Abstract
Human and chimpanzee genomes are almost identical, yet humans express higher brain capabilities. Deciphering the basis for this superiority is a long sought-after challenge. Adenosine-to-inosine (A-to-I) RNA editing is a widespread modification of the transcriptome. The editing level in humans is significantly higher compared with nonprimates, due to exceptional editing within the primate-specific Alu sequences, but the global editing level of nonhuman primates has not been studied so far. Here we report the sequencing of transcribed Alu sequences in humans, chimpanzees, and rhesus monkeys. We found that, on average, the editing level in the transcripts analyzed is higher in human brain compared with nonhuman primates, even where the genomic Alu structure is unmodified. Correlated editing is observed for pairs and triplets of specific adenosines along the Alu sequences. Moreover, new editable species-specific Alu insertions, subsequent to the human–chimpanzee split, are significantly enriched in genes related to neuronal functions and neurological diseases. The enhanced editing level in the human brain and the association with neuronal functions both hint at the possible contribution of A-to-I editing to the development of higher brain function. We show here that combinatorial editing is the most significant contributor to the transcriptome repertoire and suggest that Alu editing adapted by natural selection may therefore serve as an alternate information mechanism based on the binary A/I code.
Keywords: evolution, brain, Alu, RNA modification
Humans and chimpanzees share about 98.8% of their genome and 99.4% of their nonsynonymous DNA (1–3), but differ significantly in higher skills. Previous studies reported that the evolutionary lineage leading to humans is marked by a dramatic increase in brain size and several quantitative changes in anatomical level. It is assumed that the phenotypic difference between species can be caused by changes in protein sequences or modified regulation of gene expression. Indeed, changes in a number of proteins such as FOXP2, MCPH1, ASPM1, MAOA, AHI1, GLUT2, and MYH16 have been linked to the evolution of human-specific cognitive features (4). Several groups reported on brain-specific upregulation of gene expression along primate evolution, including genes encoding for proteins involved in neuronal function, synaptic activity, and RNA and aerobic energy metabolism (5–8). However, the majority of these interspecies expression changes were found to be neutral (9). Global analysis of alternative splicing and DNA methylation displayed differences between humans and chimpanzees (10, 11), but these changes are not sufficient to explain human-specific cognitive abilities (12). Here we show an alternative mechanism for increasing brain transcriptome diversification, which differs in humans, compared with other primates, and may provide one more possible explanation for human higher skills.
Adenosine-to-inosine (A-to-I) RNA editing is a posttranscriptional modification, altering the sequence of RNA from that encoded in the DNA. It is catalyzed by the double-stranded RNA-specific adenosine deaminase acting on RNA (ADAR) enzyme family and appears to be tissue specific with brain tissue being the most edited (13–16). The splicing and translational machineries recognize inosine (I) as guanosine (G). Therefore, the result of ADAR-mediated editing consists of genomically encoded adenosines that are read as guanosines in the RNA sequence. Many of the RNA editing targets play a central role in neurogenesis. Indeed, disruption of the editing process in lower organisms such as Caenorhabditis elegans and Drosophila melanogaster resulted in behavioral and neural defects (17, 18). Moreover, altered editing patterns in humans and mice have been linked mainly to neuropathological disorders, such as amyotrophic lateral sclerosis, epilepsy, and brain tumors (19–23).
RNA editing in humans occurs predominantly within the primate-specific Alu repetitive elements, affecting thousands of genes in tens of thousands of sites. The overwhelming majority of these sites are located in noncoding sequences (introns and UTRs) (24–27). Still, some A-to-I RNA editing occurs in coding sequences and alters the mature protein sequence and its properties. Interestingly, the level of RNA editing in humans is more than an order of magnitude higher than that in the mouse, rat, chicken, and fly (26, 28). This difference is explained by the dominance of the primate-specific Alu elements in the human transcriptome, which can generate double-stranded RNA (dsRNA) structures (29). However, the global editing level in nonhuman primates whose genomes are Alu rich has not been studied so far.
Alu sequences, present in more than a million copies per primate genome, are frequently found in gene-rich regions, generally within noncoding segments. Previous studies showed that A-to-I RNA editing of Alu elements can affect gene expression through a variety of mechanisms, including alternative splicing, mRNA stability, nuclear retention, and microRNA biogenesis and targeting (30–34). These findings led us to explore the level of RNA editing within Alu elements in primate evolution.
Results
RNA Editing Level in Primates.
To compare the editing level in different primate species we studied Alu sequences in humans (Homo sapiens), chimpanzees (Pan troglodytes), and rhesus monkeys (Macaca mulatto). Bioinformatic assessment of the global editing level in primates, using the method previously published (28), predicts a higher number of potential A-to-I RNA editing sites in humans compared with rhesus monkeys and chimpanzees (Table S1). However, the low number of available mRNA sequences for the nonhuman primates, the lack of information regarding their tissue origin and its distribution, and other potential species-dependent biases prevent us from deducing with confidence the relative enhancement of editing in humans. We therefore performed experimental analyses of editing in the three species. The Alu targets were randomly selected from the human edited gene clusters database (27) according to criteria detailed in Methods, ensuring that the genomic Alu architecture is the same in all species. Six of the 12 targets selected for fulfilling the needed requirements were successfully amplified in all samples and tissues tested (four human and four nonhuman primates) and used for the analyses (Fig. S1 and Table S2). RNA editing is known to vary among tissues, thus the comparison was made between matched tissues. To exclude the possibility of genomic polymorphism, the genomic DNA was also sequenced.
