Abstract
The quorum sensing (QS) system of Pseudomonas aeruginosa constitutes a sophisticated genome-wide gene regulatory network employing both N-acylhomoserine lactone and 2-alkyl-4-quinolone (AQ) signal molecules. AQ signalling utilizes 2-heptyl-3-hydroxy-4-quinolone (PQS) and its immediate precursor, 2-heptyl-4-quinolone (HHQ). AQ biosynthesis requires the first four genes of the pqsABCDE operon and while the biochemical function of pqsE is not known, it is required for the production of secondary metabolites such as pyocyanin. To gain insights into the relationship between the AQ stimulon, the PqsE stimulon and the regulatory function of PqsE, we constructed a pqsE inducible mutant (pqsEind) and compared the transcriptomes of the induced and uninduced states with a pqsA mutant. Of 158 genes exhibiting altered expression in the pqsA mutant, 51% were also affected in the pqsE mutant. Following induction of pqsE, 237 genes were differentially expressed compared with the wild-type strain. In the pqsEind strain, pqsA was highly expressed but following induction both pqsA expression and AQ biosynthesis were repressed, revealing a negative autoregulatory role for PqsE. Furthermore, pqsE was required for swarming motility and virulence in plant and animal infection models in the absence of AQs, while mature biofilm development required both pqsA and pqsE. Taken together these data reveal that PqsE is a key regulator within the QS circuitry facilitating the environmental adaptation of P. aeruginosa.
Introduction
Pseudomonas aeruginosa is an opportunistic pathogen capable of infecting humans, animals, plants and insects and causing both acute and chronic infections. It produces a wide variety of secondary metabolites and virulence determinants, readily forms biofilms and is naturally resistant to many antimicrobial agents. Many of the structural genes involved are under quorum sensing (QS) control and are regulated in a population density-dependent manner via both N-acylhomoserine lactone (AHL) and 2-alkyl-4-quinolone (AQ) signalling pathways. These comprise the AHL-dependent hierarchically linked las and rhl systems which are inter-connected with a third QS system that utilizes AQ signal molecules (recently reviewed by Williams and Cámara, 2009). Although P. aeruginosa produces over 50 different AQ congeners, the two major AQs which function as QS signals are 2-heptyl-3-hydroxy-4-quinolone (the ‘PseudomonasQuinolone Signal’; PQS) and its immediate precursor 2-heptyl-4-quinolone (HHQ) (Pesci et al., 1999; Déziel et al., 2004; Diggle et al., 2007). Multiple genes are required for AQ biosynthesis and signal transduction. These are mostly arranged in an operon, pqsABCDE, which is under the positive control of the transcriptional regulator, PqsR(MvfR) (Cao et al., 2001; Gallagher et al., 2002). The first four gene products of this operon are required for HHQ biosynthesis. HHQ is oxidized to PQS via the action of the putative monooxygenase PqsH, coded by the pqsH gene located some distance downstream of the pqsABCDE operon (Gallagher et al., 2002; Déziel et al., 2004).
The biosynthesis of AQs occurs via the ‘head-to-head’ condensation of anthranilate (supplied via phnAB operon or the kynABU genes) and a 3-oxo-fatty acid (Ritter and Luckner, 1971; Bredenbruch et al., 2005). Anthranilate is primed by PqsA for entry into the HHQ biosynthetic pathway (Coleman et al., 2008), whereas PqsD acts as a condensing enzyme which may either catalyse thecondensation of anthranoyl-CoA with a 3-oxo-acid, or may be involved in the formation of a 3-oxo-acid precursor (Zhang et al., 2008). PqsB and PqsC are highly homologous to 3-oxoacyl-(acyl-carrier-protein) synthases and while their precise contribution to AQ biosynthesis is not yet understood, they are likely to be involved in fatty acid recruitment and condensation (Gallagher et al., 2002).
A positive feedback loop exists within the AQ signalling pathway, because both PQS and HHQ can bind to PqsR and drive the expression of the pqsA promoter so enhancing pqsABCDE expression (McGrath et al., 2004; Wade et al., 2005; Xiao et al., 2006a,b; Diggle et al., 2007). AQ signalling is linked to AHL-dependent QS because the las system exerts positive regulatory control upon pqsR and pqsH (Déziel et al., 2004) while rhl negatively impacts on pqsABCDE and pqsR expression (McGrath et al., 2004; Wade et al., 2005; Xiao et al., 2006a). Exoproducts such as the rhamnolipids and elastase are synergistically regulated by AHL- and AQ-dependent QS (McKnight et al., 2000; Farrow et al., 2008). Furthermore, LasR has recently been shown to bind to the promoters of both pqsR and pqsA in vivo (Gilbert et al., 2009).
The fifth gene in the pqs operon, pqsE, is not required for AQ biosynthesis but instead is an effector of the AQ response (Gallagher et al., 2002; Diggle et al., 2003; Déziel et al., 2004; Farrow et al., 2008). At present the precise function of PqsE is not understood. Sequence and structural analyses indicate that PqsE belongs to the family of enzymes known as metallo-β-hydrolases, because it possesses the characteristic amino acid ‘HXHXDH’ motif of these proteins and has a metallo-β-lactamase fold with an Fe2+/Fe3+ centre in the active site (although this may not be the true cofactor) (Yu et al., 2009). Mutation of pqsE results in the reduced production of PQS-mediated exoproducts such as pyocyanin, elastase and rhamnolipid, even though the PQS and AHL levels are similar to wild-type (Gallagher et al., 2002; Diggle et al., 2003; Déziel et al., 2005; Farrow et al., 2008). PqsE was therefore termed the ‘PQS signal response’ protein. However, overexpression of PqsE enhanced pyocyanin, elastase and rhamnolipid production in the absence of PQS and PqsR (Farrow et al., 2008). Such regulation was rhl-dependent and PqsE was also shown to enhance the ability of E. coli expressing rhlR to respond to N-butanoylhomoserine lactone (C4-HSL) with respect to rhlA expression (Farrow et al., 2008).
The AQ-independent action of PqsE suggests that the primary function of PqsR in regulating the expression of key target genes is to drive the expression of pqsE. However, this does not explain why exogenous PQS but not HHQ can fully restore pyocyanin and Lectin A production in a P. aeruginosa PAO1 pqsA mutant, because both AQs drive the expression of pqsA via PqsR (Xiao et al., 2006a; Diggle et al., 2007). Given that PQS, in addition to functioning as a QS signal molecule, is an iron chelator (Bredenbruch et al., 2006; Diggle et al., 2007), a pro-oxidant and an inducer of an anti-oxidative stress response (Häussler and Becker, 2008), required for biofilm maturation (D’argenio et al., 2002; Allesen-Holm et al., 2006) and vesicle formation (Mashburn and Whiteley, 2005; Mashburn-Warren et al., 2008), it is probably involved in the regulation of both PqsE-dependent and PqsE-independent genes. Consequently, PqsE is likely to be responsible for regulating a subset of the genes controlled by the AQs. Therefore, to gain better insights into the relationship between the AQ stimulon, the PqsE stimulon and the regulatory function of PqsE, we first constructed an isopropyl β-d-l-thiogalactopyranoside (IPTG)-inducible pqsE mutant (pqsEind) to facilitate control of pqsE expression independent of the pqsA promoter. By comparing the transcriptomes of the pqsEind mutant in the absence or presence of IPTG with that of a wild-type and a pqsA mutant strain we were able to define the nature of the AQ and PqsE stimulons and their inter-relationship. Furthermore, we show that PqsE (i) is required for swarming motility and biofilm development, and (ii) restores virulence in nematode, plant and mouse infection models in a pqsA mutant strain (i.e. in the absence of AQs). PqsE was also found to control negatively its own expression through the repression of pqsA transcription, so providing an additional layer of fine-tuning regulatory sophistication to QS in P. aeruginosa.
Results
Characterization of the pqsA stimulon
To identify genes regulated by the AQs, we used high-density oligonucleotide genomic microarrays to compare the transcriptional profiles of P. aeruginosa PAO1 and an isogenic pqsA mutant which does not produce AQs (Diggle et al., 2006; Fletcher et al., 2007). Under the experimental conditions employed, the growth curves for the parent and pqsA mutant were virtually identical (data not shown). RNA for transcriptomic analysis was extracted from cells grown to an optical density at 600 nm (OD600) of 1.5, corresponding to the late exponential phase of growth. This was because reverse-transcriptase polymerase chain reaction (RT-PCR) analysis showed that transcripts for all of the genes in the pqsABCDE operon reach a maximum level of expression at this stage of growth (Appendix S1 and Fig. S1A). The results of the microarray analysis revealed that the expression of 158 genes (2.8% of all P. aeruginosa genes) is affected by the pqsA mutation at this stage of growth (full results are given in Appendix S1 and Table S1). A significant number (31%) of these have been identified previously as genes controlled by QS via the AHLs (27%; Appendix S1 and Table S1; Hentzer et al., 2003; Schuster et al., 2003; Wagner et al., 2003) or involving components of the AQ-dependent QS system (21.5%; Déziel et al., 2004; Bredenbruch et al., 2006). Considering the hierarchical organization of the QS systems of P. aeruginosa (Williams and Cámara, 2009) an overlap between the transcripts identified in these experiments and those previously reported to be controlled by the las and rhl systems was anticipated. However, our finding that many of the genes modulated by the pqsA mutation were absent from previous analyses, highlights the complexity of the reciprocal regulation occurring within the P. aeruginosa QS network (Duan and Surette, 2007; Dekimpe and Déziel, 2009).
