Abstract
This study examined the association between markers in transforming growth factor alpha (TGFA) and isolated, non-syndromic cleft lip with/without palate (CL/P) using a case–parent trio design, considering parent-of-origin effects. We also tested for gene–environmental interaction with common maternal exposures, and for gene–gene interaction using markers in TGFA and another recognized causal gene, IRF6. CL/P case–parent trios from four populations (76 from Maryland, 146 from Taiwan, 35 from Singapore, and 40 from Korea) were genotyped for 17 single nucleotide polymorphisms (SNPs) in TGFA. The transmission disequilibrium test was used to test individual SNPs, and the parent-of-origin likelihood ratio test (PO-LRT) was used to assess parent-of-origin effects. We also screened for possible gene–environment interaction using PBAT, and tested for gene–gene interaction using conditional logistic regression models. When all trios were combined, four SNPs showed significant excess maternal transmission, two of which gave significant PO-LRT values [rs3821261: P = 0.004 and OR(imprinting) = 4.17; and rs3771475: P = 0.027 and OR(imprinting) = 2.44]. Haplotype analysis of these two SNPS also supported excess maternal transmission. We saw intriguing but suggestive evidence of G × E interaction for several SNPs in TGFA when either individual SNPs or haplotypes of adjacent SNPs were considered. Thus, TGFA appears to influence risk of CL/P through unconventional means with an apparent parent-of-origin effect (excess maternal transmission) and possible interaction with maternal exposures.
Introduction
Oral clefts are one of the most common birth defects in humans and represent a significant public health problem both in terms of medical and economic burdens for affected individuals and their families. Non-syndromic cleft lip with or without palate (CL/P) is considered ‘complex’ or ‘multifactorial’ in its etiology, in that both genes and environmental risk factors control risk (Wyszynski et al. 1997; Cobourne 2004). Transforming growth factor alpha (TGFA) gene is a well-studied candidate gene for CL/P, but has shown inconsistent evidence of association with CL/P across a number of studies (Vieira 2006). Some studies have also tested for potential gene–environmental (G × E) interactions between markers in TGFA and common maternal exposures (particularly maternal smoking) (Hwang et al. 1995; Beaty et al. 1997; Shaw et al. 1998; Shaw et al. 1996; Zeiger et al. 2005). Jugessur et al. (2003a, b) raised the possibility of interaction between TGFA and MTHFR, so it would also be important to consider gene–gene interaction (G × G). Here we consider interaction between TGFA and IRF6, another frequently studied candidate gene. IRF6 on chromosome 1 has been identified as responsible for a majority of cases with van der Woude syndrome (VWS), an autosomal dominant malformation syndrome which often includes oral clefts. In addition, several studies have reported strong association between polymorphic markers with isolated non-syndromic CL/P (Zucchero et al. 2004; Park et al. 2007).
It is important to consider parent-of-origin effects when studying birth defects because maternal genotype controls the in utero environment of the developing fetus, and separating maternal genotypic effects from imprinting effects remains an important scientiffic question (Weinberg and Umbach 2005; Wilkins and Haig 2003). Maternal parent-of-origin effects have been suggested for several genes associated with non-syndromic CL/P (van Rooij et al. 2003; Rubini et al. 2005; Sull et al. 2008). However, to date no study has focused on whether TGFA gene may influence risk of CL/P through a parent-of-origin effect.
In a previous paper, we reported an association between markers in TGFA and risk of CL/P in three populations (Beaty et al. 2006). Here, we tested for association in 297 CL/P case– parent trios from 4 populations, while specifically considering parent-of-origin effects, as well as testing for gene–environmental interactions between markers in TGFA and three common maternal exposures (maternal smoking, alcohol consumption and vitamin supplementation), plus testing for interaction between markers in the TGFA and IRF6 genes.
Methods
Sample description
As part of an international study of oral clefts, we collected data on case–parent trios recruited through treatment centers in Maryland (MD) (Johns Hopkins and University of Maryland), the Chang Gung Memorial Hospital in Taiwan (TW), KK Women’s and Children’s Hospital in Singapore (SP), and Yonsei Medical Center in South Korea (KR). Research protocols were reviewed and approved by institutional review boards (IRB) at each institution. Table 1 lists the gender of all CL/P probands. The majority of cases were infants seen during a surgical or postsurgical visit. All parents of probands were unaffected in the Singapore, Taiwan, and Korean trios, but 4 parents among the 76 Maryland trios also had an oral cleft. All probands underwent clinical genetics evaluation (including assessing other congenital anomalies or major developmental delays) and were classiffied as having an isolated, non-syndromic CL/P. First-trimester maternal exposure information, including cigarette smoking, vitamin supplementation, and alcohol consumption was collected from a face-to-face interview of mothers (although our first group of trios from Taiwan had substantial rates of missing data). For maternal smoking, about 20% of mothers in Maryland reported smoking during the critical period of pregnancy (during the 3 months before conception through the first trimester). Among women in Taiwan and other Asian populations, however, smoking rates were substantially lower (about 5%). The proportion with alcohol consumption and vitamin supplementation during the first-trimester was also higher in Maryland than in the Asian populations (Table 2).
Table 1.
Gender among 297 non-syndromic cleft lip with or without cleft palate (CL/P) cases from 4 populations
| Population | CL/P cases |
||
|---|---|---|---|
| Total (n) | Male (n) | Female (n) | |
| Taiwan | 146 | 95 | 51 |
| Singapore | 35 | 24 | 11 |
| Korea | 40 | 22 | 18 |
| Maryland | 76 | 44 | 32 |
| Total | 297 | 185 | 112 |
Table 2.
Distribution of three maternal environmental factors among 297 CL/P case–parent trios from 4 populations
| Maternal smoking |
Maternal alcohol |
Maternal vitamin supplements |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Exposed | Unexp | Unknown | Exposed | Unexp | Unknown | Exposed | Unexp | Unknown | |
| TW | 5 | 82 | 59 | 1 | 86 | 59 | 27 | 58 | 61 |
| SP | 3 | 25 | 7 | 2 | 26 | 7 | 5 | 23 | 7 |
| KR | 0 | 40 | 0 | 0 | 37 | 3 | 6 | 33 | 1 |
| MD | 16 | 60 | 0 | 12 | 64 | 0 | 61 | 9 | 6 |
| Total | 24 | 207 | 66 | 15 | 213 | 69 | 99 | 123 | 75 |
SNP selection, DNA and genotyping
Single nucleotide polymorphisms (SNPs) were selected in a region surrounding TGFA on chromosome 2p13, with a goal of identifying one SNP per 5 kb of physical distance. Variants with “SNP scores” (an assessment of design quality of the Illumina assay based on a proprietary algorithm) above 0.6, high validation levels in dbSNP (this included validation levels where the submitter had validated the SNP on multiple platforms), and high heterozygosity levels (particularly in multiple populations), were given higher priority during the selection process. From 24 selected SNPs, 20 were polymorphic in all 4 populations. Two of these SNPs had low genotype call rates and one SNP deviated significantly from HWE in three of four populations, leaving only 17 SNPs with reasonable heterozygosity (Table 3).
