Abstract
Urothelial cancer has served as one of the most important sources of information about the mutational events that underlie the development of human solid maligancies. Although “field effects” that affect the entire bladder mucosa appear to initiate disease, tumors develop along two distinct biological “tracks” that present vastly different challenges for clinical management. Recent whole genome methodologies have facilitated even more rapid progress in the identification of the molecular mechanisms involved in bladder cancer initiation and progression. Specifically, whole organ mapping combined with high resolution, high throughput SNP analyses have identified a novel class of candidate tumor suppressors (“forerunner genes”) that localize near more familiar tumor suppressors but are disrupted at an earlier stage of cancer development. Furthermore, whole genome comparative genomic hybridization (CGH) and mRNA expression profiling have demonstrated that the two major subtypes of urothelial cancer (papillary/superficial and non-papillary/muscle-invasive) are truly distinct molecular entities, and in recent work our group has discovered that muscle-invasive tumors express molecular markers characteristic of a developmental process known as “epithelial-to-mesenchymal transition” (EMT). Emerging evidence indicates that urothelial cancers contain subpopulations of tumor-initiating cells (“cancer stem cells”) but the phenotypes of these cells in different tumors are heterogeneous, raising questions about whether or not the two major subtypes of cancer share a common precursor. This review will provide an overview of these new insights and discuss priorities for future investigation.
The two-track model for urothelial cancer progression
It is well recognized that human urothelial tumors develop along two major, largely independent but somewhat overlapping biological pathways, referred to as papillary and non-papillary or solid (1) (Figure 1). Papillary tumors originate from diffuse flat hyperplastic urothelial changes also referred to as low-grade intraurothelial neoplasia. They are typically of low histologic grade and grow as superficial non-invasive papillary protrusions. These tumors have a high propensity for recurrence but they practically never invade the bladder wall or metastasize. Non-papillary solid tumors evolve from severe dysplasia or carcinoma in situ (CIS) also termed high-grade intraurothelial neoplasia. The vast majority of high-grade non-papillary tumors develop in patients without a prior history of papillary tumors. High-grade solid carcinomas aggressively invade the bladder wall and may metastasize to regional lymph nodes and distant anatomic sites. Some of the low-grade superficial papillary tumors (10–15%) eventually progress to high-grade invasive carcinomas. Such progression is typically preceded by the development of CIS in the adjacent bladder mucosa or within the papillary tumor. Both types of cancer pose significant problems for public health, but for different reasons. Papillary tumors are not usually life threatening but because of their high recurrence rate, they contribute to bladder cancer’s rank as most expensive in terms of clinical management. Although about half of muscle-invasive tumors do initially respond to cisplatin-based combination chemotherapy regimens, the development of drug resistance is a major problem, and disease progression in resistant tumors is rapid and uniformly fatal. Major efforts are underway to prevent papillary tumor recurrence, to better define the biological basis of CIS and progression to muscle invasion in superficial tumors. The identification of molecular mechanisms that mediate invasion, metastasis, and resistance to therapy in muscle-invasive tumors is of prime importance for patients who present with aggressive forms of bladder cancer. It is expected that understanding the biology will lead to the identification of molecular and genetic markers that can be used to better assist in the clinical management of the disease.
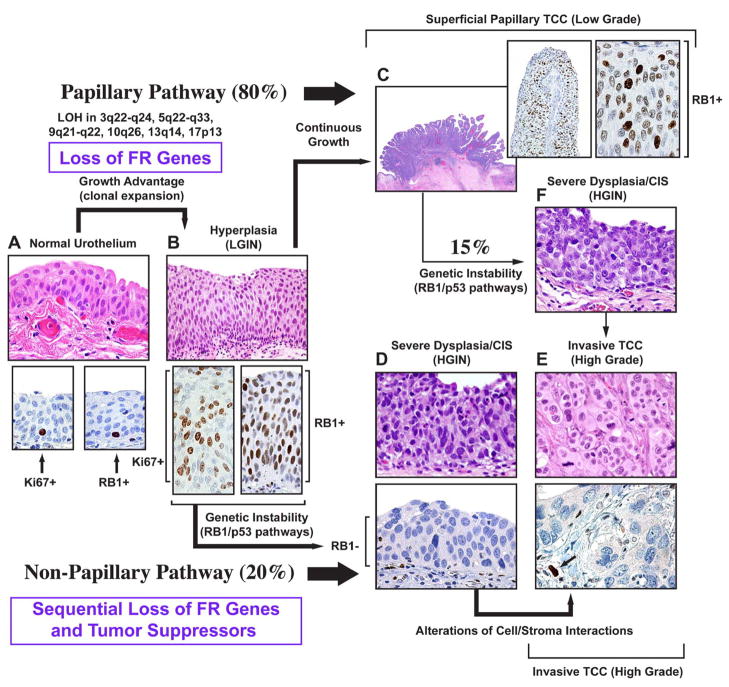
Figure 1. Dual-track concept of bladder carcinogenesis.
The expansion of a preneoplastic clone which shows minimal phenotypic deviation from the normal urothelium, is the incipient event in bladder carcinogenesis referred to as LGIN. In this phase, the loss of FR genes function provides growth advantage associated with the expansion of proliferating compartment. The proliferating cells expressing normal RB protein are seen in the entire thickness of LGIN. In contrast, normal urothelium contains only scattered proliferating cells expressing RB protein located in its basal layer. The continuous growth of LGIN leads to the development of low grade superficial papillary TCC. In the non-papillary pathway, clonal evolution results in the establishment of a successor clone with microscopic features of HGIN which often shows a loss of major tumor suppressors such as RB1 and has a high propensity for progression to an invasive high grade non papillary TCC. a, Normal urothelium (upper panel). Expression of Ki67 in proliferating basal cells of normal urothelium (lower panel, left). Expression of RB protein in peribasal cells of normal urothelium (lower panel, right). b, Urothelial hyperplasia with mild atypia referred to as LGIN (upper panel). Expression of Ki67 in the entire thickness of LGIN (lower panel, left); expression of RB protein in the entire thickness of LGIN (lower panel, right). c, Low-grade superficial TCC retaining normal expression of the RB protein: insets to c show low and high power photomicrographs illustrating the expression of normal RB protein in low grade papillary TCC. d, Severe intraurothelial dysplasia/carcinoma in situ (HGIN) (upper panel). Loss of RB protein expression in HGIN (lower panel). e, High-grade invasive nonpapillary carcinoma (upper panel). Loss of RB protein expression in high grade invasive nonpapillary TCC. Arrow shows expression of RB protein in endothelial cells adjacent to tumor which serve as an internal positive control (lower panel). f, Severe intraurothelial dysplasia/carcinoma in situ developing in bladder mucosa adjacent to a low-grade papillary tumor. It is responsible for switching the pathway and progression of some low-grade papillary tumors to high-grade invasive cancers. (Modified and reprinted with permission from T. Majewski et al. Lab Invest 88:694–721, 2008).
Field effects, forerunner genes, and the initiation of urothelial neoplasia
Epidemiological studies have clearly linked cigarette smoking to the risk of developing urothelial cancers. As is also true in the lung, tobacco carcinogens presumably cause DNA alterations throughout the entire bladder mucosa, not all of which are repaired, and one would therefore expect to observe some shared molecular alterations that contribute to most (if not all) cancers. Allelic loss on chromosome 9 is clearly one such event (2). Regions of LOH are located on both arms of the chromosome (3–8). The 9p region contains the p16/ARF locus, and studies have confirmed that p16 is frequently inactivated by LOH plus homozygous deletion or methylation independently of stage or grade in both superficial and muscle-invasive bladder cancers (9–11). The locus also contains the interferon-α (IFNα) locus (12). Given that IFNα can induce apoptosis in bladder cancer cells (13, 14), inactivation of the IFNα gene might also contribute to disease progression. Identification of the cancer-relevant genes at the 9q locus is still ongoing, but one interesting candidate is TSC1 (located at 9q.34) (15–17), which is an upstream negative regulator of the mTORC1 complex and downstream mTOR signaling (18). Overall, LOH at 9q remains one of the earliest events in bladder cancer progression identified to date (19).
Given that field effects appear to play such a crucial role in bladder cancer initiation, it seems likely that the earliest genetic alterations that mediate disease initiation would be present not just in the areas that most obviously involve cancer-related phenotypic changes but also throughout large regions of the urothelium that may be phenotypically normal. Bladder cancer progression appears to involve an accumulation of stereotyped phenotypic and molecular alterations in a manner that is similar to the more familiar progression model developed for colorectal cancers, where progression from adenoma to carcinoma involves accumulation of epigenetic and genetic damage (20). Early whole organ mapping studies demonstrated that urothelial cancers tend to be multi-focal and that regions of hyperplasia and/or atypia are found adjacent to areas containing frank cancer (21). Therefore, we have combined whole organ mapping with recombination-based marker analyses of chromosomes 1–22 to obtain a comprehensive map of the genomic alterations present in 5 cystectomy specimens (22–24). These studies identified 6 critical regions mapping to 3q22, 5q22-23, 9q21, 10q26, 13q14, and 17p13 that cooperate to drive the development of human bladder cancer. Most often patients displayed losses in 2–3 of these regions and about one fifth of cases displayed losses in all regions simultaneously.