Among the 104 genes downregulated in the pqsA mutant (Table 1 and Table S1) we found genes involved in the production of virulence determinants, such as exoproteases (aprX, aprD), the ChiC extracellular chitinolytic enzyme (chiC), the LecA lectin (lecA), and secondary metabolites such as pyocyanin (phzA1, phzC1, phzE1, phzA2, phzE2, phzC2), PA2274 (a putative flavin-dependent monooxygenase), genes involved in pyochelin biosynthesis, uptake and regulation (pchA, pchB, pchC, pchD, pchE, pchF, pchI, pchR, ftpA), pyoverdine biosynthesis (PA2412 and pvdH), in antibiotic resistance (mexG, mexH, mexI, opmD), and in biofilm development (tadA, tadZ) (Duong et al., 1992; 2001; Folders et al., 2001; Mavrodi et al., 2001; Ravel and Cornelis, 2003; Aendekerk et al., 2005; Diggle et al., 2006; Tomich et al., 2007). Many of the genes identified above have also been previously identified in microarray experiments focusing on PQS signalling via characterization of the PqsR (MvfR) regulon in P. aeruginosa PA14 (Déziel et al., 2004) and the response of the P. aeruginosa PAO1 wild type to exogenously supplied PQS (Bredenbruch et al., 2006). Similarly, we have previously reported that pqsA, lecA and the pyochelin genes are downregulated in a pqsA mutant (Diggle et al., 2007). These findings validate the array data presented here.
Table 1.
List of selected genes identified in the microarray analyses.
| PA numbera | Gene namea | wt vs pqsAb | wt vs pqsEindc | wt vs pqsEind + IPTGd | Product namea |
|---|---|---|---|---|---|
| PA0051 | phzH | −1.597 | Potential phenazine-modifying enzyme | ||
| PA0236 TR | – | 1.538 | Probable transcriptional regulator | ||
| PA0996 * | pqsA | 12.420 | 2.897 | Probable coenzyme A ligase | |
| PA0997 *** | pqsB | 7.350 | 3.801 | Homologous to beta-keto-acyl-acyl-carrier protein synthase | |
| PA0998 * | pqsC | 6.597 | 3.735 | Homologous to beta-keto-acyl-acyl-carrier protein synthase | |
| PA0999 * | pqsD | 6.466 | 2.804 | 3-Oxoacyl-[acyl-carrier-protein] synthase III | |
| PA1000 * | pqsE | 3.418 | Quinolone signal response protein | ||
| PA1001 * | phnA | 10.187 | Anthranilate synthase component I | ||
| PA1002 * | phnB | 3.462 | Anthranilate synthase component II | ||
| PA1100 | fliE | −1.570 | Flagellar hook-basal body complex protein FliE | ||
| PA1104 | fliI | −1.561 | Flagellum-specific ATP synthase FliI | ||
| PA1196 TR | – | −1.516 | Probable transcriptional regulator | ||
| PA1245 ** | aprX | 1.515 | Hypothetical protein | ||
| PA1246 | aprD | 1.624 | Alkaline protease secretion protein AprD | ||
| PA1452 | flhA | −1.509 | Flagellar biosynthesis protein FlhA | ||
| PA1603 TR | – | −1.590 | Probable transcriptional regulator | ||
| PA1899 | phzA2 | 2.556 | Probable phenazine biosynthesis protein | ||
| PA1901 | phzC2 | 2.233 | Phenazine biosynthesis protein PhzC | ||
| PA1903 | phzE2 | 2.016 | Phenazine biosynthesis protein PhzE | ||
| PA2128 | cupA1 | −2.062 | −1.581 | Fimbrial subunit CupA1 | |
| PA2147 | katE | −1.925 | Catalase HPII | ||
| PA2273 TR | soxR | 2.947 | 2.099 | Probable transcriptional regulator | |
| PA2274 *** | – | 17.122 | 3.987 | Hypothetical protein | |
| PA2300 * | chiC | 1.707 | Chitinase | ||
| PA2413 | pvdH | 1.674 | L-2,4-diaminobutyrate:2-ketoglutarate 4-aminotransferase | ||
| PA2507 | catA | −3.125 | −5.165 | Catechol 1,2-dioxygenase | |
| PA2508 | catC | −2.544 | 4.185 | −3.756 | Muconolactone delta-isomerase |
| PA2509 | catB | −3.597 | 3.577 | −6.172 | Muconate cycloisomerase I |
| PA2519 TR | xylS | −2.077 | Transcriptional regulator XylS | ||
| PA2570 * | lecA | 2.219 | LecA | ||
| PA3095 | xcpZ | −1.895 | General secretion pathway protein M | ||
| PA3096 | xcpY | −1.620 | General secretion pathway protein L | ||
| PA3098 | xcpW | −1.813 | General secretion pathway protein J | ||
| PA3099 | xcpV | −1.948 | General secretion pathway protein I | ||
| PA3102 | xcpS | −1.520 | General secretion pathway protein F | ||
| PA3718 | - | 2.242 | Probable MFS transporter | ||
| PA3875 | narG | −2.552 | Respiratory nitrate reductase alpha chain | ||
| PA3879 | narL | −2.216 | Two-component response regulator NarL | ||
| PA4205 *** | mexG | 2.944 | Hypothetical protein | ||
| PA4206 *** | mexH | 5.338 | RND efflux membrane fusion protein precursor | ||
| PA4207 * | mexI | 7.925 | 1.886 | RND efflux transporter | |
| PA4208 * | opmD | 20.813 | 6.835 | Probable outer membrane protein precursor | |
| PA4210 | phzA1 | 2.440 | 1.519 | Probable phenazine biosynthesis protein | |
| PA4212 * | phzC1 | 2.131 | Phenazine biosynthesis protein PhzC | ||
| PA4214 * | phzE1 | 1.678 | −1.629 | Phenazine biosynthesis protein PhzE | |
| PA4221 ** | fptA | 7.076 | 5.448 | 6.577 | Fe(III)-pyochelin outer membrane receptor precursor |
| PA4222 ** | pchI | 7.157 | 11.923 | 7.876 | Probable ATP-binding component of ABC transporter |
| PA4225 ** | pchF | 71.483 | 57.723 | 75.785 | Pyochelin synthetase |
| PA4226 ** | pchE | 139.217 | 94.197 | 124.376 | Dihydroaeruginoic acid synthetase |
| PA4227 **TR | pchR | 8.728 | 3.245 | 7.757 | Transcriptional regulator PchR |
| PA4228 ** | pchD | 21.066 | 16.892 | 20.643 | Pyochelin biosynthesis protein PchD |
| PA4229 ** | pchC | 1.611 | pyochelin biosynthetic protein PchC | ||
| PA4230 ** | pchB | 27.171 | 15.570 | Salicylate biosynthesis protein PchB | |
| PA4231 ** | pchA | 32.538 | 21.940 | 38.831 | Salicylate biosynthesis isochorismate synthase |
| PA4300 | tadC | −1.525 | TadC | ||
| PA4302 | tadA | 1.738 | TadA ATPase | ||
| PA4303 | tadZ | 1.946 | TadZ | ||
| PA4613 ** | katB | −2.045 | Catalase | ||
| PA4890 TR | desT | −1.595 | DesT | ||
| PA4944 | hfq | 1.742 | Hfq |
Gene number, gene name and product name are from the Pseudomonas Genome Project (http://www.pseudomonas.com). Genes previously reported to be QS-controlled are in bold (Hentzer et al., 2003; Schuster et al., 2003; Wagner et al., 2003).
indicates genes regulated by MvfR (PqsR) in Déziel and colleagues (2005);
indicate genes regulated by PQS in Bredenbruch and colleagues (2006);
known or predicted transcriptional regulator; RND, resistance-nodulation-cell division.
Fold change in gene expression of P. aeruginosa PAO1 wild type (wt) compared with P. aeruginosa PAO1 pqsA mutant (pqsA).
Fold change in gene expression of P. aeruginosa PAO1 wild type (wt) compared with P. aeruginosa PAO1 pqsEind strain (pqsEind).
Fold change in gene expression of P. aeruginosa PAO1 wild type (wt) compared with P. aeruginosa PAO1 pqsEind strain grown in presence of 1 mM IPTG (pqsEind+IPTG).
Among the 54 genes upregulated in the pqsA mutant were those involved in the catabolism of catechol and anthranilate (catA, catB, catC), in anaerobic respiration (narL, narG), and in biofilm formation (cupA1) (Table 1; Vallet et al., 2001; Oglesby et al., 2008; Toyofuku et al., 2008), suggesting that the products of the pqs operon have a negative impact on the expression of certain genes. The upregulation of the cat genes in the pqsA mutant with respect to the wild-type strain is most likely due to an accumulation of anthranilate from the inability of the pqsA mutant to convert this compound into AQs (Bredenbruch et al., 2005). The upregulation of the nar genes observed in the pqsA mutant is consistent with the negative effect exerted by PQS on denitrification in P. aeruginosa (Toyofuku et al., 2008).
The observation that genes coding for known (xylS, desT, soxR, pchR) or predicted (PA0236, PA1196, PA1603) transcriptional regulators are both up- and down-regulated in the pqsA mutant (Table 1) suggests that the AQ stimulon is enlarged through the action of multiple auxiliary regulators.
Characterization of the pqsE stimulon
The PAO1 pqsA mutant does not produce any AQs, therefore in this strain the PQS- and HHQ-dependent activation of the pqsABCDE operon is abrogated (Diggle et al., 2007). However, while the exogenous provision of either PQS or HHQ restored pqsA expression in the pqsA mutant (or in the pqsAH mutant which cannot convert exogenous HHQ to PQS; Diggle et al., 2007) only PQS fully induces the expression of PAO1 target genes such as lecA (Diggle et al., 2007). As pqsE is required for Lectin A production, these data imply that there may be differential regulation of pqsE by PQS and HHQ, i.e. exogenous PQS is capable of driving the expression of the entire pqsABCDE operon more efficiently than HHQ. To determine whether this was the case, RT-PCR was used to evaluate the levels of the pqsE transcript in pqsA and pqsAH mutants in the presence or absence of PQS or HHQ or both. Only a basal level of the pqsE gene transcript was detected in the pqsA mutant when grown in absence of HHQ or PQS (Appendix S1 and Fig. S1B). The addition of these AQs, either individually or in combination, restored the transcription of pqsE, both in the pqsA mutant and in the pqsAH double mutant, confirming that either HHQ or PQS or both can trigger the transcription of the entire pqs operon including pqsE (Appendix S1 and Fig. S1B; Diggle et al., 2007).