Table 3.
Minor allele frequencies (MAF) among parents of 297 CL/P cases from 4 populations
| No. | SNP name | Physical locationa | Minor allele | Minor allele frequency |
|||
|---|---|---|---|---|---|---|---|
| Taiwan | Singapore | Korea | Maryland | ||||
| 1 | rs473698 | 70587236 | G | 0.309 | 0.237 | 0.333 | 0.391 |
| 2 | rs4852595 | 70591414 | T | 0.134 | 0.179 | 0.184 | 0.119 |
| 3 | rs1880039 | 70598707 | C | 0.055 | 0.062 | 0.06 | 0.130 |
| 4 | rs3771516 | 70606538 | C | 0.167 | 0.193 | 0.158 | 0.088 |
| 5 | rs3755384 | 70616166 | T | 0.375 | 0.316 | 0.395 | 0.457 |
| 6 | rs1807968 | 70619916 | G | 0.375 | 0.316 | 0.395 | 0.461 |
| 7 | rs3771503 | 70623896 | T | 0.410 | 0.351 | 0.428 | 0.483 |
| 8 | rs2902345 | 70628254 | T | 0.373 | 0.328 | 0.393 | 0.443 |
| 9 | rs3755378 | 70632258 | A | 0.424 | 0.443 | 0.462 | 0.350 |
| 10 | rs3771494 | 70637007 | C | 0.211 | 0.228 | 0.184 | 0.218 |
| 11 | rs3821261 | 70640908 | G | 0.153 | 0.123 | 0.112 | 0.113 |
| 12 | rs404420 | 70645181 | T | 0.184 | 0.190 | 0.230 | 0.255 |
| 13 | rs455125 | 70658381 | C | 0.216 | 0.216 | 0.184 | 0.229 |
| 14 | rs426081 | 70662186 | C | 0.130 | 0.138 | 0.026 | 0.227 |
| 15 | rs3771475 | 70680983 | G | 0.304 | 0.319 | 0.303 | 0.123 |
| 16 | rs12473408 | 70688039 | C | 0.359 | 0.368 | 0.367 | 0.467 |
| 17 | rs11466191 | 70691736 | C | 0.290 | 0.293 | 0.253 | 0.264 |
Based on NCBI Human Genome build 35.1
Genomic DNA samples were prepared from peripheral blood by the protein precipitation method described previously (Bellus et al. 1995). DNA concentration was determined using the PicoGreen® dsDNA Quantitation Kit (Molecular Probes, Inc., Eugene OR) and all DNA samples were stored at −20°C. A 4 µg aliquot of each genomic DNA sample was dispensed into a bar-coded 96-well microtiter plate at a concentration of 100 ng/OR) and all DNA samples were stored at −20°C. A 4 µg l and geno-typed for SNP markers using the Illumina Golden-Gate™ chemistry with Sentrix® Array Matrices from the manufacturer (Oliphant et al. 2002) at the SNP Center of the Genetic Resources Core Facility (GRCF), a part of the McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins School of Medicine. Two duplicates and four CEPH controls were included on each plate to evaluate genotyping consistency within and between plates and to insure correct plate orientation. Genotypes were generated on a BeadLab 1000 system (Fan et al. 2003). No Mendelian inconsistencies were found for these 17 SNPs.
Statistical analysis
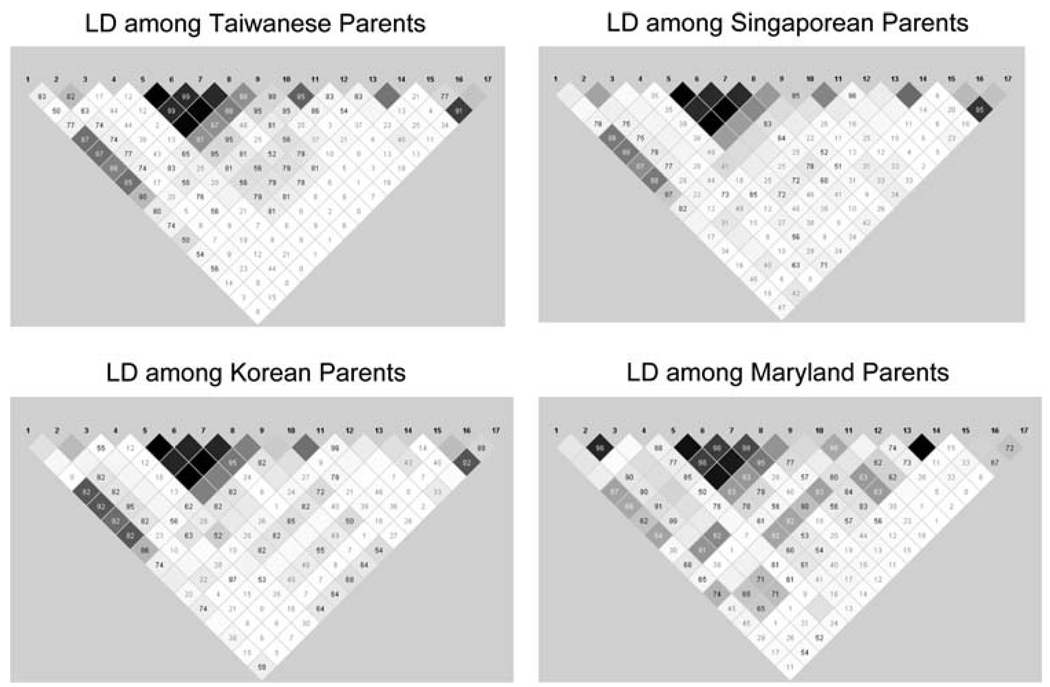
Within each population, the minor allele frequency (MAF) was computed among parents. Pairwise linkage disequilibrium (LD) was measured as r2 for all SNPs using the Haplo-view program (Barrett et al. 2005). LD blocks were identified in each population separately using pairwise LD (Fig. 1). Four blocks of LD were identified, consisting of 9, 2, 3 and 3 SNPs, respectively. Clayton’s extension of the Transmission Disequilibrium Test (TDT) incorporated into STATA 8.2 (Spielman et al. 1993; Cordell et al. 2004) was used on individual SNPs to test for evidence of linkage and LD in the combined sample of 297 CL/P trios.
