontaneous shape changes with partial motility in which PHAkt/PKB- and PTEN-enriched regions correlated with the formation and suppression of membrane protrusion, respectively. Thus, the oscillatory dynamics of the PtdIns lipids system are correlated at least in part with the spontaneous dynamics of actin-based polarity and motility.
Fig. 4.
Dynamics of the PtdIns lipid system. (A) Averaged temporal evolution dynamics of PTEN-TMR and PHAkt/PKB-EGFP obtained by principal components analysis of the traveling waves observed in the cell shown in Fig. 1. (Inset) Time trajectories of the averaged dynamics of PTEN-TMR (red) and PHAkt/PKB-EGFP (green). (B) Histograms of the oscillation periods obtained by principal components analysis of the traveling waves (n = 75 cells). (C) Spatial profiles of PTEN-TMR (red) and PHAkt/PKB-EGFP (green) along the membrane (0–2π) at a given time. The upper and lower branches of the averaged time evolution dynamics shown correspond to profiles in the yellow and blue areas, respectively. (D) Schematic illustration of wave propagation. Spatial profiles of PHAkt/PKB [PtdIns(3,4,5)P3] and PTEN are indicated as green and red bars along the membrane, respectively. At individual local regions on the membrane, the PtdIns lipid system undergoes dynamical changes in [PHAkt/PKB] and [PTEN] in accordance with the averaged oscillatory dynamics shown in A. The phases of the oscillation at individual local regions are indicated by yellow dots on the crescent-shaped traces. Arrows indicate the wave propagation direction.
The reconstructed dynamics of the PtdIns lipids system shown in Fig. 4A also reveal the spatial profiles of PTEN- and PHAkt/PKB-enriched domains along the membrane at a given time (Fig. 4C). The clockwise temporal evolutions along the crescent-shaped trace were equivalent to spatial changes along the membrane in the direction opposite that of the wave propagation. The upper and lower branches of the crescent-shaped trace correspond to the front and back regions of the PHAkt/PKB-enriched domain, respectively, and also to the back and front regions of the PTEN-enriched domain, respectively. These spatial and temporal dynamics of the PtdIns lipids system indicate that individual local regions along the membrane undergo transitions between PTEN- and PHAkt/PKB-enriched states in accordance with the crescent-shaped dynamics, whereas the membrane as a whole maintains the asymmetric spatial profile dynamically in accordance with the wave propagation (Fig. 4D). Thus, progression of the crescent-shaped dynamics at individual local regions was observed as wave propagations along the membrane as a whole. Fig. 4C also reveals that the spatial profiles of PTEN- and PHAkt/PKB-enriched domains were asymmetric along the membrane. Moreover, the time delay in the reciprocal changes between [PTEN] and [PHAkt/PKB] was revealed in the reconstruction dynamics. The [PTEN] lag was distributed over 0–40 s with a 10-s average (Fig. 1G and Fig. S4M). This time delay implies a spatial shift in the reciprocal localization between [PTEN] and [PHAkt/PKB] along the membrane (Fig. 4C). These observed asymmetries with respect to time and space are regarded as a consequence of symmetry breaking in the PtdIns lipids system.
Theoretical Model.