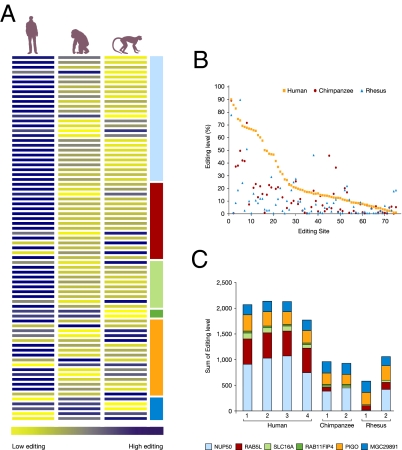
In the tested transcripts we identified higher A-to-I RNA editing, on average, in Alu elements in human vs. nonhuman primates. Fig. 1A illustrates the mean editing levels in human, chimpanzee, and rhesus cerebellum in 75 sites in the tested genes after normalization. An analysis of the actual editing level in the specific editing sites demonstrates that humans exhibit the highest editing value relative to the nonhuman primates in most sites (Fig. 1B and Table S3, Table S4, and Table S5). Comparing the normalized editing level per transcript showed a 2-fold increase in humans (P value = 1.2E-6, Mann-Whitney test; Methods). The difference between the rhesus monkey and chimpanzee was not significant (P = 0.54). Moreover, close examination of the grand total of editing levels (summed over all genes) reveals that all four individual human samples are ranked higher than the two chimpanzees and the two rhesus monkeys (P = 0.029). Fig. 1C summarizes the total editing levels for each of the different Alu targets in four human individuals relative to four nonhuman primates. Interestingly, the intraspecies variance in humans was low compared with their average editing level (gene-averaged standard deviation to average ratio, 0.12), as well as compared with the interspecies differences (gene-averaged ratio, 0.48). The differences in editing levels between chimpanzee and rhesus samples were roughly the same as the intraspecies differences of the same samples.
Fig. 1.
Higher editing level in human vs. nonhuman primates. (A) Editing levels of 75 sites in six transcripts originating from cerebellum tissues of four humans, two chimpanzees, and two rhesus monkeys were quantified after PCR amplification using the DSgene program. Average editing values were normalized (Z-score) and colored accordingly with blue-yellow gradient using the Spotfire program (Tibco). (B) Editing level per site for humans, chimpanzees, and rhesus monkeys. The human editing sites are ordered in decreasing editing levels, and the nonhuman primate editing sites are aligned, accordingly. (C) Editing levels in cerebellum tissues of eight individual primates: a total of the resulting editing level quantification in the six tested transcripts are plotted in four human, two chimpanzee, and two rhesus individuals where the bar size is proportional to the total of the editing levels in all tested sites.
We also compared coding sequence editing targets in the cerebellum of the three species. Three well-characterized coding region editing targets (CYFIP2, FLNA, and BLCAP) (35) were analyzed as previously described (20). In contrast to Alu editing, recoding RNA editing events leading to amino acid substitution appears to be gene and tissue specific; no consistent species-specific differences were observed (Table S6).
To understand the mechanism underlying increased editing in Alu sequences in humans, we examined the expression level of ADARs in the human, chimpanzee, and rhesus cerebellum using quantitative real-time PCR. Surprisingly, we found that ADARs exhibit a similar or higher expression level in the chimpanzee and rhesus brain tissues compared with the human brain (Table S7). Therefore, the differences in Alu editing levels cannot be attributed solely to the ADARs’ expression level.
Differences in noncoding RNA editing levels of a specific gene between human and nonhuman primates can be attributed to the following differences between the primate genome sequences: (i) Minor changes in the Alu DNA sequence between species, with similar genomic architecture that can affect the dsRNA structure (Fig. 2A). Such minor alterations are indeed observed in the six transcripts tested above, which contain 75 editing sites and show significantly higher editing levels in humans. Small changes in the Alu architecture may thus lead to alterations in the editing level. (ii) Inversion of one of the Alu repeats (Fig. 2B). The impact of this type of alteration was demonstrated in the RAB27A gene, where one Alu adjacent to the tested Alu is situated in opposite orientation in the human and chimpanzee, compared with the rhesus monkey (Fig. S1). This inversion may have increased the editing levels in both humans and chimpanzees (10-fold higher editing on average in humans compared with rhesus vs. only a 2-fold ratio in humans compared with chimpanzees) (Table S8). (iii) Deletion/addition of an Alu sequence (Fig. 2 C and D) might also affect RNA editing levels by modifying the dsRNA structure and stability. This type of change was demonstrated by the analysis of the MATR3 gene that fulfills our selection criteria, except that one of the rhesus Alu sequences was missing from the human and chimpanzee genomes. Indeed, MATR3 showed a higher A-to-I RNA editing level in the rhesus transcripts; see Fig. S1 and Table S4. Thus, the addition or deletion of Alu elements appears to make a major contribution to the diversity of the primate transcriptome.
Fig. 2.
