Abstract
The failure of CNS regeneration and subsequent motor and sensory loss remain major unsolved questions despite massive accumulation of experimental observations and results. The sheer volume of data and the variety of resources from which these data are generated make it difficult to integrate prior work to build new hypotheses. To address these challenges we developed a prototypic suite of computer programs to extract protein names from relevant publications and databases and associated each of them with several general categories of biological functions in nerve regeneration. To illustrate the usefulness of our data mining approach, we utilized the program output to generate a hypothesis for a biological function of CD44 interaction with osteopontin (OPN) and laminin in axon outgrowth of CNS neurons. We identified CD44 expression in retinal ganglion cells and when these neurons were plated on poly-l-lysine 3% of them initiated axon growth, on OPN 15%, on laminin-111 (1×) 41%, on laminin-111 (0.5×) 56%, and on a mixture of OPN and laminin (1×) 67% of neurons generated axon growth. With the aid of a deoxyribozyme (DNA enzyme) to CD44 that digests the target mRNA, we demonstrated that a reduction of CD44 expression led to reduced axon initiation of retinal ganglion cells on all substrates. We suggest that such an integrative, applied systems biology approach to CNS trauma will be critical to understand and ultimately overcome the failure of CNS regeneration.
Keywords: applied systems biology, axon growth, CD44, deoxyribozyme, DNA enzymes, laminin, nerve regeneration, osteopontin, text mining
The reasons for failure of axonal regeneration in the CNS are not well understood and may lie in diverse processes including apoptosis, inflammation, changes in signal transduction, cytoskeleton formation, and DNA transcription (see reviews: Goldberg and Barres 2000; Popovich 2000; Schwab 2002; Tessier-Lavigne 2002–2003, Beattie 2004). Accordingly, experiments have identified a wide array of molecules involved in CNS regeneration. For axon growth alone, such molecules include growth factors and neurotrophins (Hagg et al. 1990; Schnell et al. 1994), glial-associated inhibitory proteins (Domeniconi et al. 2002; Liu et al. 2002), extracellular matrix (ECM) molecules and their receptors (McKeon et al. 1991; Moon et al. 2001), and various downstream signaling families such as rho GTPases and transcriptional control proteins (Nikulina et al. 2004; McKerracher and Higuchi 2006).
In spite of many such molecular discoveries, an understanding of the entirety of the underlying processes leading to regeneration failure does not yet exist, and how these various protein networks interact with and influence each other remains difficult to test and thus to address experimentally. Therefore, to understand the mechanisms involved in regeneration failure, it is essential to generate a global network that spans these diverse networks (Hwang et al. 2005). To build such a global network we adopted a bioinformatics approach in the neurobiology domain in which we identified with the aid of applied systems biology the array of proteins implicated in nerve regeneration. Over 900 proteins were collected in a literature-based search, and classified accordingly to their ontology. In doing so we focused on three proteins, CD44, osteopontin (OPN), and laminin, that are expressed in neurons and glia in the CNS and PNS (Liesi 1985; Liesi and Silver 1988; Hagg et al. 1989; Cohen and Johnson 1991; Moretto et al. 1993; Morissette and Carbonetto 1995; Kaaijk et al. 1997; Alfei et al. 1999; Ichikawa et al. 2000; Jones et al. 2000; Sherman et al. 2000; Grimpe et al. 2002; Choi et al. 2004; Raivich et al. 2004; Shin and Koh 2004; Stein et al. 2004; Tuohy et al. 2004). OPN’s as well as laminin’s primary receptors are integrins (Sonnenberg et al. 1990; Tomaselli et al. 1990; Bayless et al. 1998; Yokosaki et al. 2005), and CD44’s primary ligands are hyaluronan and chondroitin sulfate proteoglycans (Alldinger et al. 2000; Fujimoto et al. 2001; Wu et al. 2005), but OPN and CD44 also interact with each other (He et al. 2006) as does laminin and CD44 (Hibino et al. 2005). OPN stimulation of CD44 induces cell migration in fibroblasts and macrophages (Weber et al. 1996). Laminin alpha 5 chain interacts with CD44 and stimulates melanoma cell growth, angiogenesis, migration, and metastasis (Hibino et al. 2005). Curiously, these two CD44 interactions have not yet been studied in the CNS. Induced to examine these molecules by our algorithmic analysis of regeneration-associated proteins, we here describe a new biological function for CD44 interactions with OPN and laminin in axon growth of retinal ganglion cells (RGCs). This study is the first approach to use information extracted from published data as a cornerstone to understanding the complex mechanisms leading to failure of regeneration after trauma to the CNS.
Materials and methods
Text mining
Our software suite runs on a PC (Dell, Round Rock, TX, USA) under SuSE Linux 9.2 (www.suse.com). This Linux distribution provided all tools for application management, development, system administration, and backups. The programs were written in Perl, and accessed a MySQL (www.mysql.com) database on the same machine as the data repository. Both Uniprot (European Bioinformatics Institute, Swiss Institute of Bioinformatics, and Georgetown University; www.ebi.uniprot.org) and PubMed (National Center for Biotechnology Information (NCBI); www.ncbi.nlm.nih.gov) records were downloaded as flatfiles and loaded into local database tables by our own import programs. The corpus for the text mining comprised the titles and abstracts of all publication records collected in PubMed classified with the Medical Subject Headings term ‘nerve regeneration’. No distinction between PNS and CNS was made.
The protein name dictionary was built dynamically at runtime from the ‘DE’ (description) field in Uniprot records. To each protein name the corresponding primary accession number was associated. Protein names were recognized by a dictionary/rule-based method similar to Egorov et al. 2004. A list of English stop words, the most frequent words used in natural language texts was obtained from MySQL v4.1, to which we added the letters from the alphabet. The example below illustrates the method’s main steps:
| ‘…detected ankyrin G, which was…’ |
get raw text instance |
| ‘detected’,‘ankyrin’, | split into words, trim non-alphanumeric characters, discard stop words, and single letters |
| ‘ankyrin’ | match as substring to name dictionary: 118 hits for ‘ankyrin’, no hit for ‘detected’ |
| …ankyrin-3lankyrin G… | use rules to generate spelling alternatives |
| …ankyrin 3lankyrin- 3lankyrin Glankyrin-G… |
match each alternative in full length to the raw text |
| ‘ …detected ankyrin G | ‘ankyrin G’ matches |
| ‘ankyrin G’, Q12955, 12507143 |
recognized protein name, primary accession number, and PubMed identifier |
| (Text sample from Kretschmer et al. 2002) | |
To remove duplicates, such as ‘glial fibrillary acidic protein’ and ‘GFAP’, the program compared the accession numbers for each name with all others in the list. Additional details and availability of the software program suite can be received from b.grimpe@ miami.edu.
In the last step we manually curated the list. A name was rejected when the name stood for: (i) no protein at all (e.g. ‘Golgi’), (ii) an entire protein class (e.g. ‘collagen’), (iii) a homonym not referring to the protein (e.g. ‘ALS’ referring to the disease ‘amyotrophic lateral sclerosis’ rather than the proteins, ‘agglutinin-like protein 2 precursor’, ‘acetylcholine receptor protein’ or ‘alpha-like growth factor-binding protein’), (iv) a protein introduced experimentally (e.g. ‘horseradish peroxidase’), or (v) a non-CNS protein (e.g. ‘hair keratin;’). Only 4.6% of all excluded proteins fell into this last category. When in doubt, we consulted the PubMed records where the name appeared. To assign functional attributes to a protein we used the ‘QuickGo’ web service (European Bioinformatics Institute; March 2006), which returns a list of Gene Ontology (www.gene-ontology.org) terms for the domains ‘molecular function’, ‘biological process’, and ‘cellular component’.
Retinal ganglion cell preparation and neurite outgrowth assay
Embryonic day 20 (E20) RGCs were purified from Sprague–Dawley rats using sequential immunopanning for Thy-1 to > 99.5% purity and cultured in defined, serum-free media as previously described in references Meyer-Franke et al. (1995) and Goldberg et al. (2002a). Circular glass coverslips (Fisher Scientific, Pittsburgh, PA, USA) were coated overnight in 24-well plates with poly-l-lysine (PLL; Sigma, St Louis, MO, USA). The next day the coverslips were rinsed three times with water. In a second overnight incubation, the coverslips were coated with OPN (0.125 µg/cm2; R&G, Minneapolis, MN, USA), laminin-111 [laminin-1 with a α1β1γ1-chain (Aumailley et al. 2005); 1 µg/cm2, Sigma] or a mixture of OPN/laminin-111 (1×) including 0.125 µg/cm2 OPN and 1 µg/cm2 laminin-111 or OPN/laminin-111 (0.5×) with 0.063 µg/cm2 OPN and 0.5 µg/cm2 laminin-111. RGCs were plated at a concentration of 2 × 103 cells/well for histology, 1.3 × 104 cells/well for RNA, and 3.3 × 105 cells/well for protein isolation. After 4 h the RGC were treated with the respective reagents.
Deoxyribozyme against CD44
The deoxyribozyme (DNA enzyme) was designed as described in Breaker and Joyce (1994), Grimpe et al. (2002), Grimpe and Silver (2004), and Grimpe et al. (2005). The sequence corresponded to the 5′ end of the rat CD44 mRNA sequence (accession number: BC061531) from nucleotide sequence 205 to 224: DNA CD44as: 5′-TACACCTGCGGGCTAGCTACAACGAAACGGCAG-3′; control DNA CD44mb: 5′-AAGTCCGTTAGCAACATCGATCGGC-TGCAGGGCAG-3′or 5′-ATTTAAGGCCGCAATATCGATCGGCGCAGGGCAG-3′. The deoxyribozyme has no homology to other mammalian sequences registered in the GenBank database of the National Institute of Health (Altshult et al. 1997). The control deoxyribozyme was designed not to bind to any mRNA and serves as a control for the presence of exogenous single-stranded DNA in a cell. Deoxyribozymes were biotinylated at the 3′ end by MWG Biotech (High Point, NC, USA). To identify deoxyribozyme uptake into RGCs, cultures were stained after 24 h with streptavidin-labeled Texas Red (1 : 200; Invitrogen, Carlsbad, CA, USA) overnight on a shaker followed by rinsing in phosphate buffered saline and mounting in GelMount (Biomed, Foster City, CA, USA).
Immunohistochemistry
Immunohistochemistry of RGCs and brains fixed in 40 mL/L paraformaldehyde and 0.25 mL/L glutaraldeyde were performed as described in Grimpe et al. (2002), Grimpe and Silver (2004), and Grimpe et al. (2005) with mouse anti-β-tubulin IgG (1 : 200; Sigma) or rabbit anti-β-tubulin IgG (1 : 500; Covance, Berkeley, CA, USA), mouse anti-OPN IgG (1 : 200; Developmental Hybridoma Bank, Iowa City, IA, USA) or rat anti-human CD44 IgG (1 : 200; Chemicon, Temecula, CA, USA) followed by goat anti-mouse, goat anti-rat Alexa 488 or goat anti-rabbit, goat anti-rat Alexa 598 secondary antibodies (1 : 500, Invitrogen), respectively with or without Triton, as described in the text.
RNA preparation and RT-PCR
Total RNA from 2.6 × 105 purified E20 RGCs was prepared following the RNAqueous®-Micro protocol for microscale RNA isolation (Ambion, Austin, TX, USA). 2.7 µg of total RNA was reverse transcribed into cDNA using the GenAmp RNA PCR Core Kit (Perkin-Elmer Life Science, Wellesley, MA, USA) in accordance with the manufacturer’s instructions. For PCR, the following primers were used: PCRCD44s: 5′-GGTGGCACACAGCTTGGGGA-3′ PCRCD44as: 5′-GGGAGGTGTTGGATGCGAGG-3′ (335 bp expected end product size). The step cycle program for the PCR was set for denaturation at 94°C for 30 s, annealing at 60°C for 45 s, and extension at 74°C for 40 s for 40 cycles. A β-actin control RT-PCR was performed as described in Grimpe et al. (2002). All RT-PCR products were separated on 1.0% agarose gels, stained with ethidium bromide, and photographed with a Polaroid camera (Kodak, Rochester, NY, USA).
