Abstract
The presence of nucleotide hybridization between the 3′ end of 16S rRNA and mRNA sequence upstream of the start codon is well known in bacteria. In this paper, we detect the presence of such hybridization sites inside the coding regions of E. coli genes, and analyze their proximity to clusters of slow-translating codons. We study this phenomenon in genes of high and low expression separately. Based on our findings, we propose an explanation for the presence of RNA hybridization within the translated regions of bacterial genes.
Keywords: codon, Shine-Dalgarno, hybridization, translation, gene expression
Introduction
During the initiation of translation in prokaryotes, the 3′ end of the 16S subunit of the ribosome is known to bind to a short stretch of nucleotides located about 5–7 bases upstream of the start codon. This purine-rich portion of mRNA, referred to as the Shine-Dalgarno (SD) sequence1 has the general form AGGAGG and is somewhat complementary (by Watson-Crick base-pairing principles) to the nucleotides at the 3′ end of the 16S rRNA. The SD sequence helps anchor the mRNA to the 16S rRNA subunit during translation initiation.2 Genes that are highly expressed in the bacterial cell have been shown to have stronger SD hybridization.3 Other studies have explored the effect of SD sequence position and mRNA structure around the start codon, and concluded that (a) the SD region needs to be at a short distance upstream of the start codon in order to enhance gene expression4 and (b) translation initiation is a rate-limiting factor for gene expression.5
As early as 1979, Sedlacek et al6 observed that nucleotide complementarity between the 16S subunit of rRNA (3′ end) and messenger RNA (mRNA) could also extend into the coding regions. By analyzing the relationship between nucleotide composition of mRNA sequences and the 16S rRNA’s 3′ end, they found complementarity throughout the translated regions, not just before the start codon. This complementarity was found to be stronger at the first and second positions of each codon in the mRNA sequence. Weaker complementarity at every third position was thought to serve the purpose of preventing the ribosome from getting stuck to the mRNA, thereby hindering the normal translation rate. Weiss et al7 also confirmed SD-like hybridization within the mRNA sequence based on a careful examination of the frameshift site in the prfB gene of E. coli. They detected SD-like base-pairing at about 5 bases upstream of the frameshift site, lending experimental evidence to the presence of rRNA-mRNA hybridization within the coding region. They made nucleotide modifications to this SD-like portion and found that the frameshift level drastically reduced, indicating the important role of this internal SD site in causing the ribosome to change its reading frame. These findings led them to conclude that the ribosome “scans the mRNA very close to the decoding sites during elongation” and raised the possibility of other functional roles for internal SD-like hybridization.7
Recently, Wen et al8 were able to present a detailed picture of ribosome translation by crystallographic techniques using optical tweezers. They found translation arrests at SD-like regions within the mRNA coding region. And upon making slight modifications to the internal SD site, the ribosomal pause was found to disappear. This study gave the clearest evidence of rRNA-mRNA hybridization during elongation. In order to find other such pause sites where there is complementarity between mRNA sequence and the 3’ end of 16S rRNA, computational methods can be used. Calculating the strength of binding between mRNA and rRNA based on the principles of thermodynamics presents a good way to capture this interaction.9,10
It is known that the process of elongation occurs at a non-uniform rate and that the speed of elongation is strongly influenced by the concentration of tRNAs.11 It has been shown that the ribosomal wait time at a slow-translating codon is roughly inversely proportional to the concentration of tRNAs that recognize that codon.12 If the stochastic search for the correct tRNA takes too long, undesirable consequences are possible. The ribosome could drop off, thereby dissociating from the mRNA before the completion of the polypeptide chain.13,14 The search for the correct tRNA isoacceptor is expected to take the longest time at codons that are recognized by lowest-concentration tRNAs,15,16 increasing the likelihood of ribosomal drop-off. This negatively impacts translation efficiency and hinders the rate of protein production. It has been shown that stable mRNA-rRNA hybridization can keep the ribosome attached to the elongating mRNA and prevent it from drop-off.8 We would therefore expect to see SD-like hybridization occuring at locations preceding slow-translating codons, atleast in highly expressed genes. This is precisely the hypothesis we propose to test in this paper.
