Abstract
B-cell chronic lymphocytic leukemia (B-CLL), the most common leukemia in the Western world, occurs in two forms, aggressive (showing for the most part high ZAP-70 expression and unmutated IgH VH) and indolent (showing low ZAP-70 expression and mutated IgH VH). We found that miR-29a is up-regulated in indolent human B-CLL as compared with aggressive B-CLL and normal CD19+ B cells. To study the role of miR-29 in B-CLL, we generated Eμ-miR-29 transgenic mice overexpressing miR-29 in mouse B cells. Flow cytometric analysis revealed a markedly expanded CD5+ population in the spleen of these mice starting at 2 mo of age, with 85% (34/40) of miR-29 transgenic mice exhibiting expanded CD5+ B-cell populations, a characteristic of B-CLL. On average, 50% of B cells in these transgenic mice were CD5 positive. At 2 y of age the mice showed significantly enlarged spleens and an increase in the CD5+ B-cell population to ∼100%. Of 20 Eμ-miR-29 transgenic mice followed to 24–26 mo of age, 4 (20%) developed frank leukemia and died of the disease. These results suggest that dysregulation of miR-29 can contribute to the pathogenesis of indolent B-CLL.
Keywords: mouse models, microRNA
Chronic lymphocytic leukemia (CLL) is the most common human leukemia, accounting for ∼30% of all cases (1), with ∼10,000 new cases observed each year in the United States. Characteristically, CLL is a disease of elderly people, with the incidence increasing linearly with each decade above age 40 y (1, 2). It is known that this disease is characterized by the clonal expansion of CD5+ B cells (2).
MicroRNAs, representing between 1% and 3% of all eukaryotic genes, are a class of endogenous noncoding RNAs, 19–25 nt in size, which regulate gene expression at the transcriptional or translational level (3). Approximately half of human microRNAs are located at fragile sites and genomic regions involved in alterations in cancers (4), and alteration of microRNA expression profiles occurs in most cancers, suggesting that individual microRNAs could function as tumor suppressors or oncogenes (5).
The 13q14 deletion is the most common CLL aberration and is detected by cytogenetic analysis in approximately half of the cases (6). Analysis of a deletion at 13q14.3 led to the discovery of two physically linked microRNAs, miR-15a and miR-16–1, as targets of these deletions (7). Consequently, miR-15a and miR-16–1 expression is reduced in the majority of CLL cases (7), and further studies indicated that miR-15a/miR-16–1 negatively regulate Bcl2 expression (8). These findings indicated that microRNAs play important roles in CLL and that down-regulation of miR-15/16 and subsequent Bcl2 up-regulation contribute to CLL pathogenesis (7). Because miR-15/16 was identified as a tumor suppressor in indolent CLL, the microRNA expression profile in CLL has been studied extensively, and a signature profile was reported describing 13 microRNAs that differentiate aggressive and indolent CLL (4).
We and others observed that miRNA-29 expression is down-regulated in aggressive CLL as compared with indolent CLL (9, 10), and several reports indicated that miR-29 might function as a tumor suppressor by targeting several oncogenes, including TCL1, MCL1, and CDK6 (9, 11, 12). On the other hand, one report showed that miR-29 expression is up-regulated in metastatic breast cancer, and a very recent study reported that miR-29 overexpression can cause acute myeloid leukemia (AML) in mice (13, 14). To clarify the role of miR-29 in B-cell leukemias, we generated transgenic mice overexpressing miR-29 in B cells and now report the phenotype of this mouse model.
Results
MiR-29 Expression in CLL and Production of the Eμ-miR-29 Transgenic Mouse Model.
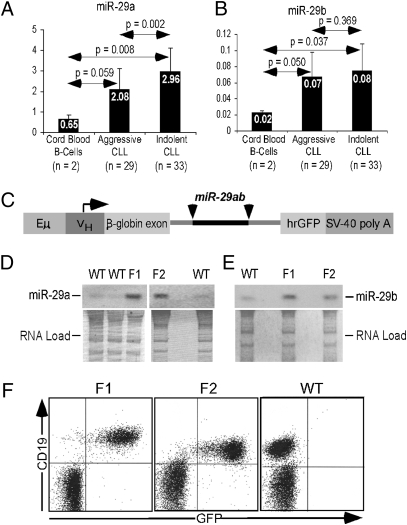
As noted above, we have reported previously that miR-29 expression is down-regulated in aggressive (compared with indolent) CLL (9, 10), but data comparing miR-29 expression in CLL and normal CD19+ B cells was not available. To determine expression levels of miR-29 in CLL and normal CD19+ B cells, we studied the expression of miR-29a and miR-29b in 29 aggressive CLL samples, 33 indolent CLL samples, and two normal CD19+ B-cell controls. Fig. 1 A and B shows real-time RT-PCR results in these samples. MiR-29a expression was 4.5-fold higher in indolent CLL than in normal CD19+ B cells, whereas aggressive CLL samples showed a 3.2-fold increase. Similarly, miR-29b expression was increased 4-fold in indolent CLL and 3.5-fold in aggressive CLL compared with normal CD19+ B cells. Both miR-29a and miR-29b were down-regulated in aggressive versus indolent CLL, confirming previous observations (9), although in the case of miR-29b this difference was not statistically significant (Fig. 1B). Interestingly, in all samples miR-29a expression level was more than 20-fold higher than that of miR-29b (Fig. 1 A and B).
