Abstract
Important biological information is encoded in the topology of biological networks. Comparative analyses of biological networks are proving to be valuable, as they can lead to transfer of knowledge between species and give deeper insights into biological function, disease, and evolution. We introduce a new method that uses the Hungarian algorithm to produce optimal global alignment between two networks using any cost function. We design a cost function based solely on network topology and use it in our network alignment. Our method can be applied to any two networks, not just biological ones, since it is based only on network topology. We use our new method to align protein-protein interaction networks of two eukaryotic species and demonstrate that our alignment exposes large and topologically complex regions of network similarity. At the same time, our alignment is biologically valid, since many of the aligned protein pairs perform the same biological function. From the alignment, we predict function of yet unannotated proteins, many of which we validate in the literature. Also, we apply our method to find topological similarities between metabolic networks of different species and build phylogenetic trees based on our network alignment score. The phylogenetic trees obtained in this way bear a striking resemblance to the ones obtained by sequence alignments. Our method detects topologically similar regions in large networks that are statistically significant. It does this independent of protein sequence or any other information external to network topology.
Keywords: network alignment, biological networks, network topology, protein function prediction, phylogeny
Introduction
Background
Increasing amounts of biological network data are becoming available, owing to advances in experimental techniques such as yeast two-hybrid assay, mass spectrometry of purified complexes, genome-wide chromatin immunoprecipitation, correlated m-RNA expression, and genetic interactions. Examples include protein-protein interaction (PPI) networks,1–11 metabolic networks,12 transcriptional regulatory networks,13 protein structure networks,14,15 and networks summarizing neuronal connectivities.16 Comparative analyses of biological networks are expected to have at least as large an impact as comparative genomics on our understanding of biology, evolution, and disease. As more biological network data is becoming available, meaningful network comparisons across species could be viewed as one of the foremost problems in evolutionary and systems biology.17 Such comparisons could guide the transfer of knowledge across species and give insights into evolutionary conservation of protein function, protein interactions, and protein complexes. They can also be used to infer phylogenetic relationships of different species based on the level of topological similarities between their molecular networks.
Exact network comparisons are computationally infeasible due to NP-completeness of the underlying subgraph isomorphism problem, which asks if one graph (or network) exists as an exact subgraph of another graph.18 Network alignment is the most common network comparison method;17 it is the more general problem of finding the best way to “fit” a graph into another graph even if the first graph is not an exact subgraph of the second one. An alignment is achieved by constructing a mapping between the nodes of networks being compared; the alignment is expected to align topologically and functionally conserved network regions. However, it is unclear how to guide the alignment process and construct such a node mapping; also, it is unclear how to measure the “goodness” of an inexact fit between the aligned networks. This is especially true for the biological networks that we consider below, due to biological variation, as well as noise in the data.19 Thus, heuristic strategies must be sought to guide the alignment process, as well as to quantify the quality of an alignment.
Analogous to sequence alignments, network alignment algorithms can be categorized as either local or global. The majority of the existing methods produce local alignments of biological networks.20–25 Such alignments typically match a small subnetwork from one network to one or more subnetworks in another network, with the hypothesis that such aligned subgraphs are conserved through evolution.21,20 In contrast to many-to-many local alignments that can be ambiguous, a global network alignment measures the overall similarity between two networks by aligning every node in the smaller network to exactly one node in the larger network, despite this possibly leading to suboptimal matchings in some local regions. Global network alignment has been applied previously in the biological networks domain,26–31 but most existing methods incorporate some a priori information external to network topology, such as sequence similarities of proteins in PPI networks,24,26,28,29,31 phylogenetic relationships between species whose networks are being aligned,22 or they use some form of learning on a set of “true” alignments.27 The best currently available global network alignment algorithm that is based solely on network topology is our recent GRAph ALigner (GRAAL),30 which uses a heuristic search strategy to quickly find approximate alignments. In contrast, here we present a more expensive search algorithm guaranteed to find optimal alignments relative to any fixed, deterministic cost function.
Our contribution
Sequences have been shown to be very valuable sources of biological information. It has already been shown that non-sequence based sources of biological information, such as the secondary or tertiary molecular structure, might be more appropriate for extracting certain types of biological knowledge than sequence-based ones.32–34 Analogously, there is important biological information that can be extracted from the topology of biological networks and that cannot easily be extracted from biological information external to network topology.35 For example, identical protein sequences can fold in different ways under different conditions, resulting in different 3-dimensional structures and functions.36–39 Since the structure of a protein is expected to define the number and type of its potential interacting partners in the PPI network, different structures would also lead to very different PPI network topologies.36–39 Moreover, entirely different sequences can produce identical protein structures.40,38 In cases where such proteins are expected to share a common function, a sequence-based function prediction would fail, whereas a network topology-based one would not. Therefore, we believe that network topology can uncover important biological information that is independent of other currently available biological information.35
Hence, unlike the previous local and global network alignment algorithms that depend only implicitly or indirectly on network topology, we introduce an algorithm called H-GRAAL (Hungarian-algorithm-based GRAAL)41 that relies solely and explicitly on a strong and direct measure of network topological similarity. As such, it is easily applicable to any type of networks, not just biological ones. In contrast to our previous greedy “seed-and-extend” approach that also relies solely on network topology,30 H-GRAAL is an optimal alignment algorithm (see below for details). Note, however, that it is trivial to include sequence or other biological information into the cost function of our method, as explained in our previous work,30 but this is out of the scope of the manuscript. Even though it is important to use all available sources of biological information to try to understand complex biological systems, it is as important to understand how much biological information can be obtained from each source of biological data individually.
We align with H-GRAAL the PPI networks of yeast and human and demonstrate that our alignment exposes topologically complex and biologically relevant regions of similarity. Since we may know a lot about some of the nodes in one network and almost nothing about topologically similar nodes in the other network, we can transfer the knowledge from one to the other to uncover new biology. Hence, we use our alignment to predict protein function of unannotated proteins in one species based on the functions of their annotated alignment partners in the other species. To demonstrate effectiveness of topological alignment, we validate a large number of our predictions in the literature.
Network alignments can also be used to measure overall similarity between networks of different species. Given a group of such biological networks, the matrix of pairwise global network similarities can be used to infer phylogenetic relationships. Thus, we apply our method to find topological similarities between metabolic networks of different species, and then build phylogenetic trees that bear a striking resemblance to the ones obtained from sequence comparisons. The significance of our method is that it uncovers large and dense optimal alignments (relative to our cost function) and extracts biologically relevant and statistically significant meaning solely from network topology, independently of any other source of biological information. Moreover, it outperforms greedy GRAAL, the current best method30 (see Results and Discussion section).
Methods
Graphlet degree vectors and signature similarities
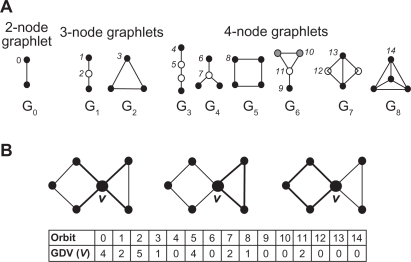
To build meaningful alignments based solely upon network topology, we match pairs of nodes from different networks using a highly constraining measure of their topological similarity, as defined by Milenković and Pržulj42 and explained below. We define a graphlet as a small, connected, induced subgraph of a larger network43,44 (Fig. 1A). An induced subgraph on a node set S ⊆ V of G is obtained by taking S and all edges of G having both end-nodes in S. For a particular node v in a network, we generalize the degree of a node, which counts the number of edges that the node touches, into the vector of graphlet degrees, counting the number of graphlets that the node touches, for all graphlets on 2 to 5 nodes (Fig. 1B). The degree of a node is the first coordinate in this vector, since an edge is the only 2-node graphlet, denoted by G0 in Figure 1A. It is important to distinguish between, for example, nodes touching a 3-node path, i.e. graphlet G1 in Figure 1A, at an end or at the middle. Hence, the notion of automorphism orbits (or just orbits, for brevity) is used: by taking into account the “symmetries” between nodes of a graphlet, there are 73 different orbits across all 2-5-node graphlets (see Figure 1A for an illustration and Przulj44 for details). We number the orbits from 0 to 72.44 The full vector of 73 coordinates is the signature of a node that describes the topology of its neighborhood and captures its interconnectivities out to a distance of 4 (see Figure 1B for an illustration and Milenković and Pržulj42 for details). This is an effective measure, since going to network distance of 4 around a node captures a large portion of network topology due to the small-world nature of many real-world networks.45 For this reason, and since the number of graphlets on n nodes increases exponentially with n, we believe that using larger graphlets would unnecessarily increase the computational complexity of the method.