If slowly-translated codons create translational problems, the ribosome must have a mechanism of stabilizing itself as it processes such codons in the highly-expressed genes. We hypothesize that the distance between SD-like hybridization (involving the ribosome and the mRNA) and slowly-translated codons plays a critical role in this process.
Methods
We downloaded the E. coli genome from NCBI GenBank (accession NC 000913.2) and extracted all the annotated gene sequences. We then used the database of E. coli gene expression (HEG)17 to select two groups of genes: (1) highly expressed genes, 243 in number (2) genes of lowest expression, 308 in number (http://genomes.urv.cat/HEG-DB/).
We calculated the hybridization free-energy signal for each of these genes based on position-wise incremental alignments between mRNA and the 3′ end of 16S rRNA. We began by aligning the two RNA strands at the position of the start codon, and estimated their hybridization free-energy using a dynamic programming approach as discussed in our earlier work.18 If no hybridization is possible, a value of zero is assigned to the free-energy estimate. Greater complementarity between the two RNA strands in consecutive positions yields higher values of free-energy, as illustrated in Figure 2.
Figure 2.
Examples of SD-like hybridization sites inside coding region of genes in E.coli, illustrating the relationship between nature of complementarity and absolute free-energy values (shown on the right side).
The 16S rRNA is then moved one base pair downstream, thereby creating a new potential hybridization site. The free-energy released from this alignment is then calculated. This procedure is repeated, each time moving the rRNA sequence in the 5′–3′ direction of the coding region, until the stop codon is encountered. The calculated series of free-energy estimates constitute our dataset of binding values which we will examine closely in order to test the proposed hypothesis.
The analysis of hybridization potentials is somewhat similar to that of Osada et al9 except that we calculate binding energy at all positions along the mRNA sequence (see Fig. 1). We found that roughly 8%–10% of the alignment positions in each gene sequence yield non-zero hybridization.
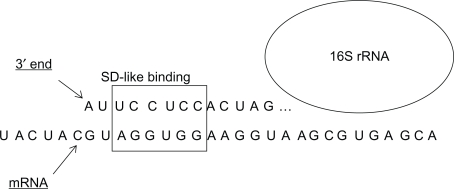
Figure 1.
Interaction between 16S rRNA and mRNA within the coding region, showing SD-like complementarity (in the region marked by a square box).
Based on experimental measurements, we selected 15 codons having the lowest tRNA concentration among all codons in E. coli, using data reported in earlier studies:12,19 UUA, AAG, AGG, UUC, AGU, UCA, CAU, UCC, CGA, CCC, CAC, AUA, ACA, CCA, CUA. The positions of these codons were marked in each examined gene sequence, and their proximity to hybridization sites was calculated. For clusters of consecutive slow-translating codons, we noted the number of such codons in each cluster (N), and the nucleotide spacing (D) between the 5′ end of such a cluster and the nearest upstream hybridization site. We then compared the values of D from highly-expressed and poorly-expressed genes to see if there is any significant difference.
Results
We observed the presence of slow-translating codons in all types of genes in E. coli, regardless of their expression level. But we found a clear difference in the proportion of such codons between genes of high and low expression level.
Since the number of slow-translating codons is higher in low-expressed genes, we found many more values for D in such genes. The values of D do not follow a normal distribution, and hence cannot be compared using a standard two-sample t-test. We therefore employed a non-parametric test of the medians in the two samples, also referred to as the Wilcoxon rank sum test.20
For every group of N slow-translating codons, we compared values of D from two distinct gene-sets: highly-expressed genes (H) and poorly-expressed genes (L). We found that the median value of D is significantly different in the two sets examined, based on the P-value (<0.01). We also found that larger groups of slow-translating codons have closer upstream hybridization sites (see Table 1). Since the sample size of the dataset decreases drastically with N, the reported P-value is somewhat unreliable for N = 4. For higher values of N, a comparison test cannot be performed since the sample size is extremely low.
Table 1.
