Abstract
Smg1 is a PI3K-related kinase (PIKK) associated with multiple cellular functions, including DNA damage responses, telomere maintenance, and nonsense-mediated mRNA decay (NMD). NMD degrades transcripts that harbor premature termination codons (PTCs) as a result of events such as mutation or alternative splicing (AS). Recognition of PTCs during NMD requires the action of the Upstream frameshift protein Upf1, which must first be phosphorylated by Smg1. However, the physiological function of mammalian Smg1 is not known. By using a gene-trap model of Smg1 deficiency, we show that this kinase is essential for mouse embryogenesis such that Smg1 loss is lethal at embryonic day 8.5. High-throughput RNA sequencing (RNA-Seq) of RNA from cells of Smg1-deficient embryos revealed that Smg1 depletion led to pronounced accumulation of PTC-containing splice variant transcripts from approximately 9% of genes predicted to contain AS events capable of eliciting NMD. Among these genes are those involved in splicing itself, as well as genes not previously known to be subject to AS-coupled NMD, including several involved in transcription, intracellular signaling, membrane dynamics, cell death, and metabolism. Our results demonstrate a critical role for Smg1 in early mouse development and link the loss of this NMD factor to major and widespread changes in the mammalian transcriptome.
Keywords: gene trap, Smg-1
Smg1 is a member of the PI3K-related kinase (PIKK) family of mammalian genes that includes ATM, ATR, the DNA-dependent protein kinase catalytic subunit, and the mammalian target of rapamycin (1, 2). These kinases play important roles in the maintenance of genomic integrity and responses to cellular stress. Like ATM, ATR, and the DNA-dependent protein kinase catalytic subunit, Smg1 is linked to DNA damage responses (3, 4). Smg1 may also function in telomere maintenance, responses to hypoxia, and TNF-α–induced apoptosis (5–7). In vitro, Smg1 has a critical role in the degradation of premature termination codon (PTC)–containing transcripts mediated by the nonsense-mediated mRNA decay (NMD) pathway (2, 3, 8). During NMD, Smg1 forms part of the Smg1-Upf1-eRF1-eRF3 surveillance complex that associates with exon junction complexes formed on spliced mRNA (9). This interaction induces the phosphorylation of Upf1 by Smg1, an indispensable step in mammalian NMD (2, 9).
NMD is a highly conserved process that removes PTC-containing transcripts so that they are not expressed as aberrant, and potentially deleterious, proteins (10). However, NMD also controls transcript abundance by regulating alternative splicing (AS) events that introduce PTCs; this process is referred to as AS-coupled NMD (11). Microarray profiling combined with bioinformatics has indicated that depletion of mammalian NMD factors substantially increases levels of predicted PTC-containing splice variants for a relatively small proportion of the total number of genes containing AS events capable of PTC introduction (12–16). To date, the genes associated with the greatest changes in PTC-containing splice variant transcripts upon NMD disruption include those encoding splicing factors or other RNA-binding/processing proteins (17–19). Increasing evidence indicates that AS-coupled NMD of splicing factor transcripts may play an important role in both autoregulation and cross-regulation of these and other RNA processing proteins.
To investigate the physiological role of Smg1, we have used a murine gene-trap model and RNA sequencing (RNA-Seq) to study the consequences of Smg1 deficiency. Smg1-deficient mice die by embryonic day 8.5 (E8.5) with marked developmental defects. Cells derived from these mutants display specific changes in their transcriptome relative to cells derived from WT embryos at the same developmental stage. Loss of Smg1 resulted in marked disruption of AS-coupled NMD in transcripts from a small subset of expressed genes. These transcripts encode splicing factors, as well as a variety of proteins involved in biological processes not previously known to be regulated by AS-coupled NMD.
Results
Characterization of Smg1-Deficient Embryonic Stem Cells.
Using data from the International Gene Trap Consortium (20), we identified an embryonic stem (ES) cell line (designated RRT449) as having a sequence tag corresponding to a putative intronic insertion of the pGT0Lxf exon trap construct into the Smg1 genomic locus. The resulting Smg1 gene-trap (Smg1gt) allele was expected to be hypomorphic as a result of fusion of exon 12 to the splice acceptor site upstream of the β-galactosidase/neomycin (β-Geo) ORF (Fig. 1A). The gene trap construct also harbors a stop codon and a premature polyadenylation signal that should abrogate expression of Smg1 exons 13 to 63. Only a relatively small N-terminal protein fragment representing aa 1 to 583 of the 3,658-aa native Smg1 protein should be produced, and this peptide should lack critical domains of Smg1, including its catalytic domain. A single GT0Lxf insertion within the Smg1 locus was confirmed by Southern blot analysis (Fig. 1B), and the presence of the hypomorphic allele was confirmed by RT-PCR detection of the corresponding fusion transcript. The gene trap ES cells were then used to produce mutant mice whose genotypes were confirmed by PCR (Fig. 1C).