Figure 1.
A) All 9 graphlets on 2, 3 and 4 nodes, denoted by G0, G1, ..., G8; they contain 15 topologically unique node types, called “automorphism orbits,” denoted by 0, 1, 2, ..., 14. In a particular graphlet, nodes belonging to the same orbit are of the same shade.44 B) An illustration of the “Graphlet Degree Vector” (GDV), or a “signature” of node v; coordinates of a GDV count how many times a node is touched by a particular automorphism orbit, such as an edge (the leftmost panel), a triangle (the middle panel), or a square (the rightmost panel). Hence, the degree is generalized to a GDV.42 The GDV of node v is presented in the table for orbits 0 to 14: v is touched by 4 edges (orbit 0), end-nodes of 2 graphlets G1 (orbit 1), etc. For an example of GDV of a node for all 73 orbits (corresponding to all 30 2–5-node graphlets), see Kuchaiev et al.30
The signature of a node provides a novel and highly constraining measure of local topology in its vicinity and comparing the signatures of two nodes provides a highly constraining measure of local topological similarity between them. The signature similarity between two nodes42 is computed as follows. For a node u ∈ G, ui denotes the ith coordinate of its signature vector, i.e. ui is the number of times node u is touched by an orbit i in G. The distance Di(u,v) between the ith orbits of nodes u and v is defined as:
where wi is the weight of orbit i that accounts for dependencies between orbits. For example, the differences in orbit 0 (i.e. in the degree) of two nodes will imply the differences in all other orbits for these nodes, since all orbits contain, i.e. “depend on”, orbit 0.42 Similarly, the differences in orbit 3 (the triangle) of two nodes will imply the differences in all other orbits that contain orbit 3, such as orbits 10–14, etc. We generalize this to all orbits, assigning higher weights to orbits that are not affected by many other orbits than to orbits that depend on many other orbits.42 By doing so, we remove the redundancy of an orbit being contained in other orbits, this being a reason why we design our own measure of similarity between graphlet degree vectors of two nodes instead of using standard metrics such as the Euclidean distance; for details, see Milenković and Pržulj.42
The total distance D(u,v) between nodes u and v is defined as:
The distance D(u,v) is in (0,1), where distance 0 means that signatures of nodes u and v are identical. Finally, the signature similarity, S(u,v), between nodes u and v is S(u,v) = 1 − D(u,v). Clearly, a higher signature similarity between two nodes corresponds to a higher topological similarity between their extended neighborhoods (out to distance 4).
H-GRAAL algorithm
Let V (G) denote the set of nodes of network G. Let G1 and G2 be networks and assume without loss of generality that |V (G1)| ≤ |V (G2)|. An alignment of G1 to G2 is a set of ordered pairs (u,v), u ∈ V (G1) and v ∈ V (G2), such that no two ordered pairs share the same G1-node or the same G2-node. We will call each such ordered pair in an alignment an aligned pair. A maximum alignment is one where every G1-node is in some aligned pair of the alignment. Unless otherwise specified, we will henceforth take “alignment” with respect to networks to mean “maximum alignment”. Note that if G2 has more nodes than G1 then for each alignment (maximum or not) at least one node in G2 will not be in any aligned pair.
Our algorithm, called H-GRAAL (Hungarian-algorithm-based GRAph ALigner), produces an alignment of minimum total cost between networks, where the total cost is summed over all aligned pairs, and the cost of each aligned pair of nodes is computed based on their node signature similarity as defined above.42 The cost of aligning two nodes u and v is modified to favor alignment of the densest parts of the networks; the cost is reduced as the degrees of both nodes increase, since higher degree nodes with similar signatures provide a tighter constraint than correspondingly similar low degree nodes; α ∈ [0, 1] is a parameter that weighs the cost-function contributions of the node signature similarity between u and v, and 1 − α weights the contribution of degrees of the nodes u and v. More specifically, if deg(u) is the degree of a node u, max_deg(G1) and max_deg(G2) are the maximum degrees of nodes in networks G1 and G2 respectively, S(u,v) is the signature similarity of nodes u and v as defined above, and α is as defined above, then the cost of aligning nodes u and v, C(u,v), is given by the following formulas:
A cost of 0 corresponds to a pair of topologically identical nodes u and v, while a cost close to 2 corresponds to a pair of topologically very different nodes.
Clearly, most nodes u and v will be of low degree, since biological networks G1 and G2 have power-law degree distributions and hence T(u,v) will be very low; this is because power-law degree distributions of G1 and G2 imply the existence of a small number of hubs (highly-linked nodes), so max_deg(G1) and max_deg(G2) will be much larger than deg(u) and deg(v) for most nodes u and v. This will give more weight to signature similarity S(u,v) even for very small α (e.g. α = 0.1). However, for α = 0, the entire weight will be given to degrees.
We use the Hungarian algorithm for minimum-weight bipartite matching41 to find an optimal alignment from G1 to G2 with respect to the cost function described above. The Hungarian algorithm is a standard polynomial-time algorithm for solving the linear assignment problem. Details of the algorithm can be found in most texts on graph algorithms e.g. West;41 an excellent outline can also be found in Mills-Tettey et al46 as a prelude to the presentation of the dynamic Hungarian algorithm. We set up a complete bipartite graph with V(G1) and V(G2) as the bipartition and each edge (u,v) from V(G1) to V(G2) is labeled with the node alignment cost between u and v. H-GRAAL then uses the Hungarian algorithm to find an alignment from G1 to G2 by minimizing the cost summed over all aligned pairs. Note that while there may be more than one optimal alignment (with the same minimum cost), H-GRAAL initially returns only one such alignment. Furthermore, the particular alignment found by H-GRAAL is highly dependent on the implementation details of the underlying Hungarian algorithm. For example, the order in which the nodes of G1 and G2 are presented to the algorithm can influence which augmenting paths are found at each stage of the algorithm, which in turn determines the ultimate matching found. While this potential variability in results returned from different implementation of the Hungarian algorithm may seem disconcerting at first, there are relatively simple and efficient measures that can be taken to learn about all possible optimal matchings, not just any one matching a particular implementation happens to return. These measures are described in the following paragraphs.
If we already have an optimal alignment A0, such as the one found from an initial invocation of H-GRAAL, a simple way to force H-GRAAL to generate another optimal alignment is to raise the alignment cost of a node-pair (u,v) in A0 to +∞ and then invoke H-GRAAL on the modified alignment problem. Artificially inflating the cost of (u,v) guarantees that the Hungarian algorithm will never select it as part of any optimal alignment, effectively removing (u,v) from consideration. We term such a cost modification of (u,v) a removal of (u,v). So any alignment (subsequent to A0) found by H-GRAAL after removing (u,v) cannot possibly contain (u,v) and must therefore be different from A0. If this subsequent invocation of H-GRAAL produces an alignment with the same cost as A0, then we have another optimal alignment to the original problem. If a costlier alignment is produced, then we need to pick another aligned pair from A0 and repeat the process. If we have picked all pairs of A0 without success, then A0 is the only optimal alignment possible.
The only problem with this simple procedure is one of efficiency. If G1 and G2 have O(n) nodes, the Hungarian algorithm can run in O(n3) time. So each invocation of H-GRAAL from scratch will take O(n3) time, which can be prohibitive for large networks, such as biological networks that we consider below. However, recent work on a dynamic version of the Hungarian algorithm allows us to solve a derived matching problem obtained by perturbing a small number of edge costs in the original problem, for which an optimal matching is already known.46 With this variant it is possible to efficiently “repair” the original matching to yield an optimal matching for the derived problem; each modified edge cost takes O(n2) time to repair, in contrast to O(n3) time if we had to start from scratch with the original Hungarian algorithm. In addition to the usual inputs to the original Hungarian algorithm, the dynamic variant takes as inputs an optimal alignment A, the final internal state of the Hungarian algorithm execution that produced A, and the set of edge costs to change. The internal state comprises, more specifically, the dual variables associated with each node in the bipartition. The dual variables are used internally by the Hungarian algorithm and their precise function need not concern us; we only need to preserve this state from one run of the algorithm to the next when using the dynamic algorithm. For our purposes, after H-GRAAL has generated the first optimal alignment, we can use the procedure described in the last paragraph, in conjunction with the dynamic Hungarian algorithm, to find subsequent optimal alignments efficiently.