Southern blot analysis
The oligodeoxynucleotide CD44 Southern blot probe (5′-GCAGAGGTCAGCTGCTTCAGTTCCTGGAGATACTGTAGCGGCC-3′) was obtained from MWG Biotech. This oligodeoxynucleotide probe was digoxigenin labeled according to the manufacture’s protocol (Roche, Indianapolis, IN, USA). The PCR product was electrophoretically separated on a 1.0% agarose gel. The gel was denatured for 15 min in 3 mol/L NaCl, 0.4 mol/L NaOH followed by a 15 min incubation in transfer solution (3 mol/L NaCl, 8 mmol/L NaOH) and blotted onto Hybond N+ (GE Health Care, Piscataway, NJ, USA) as described previously (Sambrook et al. 1989). The following day the membrane was blocked and treated according to Engler-Blum et al. (1993). As a detection reagent, CDP-Star (Roche) was used. The blots were exposed to Kodak Biomax Light Film for 1 h.
Immunoprecipitation of and western blot against CD44
Retinal ganglion cells were plated as described above, and were treated with the deoxyribozyme to CD44, control deoxyribozyme or left untreated. After 24 h the respective wells were rinsed with a Triton buffer (50 mmol/L Tris, pH 7.6, 150 mmol/L NaCl, 1 mL/L TritonX-100) as were U937 cells (1 × 106, Sheehan et al. 2004) used as positive control. Protein concentration was measured using 2-D quant kit (GE Health Care). Same amount of total protein from each treatment group was incubated for 30 min with 50 µL of streptavidin-conjugated agarose (Pierce, Rockford, IL, USA) at 4°C. Then, the supernatant was incubated sequentially with 10 µL of a rat anti-mouse CD44 monoclonal antibody or mouse anti-β-tubulin (Chemicon) overnight at 4°C on a wheel. Next day, a secondary antibody to rat biotin-labeled (Santa Cruz Biotechnology, Santa Cruz, CA, USA) or anti-mouse biotin-labeled (Invitrogen), respectively, was added. The samples were incubated again at 4°C on a wheel. After 5–6 h streptavidin-conjugated agarose was added and incubated overnight at 4°C on a wheel. Next day, the samples were centrifuged (5 min, 907 g). The supernatant was removed and the agarose was solved in non-reducing Laemmli-buffer (Laemmli 1970), boiled for 5 min and separated in a 4–14% gradient sodium dodecyl sulfate-polyacrylamid gel (Bio-Rad, Hercules, CA, USA). After blotting in 10 mmol/L Na-borate buffer overnight, the blot was incubated with the rat anti-mouse CD44 monoclonal antibody or mouse anti-β-tubulin (Chemicon) and the following day the blot was incubated with anti-rat or anti-mouse antibody conjugated to horseradish peroxidase for 3 h. The blot was developed on a Kodak Biomax Light Film after incubation with the enhanced chemiluminescence substrate solution (Pierce), for 10 s on average. The Coomassie blue gel was loaded with 6.56 µg of total protein from each RGC preparation.
Axon growth quantification
In three independent experiments, five wells of RGCs plated on OPN, laminin-111, or OPN/laminin-111 and stained with β-tubulin, as previously described, were analyzed in two ways. (i) A person not involved in the study used Stereoinvestigator (MicroBrightField, Williston, VT, USA) with a 40× magnification and a motorized stage with controlled ludl mac 500 to analyze the RGCs phenotypically. A total of 465–470 disectors on each coverslip (total area of coverslip: 11 mm2) with a counting frame area of 37.5 mm2 were used. (ii) Cells throughout the whole coverslip were characterized regarding their phenotypes such as (a) RGCs with short, multiple neurites, and a characteristic long axon; (b) RGCs with only short, multiple neurites; and (c) RGCs that had not yet extended any significant neurites. Axons were differentiated by morphology. The total number of RGCs counted on each coverslip was set to 100%, and the percentage of RGCs with at least one longer axon or only short neurites was calculated. The statistical validity of using percentages was confirmed by regression analysis (no-intercept model) of cell number versus total cells in all experimental conditions. The following three statistical tests of differences among and between experimental conditions were used: (i) Fisher’s analysis of variance (2 × 2 anova), (ii) General Linear Model procedure, and (iii) Fisher’s least significant difference test (LSD) (one-way anova). Axon lengths were measured using Neurolucida from MicroBright-Field. The analysis of mean and SD was carried out in Microsoft Excel.
Results
Text mining and gene ontology classification
We decided to collect protein names involved in regeneration processes from title and abstracts of publications for our prototypic program suite instead of using microarray or proteomics databases. This decision was based upon the existing peer-review process that is in place for publications in each journal. Proteins that have been investigated in experiments, discussed in a manuscript, and sent to any journal have to pass a rigorous review process from knowledgeable expert referees. When the manuscripts are published the investigated proteins have demonstrated their relevance in the field of ‘nerve regeneration’. This is in contrast to microarray or proteomics databases, in which many proteins have not been confirmed in experiments therefore their importance in regeneration has not been established. Although, we plan to add such microarray and proteomics data to our prototypic program suite, in the future, one has to keep in mind that these data will contain false-positive or false-negative results.
At the time of the investigation (July 2005), 12 669 records in PubMed were classified with the Medical Subject Headings term ‘nerve regeneration’. To determine the quality of this database we randomly selected 100 publications from our download. From these publications every single one dealt with regeneration of CNS neurons or PNS nerves. From all 12 669 records our text mining program suite extracted 1500 protein names; 1099 proteins remained after automated duplicate removal by accession number comparison; 942 proteins remained after removal of false-positives by manual curation, resulting in a precision of 86% of our computer program suite. To evaluate the performance of our program, we sampled 200 publications at random from the 12 669 publication records, counted proteins by hand and determined how many of them were identified by our data mining program suite. We found 98 non-unique and 71 unique protein names, of which 32 and 26, respectively, were not identified by our algorithm, giving an estimated recall of 65%. We also tested our assumption that proteins present in the chosen text corpus would be cited in the context of ‘nerve regeneration’ and are not part of our false-positive exclusion criteria (see Material and methods). We found in 200 abstracts two proteins that were introduced as part of an assay, staining, or tag, which allows an estimated error rate of 1% in our seed list.
We first asked which proteins are found most frequently in the nerve regeneration literature. We ranked the 942 unique, curated proteins by publication frequency and found that nerve growth factor topped the list with 775 publications (Table S1). Of the top 20 proteins referenced in this literature, seven were trophic factors, and three were ECM molecules (Table S1).
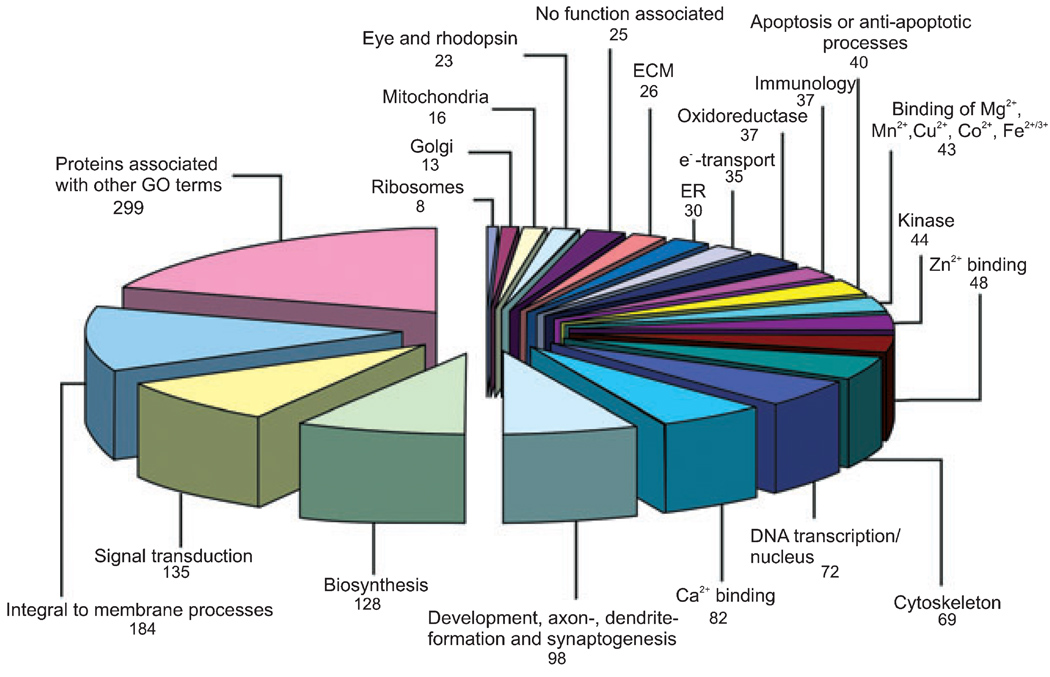
We next asked which gene ontologies these 942 proteins represented. We found that the majority of proteins identified from the regeneration literature fit into very diverse biological process classes (Fig. 1). There were only 25 proteins (2.7%) for which no classification was available in the QuickGo database, demonstrating a lag between protein identification and database completion. We identified six major categories, which are: (i) apoptosis or anti-apoptotic processes, (ii) immunology and inflammatory reactions, (iii) signal transduction, (iv) cytoskeleton and localization in the cytoplasm, (v) DNA transcription, and (vi) development, neurite-formation, and synaptogenesis that represented 32.8% of the 942 proteins, including 11 proteins from the 20 most cited proteins (Table S1). Many proteins occurred in more than one of the above-described classes; e.g. ephrin A5 is involved in development and also in signal transduction (Fig. 1). Selected well known and lesser known examples of proteins for each of these categories are shown in Tables S2– S7 with their respective Uniprot accession number and literature reference collected in our database.
Fig. 1.
Classification of the 942 proteins identified by our prototypic program suite using 20 Gene Ontology (GO-) terms. The diagram demonstrates the number of proteins present for each category. Additionally, categories including proteins which are using less common GO terms or demonstrate less esoteric characteristics and proteins for which no classification is available. As a protein may belong to several categories, the total protein sum in this diagram is 1491.
Effect of OPN on axonal outgrowth of E20 RGCs
One valuable way such database mining allows the building of new hypotheses is to focus on lesser-known or less well-studied proteins or protein interactions. By analyzing our final list of protein names generated from the prototypic program suite, we identified the commonly studied protein, laminin that appeared in our seed list on sixth position with 224 publications. On the other hand we examined proteins that appeared in one or two publications. In both attempts we tried to identify important interactions, but heretofore unknown or under-investigated in axon growth or regeneration. As the role of ECM molecules during regeneration processes is our focus, we were surprised to find an ECM molecule known from bone formation, OPN (GI: 1871123), in our ‘seed list’ of ‘nerve regeneration’ associated proteins. OPN appeared with two publications in our database. Receptors that have been studied for OPN and laminin are integrins (de Curtis et al. 1991; Duband et al. 1992; Bayless et al. 1998; Ellison et al. 1998; Clegg et al. 2000; Shin and Koh 2004), which appeared as a family in 28 publications and were thus very prevalent in our database. However, another potential receptor, CD44, previously described to interact with OPN (Weber et al. 1996) and with the laminin α5 chain (Hibino et al. 2005) in non-CNS cells were not associated with both ligands (OPN and laminin) in the CNS and no biological function in neurite outgrowth or regeneration was connected with these interactions.