Fig. 1.
MiR-29 expression in CLL and production of Eμ-miR-29 transgenic founder mice. (A) MiR-29a and (B) miR-29b expression in aggressive and indolent CLL. (C) Eμ-miR-29 construct. (D and E) Expression of (D) miR-29a and (E) miR-29b in splenic lymphocytes of Eμ-miR-29 founders. (F) Expression of GFP in splenic lymphocytes of Eμ-miR-29 founders.
Because expression levels of miR-29a and miR-29b were significantly higher in indolent CLL than in normal CD19+ B cells, we hypothesized that miR-29 could contribute to the pathogenesis of CLL. To investigate this possibility, we generated transgenic mice in which expression of the mouse miR-29a/b cluster was controlled by a VH promoter-IgH-Eμ enhancer (15), along with humanized renilla green fluorescent protein (hrGFP), and the simian virus 40 (SV40) poly(A) site. This promoter/enhancer combination drives expression of miR-29a/b in immature and mature B cells (Fig. 1C). The miR-29a/b cluster sequence was inserted within the intron of this construct (Fig. 1C). Two founders on FVB/N background, designated “F1” and “F2,” were generated and bred to establish the transgenic lines. Expression of miR-29a and miR-29b was examined by Northern blot analysis, using RNAs isolated from spleens of transgenic animals. Fig. 1 D and E shows overexpression of miR-29a and miR-29b in both transgenic lines (F1 and F2) compared with nontransgenic (WT) siblings. To confirm that the transgene is expressed in B cells, we performed flow cytometry using CD19 as a B-cell marker. Fig. 1F shows that all CD19+ cells in both transgenic lines also express GFP (F1 and F2), whereas no GFP expression was detected in WT littermates.
Eμ-miR-29 Transgenic Mice Show CLL Phenotype.
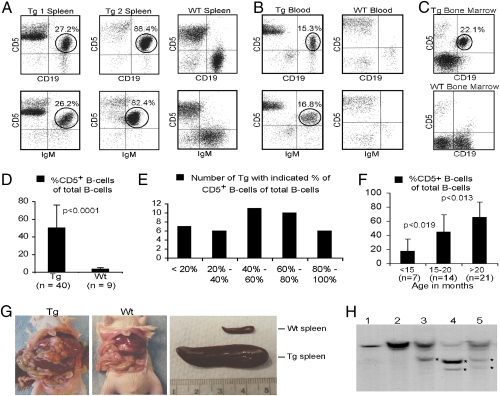
We used flow cytometry to determine the immunophenotypic profile of spleen lymphocytes from miR-29 transgenic mice. At the age of 12–24 mo, flow cytometric analysis revealed a markedly expanded CD5+ B-cell population (a characteristic of CLL) in the spleen of 34 of 40 (85%) miR-29 transgenic mice; ∼50% of B cells in these transgenic mice were CD5+. Fig. 2A (Left) shows a representative example. Although almost all spleen B cells from this animal were CD5+CD19+IgM+, these cells represented only 25–30% of all spleen lymphocytes. A more advanced CLL case is shown in Fig. 2A (Center). Almost all normal lymphocytes in the spleen of this animal were replaced by malignant CD5+CD19+IgM+ B cells. As expected, almost no CD5+CD19+IgM+ B cells were detected in spleens of WT littermates (Fig. 2A, Right). The expanded population of CD5+CD19+ B cells also was detected in peripheral blood and bone marrow from miR-29 transgenic mice, but not from WT littermates (Fig. 2, B and C). Fig. 2 D–F shows the number of animals with increased CD5+CD19+IgM+ populations in spleen. Although only 7 of 40 (17%) miR-29 transgenic mice showed 0–20% CD5+ B cells, 16 of 40 (40%) showed 60% or more CD5+CD19+IgM+ cells. In addition, miR-29 transgenic mice showed significant increases in the percentage of CD5+ splenic B cells with age (Fig. 2F). In animals younger than 15 mo, CD5+ B cells represented only ∼20% of total B cells; by 15–20 mo of age, that percentage increased to ∼40% (Fig. 2F). At the age of 20–26 mo, on average, >65% of all B cells were CD5+ (Fig. 2F). These data suggest gradual progression of indolent CLL in miR-29 transgenic mice. We followed 20 Eμ-miR-29 mice to the age of 24–26 mo. Almost all these mice showed significantly enlarged spleens, and 4 of 20 (20%) developed frank leukemia and died of disease. Fig. 2G shows a representative case of frank leukemia presenting with an enlarged spleen and liver and advanced lymphadenopathy.