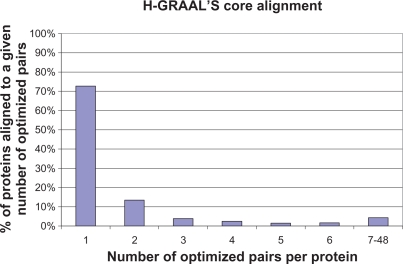
Even with the improved efficiency from using the dynamic Hungarian algorithm, network alignment problems will typically yield far too many optimal alignments to make exhaustive enumeration practical; yet it would be undeniably useful to somehow summarize information about the set of all possible optimal alignments. To this end, we say that an aligned pair is optimizing if it appears in at least one optimal alignment. The set of all optimizing aligned pairs is clearly the union of all aligned pairs from all optimal alignments, which gives a short, O(n2)-sized, synopsis of the set of all optimal alignments (Fig. 4). Furthermore, the set of optimizing pairs can be computed fairly efficiently in at worst O(n4) time. The procedure for enumerating optimizing pairs basically uses the same pair-removal trick described earlier for generating new optimal alignments. The pseudo-code for this procedure is shown below, with an explanation for each line afterwards.
Figure 4.
Statistics of H-GRAAL’s core yeast-human alignment for α = 0.5. We present the percentage of yeast proteins, out of 2,390 of them, that participate in n “optimizing pairs” (defined in Methods), for n = 1, 2, ..., 6, 7–48. Recall that an aligned pair is optimizing if it appears in at least one optimal alignment. Hence, when we examine all optimal alignments, we compute the percentage of yeast proteins that are aligned to n human proteins by optimal alignments. Around 72% of all yeast proteins have a unique human protein that they are aligned to by every optimal alignment. These yeast-human protein pairs form H-GRAAL’s core alignment.
Note that the procedure for finding optimizing aligned pairs can be embarrassingly parallelized by partitioning U amongst various processors. Each processor would have u loop over a partition Ui and the optimizing pairs can be gathered from all processors at the end.
It is clear that for each u, there will be at least one optimizing aligned pair originating from u; some u’s may be associated with several optimizing pairs. The number of optimizing pairs per u can give us an indication of how “significant” an aligned pair involving u really is. If, for example, u had 10 associated optimizing pairs, and a given optimal alignment paired u with some v, we might be inclined to view the (u,v) pairing as being rather ambiguous since there are 9 other possible candidates for v, which would be realized in some other optimal alignments. However, if (u,v) were the only optimizing pair for u, then every optimal alignment must contain (u,v), which would make it highly significant. So it is fruitful to identify the set of all such special optimizing pairs, which clearly must be a subset of every optimal alignment. We call this set the core alignment (maximum or not). For our purposes, a large core alignment is clearly desirable because it means that no matter what optimal alignment gets returned by H-GRAAL, a large proportion of the aligned pairs in that alignment are “stable” in the sense that they would still be present for a different optimal alignment, so that any one optimal alignment would be highly representative of all optimal alignments. Ideally, if all optimizing pairs belonged to the core alignment, then there would be exactly one optimal alignment.
Measures of alignment quality
Given a complete global alignment, we quantify the quality of alignments produced by our method with three scores: the edge correctness (EC), node correctness (NC), and interaction correctness (IC).30 EC is a percentage of edges in one graph that are aligned to edges in the other graph. The prerequisite to be able to measure NC and IC is to know the “true alignment,” i.e. the correct node mappings between two networks. Then, NC is a percentage of nodes in one network that are correctly aligned to nodes in the other network with respect to the true alignment. IC is the percentage of interactions that are aligned correctly with respect to the true alignment; we say that an interaction A–B is aligned correctly if two connected nodes A and B from one network are aligned with their correct alignment partners (with respect to the true alignment) and if their partners interact in the other network. Thus, IC is a stricter measure than EC, since EC does not require that the alignment partners of nodes A and B be the correct ones with respect to the true alignment.
Procedure 1.
Find all optimizing aligned pairs of nodes for networks G1 and G2 where U = V (G1), V = V (G2) and C0 is the matrix of node alignment costs between U and V.
| Procedure | Find-All-Optimizing-pairs (U, V, C0) |
|---|---|
| 1: A0, M0 ← H-GRAAL (U, V, C0) | |
| 2: Cmin ← Alignment-Cost (A0) | |
| 3: S ← ϕ | |
| 4: for allu ∈ Udo | |
| 5: A ← A0, M ← M0, C ← C0 | |
| 6: while Alignment-Cost (A) = cmindo | |
| 7: v ← A[u] | |
| 8: S ← S ∪ {v} | |
| 9: A, M ← Dynamic-H-GRAAL (U, V, C, A, M, {((u, v), +∞)}) | |
| 10: C[u][v] ← +∞ | |
| 11: end while | |
| 12: end for | |
| Line 1: | Invoke the non-dynamic, original H-GRAAL with inputs U, V and C0. Return an initial alignment A0 and the corresponding internal state M0 of the underlying Hungarian algorithm. |
| Line 2: | Compute the minimum alignment cost cmin from alignment A0. |
| Line 3: | Initialize the set S of optimizing aligned pairs to the empty set ϕ. |
| Line 4: | Loop over all nodes u in U. Procedure terminates when we have looped over all nodes in U. |
| Line 5: | Initialize working variables A, M, and C for alignment, internal state and cost matrix respectively. These change with each iteration of the while loop in line 6. |
| Line 6: | Loop while alignment A has cost Cmin. If A is costlier than Cmin, then we have found all optimizing pairs with U-node u and so we move on to the next node u from U. |
| Line 7: | v = A[u] is the node in V that was paired with u in alignment A. |
| Line 8: | Add the aligned pair (u,v) from alignment A to S. Note that we could also add the rest of the aligned pairs from A to S but that would not change the asymptotic efficiency of the procedure, which remains at O(kn2) time, where k is the number of optimizing pairs. |
| Line 9: | Invoke the dynamic version of H-GRAAL with inputs U, V, C, A, M and the set {((u,v), +∞)} consisting of one node-pair cost-change. The cost-change is for (u,v) and raises the cost to +∞. In practice, the cost is set to a sufficiently large positive number so that the Hungarian algorithm will not select the corresponding pair. Return an alignment A and the corresponding internal state M of the underlying Hungarian algorithm. Note that for a fixed u, A is guaranteed not to have a (u,v) pair for the present v and all previous v’s encountered within the while loop. |
| Line 10: | Adjust the (u,v) entry of the cost matrix C to +∞. |
In real applications, such as those involving biological networks, we do not usually know the true alignment and can therefore only measure EC. However, it is possible for two alignments to have similar ECs, one of which exposes large, dense, contiguous, and topologically complex regions of network similarity, while the other fails to do so. Thus, in addition to counting aligned edges, it is important that the aligned edges cluster together to form large and dense connected subgraphs, in order to uncover such regions of similar topology. By “dense” we mean that the aligned subgraphs share many edges among their nodes as opposed to many isolated edges. To this end, we define a common connected subgraph (CCS) as a connected subgraph (not necessarily induced) that appears in both networks. Note that it might not be clear which criterion reveals better alignment; ideally, both high EC and large and dense CCSs are desirable. Our algorithm produces both large CCSs and dense global alignments, as demonstrated below, owing probably less to the details of the algorithm and more to our strong measure of nodes’ topological network similarity.