As pqsE influences the production of secondary metabolites such as pyocyanin and the rhamnolipids (Gallagher et al., 2002; Diggle et al., 2007; Farrow et al., 2008), it was not possible to determine whether the differentially regulated genes in the pqsA stimulon were affected as a consequence of a lack of AQs or PqsE. To address this question, we constructed a P. aeruginosa strain (pqsEind) in which the chromosomal pqsE gene was placed under the control of an IPTG-inducible promoter, such that the expression of pqsE is independent from the pqsABCD genes (Fig. 1A). The levels of pqsE expression in pqsEind and wild-type strains were compared by qRT-PCR analysis at an OD600 of 0.5 (early exponential phase) and 1.5 (late exponential phase). In both cases, when grown in the absence of IPTG the pqsEind strain expressed pqsE only at a basal level, while the provision of IPTG resulted in the premature induction and overexpression of pqsE with respect to the parental strain. pqsE transcription increased 9.6-fold at an OD600 0.5, and 18.1-fold at an OD600 1.5 with respect to the wild-type strain at the same OD600 (Appendix S1 and Fig. S2). The growth curve of each of the strains evaluated was identical with or without IPTG (data not shown).
Fig. 1.
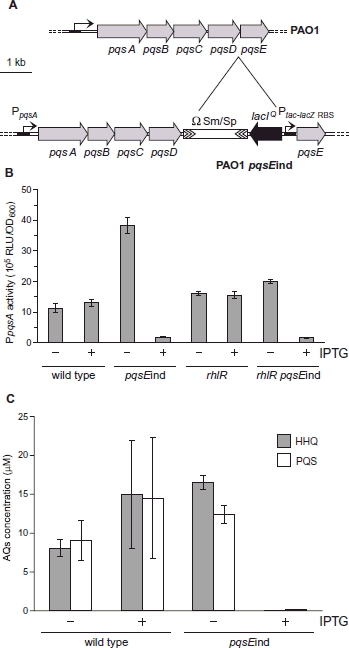
A. Schematic representation of the pqs locus in P. aeruginosa PAO1 wild type and the IPTG-inducible pqsE strain, pqsEind. The Ω interposon (SmR/SpcR) is from plasmid pHP45Ω: the lacIQ repressor is derived, together with the Ptac promoter, from plasmid pME6032. B. Activity of the PpqsA::lux promoter fusion. The activity of the PpqsA promoter was monitored during growth in PAO1 wild type, pqsEind, rhlR and pqsEind rhlR double mutants. The maximal expression levels reached during the late exponential phase of growth are shown. Where indicated (+), 1 mM IPTG was added to the growth medium. Error bars are calculated from three independent experiments. C. Concentration of HHQ (grey bars) and PQS (white bars) determined by LC-mass spectrometry in PAO1 wild type and the pqsEind mutant. The AQs were extracted from overnight cultures grown in LB broth; where indicated (+), 1 mM IPTG was added to the growth medium. The average of the results from three independent experiments is shown and error bars represent two standard deviations from the mean.
The transcriptional profiles of the PAO1 wild type and isogenic PAO1 pqsEind strains were compared in the presence and absence of 1 mM IPTG during the late exponential phase of growth (OD600 1.5). The control microarray analysis in which the wild-type PAO1 strain was grown with or without IPTG demonstrated that IPTG by itself had a negligible effect on the P. aeruginosa transcriptome (data not shown). A comparison of the parent strain and PAO1 pqsEind grown without IPTG revealed that pqsE mutation altered the expression of 57 genes (Appendix S1 and Table S1); 51% of these genes were also altered in the pqsA mutant transcriptome, indicating that the pqsE stimulon comprises a subset of the pqsA stimulon. Consequently many of the changes in gene expression noted in the pqsA transcriptome are likely to be due to a lack of pqsE expression in the pqsA mutant. However, 49% of the genes affected in the pqsEind strain were not present in the pqsA stimulon (Appendix S1 and Table S1). This finding suggests that these genes are differentially regulated by the AQs and PqsE, such that the loss of pqsE expression alone in the pqsE mutant disrupts the regulatory homeostasis so that they are apparent in the pqsE but not in the pqsA microarray, where the AQs and PqsE are absent.
PQS is a potent iron chelator and the addition of exogenous PQS to the wild type or to a pqsA mutant strongly induces genes involved in pyochelin and pyoverdin iron transport (Bredenbruch et al., 2006; Diggle et al., 2007). Conversely, compared with the wild type, mutation of either pqsA or pqsE results in the downregulation of pyochelin siderophore biosynthesis, regulatory and transport genes (pchA, pchB, pchD, pchE, pchF, pchI, pchR and ftpA; Table 1). These data suggest that the loss of PQS and its iron-chelating properties is not the sole determinant for the downregulation of the pyochelin biosynthetic pathway, and that PqsE may either play a role in iron homeostasis or in an as yet unidentified function of pyochelin unrelated to iron transport.
When compared with the wild-type parent strain, the transcriptional profile of the pqsEind strain grown in presence of 1 mM IPTG (which results in premature induction and overexpression of pqsE) revealed the differential expression of 237 genes; 133 genes were upregulated and 104 genes were downregulated. Apart from genes involved in pyocyanin and pyochelin production, in this experimental setting, PqsE also affected the expression of genes involved in type II secretion (xcpS, xcpV, xcpW, xcpY, xcpZ), flagellar biosynthesis (fliE, fliI, flhA) and the post-transcriptional regulator hfq (Table 1; Appendix S1 and Table S1). Of these, the most downregulated genes are those involved in pyochelin biosynthesis (pchA, pchD, pchE and pchF), transport (pchI, fptA) and regulation (pchR). Compared with the wild type, these genes were also downregulated in the pqsA mutant and in the pqsEind strain in the absence of IPTG. Thus either a lack of PqsE and PQS or the premature induction/overexpression of pqsE (where no PQS is produced) results in the downregulation of pyochelin genes.
The pqsABCDE operon was also present among the genes repressed as a consequence of pqsE overexpression (Table 1). This was of particular interest considering that it has previously been reported that mutation of pqsE did not affect the production of PQS (Gallagher et al., 2002). Surprisingly, the pqsE gene itself was not present among the pqsE-regulated genes. Considering that the qRT-PCR analysis performed on the same strains grown under the same experimental conditions clearly show marked changes in pqsE expression (Appendix S1 and Fig. S2), these data highlight the limitations intrinsic to high-density oligonucleotide microarrays and suggest that some genes may escape our analysis. Nevertheless, these data demonstrate that PqsE is a key player in the QS-dependent adaptive behaviour of P. aeruginosa.
PqsE can regulate pqsA expression and AQ production
To investigate further the impact of pqsE on the transcription of the pqsABCDE genes, we measured the activity of the pqsA promoter (PpqsA) via fusion with lux reporter genes in the P. aeruginosa PAO1 wild type and pqsEind strains, grown in presence and absence of IPTG. As shown in Fig. 1B, the activity of PpqsA is increased in the absence of pqsE and severely reduced when pqsE is overexpressed. The strong repression of PpqsA activity resulted in the inability of the pqsE overexpressing strain to produce HHQ and PQS (Fig. 1C) and other AQs (data not shown), while the concentrations of the AHLs, N-(3-oxododecanoyl)homoserine lactone (3-oxo-C12-HSL) and C4-HSL were unaffected (data not shown). As pqsE expression is dependent on PQS and HHQ, the ability of PqsE to reduce AQ biosynthesis through downregulation of pqsABCDE operon reveals that PqsE generates an autoregulatory negative-feedback loop.
Farrow and colleagues (2008) reported that PqsE regulates the production of pyocyanin and rhamnolipids in a RhlR/C4-HSL-dependent manner (Farrow et al., 2008). To determine whether the pqsE-dependent regulation of PpqsA is also mediated via RhlR, we determined the activity of PpqsA in a P. aeruginosa PAO1 rhlR pqsEind double mutant. Figure 1B shows that pqsE induction can still abrogate PpqsA activity in the absence of rhlR, suggesting that the mechanism of action of PqsE in this context is RhlR-independent.
PqsE is required for swarming motility and biofilm formation
As PqsE can function in an AQ-independent manner as well as modulating AQ biosynthesis, we examined the contribution of PqsE to pyocyanin and lectin production, motility and biofilm development in the pqsEind mutant and also in a pqsA pqsEind double mutant with and without induction since the latter permitted us to evaluate the impact of PqsE in the absence of AQs.
Figure 2A and B shows that neither PAO1 pqsEind nor PAO1 pqsA pqsEind produce much pyocyanin or lectin when pqsE is not induced, while IPTG-dependent pqsE overexpression results in the production of almost 2.5 times the wild-type level of pyocyanin and substantially higher levels of Lectin A in both mutants, demonstrating that PqsE controls both virulence determinants in an AQ-independent manner. Despite the differential production of pyocyanin and Lectin A, the microarray data (Table 1) did not reveal major changes in the phz or lecA transcript levels in the pqsE-inducible strain compared with the wild type. This could either reflect a possible post-transcriptional regulatory role for PqsE or highlight an experimental limitation of the microarray technique employed.
Fig. 2.