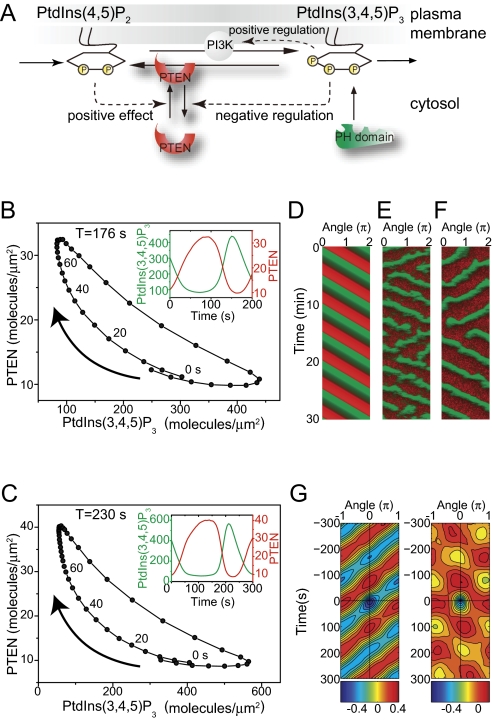
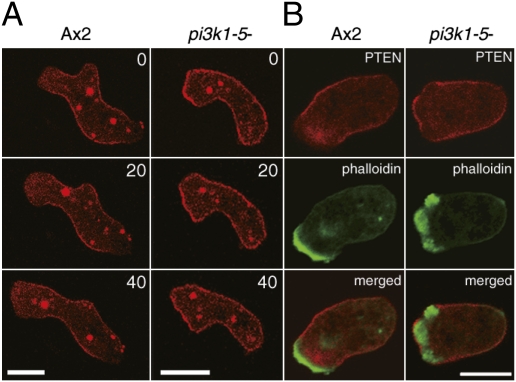
On the basis of these experimental observations, we constructed a simple reaction-diffusion model for the generation of the traveling waves in the PtdIns lipids system (Fig. 5 and SI Discussion). First, we consider the dynamics of the PtdIns lipids system at a local area along the membrane. The local reaction dynamics show a relaxation oscillation with both the reciprocal relation and time delay between [PHAkt/PKB] and [PTEN] (Fig. 4A). To reproduce the dynamics in the PtdIns lipids system with the reciprocal relation between PtdIns(3,4,5)P3 and PTEN concentrations, we assumed a PtdIns(3,4,5)P3-dependent negative regulation on PTEN recruitment to the membrane (Fig. 5A). This regulation works as a positive feedback loop on both PtdIns(3,4,5)P3 and PTEN concentrations. Such relaxation oscillators with a positive feedback loop have been found and studied in various biological systems (27). When PtdIns(3,4,5)P3 increases on the membrane, PTEN recruitment to the membrane is inhibited through negative regulation, which allows PtdIns(3,4,5)P3 to accumulate, resulting in a PtdIns(3,4,5)P3-enriched (PTEN-depleted) state. Likewise, by decreasing PtdIns(3,4,5)P3, PTEN recruitment to the membrane is accelerated, further degrading PtdIns(3,4,5)P3 and leading to a PTEN-enriched [PtdIns(3,4,5)P3-depleted] state. Thus, the assumed negative regulation can produce two stable states with rapid transitions between them. In fact, this model successfully reproduces the characteristic crescent-shaped dynamics including the time-delayed anticorrelation behavior between PtdIns(3,4,5)P3 and PTEN (Fig. 5B). Although the mechanistic basis for this negative regulation in Dictyostelium cells is not yet clear, small GTPase rhoA and its effector Rock have been suggested to be involved in the regulation of PTEN localization (28, 29). Interestingly, PTEN often showed uniform localization over the entire cell surface in pi3k1-5-null cells, but was absent from leading edge pseudopods in wild-type cells (Fig. 6A, Movie S6, and Movie S7). Furthermore, PTEN was often observed in F-actin-enriched pseudopod regions of pi3k1-5-null cells, but had an anticorrelative relationship with F-actin in wild-type cells (Fig. 6B). These observations suggest a PI3K-dependent pathway for the negative regulation of PTEN localization probably via PtdIns(3,4,5)P3. Together with a previous study that showed PTEN is excluded at regions facing a higher cAMP concentration gradient in the absence of PtdIns(3,4,5)P3 (24), these results suggest that PTEN localization is regulated by at least two mechanisms: one PtdIns(3,4,5)P3 dependent and the other PtdIns(3,4,5)P3 independent.
Fig. 5.