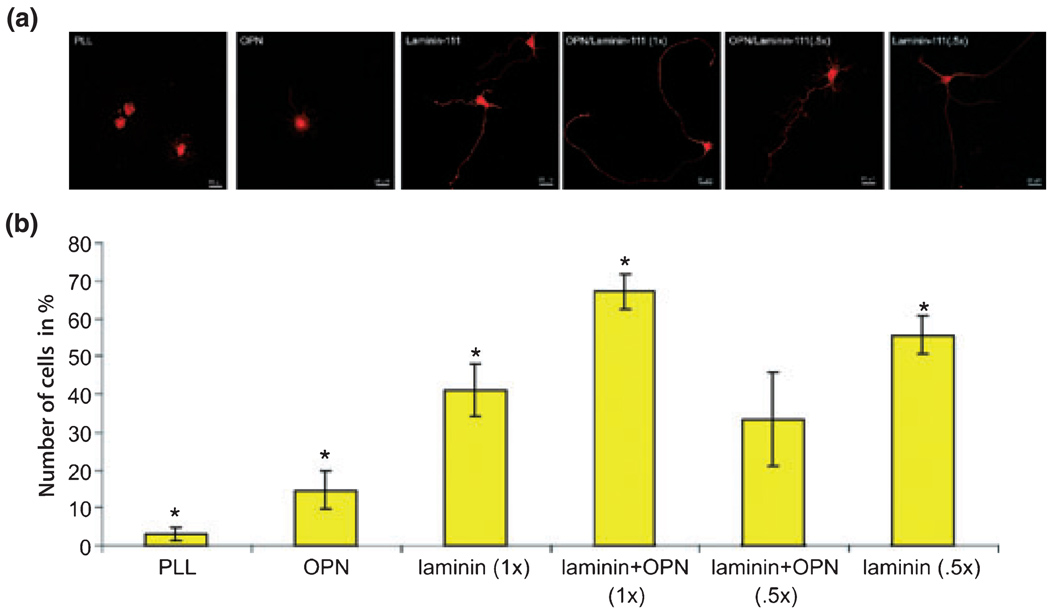
We first asked whether OPN serves as a substrate for axon growth of a CNS neuron in vitro, when compared with laminin-111 [laminin-1 with a α1β1γ1-chain (Aumailley et al. 2005)]. To address this question we chose E20 RGCs as a model system because they are commonly studied in models of in vitro survival and axon growth and because they are easily purified (Meyer-Franke et al. 1995; Goldberg et al. 2002a). This is in contrast to adult RGCs which are loosing their intrinsic ability to rapidly extend axons. We purified E20 RGCs and cultured them for 24 h in defined, serum-free media containing brain-derived neurotrophic factor, ciliary neurotrophic factor, insulin, and forskolin, which we have previously demonstrated supports maximal axon outgrowth of RGCs in culture (Goldberg et al. 2002b; Lorber et al. 2005). For these experiments we classified RGCs into three phenotypes after 24 h in culture: (i) RGCs with short, multiple neurites and a characteristic long axon; (ii) RGCs with only short, multiple neurites; and (iii) RGCs that had not yet extended any significant neurites. Poly-l-lysine, OPN, and laminin-111 were coated onto glass coverslips. We found that OPN was able to enhance axon outgrowth of E20 RGCs (15%) compared to PLL (3%), but did not support axonal extension to the same degree as laminin-111 (41%, Fig. 2). Given the weaker stimulation of axon growth by OPN compared to laminin-111, we cultured RGCs in the presence of an OPN/laminin-111 mixture. This mixture contained (i) the same concentration of both substrates as used in OPN and laminin-111 experiments alone [OPN/laminin (1×)] and (ii) half of the concentration from each substrate [OPN/laminin (0.5×)]. When OPN/laminin (1×) was used, axon initiation was found to be additive and resulted in the highest number of axon initiation on RGCs (67%). When half of the concentration of OPN and laminin-111 [OPN/laminin (0.5×)] in the mixture was used, there was a reduction in axon initiation (33%). However, half of the laminin-111 concentration [laminin (0.5×)] alone increased axon initiation (56%) compared to OPN/laminin (0.5×) and laminin (1×) but did not reach axon growth of OPN/laminin (1×). All results were statistically significant to each other with the exception of OPN/laminin (0.5×) to laminin-111 alone (Fig. 2). Thus, embryonic RGCs are responsive to OPN, OPN can support axon growth from E20 RGCs compared to PLL, but in comparison to a permissive substrate such as laminin-111 (1× and 0.5×), OPN is less stimulatory to initiate axon growth. However, when OPN with laminin-111 is provided as a substrate both are additive to axon initiation of RGCs.
Fig. 2.
RGCs plated on different substrates and characterized respectively by their axon length on these substrates. (a) Confocal images of RGCs plated on poly-l-lysine (PLL), osteopontin (OPN), laminin (1×), OPN/laminin (5×), OPN/laminin (1×), and laminin (0.5×) mixtures. (b) The percentage of RGCs that extended axons in vitro was quantified on the aforementioned substrates (5 wells/condition). RGCs adhered to all substrates; *statistically significant, p < 0.05.
Down-regulation of CD44 expression in RGCs
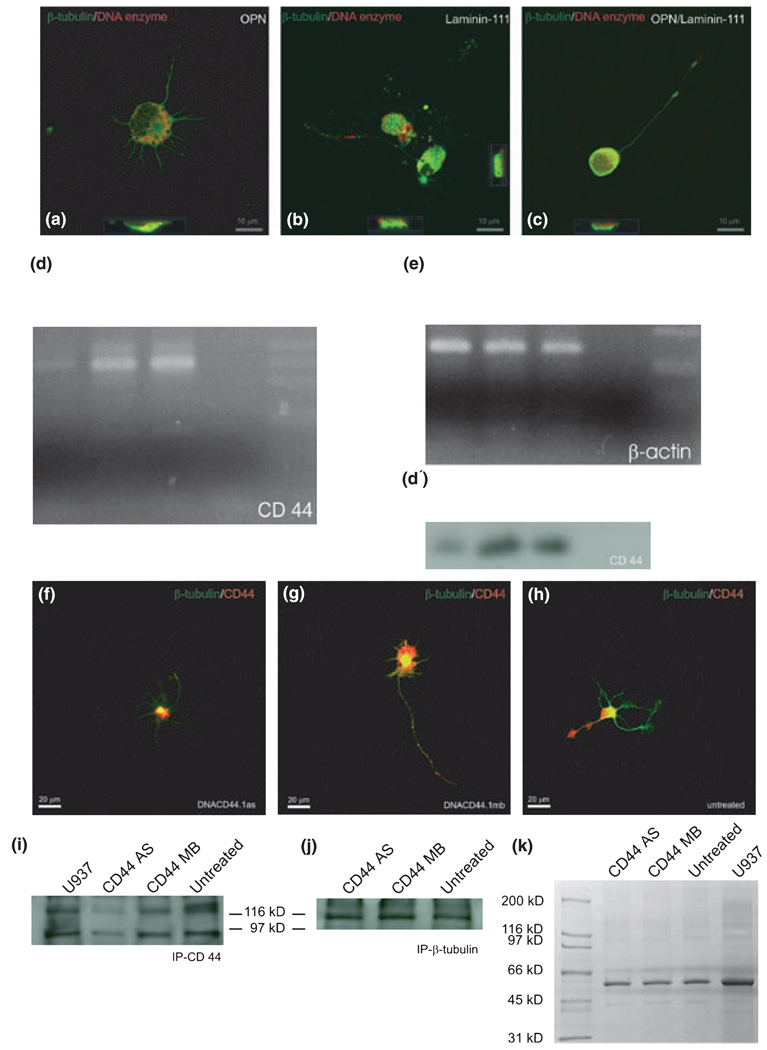
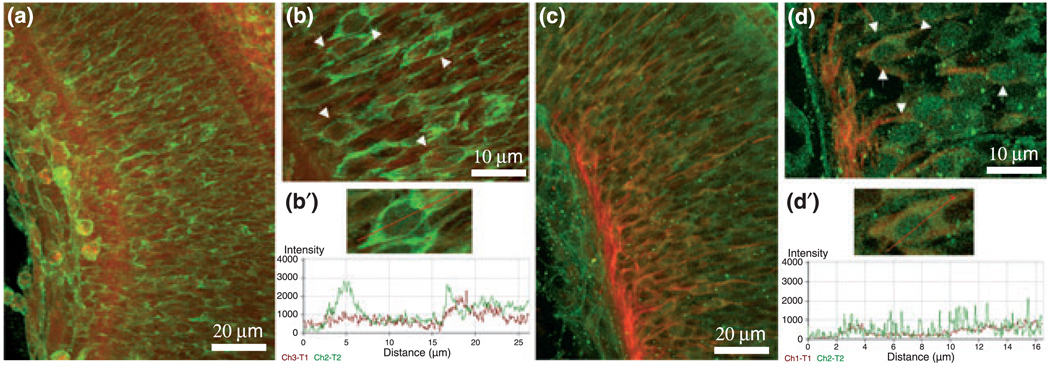
To address whether CD44 mediates the effect of OPN and laminin-111 on RGC axon growth, we first determined the proportion of RGC that express CD44 on OPN, laminin-111, and the OPN/laminin-111 mixture, which resulted in 20–30% of the cells in each of the substrate groups. Then, we interfered with CD44 expression by RGCs using a deoxyribozyme (Breaker and Joyce 1994; Grimpe et al. 2002, 2005; Grimpe and Silver 2004) targeting the CD44 mRNA. Deoxyribozymes are small catalytic DNA molecules that are able to act as enzymes by cleaving a target mRNA via their sequence-specific binding-arms. The digested pieces are released and the deoxyribozyme is able to cleave further mRNA molecules of the same gene (Breaker and Joyce 1994; Grimpe et al. 2002; Grimpe and Silver 2004). First, we confirmed that the deoxyribozyme was internalized by cultured RGCs. For this purpose we labeled the CD44 deoxyribozyme at its 5′ end with biotin and added 8 µmol/L to E20 RGCs cultured on three different substrates: OPN, laminin-111, and OPN/laminin-111 (0.5×). After 24 h in culture, RGCs were fixed and double-labeled with a Texas Red-conjugated streptavidin and counter-stained with an antibody to β-tubulin (Fig. 3). RGCs were imaged by confocal microscopy, and demonstrated double labeling for the biotin-labeled deoxyribozyme and β-tubulin in the cytoplasm in both cross-sections and the ortho-projection acquired by the laser scanning microscope (LSM) browser (Fig. 3a–c). We found that 56% of the RGCs growing on OPN, 63% growing on laminin-111, and 73% growing on OPN/laminin-111 (0.5×) internalized the biotin-labeled deoxyribozyme, demonstrated by a strong staining. Additionally, 36% of RGCs growing on OPN, 44% growing on laminin-111, and 27% plated on OPN/laminin-111 (0.5×) showed a weak staining of internalized deoxyribozyme. Only 0.2% of RGCs plated on OPN, 1.8% plated on laminin-111, and 0.34% plated on OPN/laminin-111 (0.5×) showed no deoxyribozyme uptake at all (Fig. 3a–c). Thus, the deoxyribozyme accumulated in nearly all cultured RGCs independent of substrate, and was detectable as early as 24 h.
Fig. 3.