We then used whole organ high-density SNP analyses to define the region surrounding the Rb gene (13q14) in more detail (Figure 2). The minimally deleted region associated with clonal expansion contained several putative open reading frames encoding proteins with previously uncharacterized functions (22, 23). We selected two of the genes – P2RY5 and ITM2B –for initial functional characterization in vitro. Both genes were downregulated at the mRNA level in over half of human bladder cancer cell lines in a relatively large panel. Typically this downregulation involved LOH of one allele and methylation of the other; for example, in cell lines the frequency of ITM2B methylation was approximately 40% (23). Reintroduction of either gene into bladder cancer cells that lacked it resulted in cell cycle arrest and apoptosis (23). Although the molecular mechanisms underlying these responses are still being investigated, ITM2B is homologous to the so-called BH3-only subfamily of BCL-2 proteins (25–28), and P2RY5 is a member of a subfamily of G protein-coupled receptors for lysophosphatic acid (LPA), and it has recently been implicated in genetic hair growth disorders (29–31).
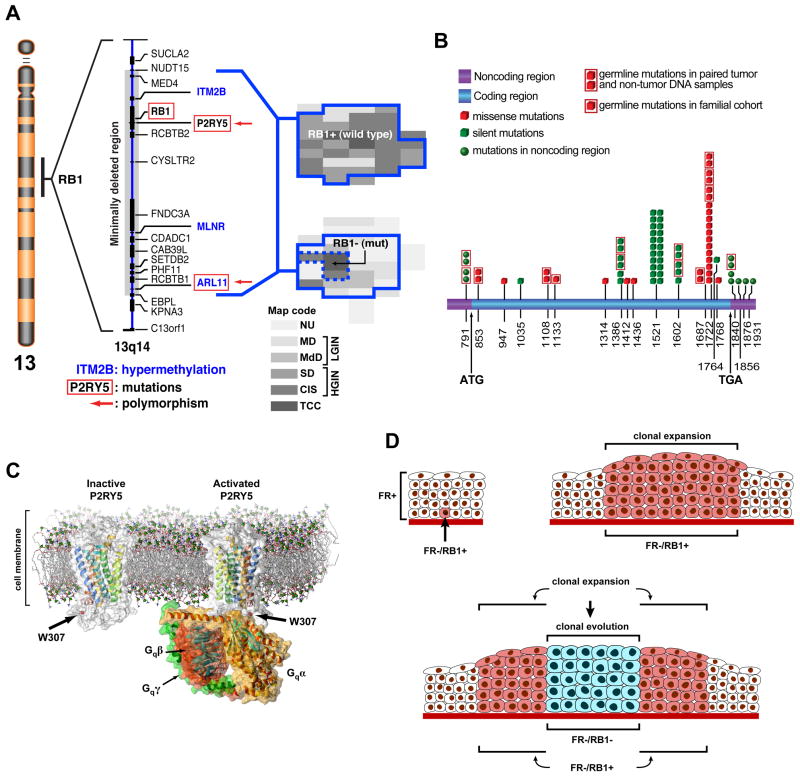
Figure 2. The concept of FR genes.
a, Chromosome 13, shown on the left, with the expanded 13q14 region flanking RB1 depicts a deleted segment associated with early expansion of in situ bladder neoplasia and containing candidate FR genes. The candidate FR genes inactivated by nucleotide substitutions (mutations or polymorphism) are depicted by red boxes. The candidate FR genes inactivated by hypermethylation are printed in blue. Histologic maps of human cystectomies with invasive bladder cancer are shown on the right. The continuous blue line depicts plaque like areas of clonal intraurothelial expansion associated with the loss of hetrozygosity within the minimal deleted region around RB1. The upper map shows a plaque-like clonal expansion involving almost the entire bladder mucosa with no inactiviation of the remaining RB1 allele. The lower map shows the inactivation of the remaining RB1 allele by a mutation restricted to an area of invasive cancer and adjacent HGIN depicted by a dashed blue line. This mutation involved codon 556 of exon 17 consisting of CGA→TGA and resulting in the change of Arg to a stop codon. b Summary of sequence analysis of P2RY5. The positions of nucleotide substitutions are shown on the full length mRNA. c, A model of inactive P2RY5 containing 7 transmembrane (H1–H7) and one cytoplasmic (H8) helix structures showing the position of polymorphism in codon 307 located within the cystoplasmic domain of the protein (left diagram) that may affect its interaction with the Gαβγ trimeric protein complex (right diagram). d, Expansion of intraurothelial neoplasia and its clonal evolution into carcinoma in situ. Inactivation of the FR genes (FR−) in a parabasal urothelial tumor initiating cell with the retention of normal RB1 expression (RB1+) (upper left panel). Clonal expansion of the FR−/RB1+ cell shown in a exhibiting minimal deviation from a normal phenotype. Typically in this phase only an increased number of urothelial cell layers can be identified in the urothelium. (upper right panel) Clonal evolution into carcinoma in situ associated with loss of RB1 function (RB1−). (bottom panel) (a, b, and c modified and reprinted with permission from S. Lee et al PNAS 104(34):13732–13737, 2007).
Interestingly, another group identified a SNP within this same genomic locus (rs2227311) that is predicted to downregulate P2RY5 gene expression and is a risk factor for developing invasive ovarian cancer (32). Similarly, a polymorphic site (G1722T) located within the P2RY5 coding sequence was detected in several bladder tumors and non-tumor DNA from the same patient. This form of the protein was defective in promoting apoptosis in vitro (22). Strikingly, in a large case-control study, 100% of smokers carrying this allele developed bladder cancer (23). We are currently characterizing the functions of several other candidate “forerunner genes” near the Rb locus that display frequent downregulation in cell lines and primary tumors, including ARL11 and GPR38.
Insights into molecular basis for the two tracks of urothelial cancer progression
Recent studies using transgenic mouse models have provided important insights into the molecular mechanisms that give rise to the two distinct cancer phenotypes observed in patients with urothelial tumors. Using a bladder-specific (uroplakin) promoter, Xue-Ru Wu’s group produced two lines of mice that closely recapitulate the two major types of cancer observed in humans. In the first, bladder-specific expression of mutant active H-ras led to the development of bladder hyperplasia/dysplasia or papillary tumors in a manner that was related to gene dosage (i.e., high copy numbers induced tumors) (33, 34). When the same promoter was used to drive expression of the SV40 large T antigen (thereby inactivating the p53 and Rb tumor suppressors), mice bearing low copy numbers developed CIS, whereas mice bearing high copy numbers developed muscle-invasive tumors with 100% penetrance (35). Analysis of the low copy number SV40 mice suggested that complete loss of p53 was associated with the development of CIS but that progression required additional events, presumably including loss of Rb function (36). Likewise, complete loss of p53 on the mutant H-ras background accelerated the development of both low-grade and high-grade superficial tumors but did not support the development of muscle-invasive disease (37). Again, additional genetic events must be present in the high copy number SV40T mice that drive progression to muscle invasion.
More recent work strongly suggests that one candidate driver of the muscle-invasive phenotype is activation of the PTEN/PI3 kinase/AKT/mTOR pathway. Abate-Shen’s group used mice bearing “floxed” p53 and PTEN genes and intravesical instillation of adenoviral Cre to conditionally eliminate the expression of these tumor suppressors in the mouse bladder (38). These mice developed early CIS that uniformly progressed to muscle-invasive and metastatic tumors. Similarly, elimination of p53 and PTEN function in human urothelial cells also made them tumorigenic, and the group’s own analysis of primary tumors from patients confirmed that PTEN disruption and AKT pathway activation are common features of muscle-invasive disease (38), consistent with previous studies (see below). The authors noted that mTOR, a downstream mediator of AKT-dependent signaling that is being evaluated as a potential therapeutic target in cancer (18), was activated in the mouse and human cancer cells (38), supporting the notion that mTOR inhibitors should be evaluated aggressively in relevant preclinical models and in human clinical trials. Curiously, another group used a different approach to induce tissue-specific loss of PTEN (in a wild-type p53 background) in the urogenital system in the mouse and showed that this resulted in the emergence of prostate but not bladder cancers, effects that were linked to differential activation of AKT (prostate-specific) versus p21 (bladder-specific)(39). Therefore, loss of p53 pathway function may be an obligate cofactor for PTEN-dependent bladder cancer initiation in the mouse.