Possible effects of Alu architecture alterations on RNA editing. Schematic representation of the genomic Alu elements’ location and orientation: Alu elements are marked as arrow-shaped boxes in the human (blue) and monkey (red) genomes. Alterations between the species are indicated in orange. (A) Minor alteration in Alu sequence between the species. (B) Inversion of one of the Alu sequences along primate evolution. (C) Deletion of Alu element along evolution. (D) Insertion of additional Alu sequence along evolution.
Increased Alu Insertions in Neuronal Genes.
Genomic comparison of Alu features revealed that the global properties of the more than one million Alu elements are highly similar for human and nonhuman primates (Fig. S2). Actually, only a few thousand new Alu insertions occurred since the split of humans and chimpanzees from their common ancestor. Interestingly, previous work showed a 3.4-fold higher number of new Alu insertions in humans compared with chimpanzees (5,530 compared with 1,642, respectively) (3, 36). Since the number of the new Alu insertions is relatively small, those Alus have minimal effect on the global trends of the Alu genomic architecture. The majority of these insertions (>95%) in both species are AluY sequences, members of the youngest and most active Alu family (3). About one-third of these insertions appear in RefSeq genes (1,477 RefSeq genes in humans compared with 497 in chimpanzees). Because Alu insertions tend to take place in Alu-rich regions, the insertion of a single Alu in the vicinity of an Alu element in reverse orientation is expected to lead to a dsRNA structure, which is the substrate for the editing-mediating enzymes. Such insertions can alter the “editing potential,” not only of the inserted Alus, but also of adjacent Alus. In humans, for example, we find that 1,601 of the 1,932 Alu insertions in RefSeq are within 2,500 bp from another reversely oriented Alu and are therefore editable. Moreover, an additional 2,084 “old” Alus are within a 2,500-bp range from a newly inserted Alu and are expected to show a modified editing pattern due to the new insertion.
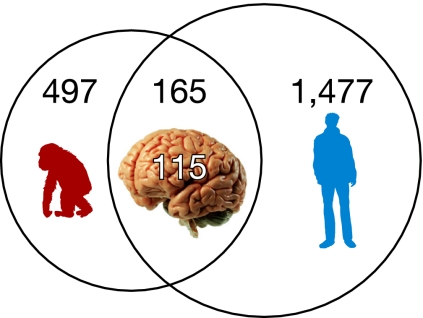
Functional clustering analysis (using Ingenuity software) of human and chimpanzee RefSeq genes harboring newly inserted Alus revealed that the nervous system development and function annotation group was significantly enriched in both humans and chimpanzees (Table 1, Dataset S1, and Dataset S2). Likewise, the cellular compartment analysis of genes containing newly inserted Alu sequences in humans and chimpanzees identified enrichment in genes coding for structures central to the nervous system with synapse and cell junction genes being the most significantly enriched (Table S9). Furthermore, enrichment analysis for disease-related genes revealed that genes with new Alu insertions are significantly associated with several disease categories (Table 2, Dataset S3, and Dataset S4). Significant enrichment was noticed for the neurological diseases category and several specific neurological and mental disorders such as bipolar affective disorder (P = 4E-45, Fisher test, Bonferroni corrected), Alzheimer's disease (P = 2E-29), amyotrophic lateral sclerosis (P = 2E-27), Parkinson's disease (P = 1E-22), and schizophrenia (P = 0.02). Additional disease categories characterized by enrichment in Alu insertions include inflammatory and cardiovascular diseases. It should be noted that the highest level of RNA editing was previously found in neuronal tissues and in immune and inflammatory cells (37). Finally, we identified 165 genes that exhibited independent new Alu insertions in both humans and chimpanzees (Fig. 3), and 115 of them (69%) were annotated with neurological function or disease (Table S10).
Table 1.
Genes harboring newly inserted Alus in humans are enriched in nervous system development function
| Function annotation | Fisher Pv | Benjamini Pv | No. of genes |
| Neurological process of axons | 1.17E-08 | 2.75E-06 | 35 |
| Guidance of axons | 7.70E-07 | 1.71E-04 | 28 |
| Neurotransmission | 6.58E-05 | 1.38E-02 | 46 |
| Development of sensory projections | 9.12E-05 | 1.77E-02 | 5 |
| Synaptic transmission | 1.23E-04 | 2.28E-02 | 43 |
| Neurological process of eukaryotic cells | 5.11E-04 | 8.08E-02 | 33 |
| Growth of neurites | 5.98E-04 | 8.95E-02 | 48 |
| Neurological process of cells | 7.39E-04 | 9.26E-02 | 36 |
| Chemorepulsion of neurons | 7.53E-04 | 9.26E-02 | 4 |
| Pathfinding of axons | 7.53E-04 | 9.26E-02 | 4 |
| Neurological process of normal cells | 8.84E-04 | 9.93E-02 | 30 |
P values calculated by Fisher's exact method are not corrected for multiple testing. Benjamini-Hochberg method was implemented to set a cumulative FDR of 0.1.
Table 2.