CD44 deoxyribozyme specifically down-regulates CD44 expression in RGCs. (a–c) Confocal images of RGCs growing for 24 h on different substrates as marked were treated with biotin-labeled deoxyribozyme to CD44 in a concentration of 8 µmol/L. Red shows the labeled deoxyribozyme localized in the cytoplasm; green is β-tubulin immunostaining. In each image, cross-sections (lower or right-hand sides of the image) demonstrate the accumulation of the deoxyribozyme in the cytoplasm (yellow/orange). (d) RT-PCR for CD44 (40 cycles) with RNA from RGCs after 24 h in culture. Reduction of CD44 mRNA after deoxyribozyme treatment against CD44 (CD44AS), as compared to control deoxyribozyme treatment (CD44MB), or to untreated cultures, was observed. (e) RT-PCR as in (d) for β-actin (25 cycles); bands demonstrate similar RNA template sample concentrations, and specificity of CD44 down-regulation from (d); MWM, molecular weight marker. (f–h) Immunostainings of RGCs to CD44 (red channel) and β-tubulin (green channel) after 24 h in culture demonstrate a reduction of CD44 after deoxyribozyme treatment to CD44 (f), compared to control deoxyribozyme treatment (g) and untreated (h) cultures. Note the strong positive CD44 staining at the short but multiple neurites of (g) and at the end-feet of the axonal growth cone in (h). (i and j) Immunoprecipitation of CD44 protein (i) or β-tubulin (j) from RGCs after treatment with 8 µmol/L of a deoxyribozyme targeting CD44 mRNA (CD33AS), 8 µmol/L control deoxyribozyme (CD44MB), or left untreated. U937 is a human leukemia monocyte lymphoma cell line, which was used as positive control, based on its known high expression of CD44. (k) Shows a Coomassie stained 4–15% SDS gel, which was loaded with crude protein extract from RGCs treated with 8 µmol/L of deoxyribozyme to CD44 (CD44AS), control deoxyribozyme (CD44MB), left untreated or as a positive control U937 cell extract. Note that the overall protein bands are equal in each lane which demonstrates that deoxyribozyme treatment has no general effect on the RGCs.
We next confirmed the specificity of the deoxyribozyme against CD44 in RGCs, using RT-PCR on total RNA isolated from deoxyribozyme- and control deoxyribozyme-treated as well as untreated RGCs cultured on OPN. RT-PCR Southern blot with a CD44-specific probe demonstrated that CD44 was down-regulated in CD44 deoxyribozyme-treated but not in mixed base (control) deoxyribozyme-treated cultures or in untreated RGCs (Fig. 3d and d′). The performed Southern blot followed by hybridization with a CD44-specific oligonucleotide probe demonstrates that the amplified product is, indeed, CD44 mRNA. A β-actin RT-PCR was utilized as an internal control to show the integrity of the RNA as well as to demonstrate that a similar amount of RNA was used in each reaction (Fig. 3e). Thus, RGCs normally express CD44 (also see below), and the CD44 deoxyribozyme specifically down-regulated CD44 expression at the mRNA level in cultured RGCs.
To demonstrate the down-regulation of the CD44 protein after 24 h of treatment with the deoxyribozyme, we performed immunoprecipitations followed by a western blot and immunostainings to CD44 and β-tubulin. In control, deoxyribozyme-treated and -untreated cultures CD44 expression was identified by immunostaining in the cell body of the RGCs, in axons and their growth cones, and in short neurites (Fig. 3f–h). In contrast deoxyribozyme-treated cultures demonstrate no immunostaining in any neurites and a greatly attenuated immunostaining in the cell body (Fig. 3f). This may reflect CD44 protein in RGC cell bodies at the time of preparation. The observation was supported by an immunoprecipitation using the same antibody against CD44 as utilized for the aforementioned immunostainings. Deoxyribozyme-treated cultures showed reduced expression of CD44, after immunopreciptation and western blot under non-reducing conditions compared to control deoxyribozyme and untreated cultures (Fig. 3i). To determine that same amount of protein was used, a β-tubulin immunoprecipitation was performed for each treatment group. Additionally, a Coomassie blue-stained polyacrylamide gel loaded with samples of each treatment group confirmed that no obvious differences exists between the various groups (Fig. 3j and k). Thus, CD44 deoxyribozyme specifically reduced CD44 protein translation in RGCs in culture.
Effect of down-regulating CD44 on axonal outgrowth of RGCs
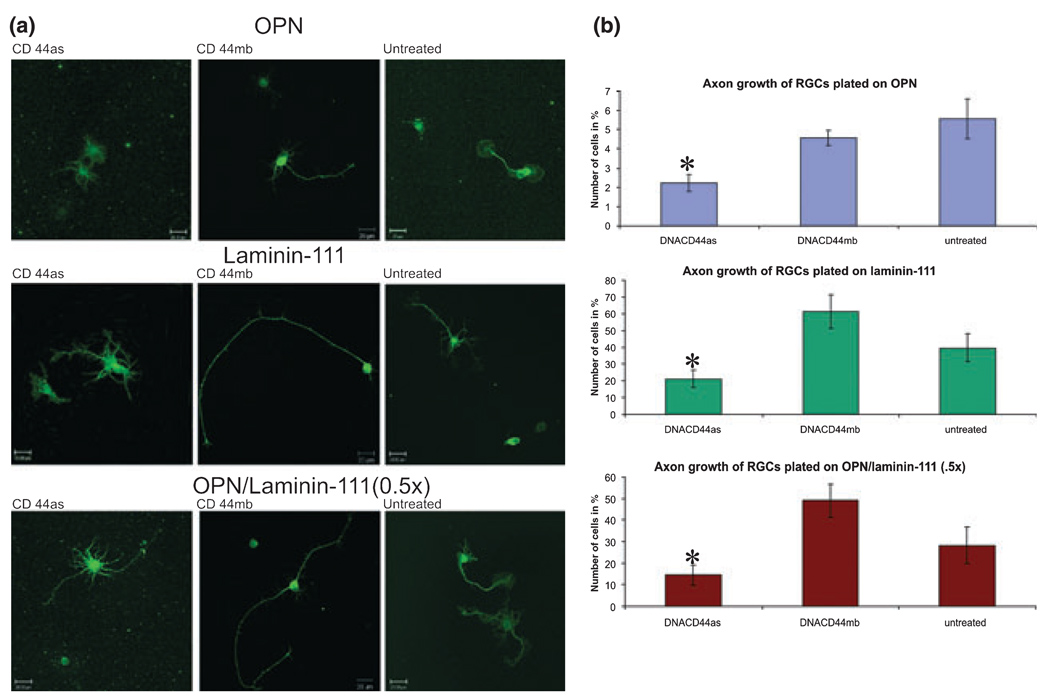
Finally, we asked whether OPN- and laminin-stimulated axon growth is dependent on CD44 expression by RGCs. We treated RGC cultures for 1 day with the CD44 deoxyribozyme, the control deoxyribozyme, or no deoxyribozyme (untreated). We found in five independent experiments that RGCs plated on OPN and treated with CD44 deoxyribozyme for 1 day did not extend as many axons compared to RGCs treated with the control deoxyribozyme or left untreated (2.22%, 4.57%, and 5.57%, respectively; Fig. 4). This difference was statistically significant as demonstrated by the Fischer least significant difference test (p ≥ 0.05). On laminin-111 as well as mixture of OPN/laminin-111 (0.5×) deoxyribozyme treatment to CD44 resulted also in inhibition of axon initiation compared to control deoxyribozyme treatment or untreated cultures [laminin-111: 21.1%, 61.35%, 39.76%; laminin-111/OPN (0.5×): 14.59%, 49.0%, 28.30%; Fig. 4]. We found that on average 15.5% of RGCs extended multiple, short neurites in any treatment/substrate group which demonstrated no statistical differences. Thus, RGC axon growth depends on CD44 expression on OPN and on laminin-111.
Fig. 4.
CD44 down-regulation decreases RGC axon initiation response. (a) Confocal images of RGCs plated on osteopontin (OPN), laminin-111, or mixture of OPN/laminin-111 (0.5×) treated with a deoxyribozyme against CD44 (as, first column), control deoxyribozyme (mb, second column), or left untreated (third column) for 1 day and immunostained for β-tubulin. (b) Quantification of the percent of RGCs that initiated axons on each substrate with deoxyribozyme or control treatments. The use of percentages were justified by increased linearly (no-intercept model) between the number of axonal cells compared to total number of cells counted. *p < 0.05 by the General Linear Model procedure (SAS Software, Cary, NC, USA), and by subsequent pair-wise tests using Fisher’s LSD-test.
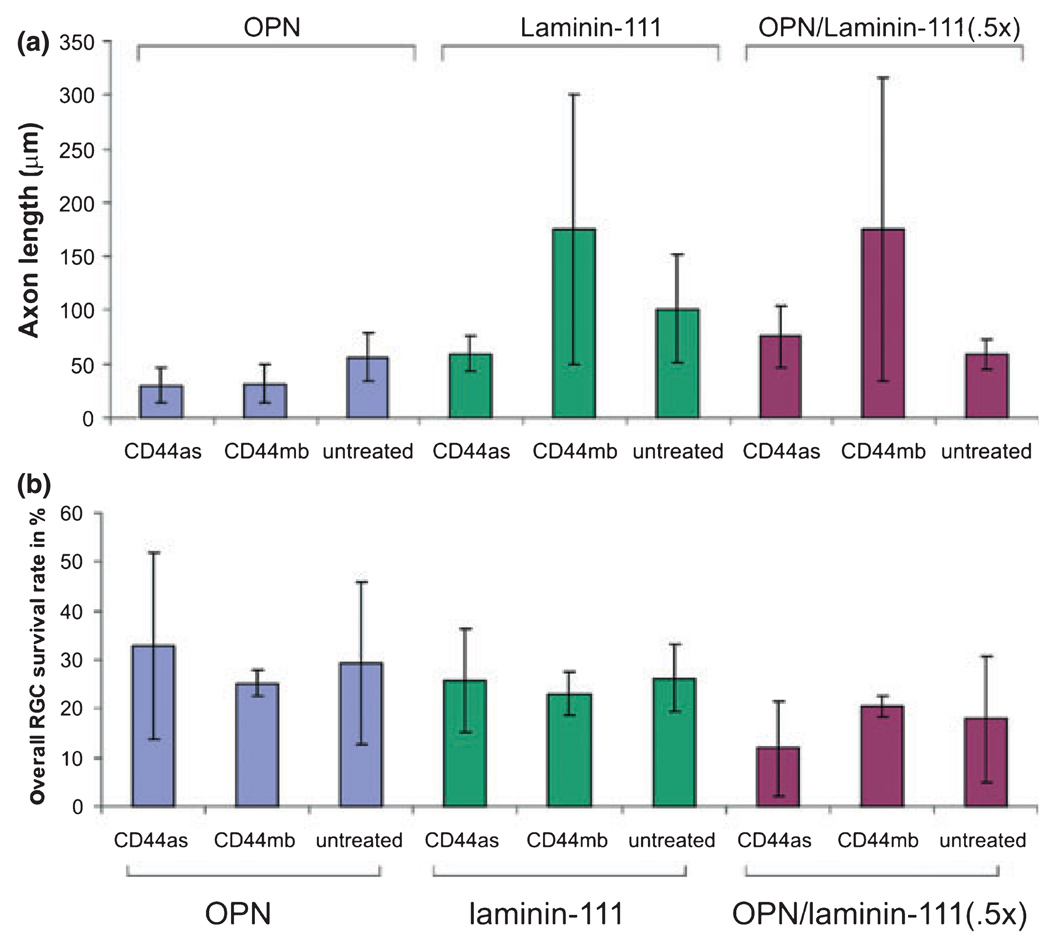
We also measured RGC axon lengths on OPN, laminin-111, and OPN/laminin-111 (0.5×) substrates, with and without CD44 deoxyribozyme down-regulation. On OPN, independent of the treatment, axons were mostly shorter and missed longer extensions. In contrast to laminin-111 and OPN/laminin-111 (0.5×) the variability in axon length was quite high because RGCs extended long and short axons. Based on the high variability in these two groups no statistical significance in axon lengths of RGCs on any substrate or in any treatment group (Fig. 5a) could be observed. Furthermore, we determined the overall survival rate of RGCs on different substrates and in each treatment group (Fig. 5b). The mean survival rate of 23.63% showed no statistical difference under any substrate or treatment conditions.