Statistical significance
Statistical significance of H-GRAAL’s alignments
Random alignment of real-world networks:
Given H-GRAAL’s alignment of two networks G1(V1, E1) and G2(V2, E2), we compute the probability of obtaining a given or better EC at random. For this purpose, an appropriate null model of random alignment is required. A random alignment is a random mapping f between nodes in two networks G1(V1, E1) and G2(V2, E2), f : V1 → V2. H-GRAAL produces global alignments, so that all nodes in the smaller network (smaller in terms of the number of nodes) are aligned with nodes in the larger network. In other words, f is defined ∀v ∈ V1. This is equivalent to aligning each edge from G1(V1, E1) with a pair of nodes (not necessarily an edge) in G2(V2, E2). Thus, we define our null model of random alignment as a random mapping g: E1 → V2 × V2. We define n1 = |V1|, n2 = |V2|, m1 = |E1|, and m2 = |E2|. We also define p = n2(n2 − 1)/2 as the number of node pairs in G2, EC = x% as the edge correctness of the given alignment, and k = [m1 × EC] = [m1 × x] as the number of edges from G1 that are aligned to edges in G2. Then, the probability P of successfully aligning k or more edges by chance (i.e. p-value) is the tail of the hypergeometric distribution:
H-GRAAL’s alignment of random model networks:
Now we describe how to estimate the statistical significance of the amount of topological similarity that H-GRAAL finds by aligning two real-world PPI networks. To do that, we need to estimate how much similarity one would expect to find with H-GRAAL between two random networks. For each of the analyzed models, we align with H-GRAAL 25 pairs of model networks of the same size as the data and average ECs over the 25 runs. Then, we apply the following form of the Vysochanskij—Petunin inequality: P(| X − μ|≥λσ)≤4/9λ2. Since the model networks that are aligned have the same number of nodes and edges as the data, it is reasonable to assume that the distribution of their ECs is unimodal. Thus, we use the Vysochanskij—Petunin inequality, since it is more precise than Chebyshev’s inequality for unimodal distributions. For more details, see Kuchaiev et al.30
Statistical significance of shared GO terms
As a biological validation of H-GRAAL’s alignments of PPI networks of different species, we find the number of aligned protein pairs sharing a Gene Ontology (GO) term.47 Next, we compute the statistical significance of this result, i.e. the probability that the same or higher number of protein pairs would share a GO term in a random alignment of our real-world networks. We use the standard model of sampling without replacement (i.e. hypergeometric distribution as described in above), where p is the number of all possible G1-to-G2 node pairs in which both proteins are annotated with at least one GO term, m2 is the number of pairs out of p pairs in which both proteins share at least one common GO term, m1 is the number of pairs in our alignment in which both proteins are annotated with at least one GO term, and k is the number of pairs out of m1 pairs in which both proteins share at least one common GO term.
Statistical significance of functional enrichments in the aligned subnetworks
As an additional biological validation of our alignments, we compute the functional enrichment of the subnetworks aligned across species. We compute the statistical significance of an enrichment, i.e. the probability that the same or higher number of proteins in a subnetwork would be involved in function F by chance, using the same formula for hypergeometric distribution described above. For this purpose, p is the number of all annotated proteins in the entire network, m2 is the number of proteins out of p proteins that have function F, m1 is the number of annotated proteins in the aligned subnetwork, and k is the number of proteins out of m1 proteins that have function F.
Statistical significance of our phylogenetic trees
We generate phylogenetic trees based on ECs produced by H-GRAAL when aligning metabolic networks of different species. To measure the probability of obtaining our trees by chance, we repeat the same procedure to generate “random trees” (defined below). Then, we compare the “distance” (defined below) between “random trees” and between our phylogenetic and “random trees.” Clearly, we expect to obtain a significantly higher distance between our phylogenetic trees and the corresponding “random trees” than between “random trees” alone. A “random tree” is created based on ECs produced by H-GRAAL when aligning model networks of the same size as the data. Since all analyzed metabolic networks are bipartite graphs, we use the bipartite random graph model with the same degree distribution as the data. Once model networks are generated for all metabolic networks, we use H-GRAAL to align all pairs of these networks and we generate the phylogenetic tree from the obtained alignment scores in the same way we generate the phylogenetic trees for real metabolic networks; we call the resulting tree the “random tree”. To take into account the randomness of the model, we repeat this procedure 30 times, resulting in 30 instances of “random trees”.
Next, we compare each pair of “random trees”. Additionally, we compare our phylogenetic trees with all 30 corresponding “random trees”. To compare two phylogenetic trees, we use the patristic distance.a After we find the average pairwise distance between all pairs of 30 “random trees” and the corresponding standard deviation, as well as the average distance between the real phylogenetic tree and the 30 “random trees”, we compute the upper bounds of p-values by using Chebyshev’s inequality:
For details, see Kuchaiev et al.30
Results and Discussion
Method validation
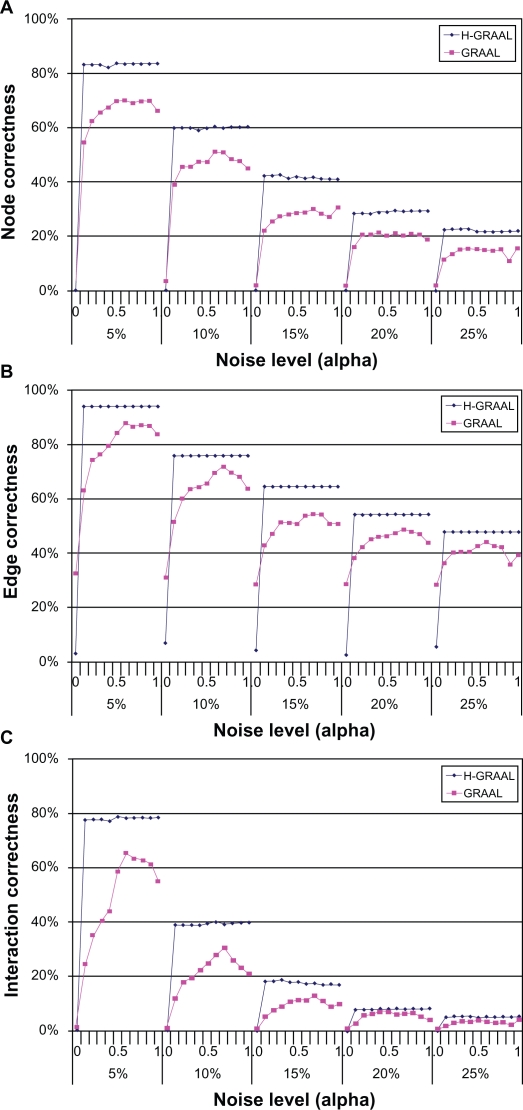
To measure the performance of H-GRAAL and benchmark it against other network alignment methods, we first analyze the largest connected component of the high-confidence yeast S. cerevisiae PPI network by Collins et al.10 We align this network with the same network augmented with interactions from the lower-confidence data set described by Collins et al.10 We analyze different noise levels, by adding 5%, 10%, 15%, 20%, and 25% of lower-confidence interactions; we add higher-confidence interactions first. For each noise level, we produce alignments by varying α from 0 to 1, in increments of 0.1 (Methods section). Since the networks being aligned are defined on the same set of nodes and differ only in the number of edges, we know the true alignment, i.e. the correct node matching. Thus, we report all three alignment quality scores: NC, EC, and IC.
Depending on the noise level, H-GRAAL achieves NC of up to 84%, EC of up to of 94%, and IC of up to 79% (Fig. 2); this demonstrates that our algorithm is capable of producing high-quality alignments with high NC, EC, and IC. Clearly, with increased level of noise, the performance deteriorates. The statistics do not differ much for α between 0.1 and 1 at the same noise level. However, the alignments are very bad for α of 0, i.e. when only node degrees are used in the cost function, without any contribution of node signatures. This indicates that node degrees alone are not an appropriate measure of network topology. The reason for very similar statistics for all non-zero values of α at a given noise level is due to the cost function giving a contribution of degree only to high-degree nodes, which are rare due to scale-free degree distributions in these networks (see Methods section).
Figure 2.
Comparison of H-GRAAL with GRAAL with respect to A) node correctness, B) edge correctness, and C) interaction correctness, for noise levels of 5%, 10%, 15%, 20%, and 25%, and for α between 0 and 1, in increments of 0.1. Note that H-GRAAL always produces better alignments than GRAAL for all values of α, and that using only degrees (α = 0) gives bad results. This tells us that graphlet-based signatures are far more valuable than a measure based on degree alone.