Fig. 1.
Generation of Smg1 gene trap mutant mice. (A) Genomic Smg1 locus (WT) and structure of the RRT449 Smg1 allele containing the pGTOLxf exon trap (Mut). Black boxes, exons; SA, splice acceptor; Beta-geo, β-Geo selection gene; pA, polyadenylation sequence. Arrows indicate forward (Fwd) and reverse (Rev) primer binding sites for PCR genotyping. (B) Single integration of pGTOLxf. Genomic DNA from Smg1+/+ and Smg1+/gt ES cells was digested with Xba1 or Sac1 and subjected to Southern blotting to detect pGTOLxf. (C) Embryonic genotyping: genomic DNA from Smg1+/+, Smg1+/gt, and Smg1gt/gt embryos was subjected to PCR to detect the WT Smg1 allele (Fwd/Rev) or the gene trap Smg1 allele (Fwd/GT-rev). (D and E) Confirmation of Smg1 deficiency and reduced Upf1 phosphorylation. (D) Protein lysates from embryonic fibroblasts derived from Smg1+/+ and Smg1gt/gt mice were analyzed by Western blotting with anti-Smg1 antibody and with anti–β-tubulin antibody as a recovery and loading control. (E) Immunoprecipitates (IP) collected by using anti-Upf1 antibody from the cell lysates were subject to sequential Western blotting with anti-Upf1 antibody or anti–phospho-S/T-Q antibody to detect phospho-Upf1. β-Tubulin loading control: phospho-specific bands in input (Left) comigrate with Upf1. For all figures, results shown are representative of at least three independent experiments.
Smg1 Deficiency Results in Embryonic Lethality.
After backcrossing for 10 generations into a C57BL6 congenic background, Smg1+/gt mice were viable and fertile, and had a normal appearance and lifespan. Intercrosses of these Smg1+/gt mice resulted in Smg1+/+ and Smg1+/gt pups at the expected Mendelian ratio but never Smg1gt/gt pups (Table 1). Smg1gt/gt embryos were present in utero until E12.5, after which only empty or debris-filled conceptuses were observed. These data indicate that Smg1 is essential for early murine embryogenesis.
Table 1.
Genotype distribution of progeny of Smg-1+/gt intercrosses by gestational age
| Stage | +/+ | +/gt | gt/gt | Ratio | χ2 value | P value |
| E 6.5–10.5 | 12 | 23 | 13 | 0.25/0.48/ 0.27 | 0.1 | >0.5 |
| E 14.5–18.5 | 7 | 6 | 0 | 0.54/0.46/0.00 | 7.6 | <0.05 |
| P ≥1 | 124 | 136 | 0 | 0.48/0.52/0.00 | 118.8 | <0.001 |
E, embryonic day; P, postnatal day; P value determined by Student t test.
We next derived murine embryonic fibroblasts (MEFs) from E8.5 Smg1+/+ (WT) and Smg1gt/gt (GT) embryos and transformed them using retroviral E1A/H-rasV12 expression to generate WT and GT transformed MEFs (tMEFs) (21). To examine Smg1 protein in these cells, we used a polyclonal antibody raised against an epitope corresponding to aa 1,623 to 1,673 of the Smg1 protein; this epitope is not present in the gene trap fusion peptide (aa 1–583). Immunoblotting readily detected the expected band of the mouse Smg1 protein (approximately 400 kDa) in WT but not GT tMEFs (Fig. 1D). Thus, expression of full-length Smg1 is suppressed by the gene trap insertion in GT tMEFs.
We also examined the phosphorylation status of Upf1, a major target of Smg1 kinase activity. Endogenous Upf1 was immunoprecipitated from lysates prepared from WT and GT tMEFs and immunoblotted with an anti–phospho-S/T-Q–specific antibody. Compared with WT, GT tMEFs showed reduced levels of an anti–phospho-S/T-Q antibody–detected band comigrating with Upf1 in both the lysate and anti-Upf1 immunoprecipitate (Fig. 1E). This result confirms that disruption of Smg1 expression in Smg1gt/gt embryos reduces the phosphorylation levels of Upf1.
Developmental Defects in Smg1gt/gt Embryos.
Examination of Smg1gt/gt embryos in utero showed that they implant, gastrulate, and begin somitogenesis before arresting at E8.5. Somites were clearly visible in both Smg1+/+ and Smg1gt/gt unturned embryos at E8.0, but the mutants were already smaller in size and lacked a developed heart field and optic pit indentation (Fig. 2A). By E10.5, the Smg1gt/gt embryos had turned but were much smaller than E10.5 Smg1+/+ embryos and did not exhibit the advanced brain and heart development characteristic of this stage (Fig. 2B). Specifically, the three primary brain vesicles did not form and heart looping and the development of a multichambered heart structure did not occur. At E12.5, Smg1gt/gt embryos showed signs of resorption but hematopoiesis apparently did initiate as pooled blood was detected in the thoracoabdominal region (Fig. 2C). However, in these embryos, no development of the vascular system was observed.