Mathematical model of PtdIns lipid traveling waves. (A) Scheme of reactions. PTEN recruitment to the membrane is negatively regulated by PtdIns(3,4,5)P3, a relationship crucial for reproducing the traveling waves observed experimentally. (B and C) Crescent-shaped dynamics of PtdIns lipids traveling waves constructed from the time series of the stochastic simulation using the same analysis performed on the experimental data. (Insets) Trajectories of the reconstructed dynamics showing reciprocal changes between PTEN and PtdIns(3,4,5)P3, irrespective of whether the Ras-PI3K-PtdIns(3,4,5)P3 positive feedback loop is included (C) or not (B) in the model. (D–F) Reproduced traveling waves of PTEN (red) and PtdIns(3,4,5)P3 (green) represented as a kymograph. Results were obtained by a deterministic simulation using a set of partial differential equations (D) or stochastic simulations (E and F) (SI Text, Table S1). The stochastic simulations were performed in the model with (F) or without (E) the feedback loop. (G) Phenotypic variability was reproduced by including stochastic noise in the model. The spatiotemporal cross-correlation function of ordered patterns between PtdIns(3,4,5)P3 and PTEN obtained from the stochastic simulation shows a traveling wave (Left) and spatiotemporal oscillation (Right).
Fig. 6.
Requirement of PtdIns(3,4,5)P3 for PTEN exclusion from the leading edge of migrating cells. (A) PTEN localization in wild-type (Left) and pi3k1-5-null (Right) cells. Numbers in the upper right of each panel are time in seconds. (Scale bar, 10 μm.) (B) Simultaneous imaging of PTEN and F-actin localization in wild-type (Left) and pi3k1-5-null (Right) cells. Images of PTEN-TMR (Top) and Alexa488-phalloidin (Middle) were obtained from the same cell and superimposed (Bottom). (Scale bar, 10 μm.)
In our model, PI3K activation is assumed to be uniform along the membrane. However, some cells exhibit RBD waves (Fig. 3 A and B), suggesting that local activation of membrane PI3K may be involved in PtdIns(3,4,5)P3/PTEN wave generation. To examine the effects of local PI3K activation on wave generation, we further included a Ras-PI3K-PtdIns(3,4,5)P3 positive feedback loop into the model (Fig. 5A). This extended model can also reproduce PtdIns(3,4,5)P3/PTEN waves with the characteristics of a relaxation oscillator (Fig. 5C). Without the positive feedback loop, PI3K activity is distributed uniformly along the membrane, whereas increasing the strength of the positive feedback loop couples PI3K activity spatiotemporally to PtdIns(3,4,5)P3/PTEN waves, suggesting that the two types of RBD dynamics observed in living cells may be derived from differences in the strength of the positive feedback loop. With this positive feedback, the wave propagation can be stabilized by modulating the oscillation period (Fig. 5 B and C). This is probably because the Ras-PI3K-PtdIns(3,4,5)P3 feedback stabilizes both the PtdIns(3,4,5)P3-enriched and the PTEN-enriched states.
We next explore the relationship between the local regions and global integration. The local reaction dynamics in individual regions interact globally through the rapid diffusion of PTEN in the cytosol. When a PtdIns(3,4,5)P3-enriched domain is formed on a membrane region, the cytosolic PTEN concentration increases to suppress new PtdIns(3,4,5)P3-enriched domains in other membrane regions (30). That is, the conservation of the total concentration of PTEN, which diffuses faster in the cytosol, affects global coupling between local regions at the membrane leading to the integration of local signaling processes to cause global polarization. Thus, the local oscillatory dynamics in individual regions can be coordinated with each other through cytosolic PTEN and thereby enable the membrane to generate traveling waves and spatiotemporal oscillations (Fig. 5 D–F).