Fig. 1.
Linkage disequilibrium as measured by r2 in TGFA among parents of CL/P children from four populations. White r2 = 0. Shades of gray 0 < r2 < 1. Black r2 = 1
Parent-of-origin analyses were conducted on the combined sample. We used the likelihood-based approach proposed by Weinberg (1999) to test for parent-of-origin effects while considering genotypes of both mother (2 parameters for maternal genetic effects) and child (2 parameters for child genetic effects). This log-linear model considers the three mating types in which the mother and father carry different numbers of variant alleles, with further stratification by the number of alleles inherited by the child. Like Weinberg, we will call this latter method the “parent-of-origin likelihood ratio test” (PO-LRT). This model considers maternally mediated in utero effects (maternal genotypic effects on the phenotype of the fetus) which could otherwise confound assessment of parent-of-origin effects, along with a separate term for imprinting (Weinberg 1999). Here imprinting reflects a differential transmission of alleles to the affected child from mothers versus fathers. This PO-LRT was executed using the LEM software (van Den Oord and Vermunt 2000).
The FAMHAP package was used to estimate haplotype frequencies and to test for excess transmission of multi-SNP haplotypes (Becker et al. 2006). The FAMHAP package calculates maximum likelihood estimates (MLEs) of haplotype frequencies (for up to 20 SNPs) in nuclear families via the expectation–maximization algorithm and is robust in handling missing SNPs (Becker and Knapp 2004). This program provides a haplotype-based test for nuclear family data. The test statistic is based on Monte-Carlo simulations where the set of transmitted and non-transmitted genotypes/haplotypes is randomly permuted for 10,000 replicates (Zhao et al. 2000; Knapp and Becker 2003). In this analysis, the chi-square statistic for marker combinations is replaced with the maximum chi-square over all haplotypes (maximum TDT statistic). The program gives an empiric P value, corrected for the multiple haplotypes being considered. Haplotype analysis was also stratified for maternal and paternal transmission for individual SNPs showing a parent-of-origin effect.
We also screened for possible gene–environment interaction between the 17 SNPs in TGFA and three common maternal exposures (maternal smoking, alcohol consumption and vitamin supplementation). In this analysis we used the strategy proposed by Vansteelandt et al. (2007) where family-based association tests are first evaluated for individual SNPs while allowing for potential gene–environment (G × E) interaction in a 2 degree of freedom (df) test, followed by a separate 1 df test for G × E interaction alone. This approach is implemented in the PBAT package (v3.6; http://www.biostat.harvard.edu/~clange/default.htm) and computes a combined score test with 2 df for main effects of genotype (G) and G × E interaction, and then using a score test with 1 df for G × E interaction alone.
We tested for possible interactions between alleles at two genes (TGFA and IRF6). We focused on SNP rs3771494 in TGFA and SNP rs2235373 in IRF6 because these SNPs gave the most significant results in the present and previous analysis (Park et al. 2007). Conditional logistic regression models were used for the gene–gene interaction test.
Results
Among these 17 SNPs, there was some variation in allele frequencies among parents from MD and the three Asian populations (Table 3). The three Asian populations (TW, SP, and KR) had very low minor allele frequencies (MAF) for SNP 3 (rs1880039) and SNP 14 (rs426081) compared to the MD trios. Pairwise LD measures across the entire gene were calculated within each population, and Fig. 1 shows similar patterns across all populations.
When individual markers were screened in the combined dataset from all four populations using the TDT without considering parent-of-origin, the odds ratio of transmission for the minor allele, OR(transmission), was significant for SNP rs3771494 (OR = 1.59, P = 0.004), SNP rs3821261 (OR = 1.56, P = 0.022), and SNP rs426081 (OR = 1.51, P = 0.030) (Table 4).
Table 4.
TDT analysis of 17 SNPs in TGFA among 297 CL/P case– parent trios ignoring parent-of-origin
| No. | SNP name | TDT |
PO-LRTa |
|||||
|---|---|---|---|---|---|---|---|---|
| T | NT | χ2 | P value | ORb | ORc | P value | ||
| 1 | rs473698 | 127 | 115 | 0.60 | 0.440 | 1.10 | 1.52 | 0.257 |
| 2 | rs4852595 | 61 | 59 | 0.33 | 0.855 | 1.03 | 0.70 | 0.389 |
| 3 | rs1880039 | 31 | 21 | 1.92 | 0.166 | 1.48 | 0.40 | 0.138 |
| 4 | rs3771516 | 60 | 60 | 0.00 | 1.000 | 1.00 | 1.98 | 0.64 |
| 5 | rs3755384 | 139 | 120 | 1.39 | 0.238 | 1.16 | 1.31 | 0.438 |
| 6 | rs1807968 | 117 | 140 | 2.06 | 0.151 | 0.84 | 1.41 | 0.329 |
| 7 | rs3771503 | 126 | 127 | 0.01 | 0.949 | 0.99 | 1.46 | 0.291 |
| 8 | rs2902345 | 134 | 119 | 0.89 | 0.346 | 1.13 | 1.30 | 0.448 |
| 9 | rs3755378 | 117 | 91 | 3.25 | 0.071 | 1.29 | 1.08 | 0.838 |
| 10 | rs3771494 | 100 | 63 | 8.40 | 0.004 | 1.59 | 1.64 | 0.205 |
| 11 | rs3821261 | 67 | 43 | 5.24 | 0.022 | 1.56 | 4.17 | 0.004 |
| 12 | rs404420 | 93 | 95 | 0.02 | 0.884 | 0.98 | 0.77 | 0.498 |
| 13 | rs455125 | 93 | 72 | 2.67 | 0.102 | 1.29 | 0.52 | 0.105 |
| 14 | rs426081 | 68 | 45 | 4.68 | 0.030 | 1.51 | 0.68 | 0.391 |
| 15 | rs3771475 | 95 | 81 | 1.11 | 0.291 | 1.17 | 2.44 | 0.027 |
| 16 | rs12473408 | 125 | 104 | 1.93 | 0.165 | 1.20 | 1.79 | 0.121 |
| 17 | rs11466191 | 100 | 86 | 1.05 | 0.305 | 1.16 | 1.97 | 0.098 |
Values given in italics indicate inferred blocks of strong linkage disequilibrium (LD)
Bold values represent results significant at the P < 0.05 level
T Transmitted, NT not transmitted, OR odds ratio, TDT transmission disequilibrium test
Parent-of-origin likelihood ratio test (PO-LRT) for a separate imprinting term
OR(transmission): Odds Ratio of transmission of minor allele
OR: Odds Ratio for imprinting effect (i.e. differential transmission from mothers vs. from fathers)
Parent-of-origin effects were investigated for all SNPs in the combined dataset (Table 4). The PO-LRTs were only significant for SNP 11 (rs3821261) [P = 0.004, OR(imprinting) = 4.17] and SNP 15 (rs3771475) [P = 0.027, OR(imprinting) = 2.44]. These significant PO-LRTs gave estimated risk ratios for an imprinting effect ranging between 2.44 and 4.17 for SNP 11 and 15, suggesting excess maternal transmission compared to paternal transmission in this region.