Fig. 5.
(a) Measurement of axon length from RGCs growing on OPN, laminin-111, and a mixture of osteopontin (OPN)/laminin-111 (0.5×) after 24 h treated with the CD44 (CD44as) and control (CD44mb) deoxyribozymes (8 µmol/L) or left untreated. (b) Demonstrates the overall survival rate of RGCs under any of our experimental conditions (substrate or treatment). No statistical difference was observed between these various conditions.
Expression of OPN and CD44 in the developing visual pathway
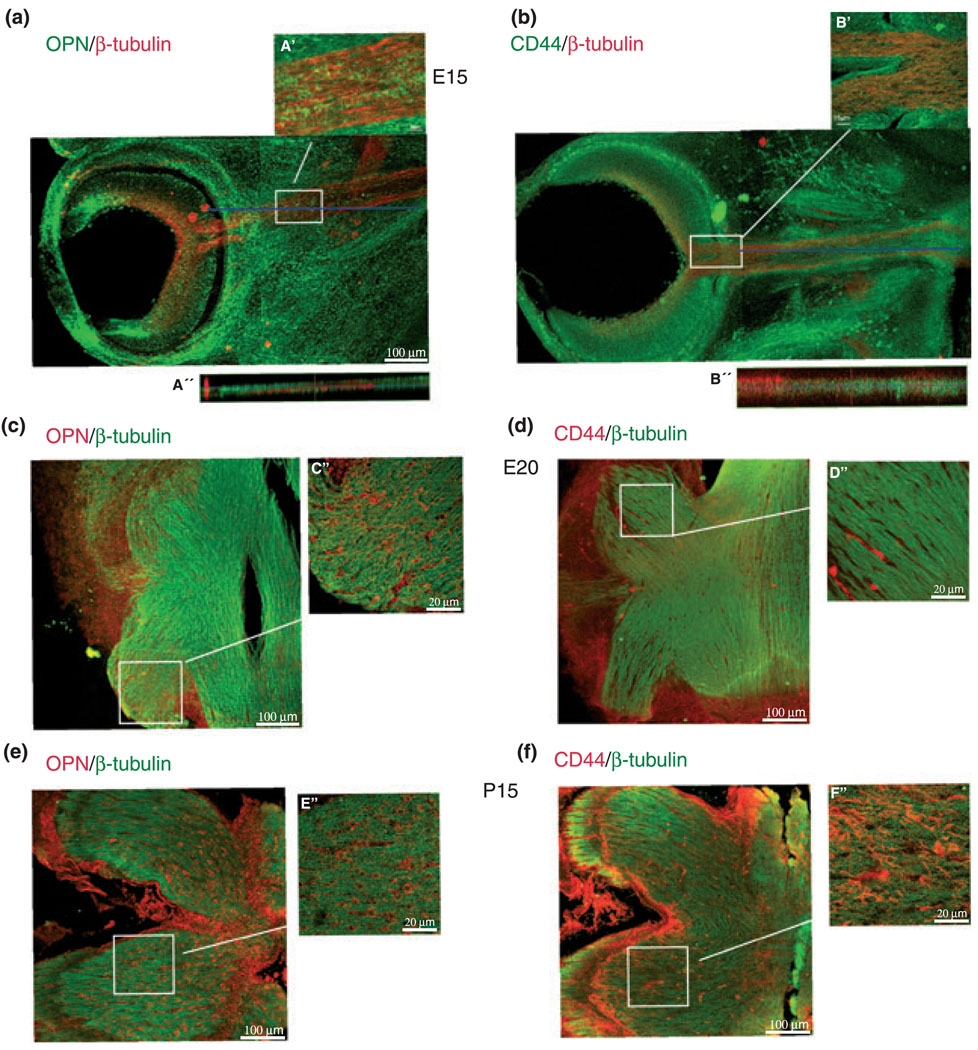
Expression of laminins in the retina and optic nerve has been demonstrated in many publications (Smalheiser et al. 1984; Hopkins et al. 1985; Cohen et al. 1987; McLoon et al. 1988; Sarthy and Fu 1990; Cohen and Johnson 1991; Morissette and Carbonetto 1995, Stier and Schlosshauser 1995; Libby et al. 2000; Aisenbrey et al. 2006). Therefore, we are focusing on the under-investigated OPN–CD44 interaction. We next explored whether the in vitro observation of OPN–CD44 interaction involved in axonal outgrowth of RGCs is potentially important in vivo by performing immunofluorescent stainings of embryonic day 15 (E15) rat retina and optic nerve, as well as E20 and postnatal day 15 (P15) optic nerve sections, with a mouse anti-rat OPN and a mouse anti-CD44 antibody (Figs 6 and 7). To identify RGC cell bodies and axons, each section was also stained with a polyclonal anti-rabbit β-tubulin antibody. We found OPN expression in the E15 retina throughout all layers (Figs 6a, 7a and a′), as well as in the E15 optic nerve (Fig. 7a and a″), consistent with prior published data (Hikita et al. 2003). We observed colocalization of OPN immunostainings with β-tubulin in a RGC in the premature ganglion cell layer of E15 rat retinas. Using ‘profile’ from the LSM software as shown in Fig. 6b′ such colocalization of OPN and β-tubulin is found at 5 µm and from 16 to 25 µm extending into the processes of the RGC. The background staining is visible throughout the first 2 µm of the y-axis (distance). In Fig. 7 a cross-section generated with the ortho-projection of the LSM browser of the confocal imaging data system through the optic nerve demonstrated OPN colocalized with β-tubulin immunostainings (Fig. 7a′). OPN localization in E20 (Fig. 6c) and P15 (Fig. 6e) optic nerve sections were found on cells that are negative for β-tubulin immunostaining.
Fig. 6.
Immunostainings of osteopontin (OPN) and CD44 in the premature E15 RGC layer of 90 µm-thick vibratome sections. (a) Confocal image of 40× magnified RGC layer immunostained for OPN (green) and β-tubulin (red). (b) 100× magnification of the premature E15 ganglion cell layer (OPN, green; β-tubulin, red). Premature ganglion cells are marked by white arrows. (b′) ‘Profile’ from the laser scanning microscope (LSM) software along the red line shows OPN protein co-expressed with β-tubulin in one RGC. ‘Profile’ uses one image from a stack of images for analysis. (c) Confocal image of 40× magnified RGC layer immunostained for CD44 (green) and β-tubulin (red) in retinas of E15 embryos. (d) Higher magnification (100×) of the premature E15 ganglion cell layer (CD44, green; β-tubulin, red) marked by white arrows. (d′) ‘Profile’ from the LSM software package shows along the red line a clustered CD44 staining in areas of β-tubulin immunostaining in one RGC.
Fig. 7.
Confocal images of sections from E15 eye and optic nerve, from E20 as well as P15 optic nerves stained with antibodies to CD44, osteopontin (OPN), and β-tubulin. (a) E15 retina and optic nerve stained with OPN (green) and β-tubulin (red). (a′) Is a higher magnification of the marked area in the optic nerve of (a). (a″) Demonstrates a co-existence of OPN and β-tubulin in the optic nerve by using the ortho-projection of the confocal imaging data coinciding with the blue line shown in (a). (b) E15 retina and optic nerve sections stained with CD44 (green) and β-tubulin (red). (b′) Is a higher magnification of the entry site of the retinal ganglion axons into the optic nerve, in which CD44 and β-tubulin co-exist. (b″) Shows the ortho-projection of the confocal imaging data from the blue line shown in (b), demonstrating that CD44 and β-tubulin are expressed throughout the optic nerve. (c–d) E20 and (e–f) P15 optic nerve section stained with β-tubulin (green) and OPN (red, c and e) or CD44 (red, d and f), respectively. (c′–f′) Are magnifications to demonstrate OPN and are not expressed from β-tubulin positive cells in the optic nerve at both ages.
CD44 immunostaining was observed as punctated or clustered staining throughout the whole retina of E15 embryos (Figs 6 and 7). In Fig. 6d′ using ‘profile’ from the LSM browser on one RGC demonstrates colocalization of CD44 with β-tubulin immunostaining. This kind of immunostaining extended into the outgrowing processes of RGCs. The first 2 µm of Fig. 6d′ show background staining followed by the colocalization of CD44 with β-tubulin immunostainings frequently above the background throughout the whole image. Further CD44 immunostaining was found in the proximal axons extending to the head (Fig. 7b″) and into the body of the optic nerve of E15 embryos (Fig. 7b′). The ortho-projection from the LSM software package (along the blue line) demonstrates CD44 immunostaining in the same area as β-tubulin (Fig. 7b′). In E20 (Fig. 7d and d′) and P15 (Fig. 7f and f′) optic nerve CD44 expression was associated with cells that are not β-tubulin positive. Thus CD44 and OPN are expressed by RGCs and surrounding cells in retina and optic nerve.
Discussion
We describe the first part of a novel bioinformatics approach, in which we identified and characterized proteins from publications classified by the term, ‘nerve regeneration’ through text mining. Interestingly but not surprising, growth factors are the most used and investigated proteins in the field of regeneration research (Table S1); however, the administration of growth factors alone has not yet led to a satisfactory cure of paralysis or recovery of function after CNS injury. Furthermore, our final protein list revealed that molecules such as tubulin, actin, acetylcholinesterase, calcitonin gene-related peptide, acetyltransferase, p75, nitric oxide synthetase, and ECM molecules have also been heavily investigated, yet no single protein has proved critical to enhancing regenerative response. Indeed, the gene ontologies of proteins studied in regeneration reflect processes from a wide range of general cell biology rather than processes specific to axon growth. For example, a high number of publications reported on proteins in the area ‘biosynthesis’ including glycolysis, β-oxidation, and Krebs cycle and many more pathways responsible to provide the cell with energy and to metabolize degradation products.
Similar systems biology approaches have been used to obtain new insights into other basic biological processes. In one example the signaling networks involved in apoptosis were investigated and revealed an autocrine loop of cytokine signaling induced by tumor necrosis factor critical for apoptosis signaling (Janes et al. 2005). In another example, a global protein network alignment between Saccharomyces cerevisiae and Helicobacter pylori identified conserved interaction pathways and cellular processes previously unknown to be associated (Kelley et al. 2003). These examples show that systems biology approaches have the potential to reveal new correlations of proteins. Therefore, we are in the process of generating a protein–protein interaction map using our final protein list to identify core networks and/or generate new hypotheses for future studies. During our computer program design we encountered several problems. The first and major problem was the use of acronyms in abstracts such as ALS. In our scientific field it stands for ‘amyotrophic lateral sclerosis’ but at the same time ALS is the abbreviation for several proteins such as ‘agglutinin-like protein 2 precursor’, ‘acetylcholine receptor protein’ or ‘alpha-like growth factor-binding protein’. The decision whether the publication refers to one of the proteins or the disease was made manually by the domain specialist. A second major problem was the high degree of semantic heterogeneity across the data sources. Names of proteins appeared in different abstracts in various notations e.g. ‘laminin 1’, ‘laminin-1’, ‘laminin1’, or ‘laminin γ1 chain’, ‘laminin-γ1 chain’, ‘laminin-gamma 1 chain’, ‘laminin gamma 1 chain’ etc. Each case of variability needs to be specifically acknowledged in our program suite. An additional problem was the identification of a protein class compared to a specific protein name. As an example in older publication ‘laminin’ refers most likely to ‘laminin-1’ because it was the only known laminin at that time. In later publications, it might refer to the whole class of laminins with 15 members (Aumailley et al. 2005). Further problems contain the ambiguous meaning of words such as ‘zinc’. The majority of readers might think immediately of the metal but there is actually a protein called ‘zinc’. We made the decision regarding the two last problems to eliminate the proteins manually by the domain specialist instead of integrating the problem in our program suite.