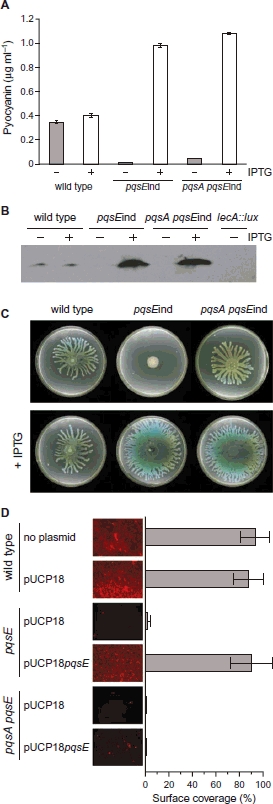
A. Pyocyanin produced by PAO1 and both pqsEind and pqsA pqsEind mutants. Bacterial cultures were grown in LB broth (grey bars) or LB broth supplemented with 1 mM IPTG (white bars), and pyocyanin was extracted after 16 h growth (early stationary phase). B. Western blot analysis of Lectin A in cell extracts of PAO1 and both pqsEind and pqsA pqsEind mutants. Proteins were extracted from cultures grown for 16 h in LB broth to an OD600 of 2.5 (early stationary phase of growth), with (+) or without (−) 1 mM IPTG. An extract from P. aeruginosa PAO1 lecA mutant (lecA::lux) was used as a negative control. C. Swarming assays performed with PAO1 and both pqsEind and pqsA pqsEind mutants in the presence or absence of 1 fmM IPTG. D. Biofilm formation on stainless steel coupons by PAO1 and both pqsE and pqsA pqsEind mutants. A representative picture of the biofilm formed by each strain is also shown. For A and D, the average of the results from three independent experiments is reported with standard deviations.
As our microarray data indicate that premature induction/overexpression of pqsE also modulates genes involved in motility such as tadC, fliE, fliI, flhA (Table 1; Tomich et al., 2007; Barken et al., 2008), we investigated the effect of pqsE on swimming, swarming and twitching motility. Twitching mainly relies on type IV pili (Mattick, 2002), while swimming motility is flagellar-mediated. Although some genes involved in flagellar motility were regulated by pqsE in the microarray experiments (fliE, fliI, flhA), agar plate motility assays indicated that pqsE did not affect swimming or twitching under the experimental conditions employed (data not shown). Conversely, the swarming plate assays demonstrated that pqsE is required for swarming, because the pqsEind mutant failed to swarm in the absence of IPTG (Fig. 2C). However, swarming was restored to wild-type levels in the pqsA pqsEind double mutant in the absence of IPTG. In both cases, the IPTG-induced expression of pqsE resulted in a ‘hyper-swarming phenotype’ (Fig. 2C). The inability of the pqsEind single mutant to swarm in the absence of IPTG implies that the AQs may inhibit swarming in the absence of PqsE.
As motility is often inversely related to biofilm formation (Verstraeten et al., 2008), we examined the effect of pqsE mutation on the ability of P. aeruginosa to form biofilms on stainless steel. As IPTG disrupts biofilm formation in P. aeruginosa (Diggle et al., 2006), we performed these experiments using isogenic PAO1 pqsE and PAO1 pqsA pqsE deletion mutants, carrying either the pUCP18 vector plasmid, or the pUCPpqsE plasmid for pqsE complementation. As shown in Fig. 2D, PqsE is required for biofilm formation, but in this case the expression of pqsE in the pqsA pqsEind mutant failed to restore biofilm production to wild-type levels. Exogenously added PQS partially restored biofilm formation (data not shown), indicating that both pqsE and PQS are required for the development of a mature biofilm.
PqsE restores virulence in the absence of AQs in nematode, plant and murine infection models
Given that PqsE restores pyocyanin and lectin production in the absence of AQs, we investigated the impact of pqsE mutation on P. aeruginosa pathogenicity using three well-established plant (lettuce leaf) and animal (Caenorhabditis elegans and mouse) experimental infection models. In these experiments, we compared the virulence of the PAO1 wild type, pqsE mutant and pqsA pqsE double mutant transformed with either the pUCP18 or pUCPpqsE vectors. The data obtained from each infection model are shown in Fig. 3. In the C. elegans model (Fig. 3A), PAO1 killed all of the nematodes after 6 days compared with ∼65% of worms fed with the PAO1 pqsA pqsE mutant. Expression of plasmid-borne pqsE in the pqsA pqsE mutant fully restored virulence. Similar results were also obtained for the pqsE mutant that was less virulent than the wild type unless complemented with pqsE (data not shown). Furthermore, C. elegans fed with the PAO1 pqsA pqsE pUCPpqsE strain exhibited symptoms of sickness, including impaired locomotion much faster (within 1 day) than was observed for C. elegans fed with the wild type (3 days). These worms also showed a significant reduction in fertility.
Fig. 3.
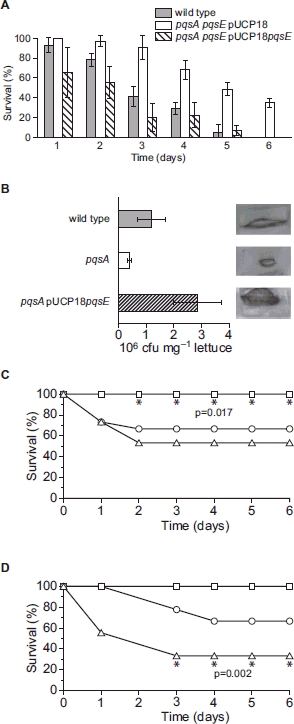
PqsE restores virulence in nematode, plant and animal infection models in the absence of AQs. A. Caenorhabditis elegans killing assay showing the percentage of nematode survival after 1–6 days of exposure to the P. aeruginosa PAO1 wild type, pqsA pqsEind mutant and pqsA pqsEind mutant transformed with the vector control, pUCP18 or pUCPpqsE respectively. The average of four independent experiments is reported with standard deviation. B. Virulence of the wild type, pqsA mutant and pqsA mutant complemented with pqsE in the lettuce leaf virulence assay. The number of bacterial cells (as colony forming units, cfu) present in 1 mg of lettuce midrib 5 days post injection is shown. Error bars were calculated from five independent experiments. A representative picture of infected midribs is also shown for each strain. C and D. Mouse acute burn wound infection showing the survival rate over time (days after burn/infection) for mice infected with (C) the P. aeruginosa wild type (▵), pqsA (□) and pqsE (○) mutants; 15 mice per mutant and (D) the P. aeruginosa pqsA mutant (□) and the pqsA mutant transformed with either pUCP18 (○) or pUCPpqsE (▵); nine mice per mutant.
When inoculated into the mid-ribs of lettuce leaves, the PAO1 pqsA mutant causes much less tissue necrosis and grows more poorly than the wild-type strain (Fig. 3B). Transformation of this mutant with plasmid-borne pqsE fully restored virulence.
In a mouse burn wound infection model, the pqsA mutant is highly attenuated compared with the pqsE mutant that in turn is less virulent than the wild type (Fig. 3C). Introduction of the pqsE-expressing plasmid into the pqsA mutant strain completely restored virulence also in this infection model (Fig. 3D).
Discussion
Pseudomonas aeruginosa employs a sophisticated multi-signal molecule QS system which operates to facilitate environmental adaptation at the population level (Fig. 4). To further refine our understanding of the individual contributions of key components of the AQ signalling pathway to P. aeruginosa physiology, we first determined the extent of the P. aeruginosa PAO1 pqsA stimulon, because this has not previously been reported. By profiling the transcripts present after maximal induction of the pqsABCDE operon, we observed that 158 genes were up- or down-regulated when the wild type was compared with the pqsA mutant strain (Table S1). However, only 18 and 21 of these genes were previously identified in transcriptome analyses of either (i) a P. aeruginosa PA14 pqsR (mvfR) mutant (Déziel et al., 2005) or (ii) P. aeruginosa PAO1 grown in the presence of exogenous PQS added at the point of inoculation (Bredenbruch et al., 2006) and compared with the corresponding wild-type strains. These variations are perhaps not particularly surprising given the differences in the strains and experimental conditions used. Although the pqsR mutant in common with the pqsA mutant is AQ-negative, it is not known whether PqsR directly drives the expression of target genes other than pqsA. Furthermore, the exogenous addition of PQS to the wild-type P. aeruginosa (which is already producing AQs in a population density dependent manner) is likely to advance and enhance PQS-dependent gene expression (Diggle et al., 2003), as well as inducing an oxidative/anti-oxidative stress response (Bredenbruch et al., 2006; Häussler and Becker, 2008). However, despite these differences, the recurrence of the same set of genes in at least two out of the three experiments, indicates that AQ-dependent QS plays a key role in regulating genes of known function including pqsABCDE, phnAB, phzABCDEFG, mexGHIompD, pchABCDEF, lecA, chiC and pvdH, as well as many others of unknown function.
Fig. 4.
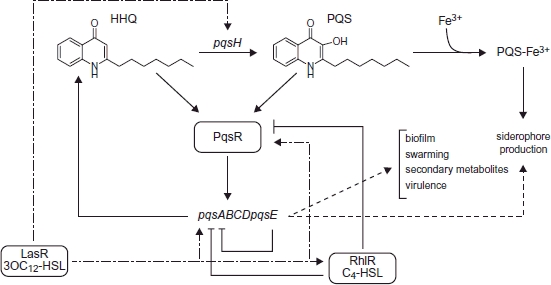
Simplified schematic representation of the AQ-dependent QS in P. aeruginosa (modified from Diggle et al., 2007). HHQ, the immediate precursor of PQS, drives the expression of the pqsABCDE operon via PqsR(MvfR) and is also converted to PQS by the action of the monooxygenase, PqsH. PQS also drives pqsABCDE expression via PqsR. PqsE positively regulates biofilm, swarming virulence and secondary metabolite gene expression but negatively regulates pqsABCDE expression. PQS also binds ferric iron which results in the induction of high affinity siderophore iron transport genes. AHL and AQ-dependent QS are linked because LasR/3-oxo-C12-HSL is required for maximal expression of pqsH and pqsR whereas pqsR and pqsABCDE are repressed by RhlR/C4-HSL.