Given the number of consecutive slow-translating codons (N), the median distance to the nearest SD-like hybridization site is shown for genes of high expression (Dh) and low expression (Dl). The P-value from Wilcoxon rank sum test and the sample sizes in each case (Sh and Sl) are also shown.
| N | Dh | Dl | P-value | Sh | Sl |
|---|---|---|---|---|---|
| 1 | 8 | 9 | 0.000000 | 5796 | 8544 |
| 2 | 6 | 9 | 0.000002 | 556 | 1669 |
| 3 | 3.5 | 9 | 0.000445 | 50 | 380 |
| 4 | 2 | 10.5 | 0.250002 | 5 | 78 |
Discussion
Our main result in this paper is that SD-like hybridization sites are found closer to larger groups of consecutive slow-translating codons. There is a clear difference between genes of high and low expression in the number of nucleotides that span the space between slow-translating codons and the nearest upstream hybridization site. This indicates two things: (a) the SD-like hybridization is preferentially closer to clusters of slow-translating codons, and (b) the proximity of such hybridization sites enables higher levels of gene expression. This can be attributed to the ability of rRNA-mRNA hybridization to latch the ribosome to the mRNA, thereby preventing it from dropping off during the time-consuming search for the tRNA isoacceptors of slow-translating codons.
Previous studies of the role of SD hybridization have examined only the 5′ region upstream of the start codon in bacterial genes.9 It has been found that translation efficiency is sensitive to the distance between the start codon and SD region21 and that there is an optimal value of 7 nucleotides for this spacing.22,23 We have examined the occurence of SD-like hybridization inside the coding regions of mRNA in E. coli, by calculating free-energy of binding between the 3′ end of the 16S rRNA and the mRNA sequence. We also examined the proximity of such hybridization sites to clusters of slow-translating codons, in an attempt to attribute a plausible role to such internal SD sites.
Using experimental methods, it has been shown that the SD interaction typically increases the rupture force of the ribosome by about 10pN.24 This leads to pauses in translation, and could serve a useful purpose if located near clusters of slow-translating codons. For such codons, the stochastic search for the correct tRNA isoacceptor takes the longest time, and can lead to undesirable consequences while the ribosome waits for the tRNA recognition to occur. The occurence of SD-like hybridization within a few nucleotides upstream of the slow-translating cluster can help prevent the ribosome from dissociating. We found stronger evidence for this mechanism in highly-expressed genes, lending support to our hypothesis.
In future studies, we are interested in exploring the nature of internal SD-like hybridization in a wide variety of bacterial species. It would be worthwhile to see if the results we found in E. coli are somewhat conserved in other species as well. It is also possible to examine the strength of hybridization (in terms of the magnitude of free-energy released) in relation to the density of slow-translating codon clusters. Such studies would shed light on critical post-transcriptional control mechanisms in bacteria.
Footnotes
Disclosures
This manuscript has been read and approved by all authors. This paper is unique and is not under consideration by any other publication and has not been puplished elsewhere. The authors and peer reviewers of this paper report no conflicts of interest. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974;71(4):1342–6. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hui A, de Boer HA. Specialized ribosome system: preferential translation of a single mrna species by a subpopulation of mutated ri-bosomes in Escherichia coli. Proc Natl Acad Sci U S A. 1987;84(14):4762–6. doi: 10.1073/pnas.84.14.4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma J, Campbell A, Karlin S. Correlations between Shine-Dalgarno sequences and gene features such as predicted expression levels and operon structures. J Bacteriol. 2002;184(20):5733–45. doi: 10.1128/JB.184.20.5733-5745.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin H, Zhao Q, Gonzalez de Valdivia EI, Ardell DH, Stenstrm M, Isaksson LA. Influences on gene expression in vivo by a Shine-Dalgarno sequence. Mol Microbiol. 2006;60(2):480–92. doi: 10.1111/j.1365-2958.2006.05110.x. [DOI] [PubMed] [Google Scholar]