Fig. 2.
Morphology and histology of Smg1gt/gt embryos. Representative age-matched littermate Smg1+/+ (Left) and Smg1gt/gt (Right) embryos are shown (N = 48). (A–C) Light microscope images of whole embryos. (D–F) Sequential sagittal sections of E12.5 embryos. Insets: High magnification images (×40). (A) Unturned E8.0 embryos are partially ensheathed by the yolk sac. Extending allantois (A) and somites (S) are visible in both genotypes, but the Smg1gt/gt embryo lacks the enlarged heart field (H) and indented optic pits (O) visible in the Smg1+/+ embryo. (B) E10.5 stage. The Smg1+/+ embryo shows visible delineation of brain ventricles (BV), brachial arches (BA), and typical heart development (H), whereas the developmentally arrested Smg1gt/gt embryo shows an abnormal heart (H) and pooled blood (Bl) in the thoracoabdominal region. (C) E12.5 stage: Smg1gt/gt embryo is much smaller than the control and shows inappropriate heart (H) formation and pooled blood (Bl) in the thoracoabdominal region. (D) H&E staining at E12.5 shows the undeveloped tissue architecture of an Smg1gt/gt embryo. (E) Reduced BrdU incorporation by a Smg1gt/gt embryo indicates a lack of cell proliferation. (F) Increased TUNEL staining by a Smg1gt/gt embryo indicates massive cell apoptosis. This embryo is dead and undergoing resorption.
Histological analyses of littermate Smg1+/+ and Smg1gt/gt embryos at E12.5 showed that the tissue architecture was severely disrupted in the mutants (Fig. 2D). Analyses of cell proliferation using BrdU incorporation confirmed that Smg1gt/gt embryos were unable to grow (Fig. 2E). Furthermore, TUNEL staining indicated that apoptosis was massive in Smg1gt/gt embryos (Fig. 2F). Thus, the embryonic lethality occurring in the absence of Smg1 is associated with profound developmental defects.
Altered Expression of PTC-Containing Splice Variants in Smg1-Deficient Cells.
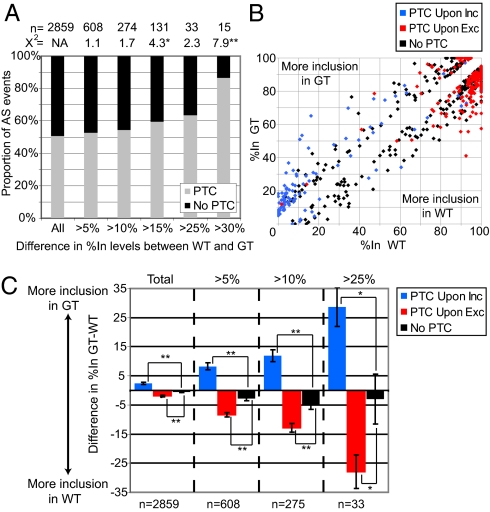
We used RNA-Seq to quantitatively compare mRNA splicing and expression patterns in primary cells derived from Smg1+/+ and Smg1gt/gt mice. PolyA+ RNA was isolated from primary MEFs derived from one E8.5 embryo/genotype and sequenced to yield approximately 56 million 50nt-reads per sample. As described in Materials and Methods, these RNA-Seq reads were mapped to a database containing more than 11,000 cassette-type AS events, of which 2,859 had sufficient read coverage in both genotypes to allow detection of differences in levels of alternatively spliced transcripts. Based on bioinformatic analysis of genome:EST/cDNA sequence alignments (13) (Materials and Methods), 51% of the 2,859 events were predicted to introduce a PTC by including an exon (i.e., PTC upon inclusion) or by excluding an exon (i.e., PTC upon exclusion). We asked whether Smg1 deficiency predominantly results in splice isoform level changes for those events predicted to introduce a PTC, as would be expected upon loss of a critical NMD factor. We grouped the 2,859 AS events according to the magnitude of the change in their percent exon inclusion (%In) level between WT and GT cells, and plotted the data according to whether the alternative splicing events were predicted to result in the incorporation of a PTC (Fig. 3A). Notably, the AS events displaying the largest %In changes were predominantly PTC-introducing AS events, consistent with an important role for Smg1 in regulating the levels of a subset of PTC-introducing alternatively spliced transcripts. For example, of 15 exons showing more than a 30%In change between WT and GT cells, 87% were PTC events, representing a significant enrichment of these events over the 51% found in the total set (P < 0.005, χ2 test). Also consistent with a critical role for Smg1 in NMD, the direction of %In changes in GT versus WT cells was consistently biased toward increased accumulation of PTC-containing transcripts in the mutant cells (Fig. 3B). More specifically, GT cells showed increased %In of alternative exons that introduce a PTC upon inclusion, and decreased %In of alternative exons that introduce a PTC upon exclusion, compared with WT cells (P < 0.005; Fig. 3C). Furthermore, the largest changes in %In between WT and GT cells were observed for events that gave rise to increased abundance of PTC-containing transcripts in GT cells.