After such clean up, we were able to identify in our list of 942 proteins the ECM molecules laminin and OPN. Laminin appears on the sixth position with 224 publications in our list of 20 most cited molecules important in ‘nerve regeneration’ (Table S1). OPN, however, appears with only two publications in this list. Because the expression of laminin in the embryonic retina and optic nerve has been described in many publications (Smalheiser et al. 1984; Hopkins et al. 1985; Cohen et al. 1987; McLoon et al. 1988; Sarthy and Fu 1990; Cohen and Johnson 1991; Morissette and Carbonetto 1995; Stier and Schlosshauser 1995; Libby et al. 2000; Aisenbrey et al. 2006) we decided to focus on OPN. OPN was originally identified in bone remodeling and was later localized to kidney, inner ear, vascular smooth muscle cells, and immune cells (Nomura et al. 1988; Butler 1989, Brown et al. 1992; Denhardt and Guo 1993, Rodan 1995; Madsen et al. 2000). It has been implicated in peripheral nerve regeneration, where it is expressed by Schwann cells, down-regulated after axon injury, and up-regulated after axon regeneration (Jander et al. 2002). In the CNS OPN deficient mice show less inflammation, greater tissue damage, and impaired locomotor recovery after contusion injury (Hashimoto et al. 2007). A potential role of OPN in myelination was supported by results in CNS oligodendrocytes, where OPN stimulates expression of myelin-basic protein (Selvaraju et al. 2004).
We asked whether OPN was able to stimulate or inhibit axon growth from CNS neurons, using E20 RGCs as a model system. We found that OPN was able to stimulate axon growth compared to PLL, but that it inhibited the growth normally stimulated by a permissive laminin-111 substrate. Our result of an inhibitory effect from OPN on axon growth of RGCs was consistent with that seen previously for dorsal root ganglion neurons in vitro (Küry et al. 2005), and with the pattern of expression after peripheral nerve injury (Nomura et al. 1988).
Furthermore, our data demonstrate by immunofluorescence in vivo that OPN might be expressed in RGCs or other retinal and optic nerve cells in the visual pathway throughout development. Previously OPN had been identified in retina although its cellular and subcellular localization has not been determined (Ju et al. 2000). OPN has also been localized in neurons of the auditory and vestibular ganglia (Davis et al. 1995), in the olfactory bulb, and in sensory nuclei of the brainstem of the mature rat (Shin et al. 1999). Thus, OPN may play a role in neurite growth throughout many areas of the CNS.
One potential receptor of OPN, CD44, was previously studied in hematopoetic cells (Weber et al. 1996). Its sequence is quite conserved when compared to other organisms utilizing BLAST. CD44 expression has been described in the CNS (Moretto et al. 1993; Kaaijk et al. 1997; Bayless et al. 1998; Alfei et al. 1999; Stein et al. 2004; Tuohy et al. 2004; Yokosaki et al. 2005; He et al. 2006), but its function in the CNS is unknown, although interactions between CD44 and various ECM components such as hyaluronan and laminin with the laminin alpha5 chain are under investigation (Skelton et al. 1998; Hibino et al. 2004). CD44 is localized to the leading edges and lamellipodia of several migrating cell types in vitro (Alfei et al. 1999; Zohar et al. 2000). As migration and axonal growth cone procession are closely related morphologically and molecularly, it makes sense to find the OPN–CD44 interaction involved in axon outgrowth of RGCs.
Reduction of CD44 mRNA using a deoxyribozyme in in vitro experiments reduced axonal outgrowth on OPN and laminin-111 when used as a substrate. Edgar et al. (1984) demonstrated that a specific domain, E8, localized at the long arm of laminin-111 consisting of the terminal globular domain and a 34 nm long rod-like element is responsible for survival and outgrowth of sympathetic neurons but not other cells. This domain might be involved in the CD44–laminin interaction observed in our experiment. Hibino et al. (2004, 2005) reported that a synthetic laminin alpha 5-chain peptide blocked the CD44 receptor expressed on melanoma cells via the glycosmainoglycans. Binding to fibroblast growth factor as well as down-stream signaling molecules involving tyrosine phosphorylation was blocked by this laminin peptide. This inhibited tumor cell migration and invasion. RGCs growing on laminin and treated with the deoxyribozyme to CD44 may have experienced the same regulatory mechanism because deoxyribozyme treatment of RGCs growing on laminin led to reduced axon initiation in our study. In the developing and adult retina, the following laminins have been identified: laminin-111 (former laminin-1), laminin-211 (former laminin-2), laminin-332 (former laminin-5), laminin-511 (former laminin-10), laminin-423 (former laminin-14), and laminin-523 (former laminin-15; McKerracher et al. 1996; Libby et al. 2000; Aumailley et al. 2005; Aisenbrey et al. 2006). Adhesion to these laminins is, in most cases, investigated via laminin-binding integrins such as α1β1, α2β1, α3β1, α4β1, α6β1, α8β1, αvβ5, and αvβ3, which were all identified in the developing retina (Clegg et al. 2000). To better understand the biological function of laminin–integrin and, in particular, laminin–CD44 interactions in regeneration processes further investigations into this area are necessary.
CD44 expression in vitro was observed in RGCs via immunofluorescence and RT-PCR (Fig. 2d and d′). This observation confirms that of Sarthy et al. (2007) who showed, for the first time, expression of enhanced green fluorescent protein under the control of the CD44 promoter in RGC of transgenic mice. Following closer characterization of the mouse retina, the authors assumed that RGC subtypes such as B1 or B3 (Sun et al. 2002) expressing CD44. Interestingly, Okamoto et al. (1999) described that the CD44 ectodomain can be proteolytically cleaved, releasing a 32 kDa soluble fragment. Could soluble CD44 in the retina or optic nerve be signaling RGCs? Soluble CD44, in a concentration of 1 ng/mL, exhibits a dose- and time-dependent neurotoxicity to a rat RGC-like cell line (RGC-5) (Choi et al. 2005). Therefore, soluble CD44 might inhibit axon growth of RGCs during development, for example when the RGC axons reach the chiasm. However, membrane-bound CD44 might have a different function as demonstrated in this publication. At the chiasm, subpopulations of RGCs may be blocked from crossing the midline by CD44-positive cells (Sretavan et al. 1994; Lin et al. 2005) but no information is given whether the membrane-bound or soluble form of CD44 is involved in this process.
Many questions regarding additional functions of CD44 interactions with OPN and laminin remain to be addressed, in particular whether they play a role in vivo in the failure of CNS axon regeneration after injury. Here, we demonstrated by describing and utilizing the first part of a new suite of programs the ability to identify novel biological functions to known protein interactions. In creating protein databases relevant to any area in biology this approach has a potential to be widely used. We applied this systems biology approach to the area of nerve regeneration and used it to predict a role of CD44 interactions with OPN and laminin in axon initiation of RGCs. By expanding the program to reveal additional novel protein interactions, we hypothesize that future applications of this data and approach will be required to overcome such complex issues as the failure of CNS regeneration.
Supplementary Material
The following supplementary material is available for this article online:
Table S1 The 20 most frequently cited proteins.
Table S2 Examples of proteins involved in apoptosis and anti-apoptotic processes.
Table S3 Examples of proteins involved in immunology and defense responses.
Table S4 Examples of proteins involved in signal transduction.
Table S5 Examples of proteins found in cytoskeleton and cytoplasm.
Table S6 Examples of proteins involved in DNA transcription and in the nucleus.
Table S7 Examples of proteins involved in development, neurite-formation and synaptogenesis.
This material is available as part of the online article from http://www.blackwell-synergy.com.
Acknowledgements
We thank John Bethea and Patrick Wood for critical reading of this manuscript, Robert Duncan for statistical evaluation, and Christian Romero and Heidrun Podini for technical assistance. Furthermore, we are grateful to Linda White for scientific input, Beata Frydel for image analysis, and William Buchser for graphical assistance. Support was provided by The Miami Project to Cure Paralysis, the Ralph Wilson Sr and Ralph Wilson Jr Medical Research Foundation, the Glaucoma Foundation (BG and JLG), The Florida State Fund (BG), the National Eye Institute [EY016790 (JLG)], National Institute of Neurological Disorder and Stroke [NS061348 (JLG) and EY014801 (Miami, JLG)] and by an unrestricted grant from Research to Prevent Blindness (Miami, JLG).
