Abstract
Activation of the cellular DNA damage response is detrimental to adenovirus (Ad) infection. Ad has therefore evolved a number of strategies to inhibit ATM- and ATR-dependent signaling pathways during infection. Recent work suggests that the Ad5 E4orf3 protein prevents ATR activation through its ability to mislocalize the MRN complex. Here we provide evidence to indicate that Ad12 has evolved a different strategy from Ad5 to inhibit ATR. We show that Ad12 utilizes a CUL2/RBX1/elongin C-containing ubiquitin ligase to promote the proteasomal degradation of the ATR activator protein topoisomerase-IIβ–binding protein 1 (TOPBP1). Ad12 also uses this complex to degrade p53 during infection, in contrast to Ad5, which requires a CUL5-based ubiquitin ligase. Although Ad12-mediated degradation of p53 is dependent upon both E1B-55K and E4orf6, Ad12-mediated degradation of TOPBP1 is solely dependent on E4orf6. We propose that Ad12 E4orf6 has two principal activities: to recruit the CUL2-based ubiquitin ligase and to act as substrate receptor for TOPBP1. In support of the idea that Ad12 E4orf6 specifically prevents ATR activation during infection by targeting TOPBP1 for degradation, we demonstrate that Ad12 E4orf6 can inhibit the ATR-dependent phosphorylation of CHK1 in response to replication stress. Taken together, these data provide insights into how Ad modulates ATR signaling pathways during infection.
Keywords: DNA damage, cullin-RING ubiquitin ligases, DNA damage, proteasome
Maintenance of genome integrity is essential for cell survival. Cells have therefore evolved a complex set of biochemical pathways to recognize and repair damaged DNA (1). In addition to recognizing cellular DNA lesions, there is increasing evidence to show that this response is also activated by viral DNA during infection, and that some DNA repair proteins can inhibit viral replication (2).
The DNA damage response is primarily controlled by two related protein kinases, ataxia-telangiectasia mutated (ATM) and ATM/Rad3-related (ATR). Both kinases have a large and partially overlapping set of substrates that includes tumor suppressor and checkpoint proteins such as p53 and BRCA1 (3). However, some proteins, such as the checkpoint effector kinases CHK1 and CHK2, are specific substrates for ATR and ATM, respectively (4). ATM predominantly responds to DNA double-strand breaks (DSBs), which are sensed by the MRE11-RAD50-NBS1 (MRN) complex (5). MRN is required for ATM activation, as it stimulates both ATM recruitment to DSBs and phosphorylation of its substrates (5, 6). In contrast, ATR becomes active in response to a much wider range of genotoxic stresses (4). This is probably accomplished by the recognition of a common signal, ssDNA, which can be generated at stalled replication forks or by enzymatic processing of DNA lesions (4). Replication protein A (RPA) binds to tracts of ssDNA and recruits ATR by interacting with its essential binding partner, ATR-interacting protein (ATRIP) (7, 8). ATR activation also requires other proteins, including topoisomerase-IIβ–binding protein 1 (TOPBP1) (9, 10). TOPBP1 contains a domain that stimulates ATR kinase activity, and so is essential for the phosphorylation of many ATR targets, including CHK1 (9, 10).
Adenovirus type 5 (Ad5) is a linear dsDNA tumor virus that provides a useful model system for studying the DNA damage response, as ATM and ATR signaling pathways are inhibited during infection (11). Ad5 E1B-55K and E4orf6 proteins prevent ATM activation by promoting the ubiquitin-dependent proteasomal degradation of MRN (11, 12). Cullin-RING ubiquitin ligases (CRLs) are multiprotein complexes required for the polyubiquitylation and proteasomal degradation of a large number of cellular proteins, to control many diverse processes such as cell cycle progression, limb patterning, and glucose sensing (13). During Ad5 infection, E1B-55K and E4orf6 associate with a CRL complex containing CUL5, RBX1, and elongins B and C, and serve to target this CRL to new substrates (14, 15). It has been suggested that E1B-55K is the substrate receptor for the complex, and E4orf6 is the adaptor that recruits the CRL as it has several motifs, termed BC boxes, that bind to the elongin B and C subunits (16, 17). As virtually all detectable Ad5 E1B-55K/E4orf6 associates with the CUL5 complex (15), it is likely that targeting cellular proteins for degradation is the major function of E1B-55K and E4orf6 during Ad5 infection (18).
E1B-55K/E4orf6–dependent degradation of MRN is sufficient to prevent ATM activation (11), and this appears to be conserved between some Ad species (19). Interestingly, however, two viral serotypes, Ad5 and Ad12, differentially regulate ATR during infection (19). The Ad5 E4orf3 protein inhibits ATR activation by relocalizing and immobilizing MRN subunits before their targeted degradation by E1B-55K/E4orf6 (20), but Ad12 E4orf3 lacks this activity (21). Yet both viruses inhibit CHK1 phosphorylation, indicating that full ATR activation is prevented by both Ad5 and Ad12, but by different strategies (19).