The results obtained in these mouse models appear to largely recapitulate what appears to happen in human superficial and muscle-invasive tumors, albeit with some caveats. For example, in one cohort of 67 tumors activating codon 12 Hras mutations were detected in almost half (40). These mutations were more common in high-grade (II or III) tumors, and 7 contained synchronous mutations at position 2719 of Hras intron D (40). This phenotype mirrored the mutations in Hras that were previously detected in the human T24 cell line, and in these tumors overall levels of Hras protein were 10-fold higher than were present in tumors that lacked the intron D mutation (40). However, in a subsequent study the frequency of activating Hras mutations was much lower (41), and an even more recent study of 35 consecutive Iranian urothelial tumors failed to identify mutations in Hras, Kras, or Nras in any of them (42), suggesting that HRas mutational frequencies vary significantly in different populations. Together, these results support the conclusion generated in the murine Hras transgenic model that Ras gene dosage is an important determinant of more aggressive biological behavior (33). However, the data do not address the issue of whether mutational activation of Hras drives the formation of superficial as opposed to muscle-invasive cancer. (Our own gene expression profiling data indicate that the T24 cell line co-clusters with muscle-invasive tumors.)
Instead, other recent work strongly supports the idea that Hras pathway activation in human papillary tumors is mediated via autocrine growth factor receptor activation. Activating mutations in the type 3 receptor for fibroblast growth factors (FGFR3) occur frequently (≥ 50%) in superficial but more rarely (≤ 20%) in muscle-invasive tumors (43–45)(P. Black et al, manuscript in preparation). The most common is a codon 7 mutation (S249C) that forms disulfide-linked homodimers that exhibit ligand-independent signaling activity (46). This mutation is found in some of the available established cell lines (i.e., UM-UC-14) and in these cells FGFR3 signaling is required for proliferation (47)(P. Black et al, manuscript in preparation). Interestingly, FGFR3 levels are elevated in superficial tumors as compared with muscle-invasive tumors overall, and a significant number of human bladder cancer cell lines that express wild-type FGFR3 are also dependent upon FGFR3 signaling for their growth (47)(P. Black et al, manuscript in preparation). Therefore, the fraction of superficial tumors that is dependent on FGFR3 signaling is probably higher than the proportion of tumors that expresses mutant FGFR3. Identifying molecular markers that can prospectively identify these tumors and targeting them with antibodies or small molecules is a high priority for clinical translation.
In the preclinical models, loss of p53 promotes CIS and contributes to progression to muscle-invasive disease (35, 38). Similarly, early work established that LOH on 17p distinguished high grade from low grade cancers (2), and many other studies have demonstrated that mutational inactivation of p53 or p53 pathway disruption (via loss of its downstream target, p21) appears to contribute to poor clinical outcome in both organ-confined and muscle-invasive disease (48–50), although as a prognostic marker its effects are stronger in lower-stage tumors (T1–T2, N0) (51, 52). An interesting exception may be found in tumors that contain exon 5 p53 mutations; in one study clinical outcomes in this group were similar to those observed in patients whose tumors expressed wild-type p53 (53).
Similarly, the phenotype of the uroplakin-SV40T transgenic mice strongly suggests that loss of Rb function also contributes to CIS and muscle invasive-disease (35). In patient tumors Rb pathway disruption occurs frequently due to mutational inactivation of Rb itself or of its upstream regulator, p16 (11, 54, 55). Predictably, loss of Rb cooperates with p53 to promote tumor recurrence and patient mortality (55). Precisely how loss of p53 and Rb promote an invasive and metastatic phenotype is not clear, since their best charactertized effects are linked to the cell cycle (and hence would be expected to be more tightly linked to grade rather than stage).
Finally, emerging data confirm that activation of the PI3 kinase/AKT/mTOR pathway is an important event in cancer progression in patients. As discussed above, loss-of-heterozygosity (LOH) involving regions on both 9p (containing the p16 locus) and 9q is extremely common in both superficial and muscle-invasive bladder cancer (56). Although a conclusive definition of the tumor suppressor(s) that are localized to this region is still lacking, the TSC1 locus is one very attractive candidate (15, 16). Interestingly, mutational inactivation of the other TSC1 allele is not a particularly common event, even in tumor subsets selected for LOH at 9q (15, 16). Therefore, it is possible that some other event suppresses TSC1 protein expression or there is a gene dosage effect such that haploinsufficiency rather than complete loss-of-function is important. Downregulation of TSC1 expression should induce downstream RAPTOR-dependent mTOR activation, consistent with the observation that mTOR is active in a subset of human tumors (38). Determining whether or not there is a direct link between downregulation of TSC1 and mTOR activation will require additional investigation.
On the other hand, and more consistent with predictions made by the preclinical models, LOH within the PTEN locus on chromosome 10 appears to be much more common in muscle-invasive as compared with superficial tumors (57–61), and a genetic signature of PTEN loss predicts a poor clinical outcome (62). PTEN clearly does regulate invasion (63), providing a molecular explanation for why loss of PTEN collaborates with p53 to promote a muscle-invasive phenotype in the preclinical models (34, 38). Other pathway defects, including activation of PI3 kinase (64) and/or AKT1 (65) may also play important roles, but their relationships to CIS and the muscle-invasive phenotypes are not as clear.
Finally, we have launched an ambitious effort to use whole genome expression profiling to directly determine whether the available preclinical models (cell lines, xenograft tumors, and transgenics) accurately recapitulate the biology of human urothelial cancers. Cross-species comparisons of gene expression confirm that the papillary tumors that arise in the Hras transgenics are highly similar to superficial (Ta and T1) lesions in patients in 3 independent datasets (66, 67)(Woonyoung Choi, manuscript in preparation). Likewise, the muscle-invasive tumors that develop in the uroplakin SV40T mouse co-cluster with muscle-invasive (T3 and T4) tumors in patients (Woonyoung Choi, manuscript in preparation). Interestingly, even the available human cell lines growing in two-dimensional culture on plastic display remarkable similarities to the two subtypes of cancer, consistent with recent gene expression profiling and whole genome CGH studies performed by Gray’s group in large panels of human breast cancer cell lines (68). In ongoing studies we are evaluating how three-dimensional culture and growth at various sites in nude mice (subcutaneous versus orthotopic) affect the similarities in gene expression between the human cell lines and primary tumors. However, the preliminary results confirm the previous conclusions based on purely histological analyses that these two transgenic mouse models are excellent surrogates for human cancer.
Epithelial-to-Mesenchymal transition and sarcomatoid transformation
Virtually all malignant epithelial neoplasms, including those arising in the bladder, may loose their original epithelial phenotype and change into mesenchymal sarcoma-like tumors during their progression. In conventional pathology, this phenomenon is referred to as sarcomatoid transformation and it was originally described over a century ago. Interestingly, the undifferentiated spindle cell sarcoma-like component shows further instability manifested by a phenotypic switch to specialized mesenchymal lineages most often osteoblastic and chondroblastic. These tumors are referred to in diagnostic pathology as sarcomatoid carcinomas and have complex morphology with juxtaposition of epithelial and mesenchymal elements. Sarcomatoid carcinomas are typically highly aggressive lethal tumors that are resistant to currently known treatment protocols. On the molecular level, they are characterized by partial or complete loss of epithelium/urothelium specific genes, complex chromosomal abnormalities and pronounced aneuploidy. Recent data show that the transition to a mesenchymal phenotype, the so called epithelial-to-mesenchmal transition (EMT), precedes the progression to microscopically recognizable sarcomatoid transformation and occurs in a subset of conventional carcinomas which retain epithelial phenotype. Currently available data support the hypothetical sequence of progression from epithelial phenotype via mesenchymal transition to sarcomatoid transformation. (Figure 3)
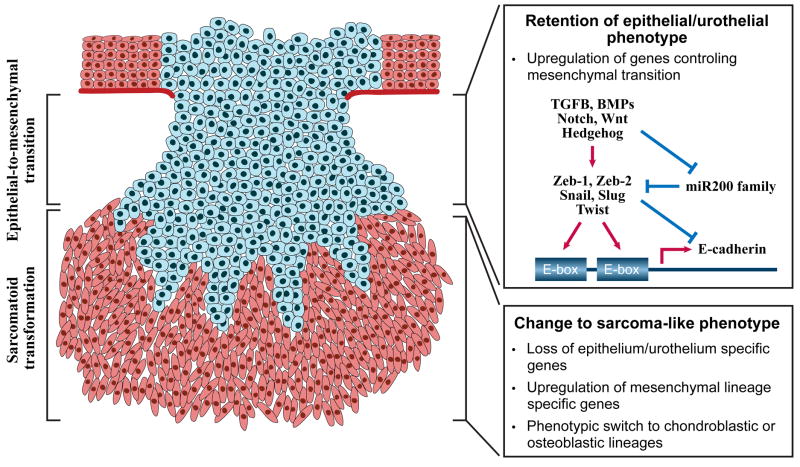
Figure 3. Molecular regulation of epithelial-to-mesenchymal transition and sarcomatoid transformation.