We compare the performance of H-GRAAL with the performance of GRAAL, a state-of-the-art algorithm for global network alignment that is based solely on network topology.30 GRAAL has already been shown to outperform all other methods capable of using only network topology as a cost function,30 including IsoRank.26 Both H-GRAAL and GRAAL are based on the same cost function, namely similarity of nodes’ graphlet degree vectors. However, GRAAL is a greedy “seed and extend” approach analogous to the popular BLAST48 algorithm for sequence alignment: it first chooses a single “seed” pair of nodes (one node from each network) with high signature similarity and then it expands the alignment radially outward around the seed as far as is practical using a greedy algorithm (see Kuchaiev et al30 for details). On the other hand, H-GRAAL finds an optimal alignment with respect to the cost function, at the expense of a longer running time. For example, H-GRAAL takes about 2 days of CPU time to produce alignment of yeast and human PPI networks described below, compared to GRAAL’s CPU time of several hours on the same hardware. Note that alignments found by H-GRAAL (or GRAAL) will not necessarily be optimal with respect to EC, even though they will be optimal with respect to the cost function.
When aligning the above mentioned data sets, H-GRAAL outperforms GRAAL for all noise levels, all α between 0.1 and 1, and with respect to all three alignment quality scores (Fig. 2). For α = 0, GRAAL’s statistics are better; this is expected, due to GRAAL’s “seed and extend” principle explained above, which results in alignments of contiguous network regions; this is not the case with H-GRAAL (see Methods section for details about H-GRAAL). However, GRAAL’s statistics for α = 0 are much worse than GRAAL’s statistics for any other analyzed α (and are thus worse than H-GRAAL’s statistics as well), again implying that node degrees are not constraining enough measure of network topology.
Pairwise alignment of PPI networks of yeast and human
Next, we apply our algorithm to pairwise alignment of PPI networks of different species. We align the yeast PPI network by Collins et al10 consisting of 16,127 interactions amongst 2,390 proteins and the human PPI network by Radivojac et al49 consisting of 41,456 interactions amongst 9,141 proteins. Due to its ease of genetic manipulation, yeast has been one of the most extensively studied organisms. Other organisms, such as fruitfly and worm, have been significantly less studied, and available networks for these organisms contain relatively higher levels of noise. Although the human PPI network is also still incomplete, studying this network is of a great interest and importance as it can give valuable insights into complex diseases. Note, however, that the analyzed human PPI network combines physical interactions from numerous data sources and thus is of large size and high coverage. Additionally, the same networks have already been analyzed with GRAAL,30 which makes the comparison of our results easier. For these reasons, we choose to align the yeast and human PPI networks.
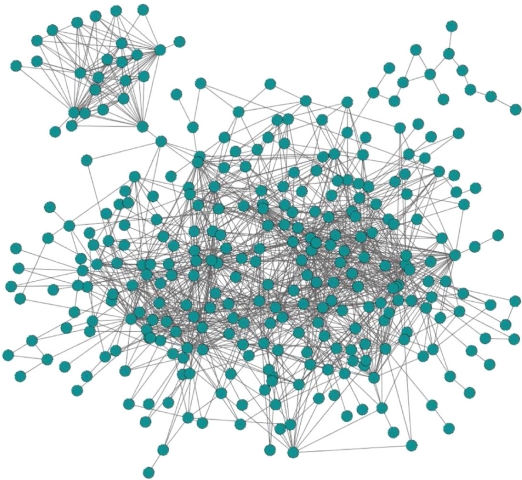
Since we are aligning two networks of different species with different sets of proteins, we do not know their “true alignment”, and therefore, we only report EC. We report an optimal yeast-human alignment produced for α of 0.5, since this α resulted in the alignment with the highest EC of 10.92% over all α between 0 and 1. Note that although we choose α so that it maximizes EC, the choice of this parameter and its influence on the resulting alignments is the subject of future research. In the analyzed optimal alignment, EC of 10.92% corresponds to aligning 16,127 * 10.92% = 1,761 edges amongst 796 proteins in the yeast network to 1,761 edges amongst 796 proteins in the human network. The aligned interactions are not randomly distributed in the yeast and human networks; instead, they form common connected subgraphs (CCSs), the largest one having 1,290 interactions amongst 317 proteins (Fig. 3), and the second largest one having 57 interactions amongst 20 proteins. Beyond these two components, there are additional 11 components on 5 or more nodes, and 166 components on 2–4 nodes.
Figure 3.
The largest common connected subgraph resulting from the alignment of the yeast and human PPI networks, consisting of 1,290 interactions amongst 317 proteins. An edge between two nodes means that an interaction exists in both species between the corresponding protein pairs. Thus, the displayed network appears, in its entirety, in the PPI networks of both species.
We find the core alignment present across all optimal alignments for α = 0.5; it is large, consisting of 1,738 out of 2,390 possible yeast-human pairs, i.e. 72.2%, of them (Fig. 4). More than 95% of the nodes in each of the two largest CCSs in the entire H-GRAAL’s analyzed optimal alignment for α = 0.5 are present in the core alignment.
Although GRAAL produces slightly higher EC of 11.72% compared to H-GRAAL when aligning the same yeast and human networks, H-GRAAL’s largest CCS with 1,290 interactions amongst 317 proteins is larger than GRAAL’s largest CCS, which has 900 edges between 267 nodes. Thus, although GRAAL aligns slightly more edges that H-GRAAL does, H-GRAAL’s alignment is more contiguous and denser. Moreover, we find that H-GRAAL’s largest CCS contains the majority of nodes from GRAAL’s first and second largest CCSs combined.
Even though the detailed node pairings are different for H-GRAAL and GRAAL, the alignments are in fact quite similar at a coarse-grained level. Indeed, although only 170/2,390 = 7.1% of aligned yeast-human pairs match exactly for the two algorithms, we find that 1,596/2,390 = 66.8% of human proteins that are in H-GRAAL’s alignment are present in GRAAL’s alignment as well; in the core part of H-GRAAL’s alignment, the same is true for 1,088/1,738 = 62.6% human proteins. (Note that since yeast is the smaller network, all yeast proteins are present in alignments produced by both methods.) Additionally, there is a significant overlap between the largest CCS of the two alignments: 222/267 = 83% of yeast proteins that were in the largest CCS in GRAAL’s alignment are also in the largest CCS of H-GRAAL’s alignment; the same is true for 175/267 = 66% of human proteins. (As mentioned above, 95% of nodes in the two largest CCSs of H-GRAAL’s alignment are in H-GRAAL’s core alignment, so the statistics for core part of H-GRAAL’s alignment are very similar.) The relatively small exact overlap of 7.1% between GRAAL’s and H-GRAAL’s alignments might not be too surprising, given that the nature of the two algorithms is different, and given that there might exist a large number of different alignments all of which may have comparable topological and biological quality. For example, it is possible that topologically identical subgraphs in two networks are aligned both by GRAAL and by H-GRAAL, but the actual aligned protein pairs differ for the two algorithms; this could happen, for example, when two cliques (complete graphs having all possible edges between the nodes) of the same size are aligned between the two networks, due to existence of multiple topologically equivalent alignments for cliques.
Statistical significance of H-GRAAL’s yeast-human alignment
First, we judge the quality of our alignment compared to a random alignment of these two particular networks. Given a random alignment of the yeast and human PPI networks, the probability of obtaining EC of 10.92% or better (p-value) is less than 7 × 10−8 (Methods section). The probability of obtaining a large CCS would be significantly smaller, so this represents a weak upper bound on our p-value.
Second, we comment on the amount of topological similarity uncovered by H-GRAAL’s alignment of yeast and human by comparing it to H-GRAAL’s alignment of random model networks. If we align with H-GRAAL networks drawn from several different random graph models50 that have the same number of nodes and edges as the yeast and human networks, we find that EC between random networks is statistically significantly lower than EC of our yeast-human alignment. We analyze the following network models: Erdös-Rényi random graphs,51 random scale-free graphs, i.e. random graphs with the same degree distribution as the data,52 3-dimensional geometric random graphs,53,43,44 scale-free gene duplication and mutation model networks,54 and geometric gene duplication and mutation model network.55 For each model, we align 25 pairs of random graphs of the same size as the data and average ECs over the 25 runs. Alignments of random graphs drawn from these models result in ECs of only 0.23 ± 0.05%, 0.23 ± 0.05%, 0.89 ± 0.1%, 2.83 ± 0.36%, and 6.25 ± 0.36%, respectively, with p-values of 9.7 × 10−6, 9.7 × 10−6, 4.5 × 10−5, 8.8 × 10−4, and 3.2 × 10−2, respectively (Methods section). Accepting geometric gene duplication and mutation model as the best available null model having the highest EC over all analyzed models, i.e. the worst case scenario for estimating the statistical significance of H-GRAAL’s yeast-human alignment, the p-value of H-GRAAL’s yeast-human alignment is 3.2 × 10−2 (Methods section). This tells us that yeast and human, two very different species, enjoy significantly more network similarity than chance would allow.