Fig. 3.
Global AS transcript changes in WT versus GT cells measured by RNA-Seq. Total of 2,859 confidently predicted AS events were used to assess changes in splice variant levels (Materials and Methods) between WT and GT cells. (A) Proportion of total events that do or do not contain a PTC for all samples (first column) or for samples that are more than 5% to more than 30% different in %In levels between WT and GT cells (subsequent columns). (B and C) Comparison of AS events grouped in three categories: those introducing a PTC as a result of exon inclusion (PTC upon inc), those introducing a PTC as a result of exon exclusion (PTC upon exc), or those not introducing a PTC (no PTC). (B) Scatter plot showing the distribution of %In levels for WT versus GT cells color-coded according to group (only events with a >5% difference in AS are shown). (C) Mean difference between WT versus GT cells in relative exon inclusion according to group for all samples (first quadrant) and for samples that are more than 5% to more than 25% different in %In between WT and GT (subsequent quadrants). Error bars indicate SEM. (*P < 0.05; **P < 0.005.)
Previous results obtained by profiling AS events following the knockdown of Upf proteins in cell culture (13–15, 22), and from analyses of NMD inhibition in Caenorhabditis elegans (16, 23), indicate that PTC containing splice variant levels of only a relatively small proportion of profiled genes are affected by NMD disruption. Consistent with these data, we found that, in the absence of Smg1, approximately 9% of predicted PTC-containing AS events displayed changes of more than 10%, and approximately 2% showed changes of more than 20%. To investigate the functions of these NMD-regulated genes, we manually annotated and applied Gene Ontology (GO) enrichment analysis (Materials and Methods) to the set of genes for which there was more than 20%-In change favoring the accumulation of PTC-containing transcripts in GT cells. As was found in previous reports on Upf factor depletion, our subset of more than 20%In change genes was significantly enriched for functions associated with RNA binding and processing and included several splicing factors. However, we also observed enrichment of genes (Flot1, Flot2) that function in the formation of lipid rafts/caveolae, and additional genes (not enriched) that function in membrane dynamics and signaling (Zyx, Cask), cell death (Bat3, Trim24), metabolism (Mgea5, Gls), and transcriptional regulation (CCAR1, Gtf2f2; Table 2, Dataset S1, and Table S1). Transcripts encoding three other transcription factors (Maz, Nfyb/NF-Y, Hsf1) were also subject to AS-coupled NMD, although they displayed a lower degree of %In change in GT versus WT cells. Thus, Smg1-dependent AS-coupled NMD appears to regulate the expression of functionally diverse genes, including splicing and transcription factors that may have additional secondary effects on gene expression (as detailed later).
Table 2.
Genes regulated by Smg1-dependent AS coupled NMD
| Function | Gene |
| Splicing | Sfrs10, Hnrnpa2b1, Srrm1, U2af1, Hnrnph1, Sf3b1, Sf3b3, Rbm5, Sf1, Sfrs11 |
| Membrane dynamics/signaling | Zyx, Flot1, Flot2, Snx13, Wdfy2, Cask, Camk2g, Pigq |
| Cell Death/DNA repair | Alkbh3, Nbr1, Trim24, Bat3 |
| Regulation of transcription | Gtf2f2, Ccar1 |
| Metabolism | Mgea5, Gls |
| Other | Rnf149, Nktr, Slc38a2, Hnrpdl |
| Unknown | Tmem183a, 1810055E12Rik, Tatdn1, Acbd5 |
Manual functional annotation of genes with AS events observed to have the largest degree of NMD regulation (>20%In change favoring accumulation of PTC transcripts in GT primary MEF). For complete list, see Dataset S1.
To assess the accuracy of the RNA-Seq read-predicted changes in splice variant levels, and to confirm that Smg1 regulates levels of PTC-containing splice variants, we performed semiquantitative RT-PCR assays on a representative subset of the genes we observed to produce NMD-regulated PTC-containing transcripts. All (N = 14) the tested events predicted by RNA-Seq to result in increased levels of PTC-containing transcripts in GT versus WT cells were validated by the RT-PCR assays (Fig. 4). Furthermore, for the analyzed events, quantification of %In changes detected in the RT-PCR assays correlated well with the changes quantified using the RNA-Seq data (Pearson r = 0.91; P < 0.0001; Fig. S1). These results validate the accuracy of our RNA-Seq–based predictions of Smg1-dependent changes in PTC-containing splice variant levels, and support the conclusion that the expression of a wider spectrum of gene functions is regulated by NMD than previously appreciated.