Haplotypes of rs3821261 and rs3771475 were analyzed using the FAMHAP program in each population. Taiwan, Singapore, and Korean parents had very similar haplotype frequencies, so all Asian trios were combined. In the combined Asian data, haploytpes T-G and G-G showed strong evidence of excess transmission to CL/P children (maximum TDT = 5.652, P = 0.054 for transmission ignoring parent-of-origin). As seen in Table 5 (maximum TDT = 7.686, P = 0.007), this is largely attributed to excess maternal transmission among these Asian trios, although the Maryland trios showed similar patterns. No haplotypes showed significant deviation from expected Mendelian transmission when transmitted from fathers.
Table 5.
Testing for excess transmission of haplotypes of SNPs rs3821261 (T/G) and rs3771475 (A/G) in TGFA in 297 CL/P case–parent trios considering maternal and paternal transmission separately (implemented with the FAMHAP program)
| Population | Haplotype | Frequency | Paternal |
Maternal |
||||
|---|---|---|---|---|---|---|---|---|
| T | NT | Maximum TDT (P value) | T | NT | Maximum TDT (P value) | |||
| Asian (221 trios) | TA | 0.612 | 53.3 | 41.6 | 2.005 (0.357) | 30.1 | 55.8 | 7.686 (0.007) |
| TG | 0.257 | 27.7 | 36.4 | 36.9 | 30.2 | |||
| GA | 0.078 | 11.2 | 18.9 | 16.4 | 7.7 | |||
| GG | 0.053 | 10.8 | 6.1 | 16.6 | 6.3 | |||
| Maryland (76 trios) | TA | 0.790 | 7.8 | 9.2 | 1.000 (1.000) | 6.5 | 13.8 | 2.652 (0.149) |
| TG | 0.114 | 2.5 | 4.5 | 7.2 | 4.5 | |||
| GA | 0.089 | 5.7 | 3.3 | 7.0 | 2.7 | |||
| GG | 0.007 | 1.0 | 0.0 | 0.3 | 0.0 | |||
Bold values represent results significant at the P < 0.05 level
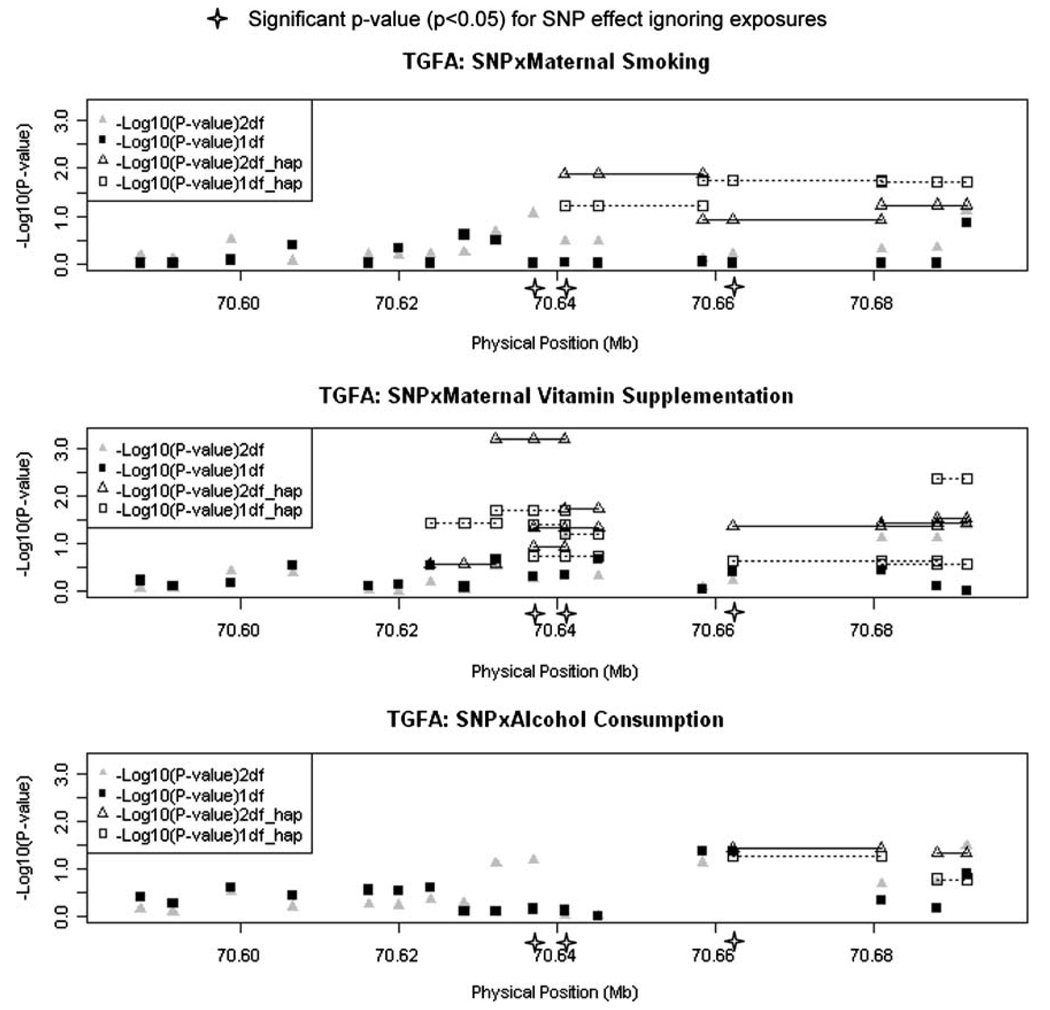
Figure 2 shows results of screening for gene–environment interaction between these 17 SNPs in TGFA and three common maternal exposures (maternal smoking, alcohol consumption and vitamin supplementation). Here −log10(P values) are plotted over physical distance for both the 2 df test of G and G × E effects considered jointly (triangles) and the 1df test for G × E effects alone (squares). A nominal 5% significance level corresponds to −log10(P value) = 1.3, a 1% significance corresponds to 2.0, etc. in this plot. The positions of the three SNPs showing marginal evidence of linkage in the presence of LD (rs3771494, rs3821261 and rs426081) when maternal exposures were ignored are noted below the X-axis, and one of these SNPs (rs3821261) showed nominal statistical significance in the score test for G × E (with 1 df) for all three maternal exposures when considered individually or in 2- and 3-SNP haplotypes. Haplotypes including rs3771494 and rs3821261 also showed statistically significant evidence of G × E interaction in the 1 df test for maternal vitamin supplementation. SNP rs455125 showed significant evidence in the 1 df test for G × E for alcohol consumption. SNP rs426081 gave nominally significant evidence of both G effects and G × E interaction in the 2 df test, and for G × E alone in the 1 df test for maternal alcohol consumption when considered alone, and for all maternal exposoures when considered as haplotypes of adjacent SNPs. SNP rs11466191 also gave significant evidence of possible interaction with maternal vitamin supplementation and alcohol consumption in the 2 df test for G and G × E (P = 0.04 and P = 0.03 for the 2 df test and P = 0.004 for the 1 df test with vitamin use). Since the marginal effect of this SNP alone (ignoring exposures) was not at all significant, this analysis illustrates the importance of considering possible G × E interaction in the etiology of CL/P.