Some conventional bladder carcinomas develop a gene expression profile characteristic of EMT but microscopically they retain a full epithelial/urothelial phenotype. Further progression to sarcomatoid transformation is associated with the partial or complete loss of epithelial phenotype and the development of mesenchymal sarcoma-type features. The hallmark feature of EMT is a loss of the homotypic adhesion molecule, E-cadherin, which is the canonical marker of an “epithelial” phenotype. E-cadherin transcription is controlled by two E-box elements located within its promoter. EMT can be induced by a variety of developmental signals, but transforming growth factor-beta (TGFb) is the best-studied stimulus. TGFβ upregulates several E-cadherin repressors (Zeb-1, Zeb-2, Snail, Twist) that inhibit E-cadherin expression by recruiting histone deacetylases (HDACs) to the E-box elements in its promoter. In addition, TGFβ downregulates expression of the miR200 family of micro-RNAs. The miR200 family interacts directly with the mRNAs encoding Zeb-1 and Zeb-2, thereby blocking their translation and promoting their degradation. In urothelial cancers expression of ΔN-p63 correlates directly with E-cadherin expression and inversely with expression of Zeb-1 and Zeb-2. Superficial urothelial tumors almost invariably display an “epithelial” phenotype, whereas muscle-invasive tumors are heterogeneous and approximately evenly divided between “epithelial” and “mesenchymal” phenotypes. Change to sarcoma-type phenotype is relatively rare and occurs in less than 10% of high grade invasive bladder cancers. Such tumors are characterized by complex chromosomal abnormalities, pronounced aneuploidy and a high degree of clinical aggressiveness.
EMT process is not cancer specific and plays an important role in tissue differentiation, organ development and wound healing (69). It is characterized by downregulation of the homotypic adhesion molecule, E-cadherin, and proteins involved in cell polarity, with parallel upregulation of fibronectin, vimentin, certain integrins, matrix metalloproteases (MMPs), and several transcriptional repressors of E-cadherin expression (Twist, Snail, Slug, Zeb-1, and Zeb-2) (69)(Figure 3). Members of the TGFβ/bone morophogenic protein (BMP) family of cytokines are the best-characterized inducers of EMT, although many inflammatory cytokines (and their transcriptional target, NFκB) and developmental signaling systems (Sonic hedgehog, Notch, and Wnt) play central roles in regulating EMT as well (69).
There is also accumulating evidence that EMT plays important roles in cancer progression and metastasis (69). E-cadherin is a metastasis suppressor in preclinical models (70) and is commonly downregulated in metastatic tumors in patients (71–73). Conversely, metastatic tumors often upregulate expression of MMPs, leading to increased invasion, migration, and angiogenesis (71, 74). Weinberg’s group recently catalyzed a resurgence in interest in the role of EMT in cancer when they showed by gene expression profiling that Twist was directly involved in metastasis in a murine breast cancer model (75, 76). Since then, the group has also defended a role for EMT in the cancer “stem cell” phenotype (77). Several years ago we performed a study to determine the significance of E-cadherin downregulation and MMP upregulation in patients with urothelial cancer (78, 79). We used immunohistochemistry to measure E-cadherin and MMP-9 expression and found that the ratio of E-cadherin to MMP9 expression predicted poor disease-specific survival in patients (78), providing the first suggestion that EMT might play a central role in progression.
We renewed our EMT studies as a result of our longstanding interest in the effects of inhibitors of the epidermal growth factor receptor (EGFR) in preclinical bladder cancer models. We developed an orthotopic (intra-bladder) xenograft model of human bladder cancer by implanting the 253J cell line into the bladder wall and “recycling” the tumor that formed 5x through the orthotopic site to enhance its tumorigenicity and invasive potential (80). The resulting model (253J B-V) is highly sensitive to small molecule EGFR inhibitors such as cetuximab and gefitinib in vitro and in vivo (81). However, over time it became clear to us that human bladder cancer cell lines display marked heterogeneity in their sensitivities to EGFR inhibition (82). To gain a better understanding of the molecular determinants of sensitivity and resistance, we screened a relatively large panel of cell lines (n = 20) for responsiveness to the EGFR inhibitors gefitinib (Iressa) and cetuximab (C225). We identified a subset of the lines (approximately 30%) that displayed strong growth inhibition when they were exposed to either agent, while the remainder of the lines was highly resistant (82, 83).
We then performed whole genome expression profiling on the lines and discovered that they naturally formed two distinct groups by unsupervised clustering (Woonyoung Choi, unpublished observations). Strikingly, all of the EGFR-sensitive lines were contained within one cluster. Analysis of the pathways that were active in the cells within this cluster revealed that they expressed high levels of E-cadherin and other “epithelial” markers (including the p53 homolog, p63) and low levels of Zeb-1, Zeb-2, and vimentin ((82, 83) and Aditi Das et al, manuscript in preparation) (Figure 3). Furthermore, knockdown of E-cadherin in the EGFR inhibitor-sensitive cells made them resistant to the drugs (83), demonstrating a cause-effect relationship between the two. Importantly, we also screened our cell lines for responsiveness to an FGFR3-selective small molecule inhibitor and discovered that all of the lines that were sensitive were resistant to Iressa but nonetheless co-clustered with the other “epithelial” lines in our panel (A. Kwan, I-L. Lee, unpublished observations). The implication of EMT in resistance to EGFR inhibitors in bladder cancer is consistent with parallel results obtained in gastrointestinal tumors and lung and head and neck cancers (84–87).
More recently we identified one of the important molecular mechanisms involved in maintaining an “epithelial” phenotype in bladder cancer cells (88). We compared whole genome miRNA expression patterns in one representative “epithelial” line (UM-UC-5) and one “mesenchymal” cell (KU7). Members of the miR200 family of micro RNAs clearly stood out as being differentially expressed in the lines, levels being high in the UM-UC-5 cells and low to undetectable in KU7 (88). An expansion of the analyses confirmed that all of the “epithelial” lines expressed high levels of miR200 family members, whereas all of the “mesenchymal” lines expressed low levels. Strikingly, reintroduction of miR200c into a representative “mesenchymal” line (UM-UC-3) reduced expression of Zeb-1 and Zeb-2, restored E-cadherin, and rendered the cells sensitive to cetuximab (88). In ongoing work we are identifying the upstream regulators that maintain the differential regulation of miR200 family expression in the “epithelial” and “mesenchymal” lines and the downstream targets of the family (including Zeb-1 and Zeb-2) that maintain EGFR inhibitor sensitivity. Our results are consistent with a wealth of emerging data implicating the miR200 family in suppression of Zeb-1/2 expression and the maintenance of the “epithelial” phenotype (89–91).
Given our historical interest in the potential role of E-cadherin in metastasis suppression, we next undertook studies to determine the relationship between EMT and the muscle-invasive phenotype in urothelial cancers (Woonyoung Choi et al, manuscript in preparation). Our analyses of previously published human gene expression profiling datasets confirmed that E-cadherin levels were higher in superficial (Ta and T1) tumors as compared to muscle-invasive (T2-4) tumors, and the opposite was true for Zeb-1 and Zeb-2. We then performed whole genome expression profiling and real-time PCR quantification of EMT-relevant markers (E-cadherin, p63, Zeb-1, Zeb-2, vimentin, MMP2, and MM9) in a new set of 103 of our own tumors for which outstanding clinical follow-up data were available. We included p63 in these analyses because it correlated closely with the “epithelial” phenotype in our cell lines, and two other groups showed that p63 tends to be downregulated in muscle-invasive disease (92–95). Our data confirmed that E-cadherin and p63 levels were decreased and Zeb-1, Zeb-2, vimentin, MMP2, and MMP9 levels were increased in the muscle-invasive tumors, consistent with the idea that EMT promotes a more invasive and metastatic phenotype. Strikingly, however, even though p63 levels were higher in Ta (superficial) tumors, we also discovered that patients with muscle-invasive tumors that expressed high levels of p63 displayed substantially reduced disease-specific survival (Woonyoung Choi et al, manuscript in preparation). Therefore, even though essentially all of the tumors that expressed Zeb-1 and Zeb-2 were muscle-invasive, tumors that displayed a more “hybrid” phenotype (muscle-invasive with retention of the “epithelial” marker, p63) appeared to possess “bad” biology. Whether or not this is related to the growing interest in p63 as a potential mediator of the epithelial “stem cell” phenotype (96–98) awaits further investigation. In addition, whether other markers of an “epithelial” phenotype (i.e., E-cadherin and miR200 family members) also identify muscle-invasive tumors with “bad” biology also requires additional experimentation.