Fig. 4.
AS-coupled NMD regulates the expression of functionally diverse genes. Alternatively spliced variant transcripts of endogenous genes in WT and GT cells were analyzed by RT-PCR. Primers pairs were designed to flank alternative exons that result in PTC introduction by exon inclusion or exon exclusion. Previously validated targets (14) are shown in regular type, and new targets identified by RNA-Seq analysis in the present study appear in bold. Numbers below gels are the mean %In for WT and GT cells as calculated by densitometric analysis of data from three independent experiments.
Altered Total mRNA Levels in the Transcriptome of Smg1-Deficient Cells.
We also used our RNA-Seq data to assess Smg1-dependent changes to total mRNA levels that might occur in addition to changes to splice variant levels. To this end, we searched for genes that displayed a more than twofold change in mRNA expression in GT versus WT cells. These changes were measured using the average read counts per kb after normalizing for overall read count differences between the genotypes. Remarkably, 19% (n = 3,396) of profiled genes displayed a more than twofold difference in total transcript levels, and 2% (n = 366) of genes showed a more than fivefold change. The genes displaying more than fivefold changes were highly enriched in the GO categories relating to development, differentiation, muscle, cytoskeleton, ECM, and calcium binding (Table S2 and Dataset S2). Thus, Smg1 directly and likely indirectly affects total mRNA levels of genes that contribute to embryogenesis. The drastic developmental defects seen in the Smg1-deficient mouse may therefore be a result of widespread changes in the transcriptome.
Discussion
NMD is a critical surveillance process that removes potentially deleterious transcripts arising from mutations and errors during gene expression. The importance of NMD has led to efforts to develop therapeutic agents that modulate this process, especially whereby suppression of a disease-related mutation that normally triggers NMD permits translation read-through and partial restoration of critical protein products (24, 25). NMD also plays an important role in the posttranscriptional regulation of numerous RNA binding and processing factors (14, 15, 18, 26). In particular, AS-coupled NMD is known to function in the autoregulation of splicing factors (17, 19). In this study, we have focused on defining the physiological role of the NMD factor Smg1, a member of the PIKK family that activates the essential NMD factor Upf1.
The gene trap ES cell line used in this study had a single integration in the Smg1 locus that suppressed Smg1 protein production and NMD. Transformed MEFs derived from GT embryos were deficient in Smg1 protein production and Upf1 phosphorylation. Whereas previous results have indicated that Smg1 knockdown results in increased sensitivity to TNF-α− or ionizing irradiation-induced apoptosis (4, 7), similar treatments did not result in a significant difference in the rate of apoptosis in GT versus WT tMEFs. This could be a result of differences in cell type, the mode of Smg1 depletion, and/or the transformation process (primary MEFs could not be collected in sufficient quantity to perform these experiments in the present study). Nevertheless, Smg1gt/gt embryos failed to reach term, indicating that one or more Smg1-dependent activities is essential for development. The timing of the gestational lethality in Smg1gt/gt embryos indicated that Smg1 is dispensable for implantation and gastrulation (E6.5) but is critical thereafter. Although the yolk sacs of Smg1gt/gt embryos appeared normal and hematopoiesis was evident, a developed heart field was conspicuously absent in Smg1gt/gt embryos after E8.0. Unsuccessful expansion and differentiation of endoderm into the lining of the heart and rudimentary blood vessels is likely the underlying cause of the developmental arrest of the Smg1gt/gt embryos at or shortly before E8.5.
The role of NMD factors in development has been investigated in other organisms, including C. elegans, Drosophila melanogaster and Danio Rerio. Taken together with our results, a picture is emerging in which NMD factors play distinct developmental roles in different species. For example, in C. elegans, the Smg1 homologue is important for NMD but dispensable for survival (27). In D. melanogaster and D. rerio, the NMD factors Upf1 and Upf2, but not Smg1, are necessary for development (28, 29). In mice, deficiency for Upf1 or Upf2 abolishes NMD and results in early embryonic lethality during the periimplantation period (30, 31). Our observation of the lethality of Smg1 gene trap embryos is therefore consistent with a requirement for NMD early during mammalian embryogenesis. The slightly later developmental arrest observed in our Smg1gt/gt mice compared with Upf protein–deficient embryos could reflect either differential requirements for these factors, or some activity of the hypomorphic Smg1 allele in the gene trap line. Nevertheless, the extensive effects of Smg1 deficiency on the transcriptomes of fibroblasts from the mutant embryos indicates that the developmental failure of Smg1gt/gt mice is most likely a result of disruption of NMD-dependent regulation of many essential genes.