Fig. 2.
Testing for main effects (G) of individual SNPs and gene–environment interaction (G × E) for three common maternal exposures in 297 CL/P case–parent trios from 4 populations. Triangles represent the 2 df test of G and G × E interaction, squares represent the 1 df test of G × E only. Haplotypes of 2- and 3-SNPs are connected by dashed lines (only nominally significant haplotypes are shown here)
The patients who received a C allele at rs3771494 in TGFA and a G allele at rs2235373 in IRF6 showed a 5.66-fold increased risk of being a case compared to those who received the T allele and A allele at these two respective SNPs, and the estimated OR for this combined effect excluded the null hypothesis (Table 6). However, the LRT formally comparing a logistic model with the G × G interaction terms to the baseline model argued this interaction was not statistically significant (P = 0.2105).
Table 6.
Odds ratio (OR) for effects of rs3771494 in TGFA and rs2235373 in IRF6 on CL/P main and combined effect
| TGFA (rs3771494) | Main effect | IRF6 (rs2235373) |
|
|---|---|---|---|
| Allele 1 (A) | Allele 2 (G) | ||
| OR (95% CI) | OR (95% CI) | ||
| 1.0 (reference) | 2.43 (1.79–3.31) (P < 0.0001) | ||
| OR (95% CI)a | Combined effect | ||
| Allele 1 (T) | 1.0 (reference) | 1.0 (reference) | 2.26 (1.61–3.18) (P < 0.0001) |
| Allele 2 (C) | 1.64 (1.18–2.27) (P = 0.003) | 1.53 (0.85–2.74) (P = 0.17) | 5.66 (3.28–9.76) (P < 0.0001) |
CI: confidence interval,
P for interaction = 0.2105
Discussion
Our study of case–parent trios from different populations (comprising a total of 297 CL/P case–parent trios) showed significant evidence of linkage and disequilibrium for three individual SNPs (rs3771494, rs3821261, and rs426081) in the TGFA gene when parent-of-origin was ignored. In screening for parent-of-origin effects, we found suggestive evidence of excess maternal transmission for at least two SNPs: rs3821261 and rs3771475. Analysis of haplotypes of these two SNPs also showed significant deviation from expected for maternal transmission, but not paternal transmission. Several SNPs in the TGFA gene also showed possible gene–environment interactions with maternal smoking and alcohol consumption, plus maternal vitamin supplementation. Lastly, we found intriguing patterns of transmission that raise the possibility of interaction between TGFA and IRF6.
Transforming growth factor alpha (TGFA) is a well-characterized mammalian growth factor (Tricoli et al. 1986), first identified as a candidate based on its expression patterns in the developing head and face. TGFA is expressed at the medial edge epithelium of fusing palatal shelves (Miettinen et al. 1999). Since the first report of an association between TGFA and oral clefts (Ardinger et al. 1989), several other case–control (Christensen and Mitchell 1996) and family-based studies (Maestri et al. 1997; Jugessur et al. 2003a) have found significant evidence of being associated with risk of oral clefts. However, other studies showed no association with risk to CL/P (Vieira 2006). The failure to detect association between oral clefts (in particularly CL/P) and TGFA could be due to both bias and genuine population diversity (Ioannidis et al. 2001). Additionally, previous case–parent trio studies did not consider parent-of-origin effects in testing for linkage in the presence of LD. Our study screened for parent-of-origin effects of individual SNPs and haplotypes and found evidence of excess maternal transmission.
Maternal genotypic effects for non-syndromic CL/P have also been reported for several other candidate genes (MTHFR, CBS, and RUNX2), but these have yet to be confirmed (Van Rooij et al. 2003; Rubini et al. 2005; Sull et al. 2008). Our results for several SNPs in TGFA (both analyzed alone and as haplotypes) showed evidence of excess maternal transmission, which could reflect an imprinting effect.
There are several examples of potential gene–environment (G × E) interactions in studies of oral clefts (Vieira 2006), but they have proven dificult to confirm across all studies. TGFA is the single most widely studied candidate gene for oral clefts, although several other candidate genes have been examined. Hwang et al. (1995) found evidence of gene–environment interaction between the Taq1 marker in TGFA and maternal smoking for cases of CP, but not CL/P cases. Infants carrying the C2 allele were more likely to have CP if the mother smoked (OR = 5.5, 95% confidence interval = 2.1–4.6). Similar patterns of interaction between TGFA and maternal smoking were found by Shaw et al. (1996), but Zeiger et al. (2005) found substantial heterogeneity among five published CP case–control studies in a meta-analysis. Shaw et al. (1998) found evidence for an interaction between this same TGFA marker and nutrient intake; where risk was highest for isolated CL/P (OR = 3.0, 95% CI = 1.4–6.6) among infants with the C2 allele whose mothers did not use vitamin supplements.