We performed both deterministic and stochastic simulations, successfully reproducing traveling waves with the characteristic relaxation oscillation dynamics (Fig. 5 D–F). In the stochastic simulation in particular, the PtdIns(3,4,5)P3-enriched domain exhibited not only continuous propagation but also the generation, separation, and fusion of other domains and also showed a tendency to reverse its propagation direction (Fig. 5 E and F). Changing parameters like the supply rate of PtdIns(4,5)P2 resulted in variations like those seen in living cells, i.e., traveling waves and spatiotemporal oscillations (Fig. 5G). These results suggest that experimentally observed phenotypic variations in the spontaneous pattern formation were generated under the influence of stochastic noise derived from the signaling components of the PtdIns lipids system as well as variations in the biochemical components of individual cells. The self-organization of the PtdIns lipids system provides the robust dynamic mechanisms necessary to establish polarity when influenced by molecular noise. In addition, molecular noise from PtdIns lipids signaling mediators can give rise to the phenotypic variability seen in PtdIns lipids dynamics. The noise-robust self-organization and noise-induced phenotypic variability in the PtdIns lipids signaling system may be the origin of the spontaneous signal generation used for the ordered patterns observed in the random cell migrations of crawling-type cells (4, 7, 31, 32).
Random cell migration in the absence of chemoattractants can be regarded as the basis for realizing directional migration in response to environmental chemoattractant signals. The self-organizing properties of the PtdIns lipids signaling system also provide important insight regarding the mechanisms for chemoattractant-induced directional migration. Eukaryotic cells, including Dictyostelium and mammalian leukocytes, can perform chemotaxis even when signaled by an input intensity approximately at the level of noise (33–35). In these cells, the PtdIns lipids system can mediate the chemotactic signals by localizing a PtdIns(3,4,5)P3-enriched domain to the membrane such that it faces a higher concentration of chemoattractants (13–17). In general, self-organization in biomolecular systems can provide robustness against molecular noise derived from the stochastic fluctuations within signaling molecules (11). In the case of directional cell migration, extracellular stimulations may modulate the self-organization dynamics within the PtdIns lipids system during stochastic signal processing against molecular noise, which in turn generates intracellular directional cues that lead to the biased random migration seen in chemotaxis.
Materials and Methods
Microscopy.
Fluorescence images were obtained by using an inverted microscope (TE2000-PFS; Nikon) equipped with a confocal unit (CSU-10; Yokogawa) through a 60× oil immersion objective lens (N.A. 1.49; Nikon). Both PHAkt/PKB-EGFP (or PI3K2-EGFP) and PTEN-TMR were excited simultaneously by 488- and 561-nm solid-state CW lasers (Sapphire 488-200 CDRH for EGFP and Compass 561-20 for TMR, respectively; Coherent). By using dual view optics (A4313; Hamamatsu Photonics), each emission was separated into two parts at 550 nm by a dichroic mirror (DML557; Asahi Spectra). The separated images were passed through an emission filter (FF01-520 for EGFP and FF01-593 for TMR; Semrock) and captured simultaneously by an EMCCD camera (DU-897 iXon+; Andor Technology). Time-lapse movies were acquired at 5-s intervals.
Cell Preparations.
D. discoideum cells were used for all experiments. Full methods and any associated references for cell preparations are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
PHAkt/PKB-EGFP was kindly provided by Y. Asano and T. Uyeda (National Institute of Advanced Industrial Science and Technology, Ibaraki, Japan). We thank P. N. Devreotes (Johns Hopkins University School of Medicine, Baltimore), R. A. Firtel (University of California, San Diego), R. R. Kay (MRC Laboratory of Molecular Biology, Cambridge, UK), and the Dicty Stock Center for providing us the PTEN gene, the GFP-RBD construct, pi3k1-5-null cells, and plc-null cells, respectively. We thank all of the members of the Stochastic Biocomputing Group at Osaka University for discussion and A. S. Mikhailov, P. Karagiannis, L. Marcucci, and T. Martin for critical reading of the manuscripts.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0908278107/-/DCSupplemental.