Our comparison of the transcriptomes of WT and GT fibroblasts demonstrated that splicing factor genes are major targets of regulation via AS-coupled NMD in vivo. These results are consistent with, and extend, previous microarray profiling and computational studies investigating PTC-containing splice variants targeted by Upf proteins (13–15, 22). Among the splicing factor genes displaying the greatest degree of Smg1-dependent AS-coupled NMD regulation in our data are Tra2b/Sfrs10, hnRNPA2/Hnrpa2b1, SRm160/Srrm1, U2AF35/U2af1, Sap155/Sf3b1, Sap130/Sf3b3, Luca15/Rbm5, Sf1, and Srsf11/Sfrs11. Five of these genes have been previously reported as AS-coupled NMD targets: Tra2b/Sfrs10 and SRm160/Srrm1 in mouse N2A cells (15) and Sap155/Sf3b1, Sf1, and Srsf11/Sfrs11 in HeLa cells (14). Some AS-coupled NMD events detected previously in HeLa or N2A cells upon knockdown of Upf proteins were not detected in the Smg1-deficient MEFs, despite the availability of sufficient RNA-Seq read coverage to detect these events. This could have resulted from one or more differences between the previous and present experiments including analysis platform, cell type, species, context (i.e., developmental stage), and/or factor dependencies. Strikingly, in our study, we found that genes with a variety of nonsplicing functions were also regulated by AS-coupled NMD, including many associated with intracellular signaling, membrane dynamics, cell death, DNA repair, transcriptional regulation, and metabolism. These results demonstrate that, in vivo, AS-coupled NMD controls genes from diverse functional categories in addition to those involved in RNA processing.
Our work has also revealed a role for Smg1 in regulating total mRNA levels of genes whose loss of function would be expected to result in the phenotypic defects of the mutant embryos. For example, genes expressed at more than 15-fold lower levels in GT versus WT cells included several that are crucial for embryonic viability: Myl1, Ptprb, Actc1, and Smyd1. Although the demise of Smg1-deficient embryos cannot be attributed to the alteration of any one particular transcript or set of transcripts, we hypothesize that disruption of the AS-coupled NMD of a specific subset of genes involved in the regulation of splicing and transcription in turn causes widespread changes in mRNA expression levels in Smg1gt/gt embryos. In particular, if loss of Smg1 alters AS-coupled NMD regulation of one (or more) transcription factors, the transcriptional regulation of many other genes with important roles in embryogenesis could be disrupted. We propose that the changes in gene regulation at the splicing and transcriptional levels induced by Smg1 deficiency likely culminated in the observed delayed development and death of the mutant embryos in utero.
Our study indicates that approximately 9% of genes with predicted PTC-introducing AS events were subject to pronounced regulation by NMD, in line with previous reports that most predicted PTC-containing transcripts are not regulated by NMD (see Results). Although this small percentage could in part reflect the sensitivity of our RNA-Seq analysis, it nevertheless supports the view that the majority of predicted PTC-containing AS transcripts are not bona fide NMD substrates. Future studies are warranted to examine how AS and NMD are coordinated in vivo to control transcript abundance, and to determine how this regulation influences normal development and pathophysiology.
Materials and Methods
Generation of Smg1gt/gt Mice and Cells.
Heterozygous gene trap ES cells (RRT449; Bay Genomics) of the 129Ola background were injected into blastocysts from C57BL/6J mice (Jackson Laboratories). Blastocysts were implanted into pseudopregnant females and resulting chimeric agouti pups with successful germline transmission of the gene trap allele were backcrossed to C57BL/6J for 10 generations. Mouse embryonic fibroblasts were derived from E8.5 embryos as described (21) and cultured in DMEM supplemented with 10% FCS.
PCR Genotyping.
Genotypes were determined using the following primers (Fig. 1A): WT Smg1 allele (forward, TTGCCAGACTTGTGGTACCGATTAT; reverse, CCACCTCTCAGAGCCCTAGCATTTA); Smg1 gene trap allele had the same forward primer plus GT-reverse GGTGTTTTAAGTGTACCCACGGTCA. PCR conditions were 40 cycles at 94 °C for 45 s, 61 °C for 30 s, and 72 °C for 1 min. Mendelian ratios were evaluated using the χ2 test, and P values for intergroup comparisons were determined using the Student t test.
Semiquantitative RT-PCR of NMD-Regulated AS.