Neurological disease enrichment in functional clustering of the genes containing new Alu insertion in humans
| Function annotation | Fisher Pv | Benjamini Pv | No. of genes |
| Neurological disorder | 7.50E-53 | 5.29E-50 | 599 |
| Bipolar affective disorder | 3.87E-48 | 2.00E-45 | 254 |
| Motor neuron disease | 1.69E-41 | 6.91E-39 | 258 |
| Progressive motor neuropathy | 2.51E-41 | 9.75E-39 | 257 |
| Neuropathy | 1.17E-40 | 4.13E-38 | 263 |
| Alzheimer's disease | 1.18E-32 | 3.38E-30 | 173 |
| Neurodegenerative disorder | 3.71E-31 | 1.03E-28 | 178 |
| Amyotrophic lateral sclerosis | 7.48E-31 | 2.00E-28 | 145 |
| Parkinson's disease | 4.36E-26 | 1.13E-23 | 141 |
| Schizophrenia | 9.02E-06 | 1.94E-03 | 61 |
| Multiple sclerosis | 1.10E-04 | 2.08E-02 | 34 |
The most significant results in neurological disease enrichment analysis are presented. (P values calculated by Fisher's exact method are not corrected for multiple testing. FDR = 0.1, Benjamini-Hochberg method).
Fig. 3.
Analysis of newly inserted Alus. Number of common human and chimpanzee genes showing new (independent) Alu element insertions. Among the 165 shared genes representing new independent Alu insertions in the human and chimpanzee, 115 are neurological function and neurological disease-associated genes.
RNA Editing and Transcriptome Complexity.
Typical editing sites are only partially edited, thus increasing transcriptome diversity to include both edited and nonedited versions. Maximum complexity is achieved when the editing level is ~50% at each site (assuming no correlations), as lower or higher values of editing reduce the potential for additional variants. We thus checked the proportion of sites containing 40–60% editing levels in all three species in the six Alu targets. Within our dataset, humans show a 1.7-fold higher number of editing sites in this range (10% of the human sites vs. 6% of the other primates; Table S11).
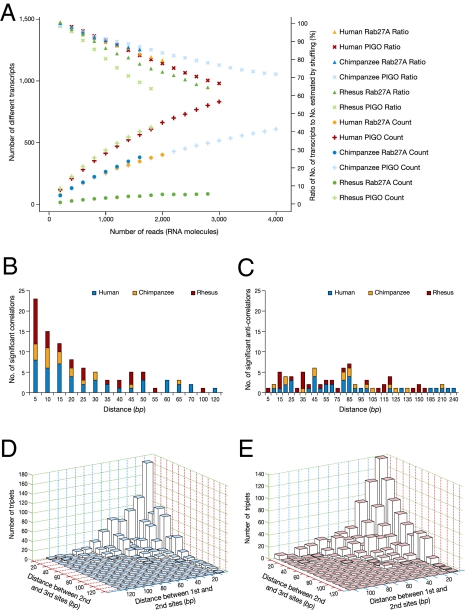
RNA editing can increase the number of different transcripts dramatically (38). For example, recent work reports on tens of different edited transcripts identified by cloning of a single Alu (39). To get a better and direct estimate of the number of combinations created by RNA editing, we generated a large number of transcripts by next-generation sequencing of two conserved Alu transcripts of human, chimpanzee, and rhesus cerebellum using the Roche 454 genome sequencer FLX system (Methods and Table S12). For this purpose, we chose two targets: the PIGO transcript, which shows a very similar ratio of an average editing level between humans and rhesus monkeys and a similar genomic architecture and the RAB27A transcript, which shows the highest difference in editing level between humans and rhesus (Table S8). Typically, edited Alu sequences exhibit a number of edited sites. Previous studies, based on traditional Sanger sequencing of a limited number of transcripts per Alu, found only ~5 editing sites per Alu on average (24–27). Looking at deep sequencing data for these two Alus, we identified 42 editing sites for the PIGO gene and 50 sites for Rab27A. We found hundreds of different variants for each of the tested targets (832 and 401 variants in humans for PIGO and RAB27A, respectively). The number of variants did not reach saturation (Fig. 4A and Dataset S5); thus the actual number of variants is expected to be even higher.
Fig. 4.
Combinatorial behavior and editing site dependencies. (A) Number of different variants as a function of the number of 454 sequencing reads. None of the graphs shows signs of saturation, indicating that the repertoire of different variants is not exhausted. The ratio of the number of different transcripts to the one expected in the absence of site–site correlations (excluding correlations resulting from the overall transcript editing level) is also shown. A ratio significantly less than 1 indicates the existence of significant dependencies between sites (see Dataset S5). (B) Number of significantly correlated and (C) anticorrelated editing-site pairs, as a function of the nucleotide distance between the sites. (D) Number of significantly triplet-correlated and (E) anticorrelated sites as a function of the nucleotide distance spanned by the triplet.
An increase of the editing level is expected to create a higher number of transcript variants. Indeed, our analysis showed that PIGO (exhibiting a similar average editing level in humans and rhesus monkeys) expresses a similar number of variants among the three species, whereas RAB27A (with a 10-fold higher editing level in humans compared with rhesus monkeys) showed an ~6-fold higher number of transcript combinations. The observation that a higher editing level contributes to increased transcriptome variability taken together with our previous analysis, which showed that the majority of tested Alu sequences had a higher editing level in humans compared with the other primates, suggests that the human transcriptome is significantly more diverse.