Fig. 2.
Eμ-miR-29 mice develop CLL. (A–C) Flow cytometric analysis of miR-29 transgenic (Tg) and control lymphocytes isolated from (A) spleen, (B) peripheral blood, and (C) bone marrow. (D–F) Analysis of CD5+ B-cell populations in miR-29 transgenic mice and WT controls. (G) Gross pathology of a representative Eμ-miR-29 transgenic mouse showing advanced CLL and a WT control of the same age. (H) Analysis of IgH gene configuration by Southern blot: spleen lymphocyte DNA isolated from five representative cases showing at least 50% CD5+CD19+ B cells. Clonal rearrangements are indicated by asterisks.
Clonal IgH gene rearrangements are typical in human CLL cases (1). These rearrangements also were observed in the Tcl1-driven mouse model of CLL (15). To determine if CD5+ B cells from Eμ-miR-29 transgenic mice show clonality, we carried out Southern blot hybridization using spleen lymphocyte DNA isolated from cases showing at least 50% CD5+CD19+IgM+ B cells. Fig. 2H shows clonal rearrangements of the IgH gene in three of five cases analyzed. These results further indicate that the expansion of CD5+ B cells in Eμ-miR-29 mice resembles human CLL.
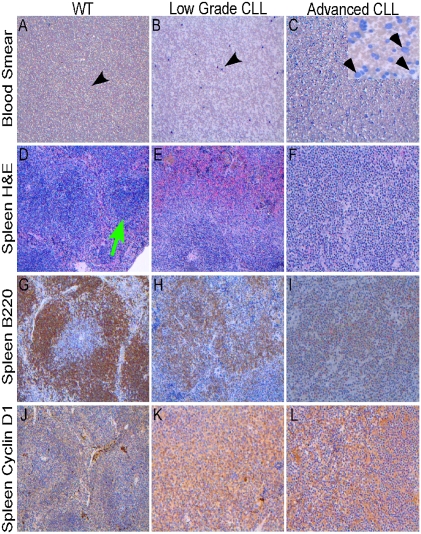
To confirm further that Eμ-miR-29 mice develop CLL-like disease, we carried out histological and immunohistological analysis. Fig. 3 A–C shows representative smears from blood of Eμ-miR-29 transgenic mice and a WT control. The smear from a WT mouse showed rare lympho-monocytes with a normal appearance (Fig. 3A). In contrast, the smear from a Eμ-miR-29 mouse with low-grade CLL exhibited an increased number of atypical lymphoid cells (Fig. 3B, black arrowheads), and the smear from a miR-29 transgenic mouse with advanced CLL presented numerous malignant lymphoid cells (Fig. 3C), including smudge cells, typical of CLL (Fig. 3C, Inset; smudge cells are indicated by arrowheads). Fig. 3 D–L shows representative histological images of Eμ-miR-29 transgenic mice and a WT control. The spleen of the WT mouse shows preserved architecture and several normal-looking lymphoid follicles (Fig. 3D, green arrow). In contrast, the spleen of a diseased miR-29 transgenic mouse with CLL exhibits distorted architecture (Fig. 3E), and the spleen of a miR-29 mouse with advanced CLL shows total obliteration of the normal architecture by malignant lymphoid proliferation (Fig. 3F). B220 staining of the same sections shows a lymphoid follicle of a WT mouse presenting a normal B-cell disposition (Fig. 3G). In contrast, transgenic spleens show lymphoid follicles in disarray because of the low-grade malignant lymphoid proliferation (Fig. 3H) or CLL with diffuse distribution of a B-cell malignant population (Fig. 3I). Fig. 3 J–L shows low expression of cyclin D1 in a WT spleen (Fig. 3J) and moderate to high cyclin D1 expression in low-grade CLL (Fig. 3K) and advanced CLL (Fig. 3L). In summary, histological and immunohistological examination confirmed that Eμ-miR-29 mice develop CLL-like disease.
Fig. 3.
Histopathological analysis of Eμ-miR-29 mice. Smudge cells indicated by arrowheads. Atypical lymphoid cells are indicated by black arrowheads. A normal lymphoid follicle is indicated by a green arrow.