Primers were designed as previously described (14) and are listed in Table S3. RT-PCR was performed by using Qiagen OneStep RT-PCR kit plus RNA extracted from primary embryonic fibroblasts using the Qiagen RNeasy kit according to the manufacturer's instructions. RT-PCR conditions were 30 cycles at 94 °C for 40 s, 60 °C for 40 s, and 72 °C for 1 min. Densitometry was performed by using the Bio-Rad Mo-lecular Imager Gel Doc XR+ system.
Immunoprecipitation and Western Blotting.
Cells were lysed with RIPA buffer (Western blot; 150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholic acid, 0.1% SDS, 50 mM Tris, pH 8.0, plus 10 μM sodium orthovanadate (Sigma) or CHAPS buffer for immunoprecipitation (100 mM NaCl, 1mM MgCl2, 0.5% CHAPS, 10 mM Tris, pH 7.5), along with phosSTOP and Protease Inhibitor Mixture (Roche). Endogenous Upf1 immunoprecipitated using 8 μg of rabbit anti-RENT1 antibody (A301-902A; Bethyl) or control rabbit IgG, crosslinked to Protein G–Sepharose beads (GE Healthcare). Anti–phospho-(Ser/Thr) no. 2851 (Cell Signaling), anti-RENT1 (as described), anti-Smg1 (A300-393A; Bethyl), and anti–β-tubulin (clone AA2; Upstate) were used for Western blotting according to standard techniques. Proteins were visualized using fluorescently labeled secondary antibodies and an Odyssey Infrared Imaging System (Li-Cor Biosciences).
Histology.
Embryos were fixed in 4% paraformaldehyde (24–48 h at 4 °C), embedded in paraffin, serially sectioned (5–7 μm), and stained with H&E according to standard methods. Whole-embryo photography was performed on previously fixed samples. For cell proliferation assays, pregnant females were injected intraperitoneally with 300 μL BrdU (10 mg/mL; cat. no. 10 280 879 001; Roche) 35 min before dissection to isolate embryos. Paraffin sections of fixed embryos were processed to detect BrdU as described (32). For apoptosis, TUNEL staining was performed using an in situ cell death detection kit (cat. no. 11684 817 910; Roche) following the manufacturer's instructions. Images were captured on a Leica MZ16F (whole-mount embryos) or a Leica DM2500 stereomicroscope (tissue sections).
mRNA Sequencing.
Illumina mRNA-Seq reads (50 nt) were first aligned to mouse genomic sequence [National Center for Biotechnology Information (NCBI) Genome Assembly build 37] using Bowtie (33), and reads mapping to multiple locations were removed. The remaining reads were mapped to RefSeq transcripts and a database of mouse splicing junctions as previously described (34), allowing as many as two mismatches/indels (insertions or deletions). The database comprised 11,423 mouse AS events identified by aligning mouse mRNA/EST sequences (NCBI UniGene build 169) to genomic sequence (NCBI Genome Assembly build 37) as described (13, 35). Searches for PTCs in AS events were performed as described (13).
GO Enrichment Profiling.
GO profiling was used to identify statistically overrepresented GO terms in our datasets. Briefly, Mouse Genome Informatics gene lists for NMD-AS events (>20%In) and mRNA expression (more than 5-fold) were profiled using corresponding full list as background with the GOstat program (36). False discovery rate (Benjamini) correction for multiple testing was used. Level 4 and higher GO hierarchy reported and the highest GOterm was used for redundant returns. P values were determined by Fisher exact test.
Calculations and Statistics.
Alternative exon inclusion levels (%In) were calculated as previously described (34). A total of 2,859 AS events with sufficient read coverage for confident prediction of alternative splicing levels (criteria described in ref. 34) were used for further analysis. The significance of differences in %In between WT and GT for PTC upon inclusion, PTC upon exclusion, and no PTC was calculated using the Wilcoxon rank-sum test. The significance of differences in PTC enrichment in AS events was determined by the χ2 test.
Supplementary Material
Acknowledgments
We thank Christine Misquitta-Ali, Arneet Saltzman, and Lynne Maquat for expert technical assistance and helpful discussions; Elize Shirdel and Sam Molyneux for help with data analysis; and Mary Saunders for scientific editing. This work was supported by grants from the Canadian Institutes of Health Research (to T.W.M. and B.J.B.), the Terry Fox Cancer Foundation (to T.W.M.), and grants from the National Cancer Institute of Canada and Genome Canada through the Ontario Genomics Institute (to B.J.B. and others).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007336107/-/DCSupplemental.