- 5.Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324(5924):255–8. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sedlcek J, Fbry M, Rychlk I, Voln D, Vtek A. The arrangement of nucleotides in the coding regions of natural templates. Mol Gen Genet. 1979;172(1):31–6. doi: 10.1007/BF00276212. [DOI] [PubMed] [Google Scholar]
- 7.Weiss RB, Dunn DM, Dahlberg AE, Atkins JF, Gesteland RF. Reading frame switch caused by base-pair formation between the 3’ end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 1988;7(5):1503–7. doi: 10.1002/j.1460-2075.1988.tb02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wen JD, Lancaster L, Hodges C, et al. Following translation by single ribosomes one codon at a time. Nature. 2008;452(7187):598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osada Y, Saito R, Tomita M. Analysis of base-pairing potentials between 16S rRNA and 5’ UTR for translation initiation in various prokaryotes. Bioinformatics. 1999;15(7–8):578–81. doi: 10.1093/bioinformatics/15.7.578. [DOI] [PubMed] [Google Scholar]
- 10.Schurr T, Nadir E, Margalit H. Identification and characterization of E.coli ribosomal binding sites by free energy computation. Nucleic Acids Res. 1993;21(17):4019–23. doi: 10.1093/nar/21.17.4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varenne S, Buc J, Lloubes R, Lazdunski C. Translation is a nonuniform process: Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J Mol Biol. 1984;180(3):549–76. doi: 10.1016/0022-2836(84)90027-5. [DOI] [PubMed] [Google Scholar]
- 12.Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol. 2009;16(3):274–80. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
- 13.Cruz-Vera LR, Magos-Castro MA, Zamora-Romo E, Guarneros G. Ribosome stalling and peptidyl-tRNA drop-off during translational delay at AGA codons. Nucleic Acids Res. 2004;32(15):4462446. doi: 10.1093/nar/gkh784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchan JR, Stansfield I. Halting a cellular production line: responses to ribosomal pausing during translation. Biol Cell. 2007;99(9):475–87. doi: 10.1042/BC20070037. [DOI] [PubMed] [Google Scholar]
- 15.Marin M. Folding at the rhythm of the rare codon beat. Biotechnol J. 2008;3(8):1047–57. doi: 10.1002/biot.200800089. [DOI] [PubMed] [Google Scholar]
- 16.Kurland CG, Ehrenberg M. Growth-optimizing accuracy of gene expression. Annu Rev Biophys Biophys Chem. 1987;16:291–317. doi: 10.1146/annurev.bb.16.060187.001451. [DOI] [PubMed] [Google Scholar]
- 17.Puigbo P, Romeu A, Garcia-Vallve S. HEG-DB: a database of predicted highly expressed genes in prokaryotic complete genomes under translational selection. Nucleic Acids Res. 2007;36:D524–D527. doi: 10.1093/nar/gkm831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ponnala L, Stomp AM, Bitzer DL, Vouk MA. Analysis of free energy signals arising from nucleotide hybridization between rrna and mrna sequences during translation in eubacteria. EURASIP J Bioinform Syst Biol. 2006;(23613) doi: 10.1155/BSB/2006/23613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong H, Nilsson L, Kurland CG. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol. 1996;260(5):649–63. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 20.Gibbons JD. In: Nonparametric Statistical Inference. 2 edition. Dekker M, editor. 1985. [Google Scholar]
- 21.Ringquist S, Shinedling S, Barrick D, et al. Translation initiation in Escherichia coli: sequences within the ribosome-binding site. Mol Microbiol. 1992;6(9):1219–29. doi: 10.1111/j.1365-2958.1992.tb01561.x. [DOI] [PubMed] [Google Scholar]
- 22.Chen H, Bjerknes M, Kumar R, Jay E. Determination of the optimal aligned spacing between the shine-dalgarno sequence and the translation initiation codon of escherichia coli mRNAs. Nucleic Acids Res. 1994;22(23):4953–4957. doi: 10.1093/nar/22.23.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444(7117):391–4. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- 24.Uemura S, Dorywalska M, Lee TH, Kim HD, Puglisi JD, Chu S. Peptide bond formation destabilizes Shine-Dalgarno interaction on the ribosome. Nature. 2007;446(7134):454–7. doi: 10.1038/nature05625. [DOI] [PubMed] [Google Scholar]