Here, we provide evidence to show that a virus can inhibit ATR signaling by specifically targeting its activator protein, TOPBP1. Ad12 E4orf6 assembles a CRL complex that targets TOPBP1 for proteasomal degradation. In contrast to previous work with Ad5, we show that this CRL contains CUL2 rather than CUL5, and that Ad12 E1B-55K is not required for TOPBP1 degradation. Rather, Ad12 E4orf6 interacts directly with TOPBP1 and is necessary and sufficient to promote its degradation. Finally, we show that, in uninfected cells exposed to replication stress, exogenous expression of Ad12 E4orf6 prevents CHK1 phosphorylation, indicating that by targeting TOPBP1 for degradation, E4orf6 can directly inhibit ATR in the absence of other viral proteins. Taken together, this study establishes Ad12 E4orf6 as a modulator of CRL-dependent ubiquitylation and a regulator of ATR/TOPBP1-dependent signaling.
Results
TOPBP1 Is Degraded in Ad12- but Not Ad5-Infected Cells.
Ad5 E1B-55K and E4orf6 proteins promote proteasomal degradation of MRN to inhibit ATM signaling (11). We reasoned that Ad12 might also degrade one or more other cellular proteins to inhibit ATR. Therefore, we infected HeLa cells with WT Ad5 or Ad12 and examined the steady-state levels of proteins involved in ATR activation at appropriate times postinfection, using MRE11 degradation by both Ad5 and Ad12 as a control (Fig. 1A). DNA ligase IV (LIG4), previously shown to be targeted for degradation in Ad5-infected cells (22), was also degraded in Ad12-infected cells, indicating that, like MRN, it may be a common target for human Ads. Further analysis revealed that ATR, ATRIP, RPA70, RPA32, and RAD9 levels remained constant in Ad5- and Ad12-infected cells. Slower migrating forms of RAD9 and RPA32 were detected in Ad12-infected cells, representing ATR-dependent phosphorylation of these proteins (19). In contrast, TOPBP1 levels were significantly reduced in Ad12- but not Ad5-infected cells, indicating that TOPBP1 may be specifically targeted for degradation by Ad12.
Fig. 1.
TOPBP1 is degraded in a proteasome-dependent manner in Ad12- but not Ad5-infected cells. (A) Effect of Ad infection on expression levels of proteins involved in ATR activation. HeLa cells were mock-infected or infected with the indicated viruses. Cells were harvested and prepared for Western blotting at the indicated time points using the appropriate antibodies. (B) Effects of the proteasome inhibitor MG132 on TOPBP1 expression levels in Ad12-infected cells. HeLa cells were mock-infected or infected with Ad12, in the presence or absence of 10 μM MG132 added 2 h after infection. Cells were harvested and prepared for Western blotting at the indicated time points, and MRE11 was used as a positive control.
Given that Ad5 promotes proteasomal degradation of MRN (12), p53 (14), and LIG4 (22), we examined whether the loss of TOPBP1 expression observed in Ad12-infected cells was also proteasome-dependent. To do this we assessed the levels of TOPBP1 and MRE11 in Ad12-infected cells treated with the proteasome inhibitor MG132. Significantly, TOPBP1 and MRE11 levels were considerably greater in Ad12-infected cells treated with MG132, than in Ad12-infected cells not treated with proteasome inhibitor (Fig. 1B). Taken together, our data show that Ad12 infection results in increased targeting of TOPBP1 to the proteasome for degradation.
TOPBP1 Localizes to Viral Replication Centers in Ad-Infected Cells.
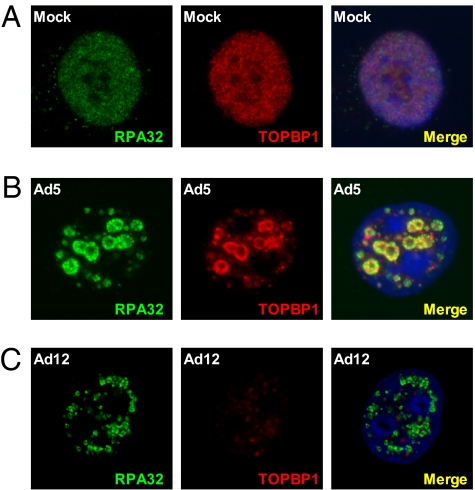
We and others have shown previously that components of the ATR pathway are recruited to Ad5 and Ad12 viral replication centers in infected cells (11, 19–21). Given the apparent differences in ATR and TOPBP1 regulation in Ad5- and Ad12-infected cells, we examined TOPBP1 localization patterns in these cells. In mock-treated cells, both RPA32 and TOPBP1 were found to be diffusely localized in the nucleoplasm, but excluded from nucleoli (Fig. 2A). However, in Ad5-infected cells, TOPBP1 was relocalized to sites of viral replication, where it colocalized with Ad5 DNA-binding protein (DBP), E1B-55K, and RPA32 at early and late times after infection (Fig. 2B and Fig. S1). Next, we determined TOPBP1 localization in Ad12-infected cells, using RPA32 as a marker of viral replication centers (21). At early times after infection, TOPBP1 was recruited to viral replication sites (Fig. S1); significantly, however, this staining pattern was lost at later times (Fig. 2C), consistent with Western blotting data shown in Fig. 1. Weak residual TOPBP1 nuclear staining was present at late times after infection, but this was not observed at viral replication centers (Fig. 2C). These results indicate that, although TOPBP1 is initially recruited to viral replication sites, it is subsequently degraded in Ad12-infected cells.