In contrast to pqsA mutants, pqsE mutants produce AQs such as PQS and HHQ but make very little pyocyanin, elastase, Lectin A or rhamnolipid (Gallagher et al., 2002; Diggle et al., 2003). This raises the question as to whether the differentially regulated genes in the pqsA mutant were affected as a consequence of a lack of AQs or PqsE. By constructing an inducible pqsE strain (pqsEind), we determined the extent of the PqsE stimulon by comparing the transcriptional profiles of the PAO1 wild-type strain with the uninduced or induced pqsEind strain, in which pqsE was not expressed or prematurely induced and overexpressed respectively. The results obtained revealed that PqsE regulates a major subset of the AQ-controlled genes, including those involved in siderophore biogenesis even in the absence of the iron-chelating properties of PQS. The observation that pyochelin genes are upregulated in the wild-type strain with respect to both the induced and the uninduced pqsEind strains suggests that a fine balancing of PQS and PqsE is required to produce physiologically profitable levels of this siderophore. It is likely that both PQS and PqsE have a positive effect on pyochelin production, as suggested by the reduced expression of genes involved in the synthesis of this siderophore in the pqsA and pqsEind strain with respect to the wild-type strain (Table 1). However, as the pyochelin genes are also downregulated when pqsE is overexpressed (pqsEind with IPTG; Table 1), this can be explained by the complete loss of PQS production in this strain (Fig. 1C).
The greater number of differentially expressed genes observed when pqsE is prematurely induced and overexpressed may be partially explained by the high level of pyocyanin produced under these conditions. This is because pyocyanin, in addition to functioning as a redox reactive virulence determinant (Lau et al., 2004), also acts as a terminal signal molecule for a subset of QS-dependent genes, including the mexGHI-ompD efflux pump and PA2274 (Dietrich et al., 2006). Pyocyanin is also capable of inducing oxidative stress, which is consistent with the upregulation of genes such as ahpF (alkylhydroperoxidase), katA and katE (catalase genes) when pqsE is induced. Interestingly, SoxR, which is required for the pyocyanin-dependent upregulation of the mexGHI-ompD pump, is downregulated in both the pqsA and pqsE mutant arrays, as well as the six genes which make up the SoxR regulon (PA2274, mexGHI-ompD and PA3718; Palma et al., 2005) (Table 1 and Table S1). Although SoxR is not a key regulatory player in the oxidative stress response of P. aeruginosa, it is essential for virulence in a mouse lung infection model (Palma et al., 2005). Given that PqsE regulates soxR and hence its regulon, this suggests that the contribution of the SoxR regulon to virulence is controlled by the AQs and PqsE.
We have shown that PqsE can repress the transcription of the pqsABCDE operon (and consequently its own transcription). This autoregulatory role of PqsE is analogous to the homeostatic effect exerted by RsaL on the production of 3-oxo-C12-HSL in P. aeruginosa. RsaL transcription is dependent on the LasR/3-oxo-C12-HSL complex. When exceeding a certain physiological concentration, RsaL binds to the rsaL-lasI bidirectional promoter repressing the transcription of both genes. This regulatory circuit generates an incoherent feed-forward loop that provides 3-oxo-C12-HSL homeostasis (Rampioni et al., 2006; 2007;). Similarly, PqsE requires the PqsR/HHQ or PqsR/PQS complex for its transcription, but when induced independently and prematurely, PqsE represses the expression of pqsABCDE, and hence AQ synthesis and its own transcription. Furthermore, in an rsaL mutant, the levels of 3-oxo-C12-HSL increase steadily whereas no AQ accumulation was observed in the pqsE mutant over the growth curve (data not shown). This difference may be ascribed to a limiting concentration of the substrates required for AQ biosynthesis, or due to the involvement of other, as yet unidentified factors regulating the transcription of the pqsABCDE operon.
While RsaL is a transcriptional regulator, PqsE is an enzyme which lacks any obvious DNA-binding domains, indicating that this protein is likely to exert its regulatory role indirectly. The structure of PqsE has been determined and the presence of the predicted metallo-β-lactamase fold with a metal centre in the active site confirmed (Yu et al., 2009). While neither the true substrate for PqsE nor the downstream signal transduction pathway has yet been identified, the purified recombinant PqsE protein is capable of slowly hydrolysing phosphodiesters, nucleic acids and thioesters (Yu et al., 2009).
Determination of the pqsE stimulon, the physiological characterization of the pqsE mutant and the consequences of expressing pqsE in a pqsA mutant demonstrate the central importance of PqsE within the AQ-signal transduction pathway and its capacity for acting independently of PQS and HHQ. While the action of PqsE in restoring pyocyanin production and rhlA expression is dependent on RhlR/C4-HSL (Farrow et al., 2008), this is not the case for the PqsE-mediated repression of pqsA, which is independent of the rhl system (Fig. 1B). Consequently PqsE may activate or repress target genes via distinct pathways.
The P. aeruginosa pqsEind mutant failed to swarm in the absence of IPTG and exhibited a hyper-swarming phenotype following induction. This result is in line with the previous finding that pqsE is involved in rhamnolipid production (Diggle et al., 2003). Surprisingly, swarming motility in the pqsA pqsEind double mutant was restored (Fig. 2C), implying a role for AQs in repressing swarming in the absence of PqsE. This phenotype may be explained by the function of PQS as an activator of the small regulatory RNA, RsmZ (S. Heeb, K.M. Righetti, M. Messina, S.A. Kuehne, C. Pustelny, S.R. Chhabra et al., unpublished data), which titrates out the RNA-binding protein RsmA (S. Heeb, K.M. Righetti, M. Messina, S.A. Kuehne, C. Pustelny, S.R. Chhabra et al., unpublished data). A PAO1 rsmA mutant is unable to swarm (Heurlier et al., 2004), and therefore mutation of pqsA (which results in the loss of PQS) would result in a reduction in RsmZ, consequently increasing the availability of RsmA and so restoring swarming motility. It is also possible that PqsE is involved in the post-transcriptional regulation of pyocyanin and Lectin A because only minor changes in phz or lecA transcript levels were noted in the pqsEind mutant in the presence or absence of IPTG despite major changes in pyocyanin and Lectin A protein levels. The presence of the post-transcriptional regulator hfq among the genes regulated by PqsE (Table 1) supports this possibility.
As well as an inability to swarm, the pqsE mutant was unable to form mature biofilms on a stainless steel substratum. However, in contrast to pyocyanin and lectin, pqsE was not sufficient to restore biofilm formation in a pqsA mutant background (Fig. 2). This is likely to be because the release of extracellular DNA which is crucial for biofilm maturation is pqsA-dependent (Allesen-Holm et al., 2006; Yang et al., 2007), and also because PQS induces oxidative DNA damage leading to DNA release and fragmentation in growing P. aeruginosa cultures (Häussler and Becker, 2008). It is therefore likely that the development of mature biofilm requires both the expression of PqsE and the production of PQS. Additionally, biofilm development is a complex pleiotropic phenotype and the impact of QS on this process is strongly dependent upon experimental conditions and growth media (Kirisits and Parsek, 2006). However, the induction of genes involved in iron sequestration observed in the pqsE mutant is indicative of iron starvation conditions, which have been reported to influence biofilm formation negatively (Banin et al., 2005), consistent with the reduced biofilm forming capacity of the pqsE mutant.
Pseudomonas aeruginosa PA14 strains carrying mutations in pqsR, pqsA, pqsB and pqsE have previously been reported to display reduced virulence in a mouse burn wound model (Déziel et al., 2005). Here we have confirmed these data for the PAO1 pqsA mutant which was completely avirulent. However, transformation of the pqsA mutant with the plasmid-borne pqsE gene fully restored virulence indicating that the AQs are dispensible for virulence. Similar results were obtained in both C. elegans and lettuce leaf infection models. Because these are acute infection models and given that PQS is essential for biofilm maturation, it is tempting to speculate that PqsE is essential for acute infections, while the AQs may play a more important role in chronic biofilm-centred infections. In this context, the autorepressive regulatory circuit generated by PqsE may play an important role in fine tuning the levels of AQ- and PqsE-regulated virulence factors.
Although the precise function of PqsE remains enigmatic, the work described here clearly demonstrates that PqsE is a global regulator within the QS network (Fig. 4) which plays a pivotal role in controlling the adaptive behaviour of P. aeruginosa.
Experimental procedures
Bacteria, growth conditions and plasmids
The bacterial strains and plasmids used are listed in Table 2. Escherichia coli and P. aeruginosa strains were routinely grown in Luria–Bertani (LB) broth. All strains were grown at 37°C in 10 ml of broth and 100 ml flasks with shaking at 200 r.p.m. The following reagents were added as required: ampicillin (Ap) 100 µg ml−1; nalidixic acid 15 µg ml−1; chloramphenicol (Cm) 30 µg ml−1 (E. coli) or 375 µg ml−1 (P. aeruginosa); tetracycline (Tc) 10 µg ml−1 (E. coli) or 200 µg ml−1 (P. aeruginosa); carbenicillin (Cb) 400 µg ml−1; streptomycin (Sm) 800 µg ml−1; IPTG 1 mM; PQS or HHQ (40 µM; synthesized as described before by Diggle et al., 2006). To select P. aeruginosa from E. coli after mating experiments, LB agar plates supplemented with nalidixic acid 15 µg ml−1 were used.
Table 2.