As noted above, only 20% of Eμ-miR-29 transgenic mice developed advanced leukemia and died from the disease. Fig. S1 shows a representative advanced case of CLL that invaded liver and kidney. Histological examination showed total obliteration of the normal spleen architecture with high expression of B220, cyclin D1, and Ki67 (Fig. S1 A–D). These B220+ malignant B cells invaded liver (Fig. S1 E–H) and kidney (Fig. S1 I–L).
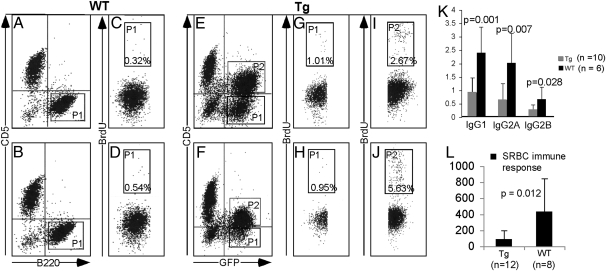
Recent investigations showed that accumulation of CLL lymphocytes can result not only from prolonged survival but also from proliferating CD5+B220+ cells originating in the bone marrow, lymph nodes, or spleen (16–18). To determine whether CLL cells from Eμ-miR-29 mice proliferate, we used cell cycle analyses based on BrdU incorporation. We assessed the proliferative capacity of B220+CD5+, as well as B220+CD5− transgenic splenic lymphocytes in comparison with WT B220+ splenic lymphocytes. Fig. 4 A–J shows that B220+CD5+ B cells from Eμ-miR-29 mice proliferate, whereas no proliferation was detected for B220+ WT lymphocytes (2.7% and 5.6% cells in S-phase for transgenic B cells versus 0.3% and 0.5% for WT B cells (Fig. 4 I and J versus C and D). Interestingly, even B220+CD5− transgenic lymphocytes showed increased proliferation compared with B220+ WT B cells, with 1.0% and 0.95% cells in S-phase versus 0.3% and 0.5% for WT B cells (Fig. 4 G and H versus C and D. These data suggest that miR-29 overexpression promotes B-cell proliferation, even in CD5− cells.
Fig. 4.
Cell-cycle analysis of leukemic cells from Eμ-miR-29 transgenic mice. (A–D) BrdU incorporation into DNA of WT B220++ B cells. (E–J) BrdU incorporation into transgenic B220+CD5+ and B220+CD5− B-cell DNA. (K) Ig levels in serum of WT and transgenic animals. (L) Levels of anti-SRBC–specific antibodies in serum of WT and transgenic animals 7 d after SRBC injection.
Human CLL is characterized by immune incompetence and progressive severe hypogammaglobulinemia that eventually develops in almost all patients (19). To determine if Eμ-miR-29 mice develop hypogammaglobulinemia, we compared levels of serum Ig in transgenic mice and in WT littermates at age ∼18 mo. Fig. 4K shows that the levels of IgG1, IgG2a, and IgG2b were decreased 2- to 4-fold in Eμ-miR-29 transgenic mice as compared with WT controls. To determine if Eμ-miR-29 mice show impaired immune response, we compared levels of anti-sheep RBC (SRBC) antibodies after injection of SRBC in miR-29 transgenic mice and WT siblings. Fig. 4L shows that serum levels of anti-SRBC antibodies were decreased ∼4-fold in serum of miR-29 transgenic mice compared with age-matched WT mice. These data clearly indicate that, as in human CLL, the CLL-like disease in Eμ-miR-29 mice is characterized by hypogammaglobulinemia and immune incompetence.