Fig. 2.
Localization of TOPBP1 and RPA32 in mock-infected cells (A), Ad5-infected cells (B), and Ad12-infected cells (C). HeLa cells were mock-infected or infected with the appropriate viruses, and fixed 24 h later for analysis by immunofluorescence and confocal microscopy.
It has previously been reported that p53 and MRN are relocalized to cytoplasmic aggresomes and nuclear track-like structures during Ad5 infection by E1B-55K and E4orf3, before degradation by E1B-55K and E4orf6 (23, 24). However, we did not observe either type of staining for TOPBP1 in Ad5- or Ad12-infected cells (Fig. S2), suggesting that TOPBP1 degradation by Ad12 may be mechanistically different from p53 and MRN degradation in Ad5-infected cells.
Elongin C and RBX1 Are Required for TOPBP1 and p53 Degradation in Ad12-Infected Cells.
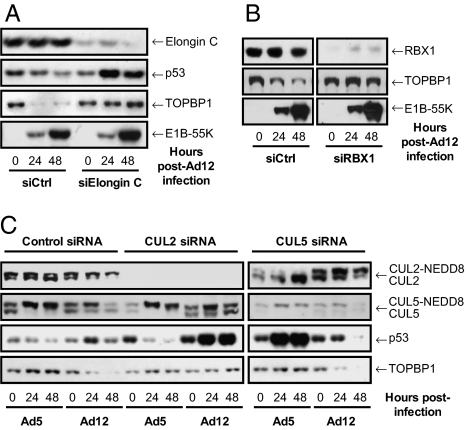
The ubiquitin ligase responsible for p53 degradation in Ad5-infected cells contains the cellular proteins CUL5, RBX1, and elongins B and C, as well as the viral E1B-55K and E4orf6 proteins (14, 15). To determine whether this complex was also required for TOPBP1 degradation in Ad12-infected cells, we initially transfected cells with nonsilencing siRNAs or siRNAs targeting elongin C, a component of both CUL2- and CUL5-based ubiquitin ligases (13). Cells depleted of elongin C were infected with Ad12, and TOPBP1 levels assessed by Western blotting, using p53 as a positive control for viral CRL activity (Fig. 3A). Elongin C knockdown resulted in the stabilization of p53 in Ad12-infected cells, probably resulting from the action of Ad E1A (25), demonstrating that the activity of the E1B-55K/E4orf6 ubiquitin ligase was compromised. Significantly, loss of elongin C also resulted in the inhibition of TOPBP1 degradation in Ad12-infected cells, indicating that, like p53, TOPBP1 is targeted for proteolysis by a CUL2- or CUL5-based ubiquitin ligase.
Fig. 3.
Elongin C (A) and RBX1 (B) are required for p53 and TOPBP1 degradation in Ad12-infected cells. (C) CUL2 is required for p53 and TOPBP1 degradation in Ad12-infected cells; CUL5 is required for p53 degradation in Ad5-infected cells. A549 cells were transfected with the indicated siRNAs, before being mock-infected or infected with Ad5 or Ad12 72 h later. Cells were harvested and prepared for Western blotting at the indicated time points using the appropriate antibodies.
We also assessed the requirement for RBX1 in promoting TOPBP1 degradation, as Ad5 E1B-55K/E4orf6 uses RBX1 to catalyze p53 polyubiquitylation (14). To do this, we transfected cells with control siRNAs and siRNAs targeting RBX1, and then infected them with Ad12. Cells were harvested as indicated and protein levels were analyzed by Western blotting. We found that RBX1 knockdown prevented TOPBP1 degradation in Ad12-infected cells (Fig. 3B); our data therefore suggest that, like Ad5, Ad12 requires a CRL to promote degradation of cellular proteins.
Ad5 and Ad12 Use Different CRLs to Degrade Cellular Proteins.
As both CUL2 and CUL5 ubiquitin ligases contain elongin C and RBX1 subunits, we wished to confirm whether Ad12 uses the same CUL5-based complex as Ad5 to direct polyubiquitylation of cellular proteins. To do this, we transfected cells with control siRNAs or siRNAs targeting CUL2 or CUL5, infected them with Ad5 or Ad12, and assessed the levels of p53 and TOPBP1 by Western blotting. As expected, in Ad5-infected cells depleted of CUL5, p53 was stabilized and TOPBP1 levels remained unaffected (Fig. 3C); CUL2 knockdown had no effect on Ad5-mediated p53 degradation. In contrast, CUL2 knockdown severely inhibited the ability of Ad12 to target both p53 and TOPBP1 for degradation, whereas CUL5 knockdown had no effect. These data indicate that Ad5 and Ad12 use different CRLs to degrade cellular proteins.