In tests for gene–environment interaction presented here, SNPs showing marginal significant evidence of gene effects ignoring exposures also gave intriguing evidence of possible interaction with common maternal exposures, when considered individually or as haplotypes. We note the exposure prevalence was low for alcohol and smoking, especially in the Asian populations and there were problems with missing data in this study. Low exposure rates and small sample sizes will become an issue for a valid evaluation of G × E interaction. Therefore, results of these tests for G × E interaction must be considered tentative. The case–parent trio design allows tests for genetic (G) and (G × E) interaction without being grossly affected by heterogeneity in genetic background (a.k.a. population stratification or confounding), but differences in exposure rates remain a source of heterogeneity. Here we found intriguing but tentative evidence of G × E interaction using PBAT in the total sample. Further analysis showed some differences between the MD and Asian trios. For example, rs3771494 showed evidence of a SNP effect ignoring all exposures in the combined sample (where 138 trios were informative for this SNP), and the 1 df score test for G × Maternal smoking was significant (P = 0.022). Among the 77 MD trios (minor allele frequency, MAF = 0.218 among parents), only 30 trios were informative, but the apparent SNP effect remained significant. Among Asian trios (MAF = 0.208) there were 108 informative trios and the SNP effect was only marginally significant (P value = 0.10), but the 1 df score test for G × Maternal vitamin use also approached signficiance (P value = 0.075) despite the lower exposure frequency. Obviously, further confirmation will be needed before firm conclusions can be drawn regarding G × E interaction, but clearly considering G × E interactions may become important in understanding the etiology of CL/P.
For non-syndormic oral clefts, gene–gene interactions have been suggested for TGFA (Vieira 2006). Interferon Regulatory factor 6 (IRF6) is a well-known candidate gene for non-syndromic CL/P, as well as being the primary causal gene for van der Woude syndrome, an autosomal dominant malformation syndrome which can include an oral cleft in carriers. Our previous study also reported several SNPs in IRF6, including rs2235373, were associated with CL/P (Park et al. 2007). Recently, IRF6-TGFA gene– gene interactions were observed in a case–parent trios study in human tooth agenesis (Vieira et al. 2007). In the present study, we tested for possible gene–gene interaction between these two genes (IRF6 and TGFA) for CL/P. Patterns consistent with G × G interaction were observed, but were not statistically significant.
Even though this candidate gene study involved a modest number of SNPs in the TGFA gene, addressing the issue of multiple comparisons is necessary before an overall statement about the significance of our findings can be made. Here we relied on a hypothesis-driven approach for 17 SNPs and conducted tests for genetic main effect, haplo-type-based test statistics, imprinting, gene–environment interaction with three exposures. SNPs in strong LD typically have highly correlated P values, adjusting significance levels via Bonferroni correction is overly conservative. Therefore, following the strategy in Sull et al. (2008), we adjusted empirical P values for the number of LD blocks, rather than the number of SNPs. In the present study, we have four LD blocks in the gene. In the second block with two SNPs (seen in Table 4), we found evidence against the null hypothesis (the empirical P value of 0.013 would still be marginally significant after correcting for the number of LD blocks). We also used haplotype-based test statistics based on permutation analysis of case–parent trio data. Salyakina et al. (2005) argue permutation tests are generally preferred over adjustments of asymptotic P values based on the estimated correlation structure among multiple markers or on conventional Bonferroni adjustment (which can be too conservative) (Nyholt 2004).
The case–parent trio design offers the advantage of testing directly for maternal versus paternal effects, and allows separating these from effects of the fetal genotype versus parental origin in a robust manner (Cordell et al. 2004; Starr et al. 2005; Sinsheimer et al. 2003). Another advantage of this design is it minimizes issues of confounding that plague traditional case–control designs. This feature permitted pooling trios from diverse populations into a combined test of allelic effects on the OR(transmission) to the affected child, while testing for parent-of-origin effects. The present study suggests maternal transmission effects for markers in TGFA and risk of non-syndromic CL/P, and we have found suggestive evidence of gene–environment interaction between markers in TGFA and maternal smoking, alcohol consumption and vitamin use, although further work is still needed to confirm its ultimate impact on risk.
Acknowledgments
This research was supported by R21-DE-013707 and R01-DE-014581 from the National Institute of Dental & Craniofacial Research, NIH R01 HL 090577, Korean Research Foundation (2005-214-E00042), and the Seoul City R&BD program (10526) in Korea. We thank all participants who donated samples for this multi-center study of oral clefts, as well as the staff at each participating site and institution. We also thank Gerald Raymond for his assistance in screening patients at Hopkins.
Footnotes
Web resources
HAPLOVIEW: http://www.broad.mit.edu/mpg/haploview/index.php/.
Online Mendelian Inheritance in Man (OMIM): http://www.ncbi.nlm.nih.gov/Omim/.
Contributor Information
Jae Woong Sull, Email: sulljw@yuhs.ac, Department of Epidemiology and Health Promotion, Institute for Health Promotion, Graduate School of Public Health, Yonsei University, Seoul, Korea.
Kung-Yee Liang, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, USA.
Jacqueline B. Hetmanski, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, USA
Tao Wu, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, USA.
Margaret Daniele Fallin, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, USA.
Roxann G. Ingersoll, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, USA Johns Hopkins School of Medicine, Baltimore, MD, USA.
Ji Wan Park, Sungkyunkwan University School of Medicine, Seoul, Korea.
Yah-Huei Wu-Chou, Chang Gung Memorial Hospital, Taoyuan, Taiwan.
Philip K. Chen, Chang Gung Memorial Hospital, Taoyuan, Taiwan
Samuel S. Chong, Department of Pediatrics, National University of Singapore, Singapore, Singapore
Felicia Cheah, Department of Pediatrics, National University of Singapore, Singapore, Singapore.
Vincent Yeow, KK Women’s and Children’s Hospital, Singapore, Singapore.
Beyoung Yun Park, Department of Plastic Surgery, College of Medicine, Yonsei University, Seoul, Korea.
Sun Ha Jee, Department of Epidemiology and Health Promotion, Institute for Health Promotion, Graduate School of Public Health, Yonsei University, Seoul, Korea; Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, USA.
Ethylin Wang Jabs, Johns Hopkins School of Medicine, Baltimore, MD, USA; Mount Sinai School of Medicine, New York, NY, USA.
Richard Redett, Johns Hopkins School of Medicine, Baltimore, MD, USA.
Alan F. Scott, Johns Hopkins School of Medicine, Baltimore, MD, USA
Terri H. Beaty, Email: tbeaty@jhsph.edu, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, USA; Department of Epidemiology, School of Public Health, Johns Hopkins University, 615 N. Wolfe St., Baltimore, MD 21205, USA.