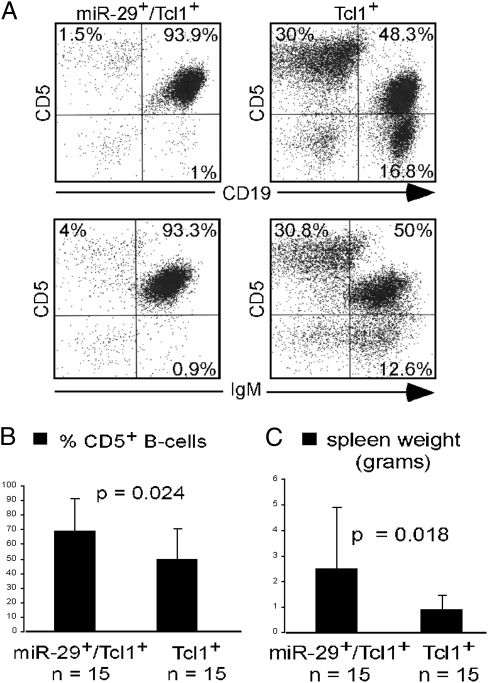
Previously, we reported development and characterization of a Tcl1-driven mouse model of CLL (15, 20, 21) and showed that miR-29 can target TCL1 expression (9). In this mouse model, the TCL1 ORF (lacking 3′ UTR) was under the control of a VH promoter-IgH-Eμ enhancer (15). Because of the absence of the 3′ UTR in the transgenic construct, miR-29 could not inhibit TCL1 expression in these mice. Eμ-TCL1 transgenic mice develop aggressive CLL, and all mice die of the disease at 12–15 mo of age (15). To determine if transgenic miR-29 expression can accelerate CLL in Eμ-TCL1 transgenic mice, we crossed Eμ-miR-29 and Eμ-TCL1 transgenic mice. Eμ-miR-29/Eμ-TCL1 mice and their Eμ-TCL1 littermates were killed at ∼8 mo of age and analyzed. Fig. 5A shows representative FACS analysis of spleen lymphocytes of these genotypes. TCL1/miR-29 double transgenic mice showed significantly increased CD5+CD19+ and CD5+IgM+ B-cell populations compared with Eμ-TCL1 mice (93.9% and 93.3% versus 48.3% and 50%). On average, Eμ-miR-29/Eμ-TCL1 mice had 40% more CD5+CD19+ splenic B cells and 3-fold increases in spleen weight compared with Eμ-TCL1 mice (Fig. 5 B and C). These data suggest that miR-29 can contribute to the pathogenesis of CLL independently of Tcl1.
Fig. 5.
Mir-29 transgene expression accelerates CLL in Eμ-TCL1 mice. (A) Flow cytometric analysis of Eμ-TCL1/Eμ-miR-29 and Eμ-TCL1 transgenic lymphocytes from spleen. (B) Percentage of CD5+ B cells in Eμ-TCL1/Eμ-miR-29 and Eμ-TCL1 transgenic spleen lymphocytes. (C) Spleen weight from Eμ-TCL1/Eμ-miR-29 and Eμ-TCL1 transgenic mice.
Analysis of miR-29 Targets.
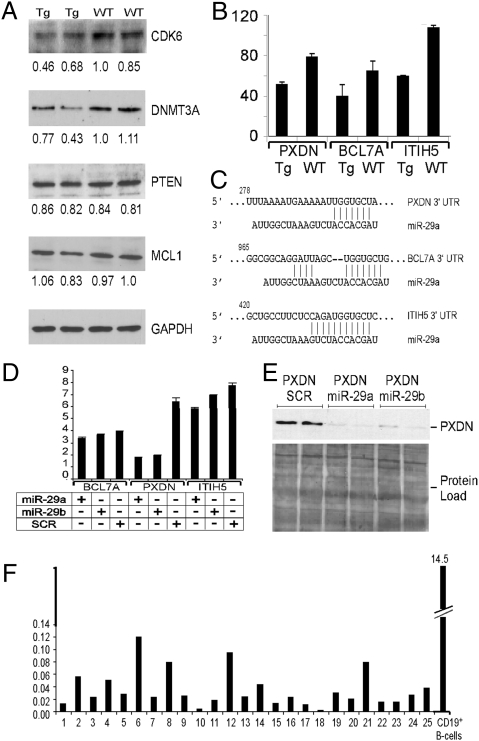
To determine whether miR-29 overexpression in mouse B cells affects expression of its targets, we analyzed expression levels of several previously reported miR-29 targets, Cdk6 (11), Mcl1 (12), and DNMT3A (22), in sorted B220+ B cells from miR-29 transgenic mice and WT controls. We found that two targets, Cdk6 and DNMT3A, are down-regulated in miR-29 transgenic mice, whereas no differences in Mcl1 and Pten were detected (Fig. 6A) [although Pten is not a proven miR-29 target, it previously was predicted to be a potential target (14)].
Fig. 6.
Analysis of miR-29 targets in Eμ-miR-29 transgenic mice. (A) Western blot analysis of Cdk6, DNMT3A, PTEN, and Mcl1 expression in CD19+ B cells of miR-29 transgenic and WT mice. (B) Microarray expression data for PXDN, BCL7A, and ITIH5 in CD19+ B cells of miR-29 transgenic and WT mice. (C) Sequence alignments of miR-29a and 3′ UTRs of PXDN, BCL7A, and ITIH5. (D) miR-29 targets PXDN but not BCL7A and ITIH5 expression in luciferase assays. (E) Effect of miR-29 on Pxdn protein expression. (F) PDXN expression in CLL.