CRLs are activated by addition of the ubiquitin-like protein NEDD8 to the Cullin subunit (13), which can be visualized by SDS/PAGE as an increase in molecular weight. Given that Ad5 and Ad12 use different CRLs, it seemed logical that CUL2 and CUL5 might be differentially neddylated during Ad5/12 infection. Indeed, when we examined the neddylation status of CUL2 and CUL5 in Ad5- and Ad12-infected cells, we observed notable differences (Fig. 3C and Fig. S3). In Ad5-infected cells, CUL5 was mostly converted into the neddylated, high molecular weight form. In contrast, CUL2 became increasingly deneddylated in Ad5-infected cells. In Ad12-infected cells, the opposite pattern of neddylation/deneddylation was observed: CUL2 became increasingly neddylated whereas CUL5 neddylation levels were not altered appreciably. These data indicate that Ad5 E4orf6 uses a neddylated CUL5-based ubiquitin ligase to degrade p53, whereas Ad12 E4orf6 uses a neddylated CUL2-based complex to degrade both p53 and TOPBP1.
Ad12 E4orf6 Is Necessary and Sufficient to Promote TOPBP1 Degradation.
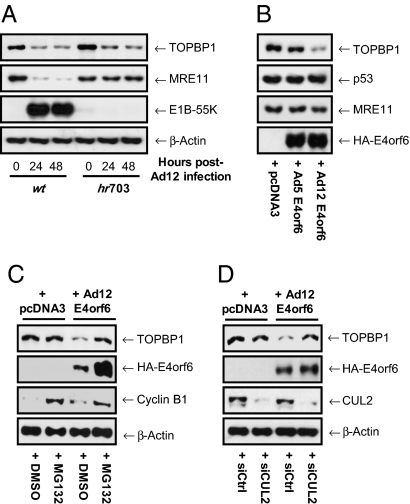
To establish whether expression of both E1B-55K and E4orf6 is needed to promote TOPBP1 degradation, we determined the requirement for E1B-55K, using the Ad12 E1B deletion mutant hr703 (26). Cells were infected with WT Ad12 or hr703, harvested at suitable times and Western blotted for TOPBP1 and MRE11. Consistent with previous reports, we found that Ad12-mediated MRE11 degradation was dependent on E1B-55K expression (Fig. 4A). However, TOPBP1 was still efficiently degraded following infection with hr703, indicating that Ad12-mediated degradation of TOPBP1 does not require E1B-55K.
Fig. 4.
(A) TOPBP1 degradation by Ad12 is independent of E1B-55K. HeLa cells were mock-infected or infected with WT Ad12 or the E1B-55K mutant virus hr703. Cells were harvested and prepared for Western blotting at the indicated times. MRE11 was used as a positive control. (B) Ad12 E4orf6 is necessary and sufficient for TOPBP1 degradation. HeLa cells were transfected with pcDNA3-Ad12-E4orf6, pcDNA3-Ad5-E4orf6, or pcDNA3 vector alone and harvested for Western blotting 24 h later. MRE11 and p53 were used as negative controls. (C) Ad12 E4orf6-mediated degradation of TOPBP1 is proteasome-dependent. HeLa cells were transfected with pcDNA3-Ad12-E4orf6 or pcDNA3 vector alone in the presence or absence of 10 μM MG132, added 6 h after transfection. Cells were harvested for Western blotting 24 h later. Cyclin B1 levels were used as a positive control for proteasome inhibition. (D) Ad12 E4orf6-mediated degradation of TOPBP1 is CUL2-dependent. HeLa cells treated with nonsilencing or CUL2 siRNAs 48 h earlier were transfected with pcDNA3-Ad12-E4orf6 or pcDNA3 vector alone, before being harvested for Western blotting 24 h later.
Given these findings, we considered the possibility that Ad12 E4orf6 alone might both recruit CRL activity and target TOPBP1 directly for degradation. To test this hypothesis, we transfected cells with plasmids expressing Ad12 E4orf6 or Ad5 E4orf6, and assessed TOPBP1 levels by Western blotting (Fig. 4B). As expected, expression of Ad5 or Ad12 E4orf6 alone had no effect on the steady-state levels of p53 or MRE11. Interestingly, and consistent with our hypothesis, Ad12 but not Ad5 E4orf6 promoted a dramatic reduction in TOPBP1 levels (Fig. 4B). Further transfection experiments with Ad12 E4orf6 and the proteasome inhibitor, MG132, showed that the Ad12 E4orf6-mediated degradation of TOPBP1 was proteasome-dependent and that Ad12 E4orf6 was itself a substrate for the proteasome (Fig. 4C). To substantiate the requirement for CUL2 in Ad12 E4orf6-mediated TOPBP1 degradation, we assessed TOPBP1 levels in cells expressing Ad12 E4orf6 that had been treated before transfection with nonsilencing siRNAs or siRNAs targeting CUL2 (Fig. 4D). In agreement with the results from our infection experiments (Fig. 3C), CUL2 knockdown negated the ability of Ad12 E4orf6 to promote TOPBP1 degradation (Fig. 4D). Taken together these results demonstrate that Ad12 E4orf6 is able to promote the CRL-mediated degradation of TOPBP1 in the absence of E1B-55K or other viral proteins.