Bacterial strains and plasmids used in this work.
| Strain/plasmid | Relevant characteristics | Source/reference |
|---|---|---|
| Strain | ||
| E. coli | ||
| DH5α | Cloning strain | Grant et al. (1990) |
| S17.1λpir | Conjugative strain for suicide plasmids. | Simon et al. (1983) |
| P. aeruginosa | ||
| PAO1 | Nottingham collection wild-type P. aeruginosa strain | |
| pqsA | pqsA mutant of PAO1 | Aendekerk et al. (2005) |
| pqsA pqsH | pqsA pqsH double mutant of PAO1 | Diggle et al. (2007) |
| lecA::lux | lecA mutant of PAO1 | Winzer et al. (2000) |
| pqsEind | PAO1 derivative in which pqsE expression is under the control of a Ptac promoter | This study |
| pqsE | pqsE in-frame deletion mutant of PAO1 | This study |
| rhlR | rhlR in-frame deletion mutant of PAO1 | This study |
| pqsA pqsEind | PAO1 pqsA mutant derivative in which pqsE expression is under the control of a Ptac promoter | This study |
| rhlR pqsEind | PAO1 rhlR mutant derivative in which pqsE expression is under the control of a Ptac promoter | This study |
| pqsA pqsE | pqsA pqsE double mutant of PAO1 | This study |
| Plasmid | ||
| pBluescript-II KS+ | Cloning vector; ColE1 replicon; ApR | Stratagene |
| pUCP18 | pUC18-derivative containing a stabilising fragment for maintenance in Pseudomonas; ApR, E. coli/CbR, P. aeruginosa. | Schweizer (1991) |
| mini-CTXlux | Promoter-probe vector containing the luxCDABE operon; TcR | Becher and Schweizer (2000) |
| pDM4 | Suicide vector; sacBR, oriR6K; CmR | Milton et al. (1996) |
| mini-CTXpqsA::lux | Plasmid to insert PpqsA-lux fusion into the chromosome of Pseudomonas; TcR | Diggle et al. (2007) |
| pDM4ΔpqsE | pDM4 derivative for pqsE in-frame deletion; CmR | This study |
| pDM4ΔrhlR | pDM4 derivative for rhlR in-frame deletion; CmR | This study |
| pDM4pqsEinda | pDM4 derivative for the generation of the pqsE-inducible strain; CmR | This study |
| pUCPpqsE | pUCP18 derivative for pqsE complementation; ApR | This study |
| pBSrhlRUp | pBluescript-II KS+ derivative containing the upstream region of rhlR; ApR | This study |
| pBSrhlRDw | pBluescript-II KS+ derivative containing the downstream region of rhlR; ApR | This study |
| pBSpqsEUp | pBluescript-II KS+ derivative containing the upstream region of pqsE; ApR | This study |
| pBSpqsEDw | pBluescript-II KS+ derivative containing the downstream region of pqsE; ApR | This study |
More details on the construction of P. aeruginosa pqsEind strain are given in Supporting Information.
DNA manipulation
Oligonucleotides used in this study are listed in Table S2 and Appendix S1. Plasmid DNA preparations, restriction enzyme digestions, agarose gel electrophoresis and ligations were performed using standard methods (Sambrook and Russell, 2001). Transformation of P. aeruginosa was carried out by electroporation as published (Farinha and Kropinski, 1990). Routine DNA sequencing was conducted in the DNA Sequencing Laboratory, Biopolymer Synthesis and Analysis Unit, Queens Medical Centre, University of Nottingham.
Construction of P. aeruginosa PAO1 mutants
The PAO1 rhlR and pqsE chromosomal deletion mutants were constructed by allelic exchange using the suicide vector pDM4ΔrhlR and pDM4ΔpqsE respectively. pDM4ΔrhlR and pDM4ΔpqsE were made as follows; using PAO1 template DNA, upstream fragments of the rhlR and pqsE genes were amplified using the primers rhlRUp1 and rhlRUp2 (for rhlR) or pqsEUp1 and pqsEUp2 (for pqsE), while downstream fragments were amplified using the primers rhlRDw1 and rhlRDw2 (for rhlR) or pqsEDw1 and pqsEDw2 (for pqsE). The resulting PCR products were cloned in pBluescript-II KS+, resulting in the plasmids pBSrhlRUp and pBSrhlRDw (for rhlR), and pBSpqsEUp and pBSpqsEDw (for pqsE). The upstream and downstream fragments of each gene were then excised with the corresponding restriction enzymes and cloned in the suicide vector pDM4 (Milton et al., 1996), resulting in the plasmids pDM4ΔrhlR and pDM4ΔpqsE. Allelic exchange in P. aeruginosa PAO1 following conjugal mating with E. coli S17-1λpir donor strains and sucrose counter-selection was performed as described by Westfall and colleagues (2004). The resulting PAO1 rhlR and PAO1 pqsE mutant strains were confirmed by PCR, sequence analysis and phenotypic assays.
The PAO1 pqsEind strain was constructed by introducing onto the chromosome the lacIQ repressor gene and the tac promoter transcribing the lacZ 5′ untranslated transcribed region and its ribosome binding site (RBS) directly upstream of the pqsE open reading frame, resulting in strong, constitutive transcription and translation only in the presence of IPTG. An ΩSmR/SpR interposon (to terminate native transcription originating upstream of pqsE) and the lacIQ repressor gene were inserted upstream of the Ptac-lacZ RBS-pqsE region (Fig. 1A). Further details are provided in Appendix S1.
To generate PAO1 rhlR pqsEind and PAO1 pqsA pqsEind double mutant strains, the pDM4pqsEind suicide vector was conjugated in the PAO1 rhlR and PAO1pqsA mutants respectively, while to generate the PAO1pqsA pqsE double mutant strain, the pDM4ΔpqsE plasmid was conjugated in the PAO1pqsA mutant. In all cases and sucrose-resistant clones were selected and verified by PCR.
Construction of pUCPpqsE
The pqsE gene was amplified from P. aeruginosa PAO1 chromosomal DNA using the primers pqsE18Fw and pqsE18RV. This PCR product was digested with SacI/XbaI and cloned into similarly digested pUCP18, resulting in the plasmid pUCPpqsE, which was verified by sequence analysis. The pUCP18 and pUCPpqsE plasmids were introduced into different P. aeruginosa PAO1 strains by electroporation (Farinha and Kropinski, 1990).
RNA extraction and expression profiling experiments
Pseudomonas aeruginosa strains were grown at 37°C in 10 ml of LB broth and 100 ml Schott Duran flasks with shaking at 200 r.p.m. Where required, LB broth was supplemented with 1 mM IPTG. RNA was extracted from each culture at an OD600 of 1.5 (late exponential phase of growth). Cells were treated with RNAprotect Bacteria Reagent (Qiagen), and total RNA extraction was performed with the RNeasy Midi Kit (Qiagen) as per the manufacturer's instructions.
For the expression profiling experiments, the microarrays were designed to contain multiple oligonucleotide probes for all the PAO1 genes including the small RNA genes and were purchased from Oxford Gene Technology (Oxford, UK). For each array, 10 µg of RNA was reverse transcribed and directly labelled with Cy5-dCTP and 2 µg of genomic DNA was directly labelled with Cy3-dCTP. Samples were hybridized onto the arrays for 16 h. Scanning of the arrays was performed using the Axon 4000B GenePix Scanner, the data extraction software used was GenePix Pro 6, both from Molecular Devices (Sunnyvale, USA). For each strain, microarray experiments were performed in triplicate and data analysis performed using GeneSpring GX10 (Agilent Technologies, Santa Clara, USA). The array data underwent Lowess normalization, the most variable 10% of data according to standard deviation within replicates was removed and genes of altered expression were determined by passing through cut-offs of both a fold change of 1.5 and a paired T-test of P = 0.05.
SDS-PAGE and immunoblotting
Pseudomonas aeruginosa cells grown in LB at 37°C overnight for 16 h to an OD600 2.5 were lysed, normalized for protein content and subjected to SDS-PAGE prior to electrophoretic transfer to nitrocellose membranes and probed with a rabbit polyclonal antibody to Lectin A as described before (Diggle et al., 2003).
Measurement of bioluminescence
The single copy fusion of the pqsA promoter to the luxCDABE genes was introduced in the different P. aeruginosa PAO1 strains by mating with the E. coli S17.1λpir carrying the mini-CTXpqsA::lux plasmid (Diggle et al., 2007). Bioluminescence was determined as a function of population density by using an automated luminometer-spectrometer (TECAN). Overnight cultures of P. aeruginosa strains carrying the chromosomal pqsA::lux fusion were diluted 1:1000 in fresh LB broth, and 0.2 ml cultures were grown at 37°C in microtitre plates. Luminescence and turbidity were determined every 30 min. Luminescence is given as relative light units divided by OD600.
Extraction and quantification of AQs
Pseudomonas aeruginosa strains were grown overnight in 10 ml of LB broth at 37°C. The AQs were extracted from 2 ml of the culture supernatant with 6 ml of acidified ethyl acetate (Diggle et al., 2003), vortexed vigorously and centrifuged at 9447 g for 5 min. The organic phase was transferred to a clean glass tube, dried to completion, resuspended in 50 µl methanol. Liquid chromatography (LC) was performed on an Agilent 1200 series HPLC with an Ascentis Express C18 150 × 2.1 mm internal diameter, 2.7 µm particle size, maintained at 50°C. The mobile phase consisted of formic acid 0.1% (v/v) in water and formic acid 0.1% (v/v) in acetonitrile run as a gradient over 20 min at a flow rate of 0.3 ml min−1. Using a Bruker HCT Plus ion trap LC-mass spectrometer in multiple reaction mode and Hystar software, ions were introduced, isolated and fragmented using positive ion electrospray. Retention times and MS2 peak spectra were matched to the 10 µM synthetic AQ standards injected at the beginning and end of each run.
Pyocyanin production, motility and biofilm assays
For pyocyanin assays, P. aeruginosa strains were grown with aeration in LB at 37°C overnight for 16 h (to early stationary phase), and pyocyanin levels were determined in the supernatants derived from 5 ml of culture, as previously described (Xu et al., 2005). For motility assays, P. aeruginosa cultures (OD600 1.0) were picked with a toothpick onto ‘Swimming Plates’ (1 g l−1 tryptone, 0.5 g l−1 yeast extract, 5 g l−1 NaCl, 3 g l−1 agar noble), or ‘Twitching Plates’ (10 g l−1 tryptone, 5 g l−1 yeast extract, 5 g l−1 NaCl, 10 g l−1 agar noble) respectively, and grown for 16 h at 37°C. For swarming assays, 2 µl of culture was spotted onto ‘Swarming Plates’ (5 g l−1 bacto agar, 8 g l−1 nutrient broth N°2, 0.5% (w/v) glucose) and grown 16 h at 37°C. Biofilm formation on stainless steel coupons was performed essentially as previously described (Diggle et al., 2006). Surface attached biofilms were stained with 0.1% (w/v) acridine orange, washed with PBS, air-dried and examined for bacterial attachment with an inverted fluorescent microscope (Nikon Eclipse TE200) using the ×40 objective lens and green filter. Ten images were collected per metal coupon using a JVC KY-F58 video camera. Sampling was conducted at random from the central portion of each coupon. Percentage surface coverage was calculated using the Lucia G/Comet software (Nikon UK). The assays were performed in triplicate for each strain.