Because Cdk6 and DNMT3 are not known to be tumor suppressors, we used Affymetrix gene expression arrays to determine potential miR-29 targets contributing to its oncogenic activity. Using microarray analysis, we compared gene expression in sorted B220+ B cells from miR-29 transgenic mice and WT controls. We then cross-referenced genes down-regulated in miR-29 transgenic B cells that had known or potential tumor suppressor function with the list of potential miR-29 targets obtained from Targetscan software (Whitehead Institute for Biomedical Research at MIT). We identified three potential targets: peroxidasin (PXDN), a p53-responsive gene down-regulated in AML (23, 24); Bcl7A, a proapoptotic gene down-regulated in T-cell lymphomas (25); and ITIH5, a member of the inter-α-trypsin inhibitor family down-regulated in breast cancer (26). Fig. 6 B and C shows the down-regulation of expression of these three genes in CD19+ B cells of miR-29 transgenic mice versus WT littermates and the alignment of miR-29a and corresponding 3′ UTRs. To determine if miR-29 indeed targets expression of PXDN, Bcl7A, and ITIH5, we inserted 3′ UTR fragments (including miR-29 homology regions) of these cDNAs downstream of the luciferase ORF into pGL3 vector, as previously described (8). HEK293 cells were cotransfected with miR-29a, miR-29b, or scrambled negative control and a pGL3 construct containing fragments of PXDN, Bcl7A, and ITIH5 cDNAs, including a region homologous to miR-29, as indicated (Fig. 6D). Expression of miR-29a or miR-29b significantly (∼3-fold) decreased luciferase expression of the construct containing the 3′ UTR of PXDN, whereas no significant effect was observed for Bcl7A and ITIH5 (Fig. 6D). Thus, we concluded that PXDN expression could be targeted by miR-29. To confirm these findings, we used full-length PXDN cDNA including 5′ and 3′ UTRs in a cytomegalovirus mammalian expression vector and investigated whether miR-29 expression affects Pdxn protein expression levels. We cotransfected this construct with miR-29a, miR-29b, or PremiR negative control (scrambled) into HEK293 cells, as indicated in Fig. 6E. These experiments revealed that coexpression of PXDN with miR-29a or miR-29b almost completely inhibited Pxdn expression (Fig. 6E). We therefore concluded that miR-29a and miR-29b target Pxdn expression at mRNA and protein levels. To determine if Pdxn could play a role in the pathogenesis of human CLL, we studied expression of PXDN in 25 human CLL samples and normal CD19+ B-cell controls. Fig. 6F shows real-time RT-PCR results in these samples. PXDN expression was drastically down-regulated (50-fold or more) in CLL samples compared with normal CD19+ B cells. These results suggest that the oncogenic role of miR-29 in B cells might be, at least in part, dependent on targeting peroxidasin.
Discussion
Although many microRNAs are up- or down-regulated in a number of solid tumors and hematological malignancies, and several are postulated to function as tumor suppressors or oncogenes (5), only three reports have used mouse models to demonstrate that dysregulation of microRNAs can cause cancer. The first report showed that overexpression of miR-155 in B cells results in pre–B-cell leukemia in mice (27). The knockout of miR-15/16 led to development of CLL with low penetrance (28, 29). Finally, a very recent report showed that overexpression of miR-29 can cause AML in mice (14). Although nearly all published reports conclude that miR-155 functions as an oncogene and that miR-15/16 is a tumor suppressor, the function of miR-29 in this respect has not been clearly defined. Previous reports showed that miR-29 inhibited tumorigenicity in lung cancer (22), its expression was down-regulated and correlated with poor prognosis in mantle cell lymphoma (11), and its expression caused apoptosis in AML (30). On the other hand, miR-29 expression was up-regulated in metastatic breast cancer (13), its overexpression in mouse myeloid cells caused AML (14), and in this report we show that miR-29 overexpression in B cells results in CLL. For AML, several reports defined miR-29 as a tumor suppressor that functions by targeting oncogenes such as MCL1 and CDK6 and promoting apoptosis in AML cells (30, 31). A very recent study reported directly opposite results, showing that miR-29 is overexpressed in some AML and that its up-regulation causes AML in a mouse model (14). Although the role of miR-29 in AML needs to be sorted out, it is likely that miR-29 can function as a tumor suppressor or oncogene depending on the cellular context.