Abbreviations used
- ALS
amyotrophic lateral sclerosis
- CD
cluster of differentiation
- ECM
extracellular matrix
- En
embryonic day
- LSM
laser scanning microscope
- OPN
osteopontin
- PLL
poly-l-lysine
- Pn
postnatal day
- RGC
retinal ganglion cell
References
- Aisenbrey S, Zhang M, Bacher D, Yee J, Brunken WJ, Hunter DD. Retinal pigment epithelial cells synthesize laminins, including lamini 5, and adhere to them through alpha3- and alpha6-containing integrins. Invest. Ophthalmol. Vis. Sci. 2006;47:5537–5544. doi: 10.1167/iovs.05-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfei L, Aita M, Caronti B, De Vita R, Margotto V, Medolago AL, Valente AM. Hyaluronate receptor CD44 is expressed by astrocytes in the adult chicken and in astrocyte cell precursors in early development of the chicken spinal cord. Eur. J. Histochem. 1999;43:29–38. [PubMed] [Google Scholar]
- Alldinger S, Fonfara S, Kremmer E, Baumgartner W. Up-regulation of the hyaluronate receptor CD44 in canine distemper demyelinated plaques. Acta Neuropathol. 2000;99:138–146. doi: 10.1007/pl00007417. [DOI] [PubMed] [Google Scholar]
- Altshult SF, Madden TL, Schaeffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumailley M, Bruckner-Tuderman L, Carter WG. A simplified laminin nomenclature. Matrix Biol. 2005;24:326–332. doi: 10.1016/j.matbio.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Bayless KJ, Meininger GA, Scholtz JM, Davis GE. Osteopontin is a ligand for the alpha4beta1 integrin. J. Cell Sci. 1998;111:1165–1174. doi: 10.1242/jcs.111.9.1165. [DOI] [PubMed] [Google Scholar]
- Beattie MS. Inflammation and apoptosis: linking therapeutic targets in spinal cord injury. Trends Mol. Med. 2004;10:580–583. doi: 10.1016/j.molmed.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Breaker RR, Joyce GF. A DNA enzyme that cleaves RNA. Chem. Biol. 1994;4:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Brown LF, Berse B, van de Water L, Papadopoulos-Sergiou A, Perruzzi CA, Manseu EJ, Dvorak DR, Senger DR. Expression and distribution of osteopontin in human tissues: widespread association with luminal epithelial surfaces. Mol. Biol. Cell. 1992;3:1169–1180. doi: 10.1091/mbc.3.10.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler WT. The nature and significant of osteopontin. Connect. Tissue Res. 1989;23:123–136. doi: 10.3109/03008208909002412. [DOI] [PubMed] [Google Scholar]
- Choi JS, Cha JH, Park HJ, Chung JW, Chun MH, Lee MY. Transient expression of osteopontin mRNA and protein in amoeboid microglia in developing rat brain. Exp. Brain Res. 2004;154:275–280. doi: 10.1007/s00221-003-1657-4. [DOI] [PubMed] [Google Scholar]
- Choi J, Miller AM, Nolan MJ, Yue BYJT, Thotz ST, Clark AF, Agarwal N, Knepper PA. Soluble CD44 is cytotoxic to trabecular meshwork and retina ganglion cells in vitro. Invest. Ophthalmol. Vis. Sci. 2005;46:214–222. doi: 10.1167/iovs.04-0765. [DOI] [PubMed] [Google Scholar]
- Clegg DO, Mullick LH, Wingerd KL, Lin H, Atienza JW, Bradshaw AD, Gervin DB, Cann GM. Adhesive events in retinal development and function: the role of integrin receptors. Results Probl. Cell Differ. 2000;31:141–156. doi: 10.1007/978-3-540-46826-4_8. [DOI] [PubMed] [Google Scholar]
- Cohen J, Johnson AR. Differential effects of laminin and merosin on neurite outgrowth by developing RGCs. J. Cell. Sci. 1991;15:1–7. doi: 10.1242/jcs.1991.supplement_15.1. [DOI] [PubMed] [Google Scholar]
- Cohen J, Burne JF, McKinlay C, Winter C. The role of laminin and the laminin/fibronectin receptor complex in the outgrowth of RGC axons. Dev. Biol. 1987;122:407–418. doi: 10.1016/0012-1606(87)90305-8. [DOI] [PubMed] [Google Scholar]
- de Curtis I, Quaranta V, Tamura RN, Reichardt LF. Laminin receptors in the retina: sequence analysis of the chick integrin α6 subunit. J. Cell Biol. 1991;113:405–416. doi: 10.1083/jcb.113.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Lopez CA, Mou K. Expression of osteopontin in the inner ear. Ann. N. Y. Acad. Sci. 1995;760:279–295. doi: 10.1111/j.1749-6632.1995.tb44638.x. [DOI] [PubMed] [Google Scholar]
- Denhardt DT, Guo X. Osteopontin: a protein with diverse functions. FASEB J. 1993;7:1475–1482. [PubMed] [Google Scholar]
- Domeniconi M, Zampieri N, Spencer T, Hilaire M, Mellado W, Chao MV, Filbin MT. MAG induces regulated intra-membrane proteolysis of the p75 neurotrophin receptor to inhibit neurite outgrowth. Neuron. 2002;46:849–855. doi: 10.1016/j.neuron.2005.05.029. [DOI] [PubMed] [Google Scholar]
- Duband LL, Belkin AM, Syfrig F, Thiery JP, Koteliansky VE. Expression of α1 integrin, a laminin-collagen receptor, during myogenesis and neurogenesis in the avian embryo. Development. 1992;116:585–600. doi: 10.1242/dev.116.3.585. [DOI] [PubMed] [Google Scholar]
- Edgar D, Timpl R, Thoenen H. The heparin-binding domain of laminin is responsible for its effects on neurite outgrowth and neuronal survival (1984) EMBO J. 1984;3:1463–1468. doi: 10.1002/j.1460-2075.1984.tb01997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egorov S, Yuryev A, Daraselia N. A simple and practical dictionary-based approach for identification of proteins in Medline abstracts. J. Am. Med. Inform. Assoc. 2004;11:174–178. doi: 10.1197/jamia.M1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison JA, Velier JJ, Spera P, Jonak ZL, Wang X, Barone FC, Feuerstein GZ. Osteopontin and its integrin receptor alpha (v) beta3 are upregulated during formation of the glial scar after focal stroke. Stroke. 1998;29:1698–1706. doi: 10.1161/01.str.29.8.1698. [DOI] [PubMed] [Google Scholar]
- Engler-Blum G, Meier M, Frank J, Miller GA. Reduction of background problems in nonradioactive Northern and Southern blot analysis enables higher sensitivity than 32P-based hybridization. Anal. Biochem. 1993;210:235–244. doi: 10.1006/abio.1993.1189. [DOI] [PubMed] [Google Scholar]
- Fujimoto T, Kawashima H, Tanaka T, Hirose M, Toyama-Sorimachi N, Matsuzawa Y, Miyasaka M. CD44 binds a chondroitin sulfate proteoglycan, aggrecan. Int. Immunol. 2001;13:359–366. doi: 10.1093/intimm/13.3.359. [DOI] [PubMed] [Google Scholar]
- Goldberg JL, Barres BA. The relationship between neuronal survival and regeneration. Annu. Rev. Neurosci. 2000;23:579–612. doi: 10.1146/annurev.neuro.23.1.579. [DOI] [PubMed] [Google Scholar]
- Goldberg JL, Espinosa JS, Xu Y, Davidson N, Kovacs GT, Barres BA. Retinal ganglion cells do not extend axons by default: promotion by neurotrophic signaling and electrical activity. Neuron. 2002a;33:689–702. doi: 10.1016/s0896-6273(02)00602-5. [DOI] [PubMed] [Google Scholar]
- Goldberg JL, Klassen MP, Hua Y, Barres BA. Amacrine-signaled loss of intrinsic axon growth ability by retinal ganglion cells. Science. 2002b;296:1860–1864. doi: 10.1126/science.1068428. [DOI] [PubMed] [Google Scholar]
- Grimpe B, Silver J. A novel DNA-enzyme reduces glycos-aminoglycan chains in the glial scar and allows microtransplanted DRG axons to regenerate beyond lesions in the spinal cord. J. Neurosci. 2004;24:1393–1397. doi: 10.1523/JNEUROSCI.4986-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimpe B, Dong S, Doller C, Temple K, Malouf AT, Silver J. The critical role of basement membrane independent laminin γ1 chain during axon regeneration in the CNS. J. Neurosci. 2002;22:3144–3160. doi: 10.1523/JNEUROSCI.22-08-03144.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimpe B, Pressman Y, Lupa MD, Horn KP, Bunge BM, Silver J. The role of proteoglycans in Schwann cell/astrocyte interactions and in regeneration failure at PNS/CNS interfaces. Mol. Cell. Neurosci. 2005;28:18–29. doi: 10.1016/j.mcn.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Hagg T, Muir D, Engvall E, Varon S, Manthorpe M. Laminin-like antigen in rat CNS neurons: distribution and changes upon brain injury and nerve growth factor treatment. Neuron. 1989;3:721–732. doi: 10.1016/0896-6273(89)90241-9. [DOI] [PubMed] [Google Scholar]
- Hagg T, Vahlsing HL, Manthorpe M, Varon S. Nerve growth factor infusion into the denervated adult rat hippocampal formation promotes its cholinergic reinnervation. J. Neurosci. 1990;10:3087–3092. doi: 10.1523/JNEUROSCI.10-09-03087.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Sun D, Rittling SR, Denhardt DT, Young W. Osteopontin-deficient mice exhibit less inflammation, greater tissue damage, and impaired locomotor recovery from spinal cord injury compared with wild-type controls. J. Neurosci. 2007;27:3603–3611. doi: 10.1523/JNEUROSCI.4805-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Mirza M, Weber GF. An osteopontin splice variant induces anchorage independence in human breast cancer cells. Oncogene. 2006;25:2192–2202. doi: 10.1038/sj.onc.1209248. [DOI] [PubMed] [Google Scholar]
- Hibino S, Shibuya M, Engbring JA, Mochizuki M, Nomizu M, Kleinman HK. Identificaiton of an active site on laminin alpha5 chain globular domain that binds to CD44 and inhibits malignancy. Cancer Res. 2004;64:4810–4816. doi: 10.1158/0008-5472.CAN-04-0129. [DOI] [PubMed] [Google Scholar]
- Hibino S, Shibuya M, Hoffman MP, Engbring JA, Hossain R, Mochizuki M, Kudoh S, Nomizu M, Kleinman HK. Laminin alpha5 chain metastasis- and angiogenesis-inhibiting peptide blocks fibroblast growth factor 2 activity by binding to the heparan sulfate chains of CD44. Cancer Res. 2005;65:10494–10501. doi: 10.1158/0008-5472.CAN-05-0314. [DOI] [PubMed] [Google Scholar]
- Hikita ST, Cann GM, Wingerd KL, Mullick LH, Wayne WC, Webb SW, Clegg DO. Integrin α4β1 (VLA-4) expression and activity in retinal and peripheral neurons. Mol. Cell. Neurosci. 2003;23:427–439. doi: 10.1016/s1044-7431(03)00065-4. [DOI] [PubMed] [Google Scholar]
- Hopkins J, Ford-Holevinski T, McCoy J, Agranoff B. Laminin and optic nerve regeneration in the goldfish. J. Neurosci. 1985;5:3030–3038. doi: 10.1523/JNEUROSCI.05-11-03030.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang D, Rust AG, Ramsey S, et al. A data integration methodology for systems biology. Proc. Natl Acad. Sci. USA. 2005;102:17296–17301. doi: 10.1073/pnas.0508647102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa H, Itota T, Nishitani Y, Tori Y, Inoue K, Sugimoto T. Osteopontin-immunoreactive primary sensory neurons in the rat spinal and trigeminal nervous system. Brain Res. 2000;863:276–281. doi: 10.1016/s0006-8993(00)02126-0. [DOI] [PubMed] [Google Scholar]
- Jander S, Bussini S, Neuen-Jacob E, Bosse F, Menge T, Muller HW, Stoll G. Osteopontin: a novel axon-regulated Schwann cell gene. J. Neurosci. Res. 2002;67:156–166. doi: 10.1002/jnr.10099. [DOI] [PubMed] [Google Scholar]
- Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science. 2005;310:1646–1653. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
- Jones LL, Liu Z, Shen J, Werner A, Kreutzberg GW, Raivich G. Regulation of the cell adhesion molecule CD44 after nerve transaction and direct trauma to the mouse brain. J. Comp. Neurol. 2000;426:468–492. doi: 10.1002/1096-9861(20001023)426:3<468::aid-cne9>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Ju WK, Kim KY, Cha JH, Kim IB, Lee MY, Oh SJ, Chung JW, Chun MH. Ganglion cells of the rat retina show osteopontin-like immunoreactivity. Brain Res. 2000;852:217–220. doi: 10.1016/s0006-8993(99)02140-x. [DOI] [PubMed] [Google Scholar]
- Kaaijk P, Pals ST, Morsink F, Bosch DA, Troost D. Differential expression of CD44 splice variants in the normal human central nervous system. J. Neuroimmunol. 1997;73:70–76. doi: 10.1016/s0165-5728(96)00167-1. [DOI] [PubMed] [Google Scholar]
- Kelley BP, Sharan R, Karp RM, Sittler T, Root DE, Stochwell BR, Ideker T. Conserved pathways within bacteria and yeast as revealed by global protein network alignment. Proc. Natl Acad. Sci. USA. 2003;100:11394–11399. doi: 10.1073/pnas.1534710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer T, Nguyen DH, Beuerman RW, Happel LT, England JD, Tiel RL, Kline DG. Painful neuromas: a potential role for a structural transmembrane protein, ankyrin G. J. Neurosurg. 2002;97:1424–1431. doi: 10.3171/jns.2002.97.6.1424. [DOI] [PubMed] [Google Scholar]