To explore the combinatorial capacity of the many editing sites within a single Alu sequence, we searched for positive and negative correlations among different sites. We thus controlled for the different levels of editing at different reads and at different sites, using a randomized matrix model with the same residual behavior. Dependencies between sites were calculated by scoring pairs or triplets of columns of the original matrices and compared with the scores of the same pairs/triplets on the randomized matrices. Previous work suggested a correlation between A-to-I editing sites on the basis of in vitro editing of synthetic substrates (40). Here we show multiple correlations and anticorrelations for editing sites (both pairs and triplets) identified for each of the endogenous targets tested (Table S13, Table S14, Table S15, Table S16, and Table S17). Figure 4 B–E demonstrates that most of the correlations were found for sites in close proximity. Yet, we also found many correlated sites located up to 244 bp apart. This pattern was found in both the human and nonhuman primates. The origin of these correlations within individual Alu sequences is not clear.
As shown in Table S18, Table S19, and Table S20, there are more than 8,000 human genes containing two inverted Alu sequences separated by up to 2,000 bp. These genes are expected to demonstrate a similar level of diversity as observed for the two tested here. Currently, alternative splicing is considered to be the main mechanism for diversity in the human transcriptome and a rough estimate of seven alternatively spliced isoforms per human gene (41) results in about 175,000 splice variants in the human transcriptome. A comparable level of diversity as a result of editing can be achieved by a mere 20 different transcripts in 8,000 edited genes. Nevertheless, the actual number of different edited transcripts is much higher, suggesting that the contribution of A-to-I RNA editing to transcriptome diversity is an order of magnitude higher than that of splicing, even though the impact of each single event is much larger for the latter.
Discussion
Human and nonhuman primates have surprisingly similar genomes, yet they differ in intellectual, cognitive, behavioral, and cultural skills. These differences are increasingly recognized by some groups to be quantitative and not qualitative, calling for a new taxonomy, grouping the chimpanzees and humans in the same genus (2). It appears that chimpanzees and bonobos are capable of informational exchange and express social, cultural, emotional, and language capabilities as well as other characteristics that were, until recently, thought to be unique to humans (42). Differences between human and nonhuman primates can be accounted for by the accumulation of alterations in DNA, RNA, and proteins. All these mechanisms may be subject to natural selection caused by climatic environmental stresses that ultimately lead to improved skills (42). However, comparing the number of coding sequence substitutions in the human and chimpanzee genomes to the genome of the most common ancestor showed no major difference in the global rate of evolution, suggesting that the difference might not lie in the coding DNA alone (2, 43). On the other hand, in contrast with the impressive DNA conservation, the brain transcriptome of humans stands out, displaying a distinctive profile of gene expression relative to nonhuman primates (5). Indeed, it was recently suggested that transcriptome modifications, such as RNA editing, contribute significantly to higher brain functions in humans (28, 44–46).
Altered Human Transcriptome Diversification Compared with Other Primates.
We and others have previously shown that the level of RNA editing in humans is significantly higher than that of several nonprimate species studied (26, 28). Most editing events in humans occur in the highly abundant repetitive Alu sequences, which comprise more than 10% of the human genome. The unique features of Alu sequences were suggested as a mechanistic explanation for the high level of editing in humans compared with organisms such as the mouse, rat, and Drosophila (29). However, the global editing level of nonhuman primates has not been studied so far. Here we provide unique experimental evidence for differences in human brain A-to-I RNA editing compared with nonhuman primates, despite similarities in the number, level of homology, and distribution of Alu elements in their genomes. Although we tested only a limited number of targets, our results suggest enhanced human transcriptome diversification compared with other primates. This finding is consistent with the hypothesized role of RNA editing in primate evolution (46). Future experiments involving transcriptome sequencing or, alternatively, measurement of global inosine level in humans, chimpanzees, and rhesus monkeys are expected to further explore this difference.
In this study we quantified the contribution of RNA editing to transcriptome complexity and diversity. Previous work, including our recent publication (39), was based on the analysis of a limited number of transcripts by cloning and sequencing. These limited studies suggested the need for high throughput analysis using next generation sequencing. Indeed, here we provide unique results of deep sequencing of Alu repeats using the 454 platform and describe the analysis of hundreds of transcripts from each target. This extended analysis indicates that RNA editing contributes to transcriptome diversity significantly more than alternative splicing, which is recognized as the central mechanism for transcriptome diversification.
Role of Natural Selection in RNA Editing.
A-to-I RNA editing is the result of enzymatic modification of RNA, replacing the genomically encoded adenosines with inosines. Inosine is recognized by the cellular machinery as guanosine. It can be argued that if there is an adaptive advantage to the presence of inosine in a specific site in the RNA molecule, natural selection will favor the genomic mutation/fixation of guanosine, which is structurally similar to inosine, in this location rather than having the disadvantageous genomic adenosine being corrected through RNA editing. Indeed, this fixation was shown to occur in a few examples in flies (47), but in many other examples, including the essential GluRB Q/R editing site, such fixation is not observed. This finding suggests that the possibility of choice imparted by editing may provide an evolutionary advantage. Similarly, we have found a few examples in which the editing of adenosine to inosine occurs in one or more of the primates analyzed, whereas in other primates guanosine is found in the DNA. Yet, in the vast majority of editing sites such fixation did not take place. When guanosine is fixated in the DNA, all RNA molecules will contain this nucleotide. Editing, on the other hand, allows for flexibility where some transcripts will contain adenosine and others will contain inosine. Tight regulation of the fraction of edited transcripts can then be recruited to fine-tune gene expression in a cell-, tissue-, function-, and developmental stage-dependent manner. Although fixation of a nucleotide in the DNA is irreversible, editing allows transient and reversible modifications of the cell transcriptome.