Ad12 E4orf6 Interacts Selectively with CUL2 and Binds Directly to TOPBP1.
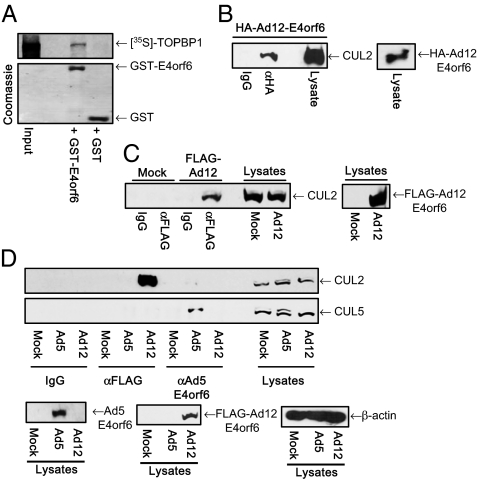
To confirm that Ad12 E4orf6 recruits CUL2 but not CUL5, and interacts with TOPBP1 directly, we assessed the ability of Ad12 E4orf6 to bind to each of these proteins. Consistent with our hypotheses, pulldown analyses showed that Ad12 E4orf6 had a greater binding capacity for CUL2 than for CUL5 (Fig. S4A), and that Ad12 E4orf6 binds to TOPBP1 directly (Fig. 5A), through at least two binding sites (Fig. S4B). To confirm the in vivo relevance of these findings, we performed immunoprecipitation (IP)–Western blot analyses. Significantly, these studies revealed that Ad12 E4orf6 associated with CUL2 in cells transfected with a plasmid expressing Ad12 E4orf6 (Fig. 5B). To address whether E4orf6 associates with CUL2 in Ad12-infected cells, we constructed an Ad12 mutant that expresses FLAG-tagged E4orf6 (FLAG-Ad12; Materials and Methods). Consistent with the transfection studies, FLAG-tagged Ad12 E4orf6 was found to associate with CUL2 in FLAG-Ad12–infected cells (Fig. 5C). To confirm whether Ad5 and Ad12 E4orf6 use distinct CRLs during infection (Fig. 3C), we determined the abilities of Ad5 E4orf6 and Ad12 E4orf6 to bind to CUL2 and CUL5 in cells infected with WT Ad5 or FLAG-Ad12. Crucially, these studies showed that Ad12 E4orf6 binds exclusively to CUL2, and that Ad5 E4orf6 binds exclusively to CUL5 (Fig. 5D). Taken together, these results indicate that Ad12 uses a CUL2-based CRL to degrade p53 and TOPBP1 during infection, whereas Ad5 uses a CUL5-based CRL to degrade p53 during infection. These data also suggest that Ad12 E4orf6 functions as a substrate receptor for TOPBP1 to recruit it directly to the CUL2-containing CRL for polyubiquitylation and degradation by the proteasome.
Fig. 5.
(A) Ad12 E4orf6 associates with TOPBP1 in vitro. GST-Ad12 E4orf6 or GST alone was incubated with [35S]-methionine-labeled TOPBP1. Following pull-downs and SDS/PAGE, radiolabeled proteins were identified by fluorography and autoradiography. (B and C) Ad12 E4orf6 associates with CUL2 in vivo. HeLa cells were transfected with pcDNA3-Ad12-E4orf6; A549 cells were either mock-infected or infected with FLAG-Ad12. Cells were harvested and subjected to IP–Western blot analyses with the appropriate antibodies 24 h later. (D) Ad5 and Ad12 E4orf6 target different Cullins in vivo. A549 cells were mock-infected or infected with WT Ad5 or FLAG-Ad12. Cells were harvested 24 h after infection and subjected to IP–Western blot analyses with the appropriate antibodies.
Ad12 E4orf6 Inhibits ATR-Dependent Phosphorylation of CHK1 in Response to Replication Stress.
We have shown previously that Ad12 infection results in partial ATR activation (19). However, despite this, CHK1 is not phosphorylated during Ad12 infection, indicating that full ATR activation is prevented. Given that TOPBP1 activates ATR in response to a variety of genotoxic stresses, it is likely that Ad12 E4orf6 inhibits the ATR-dependent phosphorylation of CHK1 in infected cells by promoting TOPBP1 degradation. To determine if Ad12 E4orf6 can similarly inhibit ATR activation in the absence of viral infection, we first transfected cells with plasmids expressing Ad12 or Ad5 E4orf6, and subsequently treated them with hydroxyurea (HU) to activate ATR and promote CHK1 phosphorylation. As anticipated, HU promoted CHK1 phosphorylation in cells expressing Ad5 E4orf6 or transfected with the empty vector (Fig. 6). Crucially, however, Ad12 E4orf6 expression significantly reduced the ATR-dependent phosphorylation of CHK1 in response to HU (Fig. 6). These data indicate that, in the absence of other viral proteins, Ad12 E4orf6 is able to inhibit ATR-dependent signaling in response to replication stress, presumably by targeting TOPBP1 for degradation.