We and others previously reported that miR-29 is down-regulated in aggressive CLL as compared with indolent CLL; we also demonstrated that miR-29 is one of the microRNAs targeting TCL1, a critical oncogene in the pathogenesis of aggressive CLL (9, 10). Here we report that miR-29 is overexpressed in indolent CLL compared with normal B cells. Because only 20% of Eμ-miR-29 transgenic mice died of leukemia in old age, but almost all mice showed expanded CD5+CD19+ B-cell populations, the phenotype of Eμ-miR-29 is similar to that of indolent CLL. Therefore up-regulation of miR-29 initiates or at least significantly contributes to the pathogenesis of indolent CLL. On the other hand, TCL1 is mostly not expressed in indolent CLL (9) and probably does not play an important role in indolent CLL. Our current hypothesis is that miR-29 overexpression is not sufficient to initiate aggressive CLL. In contrast, up-regulation of Tcl1 is a critical event in the pathogenesis of the aggressive form of CLL. Because miR-29 targets TCL1, its down-regulation in aggressive CLL (compared with the indolent form) contributes to up-regulation of Tcl1 and the development of an aggressive phenotype.
Materials and Methods
Eμ-miR-29 Transgenic Mice and Human CLL Samples.
A 1.0-kb fragment containing mouse miR-29ab cluster was cloned into the BamHI and SalI sites of the plasmid containing a mouse VH promoter (V186.2) and the IgH-Eμ enhancer (15) along with the hrGFP and the SV40 poly(A) site. The miR-29a/b cluster sequence was inserted within the intron of this construct. Transgenic mice were produced in Ohio State University transgenic mouse facility. Genotyping was performed on tail DNAs by PCR using the primers: miR29d: gct gac gtt gga gcc aca ggt aag; miR29r: aca aat tcc aaa aat gac ttc cag. Human CLL samples were obtained from the Chronic Lymphocytic Leukemia Research Consortium after informed consent was obtained from patients diagnosed with CLL. Research was performed with the approval of the Institutional Review Board of The Ohio State University. RNA extraction was carried out as previously described (32). Real-time PCR experiments were carried out using miR-29a, miR-29b, and PXDN assays for real-time PCR (Applied Biosystems) according to the manufacturer's protocol. Control human cord blood CD19+ B cells were purchased from Allcells and Lonza.
Characterization of miR-29 Transgenic Lymphocytes.
Lymphocytes from spleens and bone marrow were isolated as previously described (27). Flow cytometry measurements of SRBC immune response, Ig levels, and proliferation of B-cell populations were carried out as previously described (21). To analyze IgH gene rearrangements, Southern blot analysis of spleen lymphocyte DNA was carried out using EcoRI (Roche) digestions and mouse JH4 probe as previously described (15). For histology and immunohistochemistry, mice were necropsied, and spleens, livers, and kidneys were fixed in 10% buffered formalin, included in paraffin, and then cut in 4-μm sections as previously described (27). Sections were stained with H&E according to standard protocols (27).
Analysis of miR-29 Targets.
B cells were isolated using a B-cell isolation kit (Miltenyi Biotec) according to the manufacturer's instructions. Proteins from spleens were extracted as previously described (33). Western blot analysis was carried out using Cdk6 (H-96; Santa Cruz Biotechnology), DNMT3A (2160; Cell Signaling Technology), Pten (mmac1; Lab Vision), Mcl1 (S-19; Santa Cruz Biotechnology), Pdxn (Novus), and GAPDH (2118; Cell Signaling Technology) antibodies. For luciferase assays, fragments of PXDN, BCL7A, and ITIH5 cDNA, including regions complimentary to miR-29, were inserted into a pGL3 vector using the XbaI site immediately downstream from the stop codon of luciferase, as previously described (9). MiR-29a, miR-29b, and scrambled control RNA duplexes were purchased from Ambion. The expression construct containing full-length human PXDN was purchased from OriGene. Transfections were carried out as previously described (34).
Supplementary Material
Acknowledgments
We thank Dr. Kay Huebner for critically reviewing the manuscript and Vadim Maximov for technical assistance. The research was supported by an American Cancer Society Research Scholar grant to Y.P. and by National Institutes of Health Grant PO1-CA81534 of the Chronic Lymphocytic Leukemia Research Consortium to L.R., T.K., and C.M.C.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007186107/-/DCSupplemental.