Urothelial cancer stem cells and the two tracks of progression
Analysis of the processes involved in the development of the various cell lineages within the hematopoietic system demonstrated that they are all derived from a common progenitor, termed the hematopoietic stem cell (99). Similarly, work performed over the last 15 years strongly suggests that human tumors also contain subsets of highly tumorigenic cells that can give rise to all of the molecular heterogeneity observed in the whole tumor (99–101). Experimentally, cancer stem cells are isolated from bulk populations by sorting them on the basis of their expression of specific surface markers (99, 100, 102) and/or with functional assays (“Aldefluor” or “side population” assays, for example) (103). Like their normal counterparts, cancer stem cell biology appears to be tightly controlled via surface interactions with the other (stromal) cells and matrix proteins that are present within their “niche”(99, 101). Among these, paracrine signaling pathways that play important roles in embryonic development (Notch, Wnt, Hedgehog, BMPs/TGFβ) and integrin-matrix interactions appear to be particularly important (101).
Cells within the normal urothelium are spatially organized in a manner that correlates well with their level of differentiation (Figure 4). The single layer of basal cells located directly adjacent to the basement membrane are the least well-differentiated and they can be identified by their expression of certain cytokeratins (CK5, CK17), lack of expression of others (CK18, CK20)(104). They also express surface molecules that enable them to interact with matrix proteins (the 67 kD laminin receptor and beta integrins)(104, 105) and hyaluronic acid (CD44)(106). When pulsed with BrdU, these cells can retain label in rodents for at least a year (104), indicating that they have self-renewal capacity. Adjacent to the basal cells is a multicellular layer of “intermediate” cells that express variable levels of CD44 and CK5 and high levels of CK18 (107). The proliferative potential of these cells is more limited (108), and they resemble the partially committed, “transit amplifying” cells that have been described in other lineages (99). Finally, lining the surface is a layer of terminally differentiated “umbrella” cells that express uroplakins and cytokeratin-20 (CK20) (107). Studies in rodents have identified an essential role for ΔN-p63 in the normal development of surface epithelia, including the urothelium (109–112). Although its precise role in the maintenance of stem cell biology is still unclear, strong evidence has been advanced suggesting that it inhibits apoptosis by antagonizing p53 function (112).
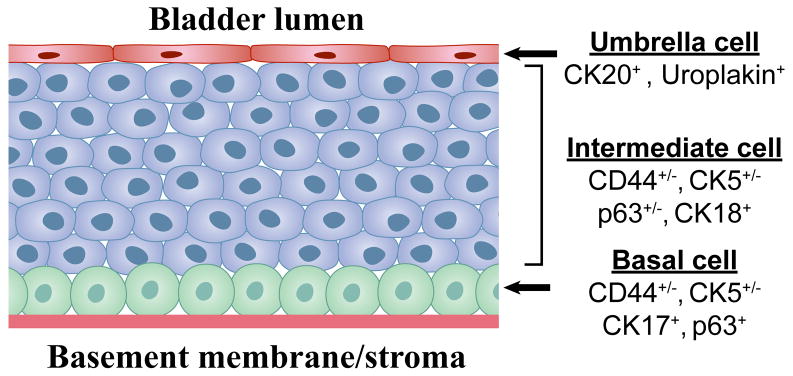
Figure 4. Differentiation-based lineage hierarchy in the normal urothelium.
The normal urothelium is comprised of 3 distinct layers. The surface epithelium consists of a single layer of terminally differentiated urothelial cells (“umbrella cells”) that express uroplakins and cytokeratin-20 (CK20) and have a low proliferative potential. It is conceivable that papillomas arise from transformation of umbrella cells, but this would require that they “de-differentiate” to reacquire self-renewal potential. Below the umbrella cells is a multi-cellular layer of intermediate cells that express CK18 and variable levels of p63, CK5, and CD44. These are most likely “transit-amplifying cells” that possess higher proliferative potential but have also begun to differentiate towards the umbrella cell phenotype. Finally, adjacent to the basement membrane is a layer of basal cells that express high levels of CK5, CD44, p63 and the 67 kD laminin receptor. This layer contains urothelial stem cells, which are tightly regulated by stromal elements (cells and matrix proteins) contained within the “niche”. Recent studies indicate that urothelial cancer stem cells share several molecular markers with the normal basal cell, including CD44, CK5, and the 67 kD laminin receptor and that these markers are upregulated at the tumor-stromal interface in xenografts.
There is currently strong interest in identifying urothelial cancer stem cells and characterizing their roles in bladder cancer progression and response to therapy. Two groups recently reported the isolation of candidate urothelial cancer stem cells from conventional and primary human xenografts (105, 106). In both cases the urothelial cancer stem cells resembled normal basal cells in terms of their expression of CK5, CK17, CD44, and the 67kD laminin receptor (105, 106). Furthermore, expression of basal markers was enriched at the edges of conventional SW780 xenografts (105), strongly suggesting that tumor-stromal interactions are critically involved in the phenotype (107). However, although most of the isolates shared certain common features (i.e., expression of the Hedgehog pathway target Gli1), analyses of stem cells from primary tumors suggested the presence of substantial inter-tumoral molecular heterogeneity (106), the origin of which is still unclear. Furthermore, whether the two “tracks” of cancer are driven by different oncogenic events (FGFR3 mutation versus p53/Rb/PTEN loss) that occur within the same target cell (presumably the basal cell), or by different oncogenic events that target different progenitors (basal versus intermediate cell)(107) remains unclear.
As introduced above, other recent work suggests that cancer stem cells possess features that are reminiscent of EMT (77). Therefore, we initiated a series of experiments to examine the relationship between the strong baseline EMT signature exhibited by our human urothelial cancer cells, their expression of stem cell markers, and their behavior in functional assays that measure “stemness” (M. Tran et al, manuscript submitted). Strikingly, the p63-positive “epithelial” lines within our panel express high levels of the markers that identify the normal basal and intermediate cells (CD44, CK5, CK17, and CK18), suggesting that EMT does not clearly correlate with “stemness” in bladder cancer cells. On the other hand, functional assays confirmed that the “mesenchymal” lines grow more readily under anchorage-independent conditions (stem cell “sphere” and soft agar assays), suggesting to us that the heterogeneity in stem cell phenotype might be linked to EMT. As discussed above, muscle-invasive tumors in patients are heterogeneous with respect to their expression of ΔN-p63 and EMT markers, and it is possible that this heterogeneity stems from differences in cell of origin and/or the transforming events that target this cell.
Conclusions and future directions
In this review we have highlighted the importance of developments in two areas (forerunner genes and EMT) as they relate to our current understanding of urothelial cancer initiation and progression. We have also attempted to provide some context with respect to other recent developments in the field, including insights into how differences in molecular genetics (FGFR3 mutations versus p53/Rb/PTEN/mTOR pathway alterations) might mediate the progression along the two independent tracks (papillary and non-papillary) of progression. There are a number of other exciting areas of research that we have not discussed in detail, including the roles of bladder cancer “stem” cells, epigenetic changes (chromatin and histone methylation/acetylation) and differential micro RNA expression (aside from the miR200 family) in cancer progression. These areas are no less important, and readers are encouraged to turn to other sources for a more complete discussion of their importance.
On the other hand, the emerging data on the roles of specific chromosomal regions (9p/q and the forerunner loci) in bladder cancer initiation raises a number of obvious questions for future research. We still do not have a comprehensive list of all of the relevant genes that are affected within each locus, or how they may potentially interact in a coordinated manner within particular biological pathways. For the somatic (as opposed to germline) changes we also do not know what caused LOH in the first place. Are these sites of chromosomal fragility and/or hypersensitivity to tobacco carcinogens and/or other agents? Finally, with respect to the forerunner genes, their biological functions need to be determined in more comprehensive studies. The observation that the first two selected for analysis (ITM2B and P2RY5) have important established physiological functions raises confidence that the others may also have important biological effects in urothelial cells.