References
- Ardinger HH, Buetow KH, Bell GI, Bardach J, VanDemark DR, Murray JC. Association of genetic variation of the transforming growth factor-alpha gene with cleft lip and palate. Am J Hum Genet. 1989;45:348–353. [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Beaty TH, Maestri NE, Hetmanski JB, Wyszynski DF, Vanderkolk CA, Simpson JC, McIntosh I, Smith EA, Zeiger JS, Raymond GV, Panny SR, Tifft CJ, Lewanda AF, Cristion CA, Wulfsberg EA. Testing for interaction between maternal smoking and TGFA genotype among oral cleft cases born in Maryland 1992– 1996. Cleft Palate Craniofac J. 1997;34:447–454. doi: 10.1597/1545-1569_1997_034_0447_tfibms_2.3.co_2. [DOI] [PubMed] [Google Scholar]
- Beaty TH, Hetmanski JB, Fallin MD, Park JW, Sull JW, McIntosh I, Liang KY, Vanderkolk CA, Redett RJ, Boyadjiev SA, Jabs EW, Chong SS, Cheah FS, Wu-Chou YH, Chen PK, Chiu YF, Yeow V, Ng IS, Cheng J, Huang S, Ye X, Wang H, Ingersoll R, Scott AF. Analysis of candidate genes on chromosome 2 in oral cleft case-parent trios from three populations. Hum Genet. 2006;120:501–518. doi: 10.1007/s00439-006-0235-9. [DOI] [PubMed] [Google Scholar]
- Becker T, Knapp M. Maximum-likelihood estimation of haplo-type frequencies in nuclear families. Genet Epidemiol. 2004;27:21–32. doi: 10.1002/gepi.10323. [DOI] [PubMed] [Google Scholar]
- Becker T, Baur MP, Knapp M. Detection of parent-of-origin effects in nuclear families using haplotype analysis. Hum Hered. 2006;62:64–76. doi: 10.1159/000095942. [DOI] [PubMed] [Google Scholar]
- Bellus GA, Hefferon TW, Ortiz de Luna RI, Hecht JT, Horton WA, Machado M, Kaitila I, McIntosh I, Francomano CA. Achondroplasia is defined by recurrent G380R mutations of FGFR3. Am J Hum Genet. 1995;56:368–373. [PMC free article] [PubMed] [Google Scholar]
- Christensen K, Mitchell LE. Familial recurrence-pattern analysis of nonsyndromic isolated cleft palate—a Danish Registry study. Am J Hum Genet. 1996;58:182–190. [PMC free article] [PubMed] [Google Scholar]
- Cobourne MT. The complex genetics of cleft lip and palate. Eur J Orthod. 2004;26:7–16. doi: 10.1093/ejo/26.1.7. [DOI] [PubMed] [Google Scholar]
- Cordell HJ, Barratt BJ, Clayton DG. Case/pseudocontrol analysis in genetic association studies: a unified framework for detection of genotype and haplotype associations, gene–gene and gene–environment interactions, and parent-of-origin effects. Genet Epidemiol. 2004;26:167–185. doi: 10.1002/gepi.10307. [DOI] [PubMed] [Google Scholar]
- Fan JB, Oliphant A, Shen R, Kermani BG, Garcia F, Gunderson KL, Hansen M, Steemers F, Butler SL, Deloukas P, Galver L, Hunt S, McBride C, Bibikova M, Rubano T, Chen J, Wickham E, Doucet D, Chang W, Campbell D, Zhang B, Kruglyak S, Bentley D, Haas J, Rigault P, Zhou L, Stuelpnagel J, Chee MS. Highly parallel SNP genotyping. Cold Spring Harb Symp Quant Biol. 2003;68:69–78. doi: 10.1101/sqb.2003.68.69. [DOI] [PubMed] [Google Scholar]
- Hwang SJ, Beaty TH, Panny SR, Street NA, Joseph JM, Gordon S, McIntosh I, Francomano CA. Association study of transforming growth factor alpha (TGF alpha) TaqI polymorphism and oral clefts: indication of gene–environment interaction in a population-based sample of infants with birth defects. Am J Epidemiol. 1995;141:629–636. doi: 10.1093/oxfordjournals.aje.a117478. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29:306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- Jugessur A, Lie RT, Wilcox AJ, Murray JC, Taylor JA, Saugstad OD, Vindenes HA, Abyholm F. Variants of developmental genes (TGFA, TGFB3, and MSX1) and their associations with orofacial clefts: a case–parent triad analysis. Genet Epidemiol. 2003a;24:230–239. doi: 10.1002/gepi.10223. [DOI] [PubMed] [Google Scholar]
- Jugessur A, Lie RT, Wilcox AJ, Murray JC, Taylor JA, Saugstad OD, Vindenes HA, Abyholm FE. Cleft palate, transforming growth factor alpha gene variants, and maternal exposures: assessing gene–environment interactions in case–parent triads. Genet Epidemiol. 2003b;25:367–374. doi: 10.1002/gepi.10268. [DOI] [PubMed] [Google Scholar]
- Knapp M, Becker T. Family-based association analysis with tightly linked markers. Hum Hered. 2003;56:2–9. doi: 10.1159/000073727. [DOI] [PubMed] [Google Scholar]
- Maestri NE, Beaty TH, Hetmanski J, Smith EA, McIntosh I, Wyszynski DF, Liang KY, Duffy DL, VanderKolk C. Application of transmission disequilibrium tests to nonsyndromic oral clefts: including candidate genes and environmental exposures in the models. Am J Med Genet. 1997;73:337–344. doi: 10.1002/(sici)1096-8628(19971219)73:3<337::aid-ajmg21>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Miettinen PJ, Chin JR, Shum L, Slavkin HC, Shuler CF, Derynck R, Werb Z. Epidermal growth factor receptor function is necessary for normal craniofacial development and palate closure. Nat Genet. 1999;22:69–73. doi: 10.1038/8773. [DOI] [PubMed] [Google Scholar]
- Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant A, Barker DL, Stuenlpnagel JR, Chee MS. BeadArray™ Technology: enabling an accurate, cost-efficient approach to high-throughput genotyping. Biotechniques. 2002;32:S56–S61. [PubMed] [Google Scholar]
- Park JW, McIntosh I, Hetmanski JB, Jabs EW, Vander Kolk CA, Wu-Chou YH, Chen PK, Chong SS, Yeow V, Jee SH, Park BY, Fallin MD, Ingersoll R, Scott AF, Beaty TH. Association between IRF6 and nonsyndromic cleft lip with or without cleft palate in four populations. Genet Med. 2007;9:219–227. doi: 10.1097/GIM.0b013e3180423cca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubini M, Brusati R, Garattini G, Magnani C, Liviero F, Bianchi F, Tarantino E, Massei A, Pollastri S, Carturan S, Amadori A, Bertagnin E, Cavallaro A, Fabiano A, Franchella A, Calzolari E. Cystathionine beta-synthase c.844ins68 gene variant and non-syndromic cleft lip and palate. Am J Med Genet A. 2005;136:368–372. doi: 10.1002/ajmg.a.30812. [DOI] [PubMed] [Google Scholar]