References
- 1.Oosawa F. Spontaneous signal generation in living cells. Bull Math Biol. 2001;63:643–654. doi: 10.1006/bulm.2001.0236. [DOI] [PubMed] [Google Scholar]
- 2.Berg HC, Brown DA. Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature. 1972;239:500–504. doi: 10.1038/239500a0. [DOI] [PubMed] [Google Scholar]
- 3.Sasaki AT, et al. G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility. J Cell Biol. 2007;178:185–191. doi: 10.1083/jcb.200611138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takagi H, Sato MJ, Yanagida T, Ueda M. Functional analysis of spontaneous cell movement under different physiological conditions. PLoS ONE. 2008;3:e2648. doi: 10.1371/journal.pone.0002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shibata T, Fujimoto K. Noisy signal amplification in ultrasensitive signal transduction. Proc Natl Acad Sci USA. 2005;102:331–336. doi: 10.1073/pnas.0403350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerisch G, Noegel AA, Schleicher M. Genetic alteration of proteins in actin-based motility systems. Annu Rev Physiol. 1991;53:607–628. doi: 10.1146/annurev.ph.53.030191.003135. [DOI] [PubMed] [Google Scholar]
- 7.Killich T, et al. The locomotion, shape and pseudopodial dynamics of unstimulated Dictyostelium cells are not random. J Cell Sci. 1993;106:1005–1013. doi: 10.1242/jcs.106.4.1005. [DOI] [PubMed] [Google Scholar]
- 8.Vicker MG. Eukaryotic cell locomotion depends on the propagation of self-organized reaction-diffusion waves and oscillations of actin filament assembly. Exp Cell Res. 2002;275:54–66. doi: 10.1006/excr.2001.5466. [DOI] [PubMed] [Google Scholar]
- 9.Gerisch G, et al. Mobile actin clusters and traveling waves in cells recovering from actin depolymerization. Biophys J. 2004;87:3493–3503. doi: 10.1529/biophysj.104.047589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiner OD, Marganski WA, Wu LF, Altschuler SJ, Kirschner MW. An actin-based wave generator organizes cell motility. PLoS Biol. 2007;5:e221. doi: 10.1371/journal.pbio.0050221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karsenti E. Self-organization in cell biology: A brief history. Nat Rev Mol Cell Biol. 2008;9:255–262. doi: 10.1038/nrm2357. [DOI] [PubMed] [Google Scholar]
- 12.Bretschneider T, et al. The three-dimensional dynamics of actin waves, a model of cytoskeletal self-organization. Biophys J. 2009;96:2888–2900. doi: 10.1016/j.bpj.2008.12.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Haastert PJ, Devreotes PN. Chemotaxis: Signalling the way forward. Nat Rev Mol Cell Biol. 2004;5:626–634. doi: 10.1038/nrm1435. [DOI] [PubMed] [Google Scholar]
- 14.Kölsch V, Charest PG, Firtel RA. The regulation of cell motility and chemotaxis by phospholipid signaling. J Cell Sci. 2008;121:551–559. doi: 10.1242/jcs.023333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parent CA, Blacklock BJ, Froehlich WM, Murphy DB, Devreotes PN. G protein signaling events are activated at the leading edge of chemotactic cells. Cell. 1998;95:81–91. doi: 10.1016/s0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- 16.Meili R, et al. Chemoattractant-mediated transient activation and membrane localization of Akt/PKB is required for efficient chemotaxis to cAMP in Dictyostelium. EMBO J. 1999;18:2092–2105. doi: 10.1093/emboj/18.8.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Postma M, et al. Sensitization of Dictyostelium chemotaxis by phosphoinositide-3-kinase-mediated self-organizing signalling patches. J Cell Sci. 2004;117:2925–2935. doi: 10.1242/jcs.01143. [DOI] [PubMed] [Google Scholar]