Biological significance of H-GRAAL’s yeast-human alignment
To quantify the biological significance of our yeast-human alignment, we count the number of aligned yeast-human protein pairs that share at least one common Gene Ontology (GO) term.47 For this analysis, we consider the complete GO annotation data set, containing all GO annotations, independent of GO evidence code. The associations between gene products and GO terms were downloaded from the Gene Ontologyb in September 2009. We do this for the entire alignment between yeast and human PPI networks, as well as for the core alignment. We also evaluate the biological quality of our alignment against GRAAL’s.
Across the entire H-GRAAL’s alignment, 45.38%, 14.54%, 4.55%, and 1.3% of aligned protein pairs share at least 1, 2, 3, and 4 common GO terms, respectively, with p-values of 4.68 × 10−8, 2 × 10−5, 8.43 × 10−5, and 4.71 × 10−2, respectively (Methods section). In GRAAL’s alignment, these percentages are 45.10%, 15.60%, 5.06%, and 2.02%. Across H-GRAAL’s core alignment, 47.4%, 16.01%, 5.21%, and 1.59% of aligned protein pairs share at least 1, 2, 3, and 4 common GO terms, respectively, with p-values of 2.77 × 10−8, 1.25 × 10−7, 3.25 × 10−6, and 7.2 × 10−3, respectively. Thus, H-GRAAL produces biologically meaningful and statistically significant alignments, and these are comparable to those of GRAAL.
We additionally validate the biological quality of H-GRAAL’s yeast-human alignment. We find that H-GRAAL aligns network regions of yeast and human in which a large and statistically significant percentage of proteins perform the same biological function in both species. Specifically, H-GRAAL aligns a 317-node subnetwork between yeast and human in which 10.2% of annotated yeast and 11.9% of annotated human proteins are involved in splicing. This result is encouraging, since splicing is known to have been conserved even between distant eukaryotes.56–58 Additionally, it aligns a 14-node subnetwork in which 92.7% of annotated yeast and 76.9% of annotated human proteins are involved in transcription. Furthermore, it aligns a 13-node subnetwork in which 92.3% of annotated yeast and 45.5% of annotated human proteins are involved in translation. Also, it aligns an additional 6-node subnetwork in which 100% of annotated yeast and 100% of annotated human proteins are involved in transcription. Finally, it aligns a 5-node subnetwork in which 100% of annotated yeast and 100% of annotated human proteins are involved in transport. The p-values for all of the above presented functional enrichments in the corresponding yeast and human subnetworks are in the 10−4 to 10−11 range (Methods section).
Application to protein function prediction
Given that we demonstrated high topological and biological quality of H-GRAAL’s yeast-human alignment, we transfer annotation between aligned proteins across the two networks. In particular, we predict from H-GRAAL’s alignment the biological characteristics (i.e. GO molecular function (MF), biological process (BP), and cellular component (CC)) of unannotated proteins based on the characteristics of their annotated aligned partners.
We make predictions with respect to two different sets of GO annotation data: the complete set described above, containing all GO annotations, independent of GO evidence codes, and a biologically-based set, containing GO annotations obtained by experimental evidence codes only.47 Many terms in the complete GO annotation data set were computationally assigned to proteins (e.g. from sequence alignments), and thus, it is biologically less confident than the biologically-based one. We identify proteins with unknown function whose aligned partners are annotated with a known MF, BP, or CC GO term, with respect to both the complete and biologically-based GO annotation data sets, and we assign all known MF, BP, or CC GO terms to the unannotated protein (see Kuchaiev et al30 for details).
With respect to the complete GO data set, we predict MF for 22 human and 299 yeast proteins, BP for 27 human and 105 yeast proteins, and CC for 37 human and 29 yeast proteins. We attempt to validate all of our predictions using the literature search and the text mining tool CiteXplorer.59 We successfully validate at least one MF, BP, and CC prediction for 44.4%, 42.9%, and 51.6% human proteins, and 49.8%, 4.7%, and 11.8% yeast proteins, respectively; by “successfully validate”, we mean that this tool finds at least one article mentioning the protein of interest in the context of at least one of our MF, BP, and CC predictions for that protein, respectively. We call the above percentages the “validation hit-rate”. In summary, we successfully validate at least one MF, BP, or CC prediction for 59% of human and 46% of yeast proteins.
With respect to the biologically-based GO data set, we predict MF for 15 human and 163 yeast proteins, BP for 22 human and 24 yeast proteins, and CC for 34 human and 15 yeast proteins. Our validation “hit-rates” with CiteXplorer for MF, BP, and CC are 25%, 23.5%, and 20.7% for human, and 55.5%, 0%, and 9.1% for yeast, respectively. Hence, in summary, we validate with CiteXplorer at least one MF, BP, or CC prediction for 29% of human and 52% of yeast proteins.
Note that, since a protein can (and is expected to) perform multiple functions, and since indications on the biological function of unannotated proteins in the literature are limited, it is possible that more of our predictions for human and yeast proteins are correct than we have been able to validate.
Our validation results are mostly better than those for GRAAL: with respect to the complete GO data set, GRAAL’s validation hit-rates for MF, BP, and CC are 34.1%, 43.4%, and 46.2% for human, and 42.1%, 3.2%, and 13% for yeast, respectively; with respect to the biologically-based GO data set, GRAAL’s validation hit-rates for MF, BP, and CC are 10%, 4.8%, and 20% for human, and 48.1%, 0%, and 0% for yeast, respectively.
Reconstruction of phylogenetic trees by aligning metabolic pathways across species
Additionally, we apply our approach to recover phylogenetic relationships between species by finding topological similarities between their metabolic networks. Although related attempts exist,60–63 they all use some biological or functional information such as sequence similarities, structural similarities, or enzyme commission numbers, to define node similarities and derive phylogenetic trees from pathways. On the other hand, we rely solely on the network topology to define node similarity, as was done with GRAAL.30 Thus, our information source is fundamentally different from the information sources used in related approaches and our algorithm recovers phylogenetic relationships (but not the evolutionary timescale of species divergence at this time) in a completely novel and independent way from all existing methods for phylogenetic recovery.
We analyze the entire metabolic networks of Eukaryotic organisms with fully sequenced genomes. There are 17 such species in the KEGG pathway database,12 seven of which are protists, six are fungi, two are plants, and two are animals. We focus on aligning protists and fungi due to lack of more data on plants and animals. Moreover, it has been shown that PPI network structure has subtle effects on the evolution of proteins and that reasonable phylogenetic inference can only be done between closely related species.64 For each of the two groups of organisms, protists and fungi, we extract the union of all metabolic pathways from KEGG. We find all-to-all pairwise H-GRAAL’s alignments between the corresponding metabolic networks, using the same α of 0.5 that we used for yeast-human alignment. We create a phylogenetic tree by using the nearest distance (single linkage) algorithmc with pairwise EC as the distance measure. We compare our results to published phylogenetic trees that contain organisms that we analyze,65–67,d as well as to topologically derived phylogenetic trees produced by GRAAL.
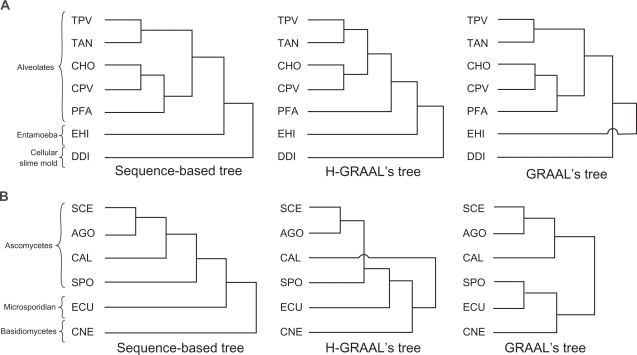
The phylogenetic tree constructed for protists using ECs produced by our method is very similar to the tree obtained from the literature,65,66 as well as to that produced by GRAAL (see Fig. 5A). Both our and GRAAL’s tree differ from the sequence-based one in α single branch: Plasmodium falciparum (PFA) is misplaced in our tree, whereas Entamoeba histolytica (EHI) (or Dictyostelium discoideum (DDI)) is misplaced in GRAAL’s tree. We can estimate the statistical significance of our tree by measuring how it compares to trees built from random networks of the same size as the metabolic networks (see Methods section and Kuchaiev et al30 for details); we find that the p-value of our tree is less than 1.6 × 10−3. Our H-GRAAL-based phylogeny reconstruction shows that the topologies of entire metabolic networks of Cryptosporidium parvum (CPV) and Cryptosporidium hominis (CHO) are very similar, since these species are grouped together in the tree. Since these organisms are two morphologically identical species of Apicomplexan protozoa with 97% genetic sequence identity, but with strikingly different hosts68 that contribute to their divergence,69,70 this validates our approach.