The basis for the difference in editing patterns between species is only starting to be elucidated. The level of expression of the editing enzymes is similar in human and nonhuman primates and therefore cannot be a major factor. On the other hand, we show that changes that occurred in the genomic architecture of Alu elements after the human–chimpanzee split, such as sequence-specific alterations, additions, deletions, and inversions may affect RNA editing. Although the rules determining editing levels in specific adenosines are not fully understood, it appears that editing in Alu sequences is not random. Editing efficiency was shown to be predetermined by the sequence and structural motifs (48) and may be adaptively channeled by natural selection. In addition, specific editing hotspots within the Alu sequence were demonstrated (27). It is possible that the future understanding of the details of structural and sequence factors affecting editability will reveal the molecular source for the differences in editing levels.
Neuronal Connection.
The global genomic distribution and architecture of the 1.4 million Alu sequences is very similar among primates. Yet 5,530 insertions of Alu elements in humans and 1,642 in chimpanzees took place after the human–chimpanzee split (36). Alu insertions, which create new editable sequences, are thus more frequent in the human genome than in the chimpanzee. Notably, we found that a significantly high fraction of new Alu insertions in both human and nonhuman primates resides in genes involved in neuronal function, such as axon guidance and neurotransmission, an observation that suggests a possible link with the evolution of the nervous system. Moreover, many genes whose structure was modified by the Alu insertion are linked to neurological and psychiatric diseases such as schizophrenia, bipolar disorder, Alzheimer's disease, and amyotrophic lateral sclerosis (ALS). Notably, diseases that have been previously associated with altered editing such as depression, ALS, epilepsy, and brain tumors are also related to the nervous system. One can suggest that the editability of brain-related transcripts provides improved regulation and fine tuning and is expected to improve neuronal functions. The price for such beneficial and intricate control may be the increased susceptibility to malfunction, resulting in disease. Taken together, these enrichment studies (Dataset S1, Dataset S2, Dataset S3, and Dataset S4) suggest that integration and fixation of Alu sequences into the primate genome may be specifically directed at genes that mediate neuronal function.
The genes modified by new editable Alu insertions include GABAergic, glutaminergic, and cholinergic receptors as well as several calcium and potassium channels. Humans and chimpanzees also show new insertions of Alu in the Neuregulin 1 gene, which was recently shown to be associated with creativity in people with high intellectual achievements (49). DSCAM, a central player in neuronal circuit formation, which was shown in Drosophila to be highly diverse due to extensive alternative splicing (50), was found here to have several new insertions in the human genome. Strikingly, although no significant alternative splicing of this gene was demonstrated in humans, there are eight Alu insertions that are expected to contribute to DSCAM transcript diversity.
RNA Editing as a Regulator of Gene Expression.
Our results suggest that editing in human vs. nonhuman primates differs in noncoding sequences, whereas there is no support for such alteration in coding regions. A-to-I RNA editing of noncoding sequences, in particular 3′ UTR regions, was shown recently to be involved in gene expression control (30, 32–34), through the anchoring of hyperedited transcripts in nuclear paraspeckles. In addition, editing affects microRNA sequences and their expression level as well as their targets (51, 52). Editing was also shown to regulate tissue-specific alternative splicing and to control exonization of intronic Alu sequences (53). It is therefore expected that the differences in editing activity may contribute to the regulation of gene expression by several complementary mechanisms.
In recent years evidence has accumulated about the abundance and importance of noncoding RNAs (ncRNAs) and their emerging role in many biological functions, particularly in the brain (45, 54), constituting a new layer of regulation and a possible mechanism for information storage. The importance of ncRNA was found to correlate with increased developmental complexity (55). RNA editing of ncRNA, in particular Alu-containing transcripts adds an additional level of complexity by contributing significantly to transcriptome diversity. We provide here evidence that this contribution may differ in the human brain compared with other primates.
RNA Editing Code.
The current extensive data demonstrate that editing of pairs and triplets of adenosine nucleotides, located either in close proximity or significantly apart, is correlated. Similarly, anticorrelation, namely, the preferred exclusion of concomitant editing of two or three adenosines, was also observed for close and distant sites. The finding of both correlation and anticorrelation of the editing of nucleotides located at variable distances from each other speaks against a simple mechanistic explanation based only on ADAR enzyme preferences or local sequence motifs. One explanation may be that editing of some sites changes the dsRNA stability and can be followed by a global change in the dsRNA structure, which is known to affect editing efficiency. These findings bring to mind information storage models. As the number of potential editing sites in each Alu-containing transcript is high, usually several dozens, the potential for combinatorial encrypted information is enormous. Binary use of A or I in millions of sites in the neural cell transcriptome can be considered equivalent to the 0’s and 1’s used for information storage and processing by computers. It is tempting to speculate that the more abundant RNA editing found in the human brain may contribute to the more advanced human capabilities such as memory, learning, and cognition. This suggestion is consistent with the hypothesis that the advantage of complex organisms lies in the development of a digital programming system based on noncoding RNA signaling (46, 56). The combinatorial posttranscriptional RNA editing of noncoding sequences may therefore contribute to higher brain functions and may play a role in the evolution of human specialization.