- Salyakina D, Seaman SR, Browning BL, Dudbridge F, Muller-Myhsok B. Evaluation of Nyholt’s procedure for multiple testing correction. Hum Hered. 2005;60:19–25. doi: 10.1159/000087540. [DOI] [PubMed] [Google Scholar]
- Shaw GM, Wasserman CR, Lammer EJ, O’Malley CD, Murray JC, Basart AM, Tolarova MM. Orofacial clefts, parental cigarette smoking, and transforming growth factor-alpha gene variants. Am J Hum Genet. 1996;58:551–561. [PMC free article] [PubMed] [Google Scholar]
- Shaw GM, Wasserman CR, Murray JC, Lammer EJ. Infant TGF-alpha genotype, orofacial clefts, and maternal periconceptional multivitamin use. Cleft Palate Craniofac J. 1998;35:366–370. doi: 10.1597/1545-1569_1998_035_0366_itagoc_2.3.co_2. [DOI] [PubMed] [Google Scholar]
- Sinsheimer JS, Palmer CG, Woodward JA. Detecting genotype combinations that increase risk for disease: maternal–fetal genotype incompatibility test. Genet Epidemiol. 2003;24:1–13. doi: 10.1002/gepi.10211. [DOI] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- Starr JR, Hsu L, Schwartz SM. Assessing maternal genetic associations: a comparison of the log-linear approach to case–parent triad data and a case–control approach. Epidemiology. 2005;16:294–303. doi: 10.1097/01.ede.0000158223.98649.eb. [DOI] [PubMed] [Google Scholar]
- Sull JW, Liang KY, Hetmanski JB, Fallin MD, Ingersoll RG, Park J, Wu-Chou YH, Chen PK, Chong SS, Cheah F, Yeow V, Park BY, Jee SH, Jabs EW, Redett R, Jung E, Ruczinski I, Scott AF, Beaty TH. Differential parental transmission of markers in RUNX2 among cleft case–parent trios from four populations. Genet Epidemiol. 2008;32:505–512. doi: 10.1002/gepi.20323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tricoli JV, Nakai H, Byers MG, Rall LB, Bell GI, Shows TB. The gene for human transforming growth factor alpha is on the short arm of chromosome 2. Cytogenet Cell Genet. 1986;42:94–98. doi: 10.1159/000132258. [DOI] [PubMed] [Google Scholar]
- van Den Oord EJ, Vermunt JK. Testing for linkage disequilibrium, maternal effects, and imprinting with (in)complete case– parent triads, by use of the computer program LEM. Am J Hum Genet. 2000;66:335–338. doi: 10.1086/302708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij IA, Vermeij-Keers C, Kluijtmans LA, Ocké MC, Zielhuis GA, Goorhuis-Brouwer SM, van der Biezen JJ, Kuijpers-Jagtman AM, Steegers-Theunissen RP. Does the interaction between maternal folate intake and the methylenetetrahydrofolate reductase polymorphisms affect the risk of cleft lip with or without cleft palate? Am J Epidemiol. 2003;157:583–591. doi: 10.1093/aje/kwg005. [DOI] [PubMed] [Google Scholar]
- Vansteelandt S, Demeo DL, Lasky-Su J, Smoller JW, Murphy AJ, Mc-Queen M, Schneiter K, Celedon JC, Weiss ST, Silverman EK, Lange C. Testing and estimating gene–environment interactions in family-based association studies. Biometrics. 2007;64:458–467. doi: 10.1111/j.1541-0420.2007.00925.x. [DOI] [PubMed] [Google Scholar]
- Vieira AR. Association between the transforming growth factor alpha gene and nonsyndromic oral clefts: a HuGE review. Am J Epidemiol. 2006;163:790–810. doi: 10.1093/aje/kwj103. [DOI] [PubMed] [Google Scholar]
- Vieira AR, Modesto A, Meira R, Barbosa AR, Lidral AC, Murray JC. Interferon regulatory factor 6 (IRF6) and fibroblast growth factor receptor 1 (FGFR1) contribute to human tooth agenesis. Am J Med Genet A. 2007;143:538–545. doi: 10.1002/ajmg.a.31620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg CR. Methods for detection of parent-of-origin effects in genetic studies of case–parents triads. Am J Hum Genet. 1999;65:229–235. doi: 10.1086/302466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg CR, Umbach DM. A hybrid design for studying genetic influences on risk of diseases with onset early in life. Am J Hum Genet. 2005;77:627–636. doi: 10.1086/496900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins JF, Haig D. What good is genomic imprinting: the function of parent-specific gene expression. Nat Rev Genet. 2003;4:359–368. doi: 10.1038/nrg1062. [DOI] [PubMed] [Google Scholar]
- Wyszynski DF, Duffy DL, Beaty TH. Maternal cigarette smoking and oral clefts: a meta-analysis. Cleft Palate Craniofac J. 1997;34:206–210. doi: 10.1597/1545-1569_1997_034_0206_mcsaoc_2.3.co_2. [DOI] [PubMed] [Google Scholar]
- Zeiger JS, Beaty TH, Liang KY. Oral clefts, maternal smoking, and TGFA: a meta-analysis of gene-environment interaction. Cleft Palate Craniofac J. 2005;42:58–63. doi: 10.1597/02-128.1. [DOI] [PubMed] [Google Scholar]
- Zhao H, Zhang S, Merikangas KR, Trixler M, Wildenauer DB, Sun F, Kidd KK. Transmission/disequilibrium tests using multiple tightly linked markers. Am J Hum Genet. 2000;67:936–946. doi: 10.1086/303073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchero TM, Cooper ME, Maher BS, Daack-Hirsch S, Nepomuceno B, Ribeiro L, Caprau D, Christensen K, Suzuki Y, Machida J, Natsume N, Yoshiura K, Vieira AR, Orioli IM, Castilla EE, Moreno L, Arcos-Burgos M, Lidral AC, Field LL, Liu YE, Ray A, Goldstein TH, Schultz RE, Shi M, Johnson MK, Kondo S, Schutte BC, Marazita ML, Murray JC. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med. 2004;351:769–780. doi: 10.1056/NEJMoa032909. [DOI] [PubMed] [Google Scholar]