- 18.Funamoto S, Milan K, Meili R, Firtel RA. Role of phosphatidylinositol 3′ kinase and a downstream pleckstrin homology domain-containing protein in controlling chemotaxis in dictyostelium. J Cell Biol. 2001;153:795–810. doi: 10.1083/jcb.153.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iijima M, Devreotes P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell. 2002;109:599–610. doi: 10.1016/s0092-8674(02)00745-6. [DOI] [PubMed] [Google Scholar]
- 20.Funamoto S, Meili R, Lee S, Parry L, Firtel RA. Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell. 2002;109:611–623. doi: 10.1016/s0092-8674(02)00755-9. [DOI] [PubMed] [Google Scholar]
- 21.Iijima M, Huang YE, Luo HR, Vazquez F, Devreotes PN. Novel mechanism of PTEN regulation by its phosphatidylinositol 4,5-bisphosphate binding motif is critical for chemotaxis. J Biol Chem. 2004;279:16606–16613. doi: 10.1074/jbc.M312098200. [DOI] [PubMed] [Google Scholar]
- 22.Brenner M, Thoms SD. Caffeine blocks activation of cyclic AMP synthesis in Dictyostelium discoideum. Dev Biol. 1984;101:136–146. doi: 10.1016/0012-1606(84)90124-6. [DOI] [PubMed] [Google Scholar]
- 23.Loovers HM, et al. Distinct roles of PI(3,4,5)P3 during chemoattractant signaling in Dictyostelium: A quantitative in vivo analysis by inhibition of PI3-kinase. Mol Biol Cell. 2006;17:1503–1513. doi: 10.1091/mbc.E05-09-0825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoeller O, Kay RR. Chemotaxis in the absence of PIP3 gradients. Curr Biol. 2007;17:813–817. doi: 10.1016/j.cub.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Sasaki AT, Chun C, Takeda K, Firtel RA. Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J Cell Biol. 2004;167:505–518. doi: 10.1083/jcb.200406177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kortholt A, King JS, Keizer-Gunnink I, Harwood AJ, Van Haastert PJ. Phospholipase C regulation of phosphatidylinositol 3,4,5-trisphosphate-mediated chemotaxis. Mol Biol Cell. 2007;18:4772–4779. doi: 10.1091/mbc.E07-05-0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murray JD. Mathematical Biology. New York; London: Springer; 2002. [Google Scholar]
- 28.Li Z, et al. Regulation of PTEN by Rho small GTPases. Nat Cell Biol. 2005;7:399–404. doi: 10.1038/ncb1236. [DOI] [PubMed] [Google Scholar]
- 29.Meili R, Sasaki AT, Firtel RA. Rho Rocks PTEN. Nat Cell Biol. 2005;7:334–335. doi: 10.1038/ncb0405-334. [DOI] [PubMed] [Google Scholar]
- 30.Gamba A, et al. Diffusion-limited phase separation in eukaryotic chemotaxis. Proc Natl Acad Sci USA. 2005;102:16927–16932. doi: 10.1073/pnas.0503974102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maeda YT, Inose J, Matsuo MY, Iwaya S, Sano M. Ordered patterns of cell shape and orientational correlation during spontaneous cell migration. PLoS ONE. 2008;3:e3734. doi: 10.1371/journal.pone.0003734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asano Y, Nagasaki A, Uyeda TQ. Correlated waves of actin filaments and PIP3 in Dictyostelium cells. Cell Motil Cytoskeleton. 2008;65:923–934. doi: 10.1002/cm.20314. [DOI] [PubMed] [Google Scholar]
- 33.Ueda M, Sako Y, Tanaka T, Devreotes P, Yanagida T. Single-molecule analysis of chemotactic signaling in Dictyostelium cells. Science. 2001;294:864–867. doi: 10.1126/science.1063951. [DOI] [PubMed] [Google Scholar]
- 34.Ueda M, Shibata T. Stochastic signal processing and transduction in chemotactic response of eukaryotic cells. Biophys J. 2007;93:11–20. doi: 10.1529/biophysj.106.100263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Haastert PJ, Postma M. Biased random walk by stochastic fluctuations of chemoattractant-receptor interactions at the lower limit of detection. Biophys J. 2007;93:1787–1796. doi: 10.1529/biophysj.107.104356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.