Methods
Selection of the Alu Targets.
Forty Alu targets were randomly selected from the human edited gene clusters database. Twelve passed the following filters: (i) the same number of Alu sequences in reverse orientation within a 4,000-bp range from the edited Alu in all three primates (a difference of 1 was allowed if the total number was higher than 3); (ii) the interspecies variability in the distance to the closest reversely oriented Alu sequence is less than 400 bp; and (iii) the similarity between the selected Alu sequence and the nearest reversely oriented sequence, as measured by BLAST identity fraction, was the same in all species (a relative difference of less than 10%); and finally, (iv) we required that there be no additional Alu sequence in the same orientation between the two inverted Alu sequences. Changes such as that in the RAB11FIP4 gene, which contains a shortened Alu (123 bp), were not considered in the analysis criteria. However, it can explain the difference in editing patterns in the rhesus monkey compared with the chimpanzee and humans, which do not have this addition and express similar editing levels.
We studied eight targets that were expressed in all tested species/tissues and could be amplified. Six of them: NUP50, RABL5, SLC16A6, PIGO, RAB11FIP4, and MGC29891 fulfill all of the above requirements. In addition, RAB27A and MATR3 that represent variation in Alu structure and orientation were also analyzed. Locations of the tested Alus and the primers used are listed in Table S5.
Additional methods are available in SI Methods.
Supplementary Material
Acknowledgments
We thank Lily Bazak for technical help. We thank the Kahn Family Foundation for their support. G.R. holds the Djerassi Chair in Oncology at the Sackler Faculty of Medicine, Tel Aviv University. E.E. and G.R. were also supported by the Israel Science Foundation (grant numbers 365/06 and 1942/08) and the Israel Ministry for Science and Technology (Scientific Infrastructure Program).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at http://star.tau.ac.il/~eli/primates/.
References
- 1.Ebersberger I, Metzler D, Schwarz C, Pääbo S. Genomewide comparison of DNA sequences between humans and chimpanzees. Am J Hum Genet. 2002;70:1490–1497. doi: 10.1086/340787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wildman DE, Uddin M, Liu G, Grossman LI, Goodman M. Implications of natural selection in shaping 99.4% nonsynonymous DNA identity between humans and chimpanzees: Enlarging genus Homo. Proc Natl Acad Sci USA. 2003;100:7181–7188. doi: 10.1073/pnas.1232172100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tarjei SM, et al. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 4.Creely H, Khaitovich P. Human brain evolution. Prog Brain Res. 2006;158:295–309. doi: 10.1016/S0079-6123(06)58014-8. [DOI] [PubMed] [Google Scholar]
- 5.Cáceres M, et al. Elevated gene expression levels distinguish human from non-human primate brains. Proc Natl Acad Sci USA. 2003;100:13030–13035. doi: 10.1073/pnas.2135499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enard W, et al. Intra- and interspecific variation in primate gene expression patterns. Science. 2002;296:340–343. doi: 10.1126/science.1068996. [DOI] [PubMed] [Google Scholar]
- 7.Preuss TM, Cáceres M, Oldham MC, Geschwind DH. Human brain evolution: Insights from microarrays. Nat Rev Genet. 2004;5:850–860. doi: 10.1038/nrg1469. [DOI] [PubMed] [Google Scholar]
- 8.Uddin M, et al. Sister grouping of chimpanzees and humans as revealed by genome-wide phylogenetic analysis of brain gene expression profiles. Proc Natl Acad Sci USA. 2004;101:2957–2962. doi: 10.1073/pnas.0308725100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khaitovich P, et al. Regional patterns of gene expression in human and chimpanzee brains. Genome Res. 2004;14:1462–1473. doi: 10.1101/gr.2538704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calarco JA, et al. Global analysis of alternative splicing differences between humans and chimpanzees. Genes Dev. 2007;21:2963–2975. doi: 10.1101/gad.1606907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enard W, et al. Differences in DNA methylation patterns between humans and chimpanzees. Curr Biol. 2004;14:R148–R149. [PubMed] [Google Scholar]
- 12.Khaitovich P, et al. Metabolic changes in schizophrenia and human brain evolution. Genome Biol. 2008;9:R124. doi: 10.1186/gb-2008-9-8-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paul MS, Bass BL. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J. 1998;17:1120–1127. doi: 10.1093/emboj/17.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu Rev Genet. 2000;34:499–531. doi: 10.1146/annurev.genet.34.1.499. [DOI] [PubMed] [Google Scholar]
- 15.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valente L, Nishikura K. ADAR gene family and A-to-I RNA editing: Diverse roles in posttranscriptional gene regulation. Prog Nucleic Acid Res Mol Biol. 2005;79:299–338. doi: 10.1016/S0079-6603(04)79006-6. [DOI] [PubMed] [Google Scholar]
- 17.Palladino MJ, Keegan LP, O'Connell MA, Reenan RA. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell. 2000;102:437–449. doi: 10.1016/s0092-8674(00)00049-0. [DOI] [PubMed] [Google Scholar]