Finally, although we were able to confirm our hypothesis that EMT is associated with the muscle-invasive “track” of urothelial cancer, we were somewhat surprised to find that the epithelial marker p63 identified the most aggressive muscle-invasive tumors. In ongoing studies we are performing functional studies to determine the potential involvement of p63 in the maintenance of bladder cancer “stem cells” and the role of EMT in the responses of cells to the platinum-based conventional therapies used in patients with muscle-invasive disease. We are also using our gene expression profiling data to determine how the muscle-invasive, p63-positive tumors can be distinguished from the papillary, p63-positive Ta lesions. Because all of the cell lines that respond to either EGFR or FGFR inhibitors fall within the “epithelial” cell line cluster, it will be important to identify markers that distinguish the EGFR- from the FGFR-dependent lines. Ultimately we are confident that by obtaining a better understanding of the biological basis of the two-track model of bladder cancer progression we will be able to develop strategies to optimize effective therapies for patients with both types of disease.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dinney CP, McConkey DJ, Millikan RE, et al. Focus on bladder cancer. Cancer Cell. 2004;6:111–6. doi: 10.1016/j.ccr.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Olumi AF, Tsai YC, Nichols PW, et al. Allelic loss of chromosome 17p distinguishes high grade from low grade transitional cell carcinomas of the bladder. Cancer Res. 1990;50:7081–3. [PubMed] [Google Scholar]
- 3.Tsai YC, Nichols PW, Hiti AL, Williams Z, Skinner DG, Jones PA. Allelic losses of chromosomes 9, 11, and 17 in human bladder cancer. Cancer Res. 1990;50:44–7. [PubMed] [Google Scholar]
- 4.Ruppert JM, Tokino K, Sidransky D. Evidence for two bladder cancer suppressor loci on human chromosome 9. Cancer Res. 1993;53:5093–5. [PubMed] [Google Scholar]
- 5.Miyao N, Tsai YC, Lerner SP, et al. Role of chromosome 9 in human bladder cancer. Cancer Res. 1993;53:4066–70. [PubMed] [Google Scholar]
- 6.Cairns P, Shaw ME, Knowles MA. Preliminary mapping of the deleted region of chromosome 9 in bladder cancer. Cancer Res. 1993;53:1230–2. [PubMed] [Google Scholar]
- 7.Keen AJ, Knowles MA. Definition of two regions of deletion on chromosome 9 in carcinoma of the bladder. Oncogene. 1994;9:2083–8. [PubMed] [Google Scholar]
- 8.Simoneau AR, Spruck CH, 3rd, Gonzalez-Zulueta M, et al. Evidence for two tumor suppressor loci associated with proximal chromosome 9p to q and distal chromosome 9q in bladder cancer and the initial screening for GAS1 and PTC mutations. Cancer Res. 1996;56:5039–43. [PubMed] [Google Scholar]
- 9.Cairns JP, Chiang PW, Ramamoorthy S, Kurnit DM, Sidransky D. A comparison between microsatellite and quantitative PCR analyses to detect frequent p16 copy number changes in primary bladder tumors. Clin Cancer Res. 1998;4:441–4. [PubMed] [Google Scholar]
- 10.Salem C, Liang G, Tsai YC, et al. Progressive increases in de novo methylation of CpG islands in bladder cancer. Cancer Res. 2000;60:2473–6. [PubMed] [Google Scholar]
- 11.Williamson MP, Elder PA, Shaw ME, Devlin J, Knowles MA. p16 (CDKN2) is a major deletion target at 9p21 in bladder cancer. Hum Mol Genet. 1995;4:1569–77. doi: 10.1093/hmg/4.9.1569. [DOI] [PubMed] [Google Scholar]
- 12.Cairns P, Tokino K, Eby Y, Sidransky D. Homozygous deletions of 9p21 in primary human bladder tumors detected by comparative multiplex polymerase chain reaction. Cancer Res. 1994;54:1422–4. [PubMed] [Google Scholar]
- 13.Papageorgiou A, Dinney CP, McConkey DJ. Interferon-alpha induces TRAIL expression and cell death via an IRF-1-dependent mechanism in human bladder cancer cells. Cancer Biol Ther. 2007;6:872–9. doi: 10.4161/cbt.6.6.4088. [DOI] [PubMed] [Google Scholar]
- 14.Papageorgiou A, Lashinger L, Millikan R, et al. Role of tumor necrosis factor-related apoptosis-inducing ligand in interferon-induced apoptosis in human bladder cancer cells. Cancer Res. 2004;64:8973–9. doi: 10.1158/0008-5472.CAN-04-1909. [DOI] [PubMed] [Google Scholar]
- 15.Hornigold N, Devlin J, Davies AM, Aveyard JS, Habuchi T, Knowles MA. Mutation of the 9q34 gene TSC1 in sporadic bladder cancer. Oncogene. 1999;18:2657–61. doi: 10.1038/sj.onc.1202854. [DOI] [PubMed] [Google Scholar]
- 16.Knowles MA, Habuchi T, Kennedy W, Cuthbert-Heavens D. Mutation spectrum of the 9q34 tuberous sclerosis gene TSC1 in transitional cell carcinoma of the bladder. Cancer Res. 2003;63:7652–6. [PubMed] [Google Scholar]
- 17.Knowles MA, Hornigold N, Pitt E. Tuberous sclerosis complex (TSC) gene involvement in sporadic tumours. Biochem Soc Trans. 2003;31:597–602. doi: 10.1042/bst0310597. [DOI] [PubMed] [Google Scholar]
- 18.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 19.Chow NH, Cairns P, Eisenberger CF, et al. Papillary urothelial hyperplasia is a clonal precursor to papillary transitional cell bladder cancer. Int J Cancer. 2000;89:514–8. [PubMed] [Google Scholar]
- 20.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 21.Koss LG, Tiamson EM, Robbins MA. Mapping cancerous and precancerous bladder changes. A study of the urothelium in ten surgically removed bladders. JAMA. 1974;227:281–6. [PubMed] [Google Scholar]
- 22.Majewski T, Lee S, Jeong J, et al. Understanding the development of human bladder cancer by using a whole-organ genomic mapping strategy. Lab Invest. 2008;88:694–721. doi: 10.1038/labinvest.2008.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S, Jeong J, Majewski T, et al. Forerunner genes contiguous to RB1 contribute to the development of in situ neoplasia. Proc Natl Acad Sci U S A. 2007;104:13732–7. doi: 10.1073/pnas.0701771104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crawford JM. The origins of bladder cancer. Lab Invest. 2008;88:686–93. doi: 10.1038/labinvest.2008.48. [DOI] [PubMed] [Google Scholar]
- 25.Deng J, Carlson N, Takeyama K, Dal Cin P, Shipp M, Letai A. BH3 profiling identifies three distinct classes of apoptotic blocks to predict response to ABT-737 and conventional chemotherapeutic agents. Cancer Cell. 2007;12:171–85. doi: 10.1016/j.ccr.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 26.Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2:183–92. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 27.Fleischer A, Rebollo A. Induction of p53-independent apoptosis by the BH3-only protein ITM2Bs. FEBS Lett. 2004;557:283–7. doi: 10.1016/s0014-5793(03)01519-9. [DOI] [PubMed] [Google Scholar]
- 28.Fleischer A, Ayllon V, Dumoutier L, Renauld JC, Rebollo A. Proapoptotic activity of ITM2B(s), a BH3-only protein induced upon IL-2-deprivation which interacts with Bcl-2. Oncogene. 2002;21:3181–9. doi: 10.1038/sj.onc.1205464. [DOI] [PubMed] [Google Scholar]
- 29.Azeem Z, Jelani M, Naz G, et al. Novel mutations in G protein-coupled receptor gene (P2RY5) in families with autosomal recessive hypotrichosis (LAH3) Hum Genet. 2008;123:515–9. doi: 10.1007/s00439-008-0507-7. [DOI] [PubMed] [Google Scholar]
- 30.Shimomura Y, Wajid M, Ishii Y, et al. Disruption of P2RY5, an orphan G protein-coupled receptor, underlies autosomal recessive woolly hair. Nat Genet. 2008;40:335–9. doi: 10.1038/ng.100. [DOI] [PubMed] [Google Scholar]
- 31.Pasternack SM, von Kugelgen I, Aboud KA, et al. G protein-coupled receptor P2Y5 and its ligand LPA are involved in maintenance of human hair growth. Nat Genet. 2008;40:329–34. doi: 10.1038/ng.84. [DOI] [PubMed] [Google Scholar]
- 32.Song H, Ramus SJ, Shadforth D, et al. Common variants in RB1 gene and risk of invasive ovarian cancer. Cancer Res. 2006;66:10220–6. doi: 10.1158/0008-5472.CAN-06-2222. [DOI] [PubMed] [Google Scholar]
- 33.Zhang ZT, Pak J, Huang HY, et al. Role of Ha-ras activation in superficial papillary pathway of urothelial tumor formation. Oncogene. 2001;20:1973–80. doi: 10.1038/sj.onc.1204315. [DOI] [PubMed] [Google Scholar]