Fig. 6.
Ad12 E4orf6 inhibits ATR signaling in response to replication stress. HeLa cells were transfected with pcDNA3-Ad12-E4orf6, pcDNA3-Ad5-E4orf6 or pcDNA3 vector alone. After 24 h, cells were mock-treated or treated with HU for the indicated times, before being harvested for Western blotting analysis.
Discussion
Ad12 E4orf6 Promotes TOPBP1 Degradation to Inhibit ATR Activation.
We have shown previously that Ad12 differentially regulates ATR during infection, as it promotes ATR-dependent phosphorylation of RPA32 and RAD9, but inhibits CHK1 phosphorylation (19). In this report, we define the mechanism by which Ad12 inhibits the ATR-dependent phosphorylation of CHK1. We show that the ATR activator protein, TOPBP1, is a unique target for proteasomal degradation in Ad12- but not Ad5-infected cells (Fig. 1). Previous work with Ad5 has shown that two viral proteins, E1B-55K and E4orf6, hijack a CUL5-based ubiquitin ligase complex to target p53, MRN, and LIG4 for degradation (12, 14, 22). In contrast, the data presented here show that Ad12 E1B-55K is not required for TOPBP1 degradation (Fig. 4A), and that Ad12 E4orf6 alone can promote TOPBP1 destruction (Fig. 4B). We have determined that Ad12 E4orf6 associates with a functionally active CUL2–RBX1–elongin B/C ubiquitin ligase complex through a direct, selective interaction with the CUL2 scaffold, and that Ad12 E4orf6 also serves as a CRL substrate receptor by recruiting TOPBP1 to the complex through direct binding (Figs. 3 and 5 and Fig. S4). Consistent with our hypothesis that Ad12 E4orf6 inhibits CHK1 phosphorylation by promoting TOPBP1 degradation, cells transfected with Ad12 E4orf6, but not Ad5 E4orf6, were unable to activate CHK1 in response to replication stress (Fig. 6). We therefore propose that Ad12 selectively negates the ATR-CHK1 signaling pathway during infection by actively promoting the CRL-mediated destruction of TOPBP1.
It is interesting to note that ATR activation is inhibited during infection not only by Ad, but also other viruses. HSV-1 ICP0, for example, promotes uncoupling of ATR and ATRIP during infection (27). It is unclear why ATR activation is detrimental to viral infection, as it does not interfere with Ad5 DNA replication (28). ATM and ATR phosphorylate many of the proteins required for checkpoint activation and DNA repair, but they also target proteins involved in RNA metabolism, including splicing (3). Ad uses the host cell transcription and splicing machinery to produce viral proteins, and a recent report indicates that ATM can inhibit protein synthesis (29). It may be that ATM and/or ATR activation inhibits late viral protein expression, which would limit the production of viral progeny; this could be a major reason why Ads have evolved to prevent activation of ATM and ATR, and will certainly require further investigation.
Ad5 and Ad12 Use Different Cullins to Degrade Cellular Targets.
It is becoming increasingly apparent that, like Ad, many other viruses target CRLs during infection. HIV, human papilloma virus, and EBV, for example, all produce proteins that recruit CRLs to target cellular proteins for degradation (30–32). However, it had not been shown that two viruses from the same family, Ad5 and Ad12, can use different CRLs to degrade host proteins. Using siRNAs targeting CRL subunits, we have determined the requirements for p53 and TOPBP1 degradation by Ad5 and Ad12. Although p53 and TOPBP1 degradation by Ad12 requires elongin C and RBX1 (Fig. 3 A and B), Ad5 specifically uses CUL5, whereas Ad12 uses CUL2 (Fig. 3C). This observation was surprising given the overall similarity between Ad5 and Ad12 E4orf6 proteins, but was supported by the altered neddylation patterns observed for CUL2 and CUL5 in infected cells, and the respective abilities of Ad5 and Ad12 proteins to bind CUL2 and CUL5 (Figs. 3C and 5). Neddylation activates CRLs (13), and our data indicate that, during Ad5 infection, CUL5 becomes increasingly neddylated whereas CUL2 is progressively deneddylated. In contrast, in Ad12-infected cells, CUL2 becomes progressively more neddylated whereas CUL5 neddylation is not altered appreciably. It will therefore be of great interest to determine how Ad regulates Cullin neddylation patterns, especially as it has recently been reported that the EBV BPLF1 protein functions as a deneddylase to regulate CRL activity in infected cells to facilitate EBV replication (33). Given that Ad5 and Ad12 E4orf6 proteins show 50% identity and 74% similarity, it would also be useful in the future to clarify what determines the Cullin-binding selectivity of Ad5 and Ad12 E4orf6 proteins, as this may shed light on how CRLs are regulated in uninfected cells.