- 18.Tonkin LA, et al. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans. EMBO J. 2002;21:6025–6035. doi: 10.1093/emboj/cdf607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maas S, Kawahara Y, Tamburro KM, Nishikura K. A-to-I RNA editing and human disease. RNA Biol. 2006;3:1–9. doi: 10.4161/rna.3.1.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paz N, et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007;17:1586–1595. doi: 10.1101/gr.6493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higuchi M, et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 22.Maas S, Patt S, Schrey M, Rich A. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc Natl Acad Sci USA. 2001;98:14687–14692. doi: 10.1073/pnas.251531398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawahara Y, et al. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004;427:801. doi: 10.1038/427801a. [DOI] [PubMed] [Google Scholar]
- 24.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blow M, Futreal PA, Wooster R, Stratton MR. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim DD, et al. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levanon EY, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 28.Eisenberg E, et al. Is abundant A-to-I RNA editing primate-specific? Trends Genet. 2005;21:77–81. doi: 10.1016/j.tig.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 29.Neeman Y, Levanon EY, Jantsch MF, Eisenberg E. RNA editing level in the mouse is determined by the genomic repeat repertoire. RNA. 2006;12:1802–1809. doi: 10.1261/rna.165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen LL, Carmichael GG. Gene regulation by SINES and inosines: Biological consequences of A-to-I editing of Alu element inverted repeats. Cell Cycle. 2008;7:3294–3301. doi: 10.4161/cc.7.21.6927. [DOI] [PubMed] [Google Scholar]
- 31.Nishikura K. Editor meets silencer: Crosstalk between RNA editing and RNA interference. Nat Rev Mol Cell Biol. 2006;7:919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: Functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–1705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prasanth KV, et al. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 35.Levanon EY, et al. Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res. 2005;33:1162–1168. doi: 10.1093/nar/gki239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mills RE, et al. Recently mobilized transposons in the human and chimpanzee genomes. Am J Hum Genet. 2006;78:671–679. doi: 10.1086/501028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang JH, et al. Widespread inosine-containing mRNA in lymphocytes regulated by ADAR1 in response to inflammation. Immunology. 2003;109:15–23. doi: 10.1046/j.1365-2567.2003.01598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keegan LP, Gallo A, O'Connell MA. The many roles of an RNA editor. Nat Rev Genet. 2001;2:869–878. doi: 10.1038/35098584. [DOI] [PubMed] [Google Scholar]
- 39.Barak M, et al. Evidence for large diversity in the human transcriptome created by Alu RNA editing. Nucleic Acids Res. 2009;37:6905–6915. doi: 10.1093/nar/gkp729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koeris M, Funke L, Shrestha J, Rich A, Maas S. Modulation of ADAR1 editing activity by Z-RNA in vitro. Nucleic Acids Res. 2005;33:5362–5370. doi: 10.1093/nar/gki849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 42.Roffman I, Nevo E. Can chimpanzee biology highlight human origin and evolution? Rambam Maimonides Med J. 2010;1:e0009. doi: 10.5041/RMMJ.10009. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clark AG, et al. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- 44.Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- 45.Gommans WM, Mullen SP, Maas S. RNA editing: A driving force for adaptive evolution? Bioessays. 2009;31:1137–1145. doi: 10.1002/bies.200900045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mattick JS. Deconstructing the dogma: A new view of the evolution and genetic programming of complex organisms. Ann N Y Acad Sci. 2009;1178:29–46. doi: 10.1111/j.1749-6632.2009.04991.x. [DOI] [PubMed] [Google Scholar]
- 47.Tian N, Wu X, Zhang Y, Jin Y. A-to-I editing sites are a genomically encoded G: Implications for the evolutionary significance and identification of novel editing sites. RNA. 2008;14:211–216. doi: 10.1261/rna.797108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lehmann KA, Bass BL. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry. 2000;39:12875–12884. doi: 10.1021/bi001383g. [DOI] [PubMed] [Google Scholar]
- 49.Kéri S. Genes for psychosis and creativity: A promoter polymorphism of the neuregulin 1 gene is related to creativity in people with high intellectual achievement. Psychol Sci. 2009;20:1070–1073. doi: 10.1111/j.1467-9280.2009.02398.x. [DOI] [PubMed] [Google Scholar]
- 50.Schmucker D, Chen B. Dscam and DSCAM: Complex genes in simple animals, complex animals yet simple genes. Genes Dev. 2009;23:147–156. doi: 10.1101/gad.1752909. [DOI] [PubMed] [Google Scholar]
- 51.Borchert GM, et al. Adenosine deamination in human transcripts generates novel microRNA binding sites. Hum Mol Genet. 2009;18:4801–4807. doi: 10.1093/hmg/ddp443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang W, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lev-Maor G, et al. RNA-editing-mediated exon evolution. Genome Biol. 2007;8:R29. doi: 10.1186/gb-2007-8-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mattick JS, Mehler MF. RNA editing, DNA recoding and the evolution of human cognition. Trends Neurosci. 2008;31:227–233. doi: 10.1016/j.tins.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 55.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mattick JS. RNA regulation: A new genetics? Nat Rev Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.