- 34.Mo L, Zheng X, Huang HY, et al. Hyperactivation of Ha-ras oncogene, but not Ink4a/Arf deficiency, triggers bladder tumorigenesis. J Clin Invest. 2007;117:314–25. doi: 10.1172/JCI30062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang ZT, Pak J, Shapiro E, Sun TT, Wu XR. Urothelium-specific expression of an oncogene in transgenic mice induced the formation of carcinoma in situ and invasive transitional cell carcinoma. Cancer Res. 1999;59:3512–7. [PubMed] [Google Scholar]
- 36.Cheng J, Huang H, Pak J, et al. Allelic loss of p53 gene is associated with genesis and maintenance, but not invasion, of mouse carcinoma in situ of the bladder. Cancer Res. 2003;63:179–85. [PubMed] [Google Scholar]
- 37.Gao J, Huang HY, Pak J, et al. p53 deficiency provokes urothelial proliferation and synergizes with activated Ha-ras in promoting urothelial tumorigenesis. Oncogene. 2004;23:687–96. doi: 10.1038/sj.onc.1207169. [DOI] [PubMed] [Google Scholar]
- 38.Puzio-Kuter AM, Castillo-Martin M, Kinkade CW, et al. Inactivation of p53 and Pten promotes invasive bladder cancer. Genes Dev. 2009;23:675–80. doi: 10.1101/gad.1772909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoo LI, Liu DW, Le Vu S, Bronson RT, Wu H, Yuan J. Pten deficiency activates distinct downstream signaling pathways in a tissue-specific manner. Cancer Res. 2006;66:1929–39. doi: 10.1158/0008-5472.CAN-05-1986. [DOI] [PubMed] [Google Scholar]
- 40.Czerniak B, Cohen GL, Etkind P, et al. Concurrent mutations of coding and regulatory sequences of the Ha-ras gene in urinary bladder carcinomas. Hum Pathol. 1992;23:1199–204. doi: 10.1016/0046-8177(92)90285-b. [DOI] [PubMed] [Google Scholar]
- 41.Knowles MA, Williamson M. Mutation of H-ras is infrequent in bladder cancer: confirmation by single-strand conformation polymorphism analysis, designed restriction fragment length polymorphisms, and direct sequencing. Cancer Res. 1993;53:133–9. [PubMed] [Google Scholar]
- 42.Karimianpour N, Mousavi-Shafaei P, Ziaee AA, et al. Mutations of RAS gene family in specimens of bladder cancer. Urol J. 2008;5:237–42. [PubMed] [Google Scholar]
- 43.Sibley K, Cuthbert-Heavens D, Knowles MA. Loss of heterozygosity at 4p16.3 and mutation of FGFR3 in transitional cell carcinoma. Oncogene. 2001;20:686–91. doi: 10.1038/sj.onc.1204110. [DOI] [PubMed] [Google Scholar]
- 44.Sibley K, Stern P, Knowles MA. Frequency of fibroblast growth factor receptor 3 mutations in sporadic tumours. Oncogene. 2001;20:4416–8. doi: 10.1038/sj.onc.1204543. [DOI] [PubMed] [Google Scholar]
- 45.Knowles MA. Role of FGFR3 in urothelial cell carcinoma: biomarker and potential therapeutic target. World J Urol. 2007;25:581–93. doi: 10.1007/s00345-007-0213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomlinson DC, Hurst CD, Knowles MA. Knockdown by shRNA identifies S249C mutant FGFR3 as a potential therapeutic target in bladder cancer. Oncogene. 2007;26:5889–99. doi: 10.1038/sj.onc.1210399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qing J, Du X, Chen Y, et al. Antibody-based targeting of FGFR3 in bladder carcinoma and t(4;14)-positive multiple myeloma in mice. J Clin Invest. 2009;119:1216–29. doi: 10.1172/JCI38017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Esrig D, Elmajian D, Groshen S, et al. Accumulation of nuclear p53 and tumor progression in bladder cancer. N Engl J Med. 1994;331:1259–64. doi: 10.1056/NEJM199411103311903. [DOI] [PubMed] [Google Scholar]
- 49.Esrig D, Spruck CH, 3rd, Nichols PW, et al. p53 nuclear protein accumulation correlates with mutations in the p53 gene, tumor grade, and stage in bladder cancer. Am J Pathol. 1993;143:1389–97. [PMC free article] [PubMed] [Google Scholar]
- 50.Chatterjee SJ, Datar R, Youssefzadeh D, et al. Combined effects of p53, p21, and pRb expression in the progression of bladder transitional cell carcinoma. J Clin Oncol. 2004;22:1007–13. doi: 10.1200/JCO.2004.05.174. [DOI] [PubMed] [Google Scholar]
- 51.Shariat SF, Bolenz C, Karakiewicz PI, et al. p53 expression in patients with advanced urothelial cancer of the urinary bladder. BJU Int. 2009 doi: 10.1111/j.1464-410X.2009.08742.x. [DOI] [PubMed] [Google Scholar]
- 52.Shariat SF, Lotan Y, Karakiewicz PI, et al. p53 predictive value for pT1–2 N0 disease at radical cystectomy. J Urol. 2009;182:907–13. doi: 10.1016/j.juro.2009.05.024. [DOI] [PubMed] [Google Scholar]
- 53.George B, Datar RH, Wu L, et al. p53 gene and protein status: the role of p53 alterations in predicting outcome in patients with bladder cancer. J Clin Oncol. 2007;25:5352–8. doi: 10.1200/JCO.2006.10.4125. [DOI] [PubMed] [Google Scholar]
- 54.Xu HJ, Cairns P, Hu SX, Knowles MA, Benedict WF. Loss of RB protein expression in primary bladder cancer correlates with loss of heterozygosity at the RB locus and tumor progression. Int J Cancer. 1993;53:781–4. doi: 10.1002/ijc.2910530513. [DOI] [PubMed] [Google Scholar]
- 55.Cote RJ, Dunn MD, Chatterjee SJ, et al. Elevated and absent pRb expression is associated with bladder cancer progression and has cooperative effects with p53. Cancer Res. 1998;58:1090–4. [PubMed] [Google Scholar]
- 56.Cairns P, Shaw ME, Knowles MA. Initiation of bladder cancer may involve deletion of a tumour-suppressor gene on chromosome 9. Oncogene. 1993;8:1083–5. [PubMed] [Google Scholar]
- 57.Cairns P, Evron E, Okami K, et al. Point mutation and homozygous deletion of PTEN/MMAC1 in primary bladder cancers. Oncogene. 1998;16:3215–8. doi: 10.1038/sj.onc.1201855. [DOI] [PubMed] [Google Scholar]
- 58.Teng DH, Hu R, Lin H, et al. MMAC1/PTEN mutations in primary tumor specimens and tumor cell lines. Cancer Res. 1997;57:5221–5. [PubMed] [Google Scholar]
- 59.Aveyard JS, Skilleter A, Habuchi T, Knowles MA. Somatic mutation of PTEN in bladder carcinoma. Br J Cancer. 1999;80:904–8. doi: 10.1038/sj.bjc.6690439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang DS, Rieger-Christ K, Latini JM, et al. Molecular analysis of PTEN and MXI1 in primary bladder carcinoma. Int J Cancer. 2000;88:620–5. doi: 10.1002/1097-0215(20001115)88:4<620::aid-ijc16>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 61.Harris LD, De La Cerda J, Tuziak T, et al. Analysis of the expression of biomarkers in urinary bladder cancer using a tissue microarray. Mol Carcinog. 2008;47:678–85. doi: 10.1002/mc.20420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saal LH, Johansson P, Holm K, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci U S A. 2007;104:7564–9. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gildea JJ, Herlevsen M, Harding MA, et al. PTEN can inhibit in vitro organotypic and in vivo orthotopic invasion of human bladder cancer cells even in the absence of its lipid phosphatase activity. Oncogene. 2004;23:6788–97. doi: 10.1038/sj.onc.1207599. [DOI] [PubMed] [Google Scholar]
- 64.Platt FM, Hurst CD, Taylor CF, Gregory WM, Harnden P, Knowles MA. Spectrum of phosphatidylinositol 3-kinase pathway gene alterations in bladder cancer. Clin Cancer Res. 2009;15:6008–17. doi: 10.1158/1078-0432.CCR-09-0898. [DOI] [PubMed] [Google Scholar]
- 65.Askham JM, Platt F, Chambers PA, Snowden H, Taylor CF, Knowles MA. AKT1 mutations in bladder cancer: identification of a novel oncogenic mutation that can co-operate with E17K. Oncogene. 2009 doi: 10.1038/onc.2009.315. [DOI] [PubMed] [Google Scholar]
- 66.Blaveri E, Simko JP, Korkola JE, et al. Bladder cancer outcome and subtype classification by gene expression. Clin Cancer Res. 2005;11:4044–55. doi: 10.1158/1078-0432.CCR-04-2409. [DOI] [PubMed] [Google Scholar]
- 67.Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J Clin Oncol. 2006;24:778–89. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- 68.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–28. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 70.Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68:3645–54. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 71.Slaton JW, Inoue K, Perrotte P, et al. Expression levels of genes that regulate metastasis and angiogenesis correlate with advanced pathological stage of renal cell carcinoma. Am J Pathol. 2001;158:735–43. doi: 10.1016/S0002-9440(10)64016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anzai H, Kitadai Y, Bucana CD, Sanchez R, Omoto R, Fidler IJ. Expression of metastasis-related genes in surgical specimens of human gastric cancer can predict disease recurrence. Eur J Cancer. 1998;34:558–65. doi: 10.1016/s0959-8049(97)10075-2. [DOI] [PubMed] [Google Scholar]