The work presented here is important in highlighting that closely related human Ad species have adopted different strategies to counteract host cell DNA damage signaling pathways activated during infection. This work also establishes that Ad12 E4orf6, at least, has functions independent of Ad12 E1B-55K, which may have important ramifications for understanding the role of E4orf6 during viral infection and the processes of Ad-mediated cellular transformation and oncogenesis. It is becoming increasingly clear that viruses have evolved to target common host cell intracellular signaling pathways through divergent mechanisms during their infectious life cycle, to neutralize host-defense processes and promote viral replication. Investigating the processes by which different viruses modulate these pathways will be important in gaining a more complete understanding of how these pathways operate at the molecular level.
Materials and Methods
Rabbit polyclonal antibody raised against TOPBP1 has been described previously (34). Further details of antibodies, Western blotting, and immunoprecipitation and immunofluorescence techniques can be found in SI Materials and Methods.
Cells and Viruses.
A549 and HeLa cells were used interchangeably throughout this study as reliable cell models for studying adenovirus infection and viral gene function. Cells were maintained in DMEM supplemented with 8% FCS and 2 mM L-glutamine (Invitrogen) and cultured at 37 °C in humidified incubators supplied with 5% CO2. WT human adenoviruses Ad5 and Ad12 were obtained from the American Tissue Culture Collection. The Ad12 E1B mutant hr703 has been described previously (26). Infections were carried out at a multiplicity of infection of 25. Where appropriate, medium was supplemented with MG132 or HU (both from Sigma-Aldrich) at the indicated concentration.
Plasmids and Transfections.
CUL2-, CUL5-, and Myc-TOPBP1-pcDNA3 vectors were gifts from P. Branton (McGill University, Montreal) and J. Chen (University of Texas M.D. Anderson Cancer Center, Houston). HA-E4orf6 and Gal4-DBD-TOPBP1 plasmids have been described previously (35, 36). Ad12 E4orf6 was cloned into pGEX-4T-1 and expressed in BL21 (Stratagene) for GST-fusion protein production. Plasmids were transfected into HeLa cells using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen).
Generation of FLAG-Ad12.
A FLAG tag was introduced into Ad12 E4orf6, following its ATG start codon, by insertion PCR using the QuikChange in vitro mutagenesis kit (Stratagene), a bacmid template encompassing the E1 (nucleotides 1–5,930) and E4 (nucleotides 29,514–34,125) regions of the Ad12 genome (Ad12pPG-S1-SwaI E1-E4-Box), and the following oligonucleotide primers: CGCTCGCAAGTCTGTTGTTTACGATG-GACTACAAGGACGACGATGACAAG-CAGCGCGACAGACGGTATCGCTACAG (forward) and CTGTAGCGATACCGTCTGTCGCGCTG-CTTGTCATCGTCGTCCTTGTAGTC-CATCGTAAACAACAGACTTGCGAGCG (reverse). Following the generation of the resultant Ad12pPG-S1-SwaI E1-FLAG-E4orf6 construct, the central region of the Ad12 genome (nucleotides 5,391–29,513) was reintroduced into the Ad12pPG-S1-SwaI E1-FLAG-E4orf6 construct, using a unique CsiI cloning site, to generate a full-length Ad12 genomic mutant with FLAG-tagged E4orf6 (FLAG-Ad12pPG-S1-SwaI). To generate FLAG-Ad12 we performed a SwaI digest and transfected the linearized bacmid in 2E2-cells (37) to generate virus particles. Virus was propagated by several rounds of infection and titered by plaque assay.
GST Pull-Down Assays.
In vitro GST pull-down assays were performed as described previously (19).
RNA Interference.
siRNAs used in this study targeting CUL2, CUL5, elongin C, and RBX1 were SMARTpools purchased from Dharmacon. siRNAs were transfected using Oligofectamine (Invitrogen) as described previously (19).
Supplementary Material
Acknowledgments
We thank Philip Branton (McGill University, Montreal), Junjie Chen (The University of Texas M.D. Anderson Cancer Center, Houston), and Paul Freimuth (Brookhaven National Laboratory, Upton, NY) for reagents. This work was funded by the University of Birmingham College of Medical and Dental Sciences. The Heinrich-Pette Institute is supported by the Freie und Hansestadt Hamburg and the Bundesministerium für Gesundheit.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0914605107/-/DCSupplemental.