References
- 1.Lovejoy CA, Cortez D. Common mechanisms of PIKK regulation. DNA Repair (Amst) 2009;8:1004–1008. doi: 10.1016/j.dnarep.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denning G, Jamieson L, Maquat LE, Thompson EA, Fields AP. Cloning of a novel phosphatidylinositol kinase-related kinase: Characterization of the human SMG-1 RNA surveillance protein. J Biol Chem. 2001;276:22709–22714. doi: 10.1074/jbc.C100144200. [DOI] [PubMed] [Google Scholar]
- 3.Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001;15:2215–2228. doi: 10.1101/gad.913001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brumbaugh KM, et al. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol Cell. 2004;14:585–598. doi: 10.1016/j.molcel.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Azzalin CM, et al. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 6.Chen RQ, et al. Kinome siRNA screen identifies SMG-1 as a negative regulator of hypoxia-inducible factor-1alpha in hypoxia. J Biol Chem. 2009;284:16752–16758. doi: 10.1074/jbc.M109.014316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliveira V, et al. A protective role for the human SMG-1 kinase against tumor necrosis factor-alpha-induced apoptosis. J Biol Chem. 2008;283:13174–13184. doi: 10.1074/jbc.M708008200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamashita A, Kashima I, Ohno S. The role of SMG-1 in nonsense-mediated mRNA decay. Biochim Biophys Acta. 2005;1754:305–315. doi: 10.1016/j.bbapap.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Kashima I, et al. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isken O, Maquat LE. Quality control of eukaryotic mRNA: Safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 11.Weischenfeldt J, Lykke-Andersen J, Porse B. Messenger RNA surveillance: Neutralizing natural nonsense. Curr Biol. 2005;15:R559–R562. doi: 10.1016/j.cub.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 13.Pan Q, et al. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes Dev. 2006;20:153–158. doi: 10.1101/gad.1382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saltzman AL, et al. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol Cell Biol. 2008;28:4320–4330. doi: 10.1128/MCB.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ni JZ, et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barberan-Soler S, Zahler AM. Alternative splicing regulation during C. elegans development: splicing factors as regulated targets. PLoS Genet. 2008;4:e1000001. doi: 10.1371/journal.pgen.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGlincy NJ, Smith CW. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem Sci. 2008;33:385–393. doi: 10.1016/j.tibs.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Spellman R, Llorian M, Smith CW. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol Cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lareau LF, et al. The Coupling of Alternative Splicing and Nonsense-Mediated mRNA Decay. In: Blencowe BJ, Graveley BR, editors. Alternative Splicing in the Postgenomic Era. Austin: Landes Bioscience; 2007. pp. 190–211. [DOI] [PubMed] [Google Scholar]
- 20.Nord AS, et al. The International Gene Trap Consortium website: A portal to all publicly available gene trap cell lines in mouse. Nucleic Acids Res. 2006;34(database issue):D642–D648. doi: 10.1093/nar/gkj097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hakem A, et al. RhoC is dispensable for embryogenesis and tumor initiation but essential for metastasis. Genes Dev. 2005;19:1974–1979. doi: 10.1101/gad.1310805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen KD, et al. Genome-wide identification of alternative splice forms down-regulated by nonsense-mediated mRNA decay in Drosophila. PLoS Genet. 2009;5:e1000525. doi: 10.1371/journal.pgen.1000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramani AK, et al. High resolution transcriptome maps for wild-type and nonsense-mediated decay-defective Caenorhabditis elegans. Genome Biol. 2009;10:R101. doi: 10.1186/gb-2009-10-9-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.James PD, et al. Aminoglycoside suppression of nonsense mutations in severe hemophilia. Blood. 2005;106:3043–3048. doi: 10.1182/blood-2005-03-1307. [DOI] [PubMed] [Google Scholar]
- 25.Wilschanski M, et al. Gentamicin-induced correction of CFTR function in patients with cystic fibrosis and CFTR stop mutations. N Engl J Med. 2003;349:1433–1441. doi: 10.1056/NEJMoa022170. [DOI] [PubMed] [Google Scholar]
- 26.Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 27.Grimson A, O'Connor S, Newman CL, Anderson P. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA decay in Caenorhabditis elegans. Mol Cell Biol. 2004;24:7483–7490. doi: 10.1128/MCB.24.17.7483-7490.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metzstein MM, Krasnow MA. Functions of the nonsense-mediated mRNA decay pathway in Drosophila development. PLoS Genet. 2006;2:e180. doi: 10.1371/journal.pgen.0020180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wittkopp N, et al. Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol Cell Biol. 2009;29:3517–3528. doi: 10.1128/MCB.00177-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Medghalchi SM, et al. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet. 2001;10:99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- 31.Weischenfeldt J, et al. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 2008;22:1381–1396. doi: 10.1101/gad.468808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hao Z, et al. Specific ablation of the apoptotic functions of cytochrome C reveals a differential requirement for cytochrome C and Apaf-1 in apoptosis. Cell. 2005;121:579–591. doi: 10.1016/j.cell.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 33.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 35.Pan Q, et al. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet. 2005;21:73–77. doi: 10.1016/j.tig.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 36.Beissbarth T, Speed TP. GOstat: Find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.