- 73.Kitadai Y, Ellis LM, Takahashi Y, et al. Multiparametric in situ messenger RNA hybridization analysis to detect metastasis-related genes in surgical specimens of human colon carcinomas. Clin Cancer Res. 1995;1:1095–102. [PubMed] [Google Scholar]
- 74.Greene GF, Kitadai Y, Pettaway CA, von Eschenbach AC, Bucana CD, Fidler IJ. Correlation of metastasis-related gene expression with metastatic potential in human prostate carcinoma cells implanted in nude mice using an in situ messenger RNA hybridization technique. Am J Pathol. 1997;150:1571–82. [PMC free article] [PubMed] [Google Scholar]
- 75.Yang J, Mani SA, Weinberg RA. Exploring a new twist on tumor metastasis. Cancer Res. 2006;66:4549–52. doi: 10.1158/0008-5472.CAN-05-3850. [DOI] [PubMed] [Google Scholar]
- 76.Yang J, Mani SA, Donaher JL, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 77.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Slaton JW, Millikan R, Inoue K, et al. Correlation of metastasis related gene expression and relapse-free survival in patients with locally advanced bladder cancer treated with cystectomy and chemotherapy. J Urol. 2004;171:570–4. doi: 10.1097/01.ju.0000108845.91485.20. [DOI] [PubMed] [Google Scholar]
- 79.Inoue K, Kamada M, Slaton JW, et al. The prognostic value of angiogenesis and metastasis-related genes for progression of transitional cell carcinoma of the renal pelvis and ureter. Clin Cancer Res. 2002;8:1863–70. [PubMed] [Google Scholar]
- 80.Dinney CP, Fishbeck R, Singh RK, et al. Isolation and characterization of metastatic variants from human transitional cell carcinoma passaged by orthotopic implantation in athymic nude mice. J Urol. 1995;154:1532–8. [PubMed] [Google Scholar]
- 81.Perrotte P, Matsumoto T, Inoue K, et al. Anti-epidermal growth factor receptor antibody C225 inhibits angiogenesis in human transitional cell carcinoma growing orthotopically in nude mice. Clin Cancer Res. 1999;5:257–65. [PubMed] [Google Scholar]
- 82.Shrader M, Pino MS, Brown G, et al. Molecular correlates of gefitinib responsiveness in human bladder cancer cells. Mol Cancer Ther. 2007;6:277–85. doi: 10.1158/1535-7163.MCT-06-0513. [DOI] [PubMed] [Google Scholar]
- 83.Black PC, Brown GA, Inamoto T, et al. Sensitivity to epidermal growth factor receptor inhibitor requires E-cadherin expression in urothelial carcinoma cells. Clin Cancer Res. 2008;14:1478–86. doi: 10.1158/1078-0432.CCR-07-1593. [DOI] [PubMed] [Google Scholar]
- 84.Thomson S, Buck E, Petti F, et al. Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer Res. 2005;65:9455–62. doi: 10.1158/0008-5472.CAN-05-1058. [DOI] [PubMed] [Google Scholar]
- 85.Witta SE, Gemmill RM, Hirsch FR, et al. Restoring E-cadherin expression increases sensitivity to epidermal growth factor receptor inhibitors in lung cancer cell lines. Cancer Res. 2006;66:944–50. doi: 10.1158/0008-5472.CAN-05-1988. [DOI] [PubMed] [Google Scholar]
- 86.Yauch RL, Januario T, Eberhard DA, et al. Epithelial versus mesenchymal phenotype determines in vitro sensitivity and predicts clinical activity of erlotinib in lung cancer patients. Clin Cancer Res. 2005;11:8686–98. doi: 10.1158/1078-0432.CCR-05-1492. [DOI] [PubMed] [Google Scholar]
- 87.Haddad Y, Choi W, McConkey DJ. Delta-crystallin enhancer binding factor 1 controls the epithelial to mesenchymal transition phenotype and resistance to the epidermal growth factor receptor inhibitor erlotinib in human head and neck squamous cell carcinoma lines. Clin Cancer Res. 2009;15:532–42. doi: 10.1158/1078-0432.CCR-08-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Adam L, Zhong M, Choi W, et al. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res. 2009;15:5060–72. doi: 10.1158/1078-0432.CCR-08-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008 doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 91.Hurteau GJ, Carlson JA, Spivack SD, Brock GJ. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 2007;67:7972–6. doi: 10.1158/0008-5472.CAN-07-1058. [DOI] [PubMed] [Google Scholar]
- 92.Di Como CJ, Urist MJ, Babayan I, et al. p63 expression profiles in human normal and tumor tissues. Clin Cancer Res. 2002;8:494–501. [PubMed] [Google Scholar]
- 93.Urist MJ, Di Como CJ, Lu ML, et al. Loss of p63 expression is associated with tumor progression in bladder cancer. Am J Pathol. 2002;161:1199–206. doi: 10.1016/S0002-9440(10)64396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Koga F, Kawakami S, Fujii Y, et al. Impaired p63 expression associates with poor prognosis and uroplakin III expression in invasive urothelial carcinoma of the bladder. Clin Cancer Res. 2003;9:5501–7. [PubMed] [Google Scholar]
- 95.Koga F, Kawakami S, Kumagai J, et al. Impaired Delta Np63 expression associates with reduced beta-catenin and aggressive phenotypes of urothelial neoplasms. Br J Cancer. 2003;88:740–7. doi: 10.1038/sj.bjc.6600764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Signoretti S, Pires MM, Lindauer M, et al. p63 regulates commitment to the prostate cell lineage. Proc Natl Acad Sci U S A. 2005;102:11355–60. doi: 10.1073/pnas.0500165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Senoo M, Pinto F, Crum CP, McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–36. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 98.Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 2008;452:225–9. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 100.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–8. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 101.He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Biol. 2009;25:377–406. doi: 10.1146/annurev.cellbio.042308.113248. [DOI] [PubMed] [Google Scholar]
- 102.Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 2009;23:2563–77. doi: 10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kakarala M, Wicha MS. Implications of the cancer stem-cell hypothesis for breast cancer prevention and therapy. J Clin Oncol. 2008;26:2813–20. doi: 10.1200/JCO.2008.16.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kurzrock EA, Lieu DK, Degraffenried LA, Chan CW, Isseroff RR. Label-retaining cells of the bladder: candidate urothelial stem cells. Am J Physiol Renal Physiol. 2008;294:F1415–21. doi: 10.1152/ajprenal.00533.2007. [DOI] [PubMed] [Google Scholar]
- 105.He X, Marchionni L, Hansel DE, et al. Differentiation of a highly tumorigenic basal cell compartment in urothelial carcinoma. Stem Cells. 2009;27:1487–95. doi: 10.1002/stem.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chan KS, Espinosa I, Chao M, et al. Identification, molecular characterization, clinical prognosis, and therapeutic targeting of human bladder tumor-initiating cells. Proc Natl Acad Sci U S A. 2009;106:14016–21. doi: 10.1073/pnas.0906549106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Brandt WD, Matsui W, Rosenberg JE, et al. Urothelial carcinoma: stem cells on the edge. Cancer Metastasis Rev. 2009;28:291–304. doi: 10.1007/s10555-009-9187-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Farsund T. Cell kinetics of mouse urinary bladder epithelium. II. Changes in proliferation and nuclear DNA content during necrosis regeneration, and hyperplasia caused by a single dose of cyclophosphamide Virchows. Arch B Cell Pathol. 1976;21:279–98. [PubMed] [Google Scholar]
- 109.Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–13. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 110.Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 2004;18:126–31. doi: 10.1101/gad.1165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McKeon F. p63 and the epithelial stem cell: more than status quo? Genes Dev. 2004;18:465–9. doi: 10.1101/gad.1190504. [DOI] [PubMed] [Google Scholar]
- 112.Cheng W, Jacobs WB, Zhang JJ, et al. DeltaNp63 plays an anti-apoptotic role in ventral bladder development. Development. 2006;133:4783–92. doi: 10.1242/dev.02621. [DOI] [PubMed] [Google Scholar]