References
- 1.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaurushiya MS, Weitzman MD. Viral manipulation of DNA repair and cell cycle checkpoints. DNA Repair (Amst) 2009;8:1166–1176. doi: 10.1016/j.dnarep.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 4.Cimprich KA, Cortez D. ATR: An essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee J-H, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 6.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 7.Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: Partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- 8.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 9.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 10.Liu S, et al. Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol Cell Biol. 2006;26:6056–6064. doi: 10.1128/MCB.00492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carson CT, et al. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 2003;22:6610–6620. doi: 10.1093/emboj/cdg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stracker TH, Carson CT, Weitzman MD. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature. 2002;418:348–352. doi: 10.1038/nature00863. [DOI] [PubMed] [Google Scholar]
- 13.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 14.Querido E, et al. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 2001;15:3104–3117. doi: 10.1101/gad.926401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harada JN, Shevchenko A, Shevchenko A, Pallas DC, Berk AJ. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J Virol. 2002;76:9194–9206. doi: 10.1128/JVI.76.18.9194-9206.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanchette P, et al. Both BC-box motifs of adenovirus protein E4orf6 are required to efficiently assemble an E3 ligase complex that degrades p53. Mol Cell Biol. 2004;24:9619–9629. doi: 10.1128/MCB.24.21.9619-9629.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng CY, Blanchette P, Branton PE. The adenovirus E4orf6 E3 ubiquitin ligase complex assembles in a novel fashion. Virology. 2007;364:36–44. doi: 10.1016/j.virol.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 18.Blackford AN, Grand RJA. Adenovirus E1B 55-kilodalton protein: Multiple roles in viral infection and cell transformation. J Virol. 2009;83:4000–4012. doi: 10.1128/JVI.02417-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blackford AN, et al. A role for E1B-AP5 in ATR signaling pathways during adenovirus infection. J Virol. 2008;82:7640–7652. doi: 10.1128/JVI.00170-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carson CT, et al. Mislocalization of the MRN complex prevents ATR signaling during adenovirus infection. EMBO J. 2009;28:652–662. doi: 10.1038/emboj.2009.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stracker TH, et al. Serotype-specific reorganization of the Mre11 complex by adenoviral E4orf3 proteins. J Virol. 2005;79:6664–6673. doi: 10.1128/JVI.79.11.6664-6673.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baker A, Rohleder KJ, Hanakahi LA, Ketner G. Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J Virol. 2007;81:7034–7040. doi: 10.1128/JVI.00029-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans JD, Hearing P. Relocalization of the Mre11-Rad50-Nbs1 complex by the adenovirus E4 ORF3 protein is required for viral replication. J Virol. 2005;79:6207–6215. doi: 10.1128/JVI.79.10.6207-6215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Shevchenko A, Shevchenko A, Berk AJ. Adenovirus exploits the cellular aggresome response to accelerate inactivation of the MRN complex. J Virol. 2005;79:14004–14016. doi: 10.1128/JVI.79.22.14004-14016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grand RJ, Grant ML, Gallimore PH. Enhanced expression of p53 in human cells infected with mutant adenoviruses. Virology. 1994;203:229–240. doi: 10.1006/viro.1994.1480. [DOI] [PubMed] [Google Scholar]
- 26.Byrd PJ, Grand RJ, Breiding D, Williams JF, Gallimore PH. Host range mutants of adenovirus type 12 E1 defective for lytic infection, transformation, and oncogenicity. Virology. 1988;163:155–165. doi: 10.1016/0042-6822(88)90242-5. [DOI] [PubMed] [Google Scholar]
- 27.Wilkinson DE, Weller SK. Herpes simplex virus type I disrupts the ATR-dependent DNA-damage response during lytic infection. J Cell Sci. 2006;119:2695–2703. doi: 10.1242/jcs.02981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lakdawala SS, et al. Differential requirements of the C terminus of Nbs1 in suppressing adenovirus DNA replication and promoting concatemer formation. J Virol. 2008;82:8362–8372. doi: 10.1128/JVI.00900-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Braunstein S, Badura ML, Xi Q, Formenti SC, Schneider RJ. Regulation of protein synthesis by ionizing radiation. Mol Cell Biol. 2009;29:5645–5656. doi: 10.1128/MCB.00711-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu X, et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 31.Huh K, et al. Human papillomavirus type 16 E7 oncoprotein associates with the cullin 2 ubiquitin ligase complex, which contributes to degradation of the retinoblastoma tumor suppressor. J Virol. 2007;81:9737–9747. doi: 10.1128/JVI.00881-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato Y, et al. Degradation of phosphorylated p53 by viral protein-ECS E3 ligase complex. PLoS Pathog. 2009;5:e1000530. doi: 10.1371/journal.ppat.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gastaldello S, et al. A deneddylase encoded by Epstein-Barr virus promotes viral DNA replication by regulating the activity of cullin-RING ligases. Nat Cell Biol. 2010;12:351–361. doi: 10.1038/ncb2035. [DOI] [PubMed] [Google Scholar]
- 34.Boner W, et al. A Functional interaction between the human papillomavirus 16 transcription/replication factor E2 and the DNA damage response protein TopBP1. J Biol Chem. 2002;277:22297–22303. doi: 10.1074/jbc.M202163200. [DOI] [PubMed] [Google Scholar]
- 35.Rubenwolf S, Schütt H, Nevels M, Wolf H, Dobner T. Structural analysis of the adenovirus type 5 E1B 55-kilodalton-E4orf6 protein complex. J Virol. 1997;71:1115–1123. doi: 10.1128/jvi.71.2.1115-1123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wright RH, Dornan ES, Donaldson MM, Morgan IM. TopBP1 contains a transcriptional activation domain suppressed by two adjacent BRCT domains. Biochem J. 2006;400:573–582. doi: 10.1042/BJ20060831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Catalucci D, et al. An adenovirus type 5 (Ad5) amplicon-based packaging cell line for production of high-capacity helper-independent deltaE1-E2-E3-E4 Ad5 vectors. J Virol. 2005;79:6400–6409. doi: 10.1128/JVI.79.10.6400-6409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.