Virulence assays
C. elegans virulence assays were performed essentially as described by Papaioannou and colleagues (2009). Caenorhabditis elegans was incubated on bacterial lawns at 21°C on nematode growth medium (Stiernagle, 2006) and scored for live worms every day. For statistical purposes, four replicates with 10 worms for each strain were performed, and four replicates per trial were carried out with E. coli OP50 as negative control.
For the lettuce leaf virulence assay, 10 µl aliquots of bacterial cultures resuspended to an A600 of 0.1 in 10 mM MgSO4 were injected into the midribs of fresh Romaine lettuce leaves, and incubated for 2–5 days as previously described (Starkey and Rahme, 2009). Lettuce leaves were monitored for the appearance of soft-rot symptoms and the numbers of bacterial cells in the midrib were determined after a defined incubation period.
The mouse acute burn wound model described by Schaber and colleagues (2007) and by Rumbaugh and colleagues (2009) was carried out as follows. Female, Swiss Webster, mice were obtained from Charles River Laboratories (Wilmington, MA, USA). Mice used in experiments were 6–8 weeks old and weighed 20–25 g. Mice were anaesthetized, their backs were shaved and an acute scald wound induced as previously described, with infection doses of 102 bacteria. Mice were housed and studied under protocols approved by the Institutional Animal Care and Use Committee in the animal facility of TTUHSC (Lubbock, TX).
Acknowledgments
This work was supported by research grants from the Biological and Biotechnological Sciences Research Council U.K. (BBF0143921), the European Union (FP6 Marie Curie EST project ANTIBIOTARGET; MEST-CT-2005-020278) and the American Diabetes Association (KPR). We thank Siri Ram Chhabra and Alex Truman for AQ synthesis.
Supporting information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. RT-PCR analysis of the pqsABCDE operon.
A. Amplification of the individual pqs genes in PAO1. cDNA was synthesized from RNA extracted from P. aeruginosa PAO1 at various stages of growth (as determined by OD600). The corresponding RNA was amplified in parallel as a negative control, and the P. aeruginosa PAO1 genomic DNA was used as a positive control.
B. Amplification of the pqsE gene in PAO1 pqsA and PAO1 pqsA pqsH mutants. cDNA was synthesized from RNA extracted from cells grown to OD600 1.5. Where indicated (+), strains were grown in presence of 40 μM HHQ and/or PQS. The corresponding RNA was amplified in parallel as a negative control, and the P. aeruginosa PAO1 genomic DNA was used as a positive control.
Fig. S2. Analysis of pqsE expression as a function of growth in the pqsEind strain. The fold change in pqsE transcript levels in the pqsEind strain grown to early exponential phase (OD600 = 0.5) and late exponential phase (OD600 = 1.5) was determined by qRT-PCR in the presence of absence of IPTG. The relative expression of pqsE is compared with that of the wild type at an OD600 of 0.5. Where indicated, IPTG (1 mM) was added to the growth medium.
Table S1. Genes regulated in the microarray experiments.
Table S2. Primers used in this work.
Appendix S1. Experimental procedures.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Aendekerk S, Diggle SP, Song Z, Høiby N, Cornelis P, Williams P, Cámara M. The MexGHI-OpmD multidrug efflux pump controls growth, antibiotic susceptibility and virulence in Pseudomonas aeruginosa via 4-quinolone-dependent cell-to-cell communication. Microbiology. 2005;151:1113–1125. doi: 10.1099/mic.0.27631-0. [DOI] [PubMed] [Google Scholar]
- Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, et al. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol. 2006;59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- Banin E, Vasil ML, Greenberg EP. Iron and Pseudomonas aeruginosa biofilm formation. Proc Natl Acad Sci USA. 2005;102:11076–11181. doi: 10.1073/pnas.0504266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barken KB, Pamp SJ, Yang L, Gjermansen M, Bertrand JJ, Klausen M, et al. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ Microbiol. 2008;10:2331–2343. doi: 10.1111/j.1462-2920.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- Becher A, Schweizer HP. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques. 2000;29:948–950. doi: 10.2144/00295bm04. [DOI] [PubMed] [Google Scholar]
- Bredenbruch F, Nimtz M, Wray V, Morr M, Müller R, Häussler S. Biosynthetic pathway of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines. J Bacteriol. 2005;187:3630–3635. doi: 10.1128/JB.187.11.3630-3635.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenbruch F, Geffers R, Nimtz M, Buer J, Häussler S. The Pseudomonas aeruginosa quinolone signal (PQS) has an iron-chelating activity. Environ Microbiol. 2006;8:1318–1329. doi: 10.1111/j.1462-2920.2006.01025.x. [DOI] [PubMed] [Google Scholar]
- Cao H, Krishnan G, Goumnerov B, Tsongalis J, Tompkins R, Rahme LG. A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc Natl Acad Sci USA. 2001;98:14613–14618. doi: 10.1073/pnas.251465298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JP, Hudson LL, McKnight SL, Farrow JM, 3rd, Calfee MW, Lindsey CA, Pesci EC. Pseudomonas aeruginosa PqsA is an anthranilate-coenzyme A ligase. J Bacteriol. 2008;190:1247–1255. doi: 10.1128/JB.01140-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Argenio DA, Calfee MW, Rainey PB, Pesci EC. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J Bacteriol. 2002;184:6481–6489. doi: 10.1128/JB.184.23.6481-6489.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekimpe V, Déziel E. Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR-specific factors. Microbiology. 2009;155:712–723. doi: 10.1099/mic.0.022764-0. [DOI] [PubMed] [Google Scholar]
- Déziel E, Lépine F, Milot S, He J, Mindrinos MN, Tompkins RG, Rahme LG. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci USA. 2004;101:1339–1344. doi: 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déziel E, Gopalan S, Tampakaki AP, Lépine F, Padfield KE, Saucier M, et al. The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-l-homoserine lactones. Mol Microbiol. 2005;55:998–1014. doi: 10.1111/j.1365-2958.2004.04448.x. [DOI] [PubMed] [Google Scholar]
- Dietrich LE, Price-Whelan A, Petersen A, Whiteley M, Newman DK. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol Microbiol. 2006;61:1308–1321. doi: 10.1111/j.1365-2958.2006.05306.x. [DOI] [PubMed] [Google Scholar]
- Diggle SP, Winzer K, Chhabra SR, Worrall KE, Cámara M, Williams P. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol Microbiol. 2003;50:29–43. doi: 10.1046/j.1365-2958.2003.03672.x. [DOI] [PubMed] [Google Scholar]
- Diggle SP, Stacey RE, Dodd C, Cámara M, Williams P, Winzer K. The galactophilic lectin, LecA, contributes to biofilm development in Pseudomonas aeruginosa. Environ Microbiol. 2006;8:1095–1104. doi: 10.1111/j.1462-2920.2006.001001.x. [DOI] [PubMed] [Google Scholar]
- Diggle SP, Matthijs S, Wright VJ, Fletcher MP, Chhabra SR, Lamont IL, et al. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem Biol. 2007;14:87–96. doi: 10.1016/j.chembiol.2006.11.014. [DOI] [PubMed] [Google Scholar]
- Duan K, Surette MG. Environmental regulation of Pseudomonas aeruginosa PAO1 Las and Rhl quorum-sensing systems. J Bacteriol. 2007;189:4827–4836. doi: 10.1128/JB.00043-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong F, Lazdunski A, Cami B, Murgier M. Sequence of a cluster of genes controlling synthesis and secretion of alkaline protease in Pseudomonas aeruginosa: relationships to other secretory pathways. Gene. 1992;121:47–54. doi: 10.1016/0378-1119(92)90160-q. [DOI] [PubMed] [Google Scholar]
- Duong F, Bonnet E, Géli V, Lazdunski A, Murgier M, Filloux A. The AprX protein of Pseudomonas aeruginosa: a new substrate for the Apr type I secretion system. Gene. 2001;262:147–153. doi: 10.1016/s0378-1119(00)00541-2. [DOI] [PubMed] [Google Scholar]
- Farinha MA, Kropinski AM. High efficiency electroporation of Pseudomonas aeruginosa using frozen cell suspensions. FEMS Microbiol Lett. 1990;58:221–225. doi: 10.1111/j.1574-6968.1990.tb13982.x. [DOI] [PubMed] [Google Scholar]
- Farrow JM, Sund ZM, Ellison ML, Wade DS, Coleman JP, Pesci EC. PqsE functions independently of PqsR-Pseudomonas quinolone signal and enhances the rhl quorum-sensing system. J Bacteriol. 2008;190:7043–7051. doi: 10.1128/JB.00753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher MP, Diggle SP, Crusz S, Chhabra SR, Cámara M, Williams P. A dual biosensor for 2-alkyl-4-quinolone quorum-sensing signal molecules. Environ Microbiol. 2007;9:2683–2693. doi: 10.1111/j.1462-2920.2007.01380.x. [DOI] [PubMed] [Google Scholar]