References
- 1.Sgambati M, Linet M, Devesa S. Chronic lymphocytic leukemia, epidemiological, familial, and genetic aspects. In: Cheson B, editor. Chronic Lymphocytic Leukemias. 2nd Ed. New York: Marcel Dekker, Inc.; 2001. pp. 33–62. [Google Scholar]
- 2.Bullrich F, Croce C. Molecular biology of chronic lymphocytic leukemia. In: Cheson B, editor. Chronic Lymphocytic Leukemias. 2nd Ed. New York: Marcel Dekker, Inc.; 2001. pp. 9–32. [Google Scholar]
- 3.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Calin GA, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Döhner H, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 7.Calin GA, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cimmino A, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pekarsky Y, et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66:11590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 10.Herling M, et al. TCL1 shows a regulated expression pattern in chronic lymphocytic leukemia that correlates with molecular subtypes and proliferative state. Leukemia. 2006;20:280–285. doi: 10.1038/sj.leu.2404017. [DOI] [PubMed] [Google Scholar]
- 11.Zhao JJ, et al. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood. 2010;115:2630–2639. doi: 10.1182/blood-2009-09-243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gebeshuber CA, Zatloukal K, Martinez J. miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep. 2009;10:400–405. doi: 10.1038/embor.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han YC, et al. microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J Exp Med. 2010;207:475–489. doi: 10.1084/jem.20090831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bichi R, et al. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc Natl Acad Sci USA. 2002;99:6955–6960. doi: 10.1073/pnas.102181599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiorazzi N. Cell proliferation and death: Forgotten features of chronic lymphocytic leukemia B cells. Best Pract Res Clin Haematol. 2007;20:399–413. doi: 10.1016/j.beha.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Messmer BT, et al. In vivo measurements document the dynamic cellular kinetics of chronic lymphocytic leukemia B cells. J Clin Invest. 2005;115:755–764. doi: 10.1172/JCI23409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sieklucka M, et al. Apoptosis in B-CLL: The relationship between higher ex vivo spontaneous apoptosis before treatment in III-IV Rai stage patients and poor outcome. Oncol Rep. 2008;19:1611–1620. [PubMed] [Google Scholar]
- 19.Caligaris-Cappio F, Hamblin TJ. B-cell chronic lymphocytic leukemia: A bird of a different feather. J Clin Oncol. 1999;17:399–408. doi: 10.1200/JCO.1999.17.1.399. [DOI] [PubMed] [Google Scholar]
- 20.Zanesi N, et al. Effect of rapamycin on mouse chronic lymphocytic leukemia and the development of nonhematopoietic malignancies in Emu-TCL1 transgenic mice. Cancer Res. 2006;66:915–920. doi: 10.1158/0008-5472.CAN-05-3426. [DOI] [PubMed] [Google Scholar]
- 21.Efanov A, et al. CD5+CD23+ leukemic cell populations in TCL1 transgenic mice show significantly increased proliferation and Akt phosphorylation. Leukemia. 2010;24:970–975. doi: 10.1038/leu.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fabbri M, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horikoshi N, Cong J, Kley N, Shenk T. Isolation of differentially expressed cDNAs from p53-dependent apoptotic cells: Activation of the human homologue of the Drosophila peroxidasin gene. Biochem Biophys Res Commun. 1999;261:864–869. doi: 10.1006/bbrc.1999.1123. [DOI] [PubMed] [Google Scholar]
- 24.Desmond JC, et al. Discovery of epigenetically silenced genes in acute myeloid leukemias. Leukemia. 2007;21:1026–1034. doi: 10.1038/sj.leu.2404611. [DOI] [PubMed] [Google Scholar]
- 25.van Doorn R, et al. Epigenetic profiling of cutaneous T-cell lymphoma: Promoter hypermethylation of multiple tumor suppressor genes including BCL7a, PTPRG, and p73. J Clin Oncol. 2005;23:3886–3896. doi: 10.1200/JCO.2005.11.353. [DOI] [PubMed] [Google Scholar]
- 26.Himmelfarb M, et al. ITIH5, a novel member of the inter-alpha-trypsin inhibitor heavy chain family is downregulated in breast cancer. Cancer Lett. 2004;204:69–77. doi: 10.1016/j.canlet.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 27.Costinean S, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klein U, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 29.Zanesi N, Pekarsky Y, Trapasso F, Calin G, Croce C. MicroRNAs in mouse models of lymphoid malignancies. J Nucleic Acids Invest. 2010;1:36–40. doi: 10.4081/jnai.2010.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garzon R, et al. MicroRNA 29b functions in acute myeloid leukemia. Blood. 2009;114:5331–5341. doi: 10.1182/blood-2009-03-211938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garzon R, et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci USA. 2008;105:3945–3950. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palamarchuk A, et al. 13q14 deletions in CLL involve cooperating tumor suppressors. Blood. 2010;115:3916–3922. doi: 10.1182/blood-2009-10-249367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palamarchuk A, et al. Tal1 transgenic expression reveals absence of B lymphocytes. Cancer Res. 2006;66:6014–6017. doi: 10.1158/0008-5472.CAN-06-0937. [DOI] [PubMed] [Google Scholar]
- 34.Pekarsky Y, et al. Tcl1 functions as a transcriptional regulator and is directly involved in the pathogenesis of CLL. Proc Natl Acad Sci USA. 2008;105:19643–19648. doi: 10.1073/pnas.0810965105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.