- Küry P, Zickler P, Stoll G, Hartung HP, Jander S. Osteopontin, a macrophage-derived matricellular gylcoprotein, inhibits axon outgrowth. FASEB J. 2005;19:398–400. doi: 10.1096/fj.04-1777fje. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Libby RT, Champliaud MF, Claudepierre T, Xu Y, Gibbons EP, Koch M, Burgeson RE, Hunter DD, Brunken WJ. Laminin expression in adult and developing retinae: evidence of two novel CNS laminins. J. Neurosci. 2000;20:6517–6528. doi: 10.1523/JNEUROSCI.20-17-06517.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesi P. Laminin-immunoreactive glia distinguish regenerative adult CNS systems from non-regenerative ones. EMBO J. 1985;4:2505–2511. doi: 10.1002/j.1460-2075.1985.tb03963.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesi P, Silver J. Is astrocyte laminin involved in axon guidance in the mammalian CNS? Dev. Biol. 1988;130:774–785. doi: 10.1016/0012-1606(88)90366-1. [DOI] [PubMed] [Google Scholar]
- Lin L, Cheung AW, Chan SO. Chiasmatic neurons in the ventral diencephalon of mouse embryos- changes in arrangement and heterogeneity in surface antigen expression. Brain Res. Dev. Brain Res. 2005;158:1–12. doi: 10.1016/j.devbrainres.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Liu BP, Fournier A, GrandPre T, Strittmatter SM. Myelin-associated glycoprotein as a functional ligand for the Nogo-66 receptor. Science. 2002;297:1190–1193. doi: 10.1126/science.1073031. [DOI] [PubMed] [Google Scholar]
- Lorber B, Berry M, Logan A. Lens injury stimulates adult mouse retinal ganglion cell axon regeneration via both macrophage- and lens-derived factors. Eur. J. Neurosci. 2005;21:2029–2034. doi: 10.1111/j.1460-9568.2005.04034.x. [DOI] [PubMed] [Google Scholar]
- Madsen JE, Hukkanen M, Aspenberg P, Polak J, Nordsletten L. Time-dependent sensory nerve ingrowth into a bone conduction chamber. Acta Orthop. Scand. 2000;71:74–79. doi: 10.1080/00016470052943946. [DOI] [PubMed] [Google Scholar]
- McKeon RJ, Schreiber RC, Rudge JS, Silver J. Reduction of neurite outgrowth in a model of glial scarring following CNS injury is correlated with the expression of inhibitory molecules on reactive astrocytes. J. Neurosci. 1991;11:3398–3411. doi: 10.1523/JNEUROSCI.11-11-03398.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKerracher L, Higuchi H. Targeting Rho to stimulate repair after spinal cord injury. J. Neurotrauma. 2006;23:309–317. doi: 10.1089/neu.2006.23.309. [DOI] [PubMed] [Google Scholar]
- McKerracher L, Chamoux M, Arregui CO. Role of laminin and integrin interaction in growth cone guidance. Mol. Neurobiol. 1996;12:95–116. doi: 10.1007/BF02740648. [DOI] [PubMed] [Google Scholar]
- McLoon SC, McLoon LK, Palm SI, Furcht L. Transient expression of laminin in the optic nerve of the developing rat. J. Neurosci. 1988;8:1981–1990. doi: 10.1523/JNEUROSCI.08-06-01981.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Franke A, Kaplan MR, Pfrieger FW, Barres BA. Characterization of the signaling interactions that promote the survival and growth of developing retinal ganglion cells in culture. Neuron. 1995;15:805–819. doi: 10.1016/0896-6273(95)90172-8. [DOI] [PubMed] [Google Scholar]
- Moon LD, Asher RA, Rhodes KE, Fawcett JW. Regeneration of CNS axons back to their target following treatment of adult rat brain with chondroitinase ABC. Nat. Neurosci. 2001;4:465–466. doi: 10.1038/87415. [DOI] [PubMed] [Google Scholar]
- Moretto G, Xu RY, Kim SU. CD44 expression in human astrocytes and oligodendrocytes in culture. Neuropathol. Exp. Neurol. 1993;52:419–423. doi: 10.1097/00005072-199307000-00009. [DOI] [PubMed] [Google Scholar]
- Morissette N, Carbonetto S. Laminin α2 chain (M chain) is found within the pathway of avian and murine retinal projections. J. Neurosci. 1995;15:8067–8082. doi: 10.1523/JNEUROSCI.15-12-08067.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikulina E, Tidwell JL, Dai HN, Bregman BS, Filbin MT. The phosphodiesterase inhibitor rolipram delivered after a spinal cord lesion promotes axonal regeneration and functional recovery. Proc. Natl Acad. Sci. USA. 2004;101:8786–8790. doi: 10.1073/pnas.0402595101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura S, Will AJ, Edwards DR, Heath JK, Hogan BL. Developmental expression of 2ar (osteopontin) and SPARC (osteonectin) RNA as revealed by in situ hybridization. J. Cell Biol. 1988;106:441–450. doi: 10.1083/jcb.106.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto I, Kawano Y, Matsumoto M, Suga M, Kaibuchi K, Ando M, Saya H. Regulated CD44 cleavage under the control of protein kinase C, calcium influx, and the Rho family of small G proteins. J. Biol. Chem. 1999;274:25525–25534. doi: 10.1074/jbc.274.36.25525. [DOI] [PubMed] [Google Scholar]
- Popovich PG. Immunological regulation of neuronal degeneration and regeneration in the injured spinal cord. Prog. Brain Res. 2000;128:43–58. doi: 10.1016/S0079-6123(00)28006-0. [DOI] [PubMed] [Google Scholar]
- Raivich G, Bohatschek M, Da Costa C, et al. The AP1 transcription factor c-Jun is required for efficient axonal regeneration. Neuron. 2004;43:57–67. doi: 10.1016/j.neuron.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Rodan GA. Osteopontin overview. Ann. N. Y. Acad. Sci. 1995;760:1–5. doi: 10.1111/j.1749-6632.1995.tb44614.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Sarthy PV, Fu M. Localization of laminin B1 mRNA in RGCs by in situ hybridization. J. Cell Biol. 1990;10:2099–2108. doi: 10.1083/jcb.110.6.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarthy V, Hoshi H, Mills S, Dudley VJ. Characterization of green fluorescent protein-expressing retinal ganglion cells in CD44-transgenic mice. Neuroscience. 2007;144:1087–1093. doi: 10.1016/j.neuroscience.2006.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell L, Schneider R, Kolbeck R, Barde YA, Schwab ME. Neurotrophin-3 enhances sprouting of corticospinal tract during development and after adult spinal cord lesion. Nature. 1994;367:170–173. doi: 10.1038/367170a0. [DOI] [PubMed] [Google Scholar]
- Schwab ME. Repairing the injured spinal cord. Science. 2002;295:1029–1031. doi: 10.1126/science.1067840. [DOI] [PubMed] [Google Scholar]
- Selvaraju R, Bernasconi L, Losberger C, et al. Osteopontin is upregulated during in vivo demyelination and remyelination and enhances myelin formation in vitro. Mol. Cell. Neurosci. 2004;25:707–721. doi: 10.1016/j.mcn.2003.12.014. [DOI] [PubMed] [Google Scholar]
- Sheehan KM, DeLott LB, West RA, Bonnema JD, DeHeer DH. Hyaluronic acid of high molecular weight inhibits proliferation and induces cell death in U937 macrophage cells. Life Sci. 2004;75:3087–3102. doi: 10.1016/j.lfs.2004.02.038. [DOI] [PubMed] [Google Scholar]
- Sherman LS, Rizvi TA, Karyala S, Ratner N. CD44 enhances neuregulin signaling by Schwann cells. J. Cell Biol. 2000;150:1071–1084. doi: 10.1083/jcb.150.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin T, Koh CS. Immunohistochemical detection of osteopontin in the spinal cord of mice with Theiler’s murine encephalomyelitis virus-induced demyelinating disease. Neurosci. Lett. 2004;356:72–74. doi: 10.1016/j.neulet.2003.11.023. [DOI] [PubMed] [Google Scholar]
- Shin SL, Cha JH, Chun MH, Chung JW, Lee MY. Expression of ostepontin mRNA in the adult rat brain. Neurosci. Lett. 1999;273:73–76. doi: 10.1016/s0304-3940(99)00516-9. [DOI] [PubMed] [Google Scholar]
- Skelton TP, Zeng C, Nocks A, Stamenkovic I. Glycosylation provides both stimulatory and inhibitory effects on cell surface and soluble CD44 binding to hyaluronan. J. Cell Biol. 1998;140:431–446. doi: 10.1083/jcb.140.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR, Crain SM, Reid LM. Laminin as a substrate for retinal axons in vitro. Dev. Brain Res. 1984;12:136–140. doi: 10.1016/0165-3806(84)90184-6. [DOI] [PubMed] [Google Scholar]
- Sonnenberg A, Linders CJT, Modderman PW, Damsky CH, Aumailley M, Timpl R. Integrin recognition of different cell binding fragments of laminin (P1, E3, E8) and evidence that α6β1 functions as a major receptor for fragment E8. J. Cell Biol. 1990;110:2145–2155. doi: 10.1083/jcb.110.6.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sretavan DW, Feng L, Pure E, Reichardt LF. embryonic neurons of the developing optic chiasm express L1 and CD44, cell surface molecules with opposite effects on retinal axon growth. Neuron. 1994;12:957–975. doi: 10.1016/0896-6273(94)90307-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein VM, Czub M, Schreiner N, Moore PF, Vandevelde M, Zurbriggen A, Tipold A. Microglial cell activation in demyelinating canine distemper lesions. J. Neuroimmunol. 2004;153:122–131. doi: 10.1016/j.jneuroim.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Stier H, Schlosshauser B. Axonal guidance in the chicken retina. Development. 1995;121:1443–1454. doi: 10.1242/dev.121.5.1443. [DOI] [PubMed] [Google Scholar]
- Sun W, Li N, He S. Large-scale morphological survey of mouse retinal ganglion cells. J. Comp. Neurol. 2002;451:115–126. doi: 10.1002/cne.10323. [DOI] [PubMed] [Google Scholar]
- Tessier-Lavigne M. Wiring the brain: the logic and molecular mechanisms of axon guidance and regeneration. Harvey Lect. 2002–2003;98:103–143. [PubMed] [Google Scholar]
- Tomaselli K, Hall DE, Flier LA, Gehlsen KR, Turner DC, Carbonetto S, Reichardt LF. A neuronal cell line (PC12) expresses two β1-class integrins—α1β1 and α3β1—that recognize different outgrowth-promoting domains in laminin. Neuron. 1990;5:651–662. doi: 10.1016/0896-6273(90)90219-6. [DOI] [PubMed] [Google Scholar]
- Tuohy TM, Wallingford N, Liu Y, Chan FH, Rizvi T, Xing R, Bebo B, Rao MS, Sherman LS. CD44 overexpression by oligodendrocytes: a novel model of inflammation-independent demyelination and dysmyelination. Glia. 2004;47:335–345. doi: 10.1002/glia.20042. [DOI] [PubMed] [Google Scholar]
- Weber GF, Ashkar S, Glimcher MJ, Cantor H. Receptor-ligand interaction between CD44 and osteopontin (Eta-1) Science. 1996;271:509–512. doi: 10.1126/science.271.5248.509. [DOI] [PubMed] [Google Scholar]
- Wu YJ, La Pierre DP, Wu J, Yee AJ, Yang BB. The interaction of versican with its binding partner. Cell Res. 2005;15:483–494. doi: 10.1038/sj.cr.7290318. [DOI] [PubMed] [Google Scholar]
- Yokosaki Y, Tanaka K, Higashikawa F, Yamashita K, Eboshida A. Distinct structural requirements for binding of the integrins alphavbeta6, alphavbeta3, alphavbeta5, alpha5beta1 and alpha9-beta1 to osteopontin. Matrix Biol. 2005;24:418–427. doi: 10.1016/j.matbio.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Zohar R, Suzuki N, Suzuki K, Arora P, Glogauer M, McCullock CA, Sodek J. Intracellular osteopontin is an integral component of the CD44-ERM complex involved in cell migration. J. Cell. Physiol. 2000;184:118–130. doi: 10.1002/(SICI)1097-4652(200007)184:1<118::AID-JCP13>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following supplementary material is available for this article online:
Table S1 The 20 most frequently cited proteins.
Table S2 Examples of proteins involved in apoptosis and anti-apoptotic processes.
Table S3 Examples of proteins involved in immunology and defense responses.
Table S4 Examples of proteins involved in signal transduction.
Table S5 Examples of proteins found in cytoskeleton and cytoplasm.
Table S6 Examples of proteins involved in DNA transcription and in the nucleus.
Table S7 Examples of proteins involved in development, neurite-formation and synaptogenesis.
This material is available as part of the online article from http://www.blackwell-synergy.com.