Figure 5.
A) Comparison of the phylogenetic trees for protists obtained by genetic sequence alignments (left), H-GRAAL’s metabolic network alignments (middle), and GRAAL’s metabolic network alignments (right). The following abbreviations are used for species: CHO—Cryptosporidium hominis, DDI—Dictyostelium discoideum, CPV—Cryptosporidium parvum, PFA—Plasmodium falciparum, EHI—Entamoeba histolytica, TAN—Theileria annulata, TPV—Theileria parva; the species are grouped into “Alveolates”, “Entamoeba”, and “Cellular Slime mold” classes. B) Comparison of the phylogenetic trees for fungi obtained by genetic sequence alignments (left), H-GRAAL’s metabolic network alignments (middle), and GRAAL’s metabolic network alignments (right). The following abbreviations are used for species: AGO—Ashbya gossypii (Eremothecium gossypii), CAL—Candida albicans, CNE—Cryptococcus neoformans, ECU—Encephalitozoon cuniculi, SCE—Saccharomyces cerevisiae, SPO—Schizosaccharomyces pombe; the species are grouped into “Ascomycetes”, “Microsporidian”, and “Basidiomycetes” classes.
Given that H-GRAAL’s phylogenetic tree, as well as GRAAL’s tree, is slightly different from the sequence-based one, and given that H-GRAAL’s tree is slightly different from GRAAL’s one, there is no reason to believe that the sequence-based tree or GRAAL’s one should a priori be considered the correct one. Sequence-based phylogenetic trees are built based on multiple alignment of gene sequences and whole genome alignments. Multiple alignments can suffer from a number of problems: they can be misleading due to gene rearrangements, inversions, transpositions, and translocations that occur at the substring level. Also, different species might have an unequal number of genes or genomes of vastly different lengths. Furthermore, whole genome phylogenetic analyses can be misleading due to noncontiguous copies of a gene or non-decisive gene order.71 Moreover, the trees are built incrementally from smaller pieces that are “patched” together probabilistically,65 so probabilistic errors in the tree are expected. H-GRAAL’s and GRAAL’s trees suffer from none of these problems, but they may suffer from other problems, such as noise and incompleteness of PPI networks.
We also construct a phylogenetic tree for fungi. Our tree for fungi is much more similar to the sequence-based one than the tree produced by GRAAL (see Fig. 5B): unlike in GRAAL’s fungi tree, only a single branch, Candida albicans (CAL), is misplaced in our tree compared to the sequence-based one. The p-value of our phylogenetic tree for fungi is less than 4.5 × 10−3. We can see that Encephalitozoon cuniculi (ECU) of the Microsporidia group is grouped next to Schizosaccharomyces pombe (SPO) of the Ascomycetes group. This result is encouraging since it has been shown that Microsporidia consistently falls not only within fungal diversification but also close to Ascomycetes,67 even though for a long time it has been difficult to resolve the evolutionary relationship between the microsporidia and other eukaryotes.
Note that in addition to metabolic networks of protists and fungi, our method might need to be tested on other data sets before it can be widely used for phylogenetic tree reconstruction. Also note that some parts of the metabolic networks that we analyze are derived experimentally, while others are obtained by network reconstructions based on orthology relationships between species. These orthology relationships are based on alignments of protein (enzyme) sequences. Therefore, the striking resemblance between our phylogenetic trees and sequence-based ones explicitly validates our method. Furthermore, given that different source of sequence data is used for reconstructing phylogenetic trees in the literature (alignments of mitochondrial proteins or ribosomal RNA are used) and for reconstructing metabolic networks (protein sequences of enzymes are used),30 the phylogenetic trees obtained from our network alignments might already be viewed as new and independent sources of phylogenetic information. Our results will only gain in biological significance when purely experimentally obtained networks become available.
Comparison with other methods
The current best global alignment algorithm is GRAAL which has been shown to produce by far the most complete topological alignments of biological networks to date that are statistically significant and biologically valid.30 Therefore, since GRAAL has been shown to outperform other network alignment methods,30 we compare the results of H-GRAAL’s alignment only to those of GRAAL. We demonstrate that H-GRAAL’s alignments are comparable or superior to those of GRAAL. First, H-GRAAL outperforms GRAAL when aligning high-confidence to high- + lower-confidence yeast PPI networks10 with respect to all three NC, EC, and IC, all non-zero α, and all noise levels (see Fig. 2). Second, although H-GRAAL’s yeast-human alignment has slightly lower EC than GRAAL’s, it is more contiguous and denser, with bigger largest CCS. Third, H-GRAAL produces biologically meaningful and statistically significant alignments, which are comparable to those of GRAAL. Fourth, with respect to protein function prediction by H-GRAAL and GRAAL, literature validation hit-rates are higher for H-GRAAL’s predictions. Finally, H-GRAAL’s fungi phylogenetic tree is much more similar to the literature-based one than GRAAL’s fungi tree (Fig. 5B). This is likely due to the fact that H-GRAAL produces an optimal alignment with respect to the cost function, whereas GRAAL does not. However, it does so at the expense of running time (e.g. H-GRAAL takes about 2 days to align yeast and human PPI networks analyzed in this study, compared to several hours that GRAAL takes to align them on the same hardware; also see Methods).
Acknowledgments
This project was supported by the NSF CAREER IIS-0644424 grant.