- Folders J, Algra J, Roelofs MS, van Loon LC, Tommassen J, Bitter W. Characterization of Pseudomonas aeruginosa chitinase, a gradually secreted protein. J Bacteriol. 2001;183:7044–7052. doi: 10.1128/JB.183.24.7044-7052.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher LA, McKnight SL, Kuznetsova MS, Pesci EC, Manoil C. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J Bacteriol. 2002;184:6472–6480. doi: 10.1128/JB.184.23.6472-6480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert KB, Kim TH, Gupta R, Greenberg EP, Schuster M. Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol Microbiol. 2009;73:1072–1085. doi: 10.1111/j.1365-2958.2009.06832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SG, Jessee J, Bloom FR, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häussler S, Becker T. The pseudomonas quinolone signal (PQS) balances life and death in Pseudomonas aeruginosa populations. PLoS Pathog. 2008;4:e1000166. doi: 10.1371/journal.ppat.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentzer M, Wu H, Andersen JB, Riedel K, Rasmussen TB, Bagge N, et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 2003;22:3803–3815. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurlier K, Williams F, Heeb S, Dormond C, Pessi G, Camara M, et al. Positive control of swarming and lipase production by the post-transcriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J Bacteriol. 2004;186:2936–2945. doi: 10.1128/JB.186.10.2936-2945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisits MJ, Parsek MR. Does Pseudomonas aeruginosa use intercellular signalling to build biofilm communities? Cell Microbiol. 2006;8:1841–1849. doi: 10.1111/j.1462-5822.2006.00817.x. [DOI] [PubMed] [Google Scholar]
- Lau GW, Hassett DJ, ran H, Kong F. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Microbiol. 2004;10:599–606. doi: 10.1016/j.molmed.2004.10.002. [DOI] [PubMed] [Google Scholar]
- McGrath S, Wade DS, Pesci EC. Dueling quorum sensing systems in Pseudomonas aeruginosa control the production of the Pseudomonas quinolone signal (PQS) FEMS Microbiol Lett. 2004;230:27–34. doi: 10.1016/S0378-1097(03)00849-8. [DOI] [PubMed] [Google Scholar]
- McKnight SL, Iglewski BH, Pesci EC. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 2000;182:2702–2708. doi: 10.1128/jb.182.10.2702-2708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn LM, Whiteley M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature. 2005;437:422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- Mashburn-Warren L, Howe J, Garidel P, Richter W, Steiniger F, Roessle M, et al. Interaction of quorum signals with outer membrane lipids: insights into prokaryotic membrane vesicle formation. Mol Microbiol. 2008;69:491–502. doi: 10.1111/j.1365-2958.2008.06302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol. 2001;183:6454–6465. doi: 10.1128/JB.183.21.6454-6465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milton DL, O'Toole R, Horstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. J Bacteriol. 1996;178:1310–1319. doi: 10.1128/jb.178.5.1310-1319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oglesby AG, Farrow JM, 3rd, Lee JH, Tomaras AP, Greenberg EP, Pesci EC, Vasil ML. The influence of iron on Pseudomonas aeruginosa physiology: a regulatory link between iron and quorum sensing. J Biol Chem. 2008;283:15558–15567. doi: 10.1074/jbc.M707840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma M, Zurita J, Ferreras JA, Worgall S, Larone DH, Shi L, et al. Pseudomonas aeruginosa SoxR does not conform to the archetypal paradigm for SoxR-dependent regulation of the bacterial oxidative stress adaptive response. Infect Immun. 2005;73:2958–2966. doi: 10.1128/IAI.73.5.2958-2966.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaioannou E, Wahjudi M, Nadal-Jimenez P, Koch G, Setroikromo R, Quax WJ. Quorum-quenching acylase reduces the virulence of Pseudomonas aeruginosa in a Caenorhabditis elegans infection model. Antimicrob Agents Chemother. 2009;53:4891–4897. doi: 10.1128/AAC.00380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesci EC, Milbank JBJ, Pearson JP, McKnight S, Kende AS, Greenberg EP, Iglewski BH. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:11229–11234. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampioni G, Bertani I, Zennaro E, Polticelli F, Venturi V, Leoni L. The quorum-sensing negative regulator RsaL of Pseudomonas aeruginosa binds to the lasI promoter. J Bacteriol. 2006;188:815–819. doi: 10.1128/JB.188.2.815-819.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampioni G, Schuster M, Greenberg EP, Bertani I, Grasso M, Venturi V, et al. RsaL provides quorum sensing homeostasis and functions as a global regulator of gene expression in Pseudomonas aeruginosa. Mol Microbiol. 2007;66:1557–1565. doi: 10.1111/j.1365-2958.2007.06029.x. [DOI] [PubMed] [Google Scholar]
- Ravel J, Cornelis P. Genomics of pyoverdine-mediated iron uptake in pseudomonads. Trends Microbiol. 2003;11:195–200. doi: 10.1016/s0966-842x(03)00076-3. [DOI] [PubMed] [Google Scholar]
- Ritter C, Luckner M. Zur Biosynthese der 2-n-Alkyl-4-hydroxychinolinderivate (Pseudane) bei Pseudomonas aeruginosa. Eur J Biochem. 1971;18:391–400. doi: 10.1111/j.1432-1033.1971.tb01255.x. [DOI] [PubMed] [Google Scholar]
- Rumbaugh KP, Diggle SP, Watters CM, Ross-Gillespie A, Griffin AS, West SA. Quorum sensing and the social evolution of bacterial virulence. Curr Biol. 2009;19:341–345. doi: 10.1016/j.cub.2009.01.050. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schaber JA, Triffo WJ, Suh SJ, Oliver JW, Hastert MC, Griswold JA, et al. Pseudomonas aeruginosa forms biofilms in acute infection independent of cell-to-cell signaling. Infect Immun. 2007;75:3715–3721. doi: 10.1128/IAI.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M, Lohstroh CP, Ogi T, Greemberg EP. Identification, timing and signal specifity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer HP. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene. 1991;97:109–121. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic-engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology. 1983;1:784–791. [Google Scholar]
- Starkey M, Rahme LG. Modeling Pseudomonas aeruginosa pathogenesis in plant hosts. Nat Protoc. 2009;4:117–124. doi: 10.1038/nprot.2008.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T. Maintenance of C. elegans. WormBook. 2006;11:1–11. doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomich M, Placet PJ, Figurski DH. The tad locus: postcards from the widespread colonization island. Nat Rev Microbiol. 2007;5:363–375. doi: 10.1038/nrmicro1636. [DOI] [PubMed] [Google Scholar]
- Toyofuku M, Nomura N, Kuno E, Tashiro Y, Nakajima T, Uchiyama H. Influence of the Pseudomonas quinolone signal on denitrification in Pseudomonas aeruginosa. J Bacteriol. 2008;190:7947–7956. doi: 10.1128/JB.00968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet I, Olson JW, Lory S, Lazdunski A, Filloux A. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc Natl Acad Sci USA. 2001;98:6911–6916. doi: 10.1073/pnas.111551898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraeten N, Braeken K, Debkumari B, Fauvart M, Fransaer J, Vermant J, Michiels J. Living on a surface: swarming and biofilm formation. Trends Microbiol. 2008;16:496–506. doi: 10.1016/j.tim.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Wade DS, Calfee MW, Rocha ER, Ling EA, Engstrom E, Coleman JP, Pesci EC. Regulation of Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. J Bacteriol. 2005;187:4372–4380. doi: 10.1128/JB.187.13.4372-4380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewsky BH. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol. 2003;185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall LW, Luna AM, San Francisco M, Diggle SP, Worrall KE, Williams P, et al. The Pseudomonas aeruginosa global regulator MvaT specifically binds to the ptxS upstream region and enhances ptxS expression. Microbiology. 2004;150:3797–3806. doi: 10.1099/mic.0.27270-0. [DOI] [PubMed] [Google Scholar]
- Williams P, Cámara M. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr Opin Microbiol. 2009;12:182–191. doi: 10.1016/j.mib.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Winzer K, Falconer C, Garber NC, Diggle SP, Cámara M, Williams P. The Pseudomonas aeruginosa lectins PA-IL and PA-IIL are controlled by quorum sensing and by RpoS. J Bacteriol. 2000;182:6401–6411. doi: 10.1128/jb.182.22.6401-6411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G, Déziel E, He J, Lépine F, Lesic B, Castonguay MH, et al. MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol Microbiol. 2006a;62:1689–1699. doi: 10.1111/j.1365-2958.2006.05462.x. [DOI] [PubMed] [Google Scholar]
- Xiao G, He J, Rahme LG. Mutation analysis of the Pseudomonas aeruginosa mvfR and pqsABCDE gene promoters demonstrates complex quorum-sensing circuitry. Microbiology. 2006b;152:1679–1686. doi: 10.1099/mic.0.28605-0. [DOI] [PubMed] [Google Scholar]
- Xu H, Lin W, Xia H, Xu S, Li Y, Yao H, et al. Influence of ptsP gene on pyocyanin production in Pseudomonas aeruginosa. FEMS Microbiol Lett. 2005;253:103–109. doi: 10.1016/j.femsle.2005.09.027. [DOI] [PubMed] [Google Scholar]
- Yang L, Barken KB, Skindersoe ME, Christensen AB, Givskov M, Tolker-Nielsen T. Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology. 2007;153:1318–1328. doi: 10.1099/mic.0.2006/004911-0. [DOI] [PubMed] [Google Scholar]
- Yu S, Jensen V, Seeliger J, Feldmann I, Weber S, Schleicher E, et al. Structure elucidation and preliminary assessment of hydrolase activity of PqsE, the Pseudomonas quinolone signal (PQS) response protein. Biochemistry. 2009;48:10298–10307. doi: 10.1021/bi900123j. [DOI] [PubMed] [Google Scholar]
- Zhang YM, Frank MW, Zhu K, Mayasundari A, Rock CO. PqsD is responsible for the synthesis of 2,4-dihydroxyquinoline, an extracellular metabolite produced by Pseudomonas aeruginosa. J Biol Chem. 2008;283:28788–28794. doi: 10.1074/jbc.M804555200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