Footnotes
Disclosures
This manuscript has been read and approved by all authors. This paper is unique and is not under consideration by any other publication and has not been published elsewhere. The authors and peer reviewers of this paper report no conflicts of interest. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.Ito T, et al. Toward a protein-protein interaction map of the budding yeast: A comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc Natl Acad Sci U S A. 2000;97(3):1143–7. doi: 10.1073/pnas.97.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uetz P, et al. A comprehensive analysis of protein-protein interactions in saccharomyces cerevisiae. Nature. 2000;403:623–7. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 3.Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415(6868):141–7. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 4.Ho Y, et al. Systematic identification of protein complexes in saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415(6868):180–3. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 5.Giot L, et al. A protein interaction map of drosophila melanogaster. Science. 2003;302(5651):1727–36. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 6.Li S, et al. A map of the interactome network of the metazoan c. elegans. Science. 2004;303:540–3. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stelzl U, et al. A human protein-protein interaction network: A resource for annotating the proteome. Cell. 2005;122:957–68. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 8.Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–78. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 9.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–43. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 10.Collins SR, et al. Toward a comprehensive atlas of the physical interactome of saccharomyces cerevisiae. Molecular and Cellular Proteomics. 2008;6(3):439–50. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Simonis N, et al. Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nature Methods. 2009;6(1):47–54. doi: 10.1038/nmeth.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanehisa M, Goto S. Kegg: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davidson EH, et al. A genomic regulatory network for development. Science. 2006;295(5560):1669–78. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- 14.Berman HM, Westbrook J, Feng Z, et al. The protein data bank. Nucleic Acids Research. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milenković T, Filippis I, Lappe M, Pržulj N. Optimized null model for protein structure networks. PLoS ONE. 2009;4(6):e5967. doi: 10.1371/journal.pone.0005967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White J, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode c elegans. Philosophical Transactions of the Royal Society of London—Series B: Biological Sciences. 1986;314:1340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 17.Sharan R, Ideker T. Modeling cellular machinery through biological network comparison. Nat Biotech. 2006 Apr;24(4):427–33. doi: 10.1038/nbt1196. [DOI] [PubMed] [Google Scholar]
- 18.Cook SA. Proc 3rd Ann ACM Symp on Theory of Computing: 1971; New York. Association for Computing Machinery; 1971. The complexity of theorem-proving procedures; pp. 151–58. [Google Scholar]
- 19.Venkatesan K, et al. An empirical framework for binary interactome mapping. Nature Methods. 2009;6(1):83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelley BP, Bingbing Y, Lewitter F, Sharan R, Stockwell BR, Ideker T. PathBLAST: a tool for alignment of protein interaction networks. Nucl Acids Res. 2004;32:83–8. doi: 10.1093/nar/gkh411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berg J, Lassig M. Local graph alignment and motif search in biological networks. PNAS. 2004;101:14689–94. doi: 10.1073/pnas.0305199101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flannick J, Novak A, Srinivasan BS, McAdams HH, Batzoglou S. Graemlin: General and robust alignment of multiple large interaction networks. Genome Res. 2006;16(9):1169–81. doi: 10.1101/gr.5235706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang Z, Xu M, Teng M, Niu L. NetAlign: a web-based tool for comparison of protein interaction networks. Bioinformatics. 2006;22(17):2175–7. doi: 10.1093/bioinformatics/btl287. [DOI] [PubMed] [Google Scholar]
- 24.Berg J, Lassig M. Cross-species analysis of biological networks by Bayesian alignment. Proceedings of the National Academy of Sciences. 2006;103(29):10967–72. doi: 10.1073/pnas.0602294103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cootes AP, Muggleton SH, Sternberg MJ. The identification of similarities between biological networks: Application to the metabolome and interactome. Journal of Molecular Biology. 2007;369(4):1126–39. doi: 10.1016/j.jmb.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 26.Singh R, Xu J, Berger B. Research in Computational Molecular Biology. Springer; 2007. Pairwise global alignment of protein interaction networks by matching neighborhood topology; pp. 16–31. [Google Scholar]
- 27.Flannick J, Novak AF, Do CB, Srinivasan BS, Batzoglou S. Automatic parameter learning for multiple network alignment. RECOMB. 2008:214–231. doi: 10.1089/cmb.2009.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaslavskiy M, Bach F, Vert JP. Global alignment of protein-protein interaction networks by graph matching methods. Bioinformatics. 2009;25(12):i259–67. doi: 10.1093/bioinformatics/btp196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao C, Lu K, Baym M, Singh R, Berger B. Isorankn: spectral methods for global alignment of multiple protein networks. Bioinformatics. 2009;25(12):i253–8. doi: 10.1093/bioinformatics/btp203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuchaiev O, Milenković T, Memisević V, Hayes W, Pržulj N. Journal of the Royal Society Interface. 2010. Topological network alignment uncovers biological function and phylogeny. doi: 10.1098/rsif.2010.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chindelevitch CS, an Liao L, Berger B. Local optimization for global alignment of protein interaction networks. Pacific Symposium on Biocomputing. 2010;15:123–32. doi: 10.1142/9789814295291_0015. [DOI] [PubMed] [Google Scholar]
- 32.Gautheret D, Major F, Cedergren R. Pattern searching/alignment with rna primary and secondary structures: an effective descriptor for trna. Comput Appl Biosci. 1990;6(4):325–31. doi: 10.1093/bioinformatics/6.4.325. [DOI] [PubMed] [Google Scholar]
- 33.Woese CR, Gutell R, Gupta R, Noller HF. Detailed analysis of the higher-order structure of 16 s-like ribosomal ribonucleic acids. Microbiological reviews. 1983;47(4):621–69. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webb CH, Riccitelli NJ, Ruminski DJ, Lupták A. Widespread occurrence of self-cleaving ribozymes. Science (New York, N.Y.) 2009;326(5955):953. doi: 10.1126/science.1178084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Memisević V, Milenković T, Pržulj N. Complementarity of network and sequence structure in homologous proteins. Journal of Integrative Bioinformatics. 2010;7(3) doi: 10.2390/biecoll-jib-2010-135. [DOI] [PubMed] [Google Scholar]
- 36.Komili S, Farny NG, Roth FP, Silver PA. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131(3):557–71. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watson JD, Laskowski RA, Thornton JM. Predicting protein function from sequence and structural data. Current opinion in structural biology. 2005;15(3):275–84. doi: 10.1016/j.sbi.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 38.Whisstock JC, Lesk AM. Prediction of protein function from protein sequence and structure. Q Rev Biophys. 2003;36(3):307–40. doi: 10.1017/s0033583503003901. [DOI] [PubMed] [Google Scholar]
- 39.Kosloff M, Kolodny R. Sequence-similar, structure-dissimilar protein pairs in the pdb. Proteins. 2008;71(2):891–902. doi: 10.1002/prot.21770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laurents DV, Subbiah S, Levitt M. Different protein sequences can give rise to highly similar folds through different stabilizing interactions. Protein Sci. 1994;3(11):1938–44. doi: 10.1002/pro.5560031105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.West DB. Introduction to Graph Theory. 2nd edition. Prentice Hall; Upper Saddle River, NJ.: 2001. [Google Scholar]
- 42.Milenković T, Pržulj N. Uncovering biological network function via graphlet degree signatures. Cancer Informatics. 2008;6:257–73. [PMC free article] [PubMed] [Google Scholar]
- 43.Pržulj N, Corneil DG, Jurisica I. Modeling interactome: Scale-free or geometric? Bioinformatics. 2004;20(18):3508–15. doi: 10.1093/bioinformatics/bth436. [DOI] [PubMed] [Google Scholar]
- 44.Pržulj N. Biological network comparison using graphlet degree distribution. Bioinformatics. 2007;23:e177–83. doi: 10.1093/bioinformatics/btl301. [DOI] [PubMed] [Google Scholar]
- 45.Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393:440–2. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 46.Mills-Tettey GA, Stentz A, Dias MB. Technical Report CMU-RI-TR-07-27. Robotics Institute; Pittsburgh, PA: Jul, 2007. The dynamic Hungarian algorithm for the assignment problem with changing costs. [Google Scholar]
- 47.The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altschul SF, Gish W, Miller W, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 49.Radivojac P, Peng K, Clark WT, et al. An integrated approach to inferring gene-disease associations in humans. Proteins. 2008;72(3):1030–7. doi: 10.1002/prot.21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Milenković T, Lai J, Pržulj N. Graphcrunch: a tool for large network analyses. BMC Bioinformatics. 2008;9(70) doi: 10.1186/1471-2105-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Erdös P, Rényi A. On random graphs. Publicationes Mathematicae. 1959;6:290–7. [Google Scholar]
- 52.Molloy M, Reed B. A critical point of random graphs with a given degree sequence. Random Structures and Algorithms. 1995;6:161–80. [Google Scholar]
- 53.Penrose M. Geometric Random Graphs. Oxford University Press; 2003. [Google Scholar]
- 54.Vazquez A, Flammini A, Maritan A, Vespignani A. Modeling of protein interaction networks. ComPlexUs. 2003;1:38–44. doi: 10.1038/nbt825. [DOI] [PubMed] [Google Scholar]
- 55.Pržulj N, Kuchaiev O, Stevanović A, Hayes W. Geometric evolutionary dynamics of protein interaction networks. Proceedings of the 2010 Pacific Symposium on Biocomputing (PSB), Big Island, Hawaii. 2010 Jan 4–8;:178–89. doi: 10.1142/9789814295291_0020. [DOI] [PubMed] [Google Scholar]
- 56.Collins L, Penny D. Complex spliceosomal organization ancestral to extant eukaryotes. Molecular Biology and Evolution. 2005;22(4):1053–66. doi: 10.1093/molbev/msi091. [DOI] [PubMed] [Google Scholar]
- 57.Wentz-Hunter K, Potashkin J. The evolutionary conservation of the splicing apparatus between fission yeast and man. Nucleic Acids Symp. 1995;33:226–8. [PubMed] [Google Scholar]
- 58.Lorkovic ZJ, Lehner R, Forstner C, Barta A. Evolutionary conservation of minor u12-type spliceosome between plants and humans. RNA. 2005;11(7):1095–107. doi: 10.1261/rna.2440305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Labarga A, Valentin F, Andersson M, Lopez R. Web services at the european bioinformatics institute. Nucleic Acids Research. 2007;35(Web Server issue):W6–11. doi: 10.1093/nar/gkm291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suthram S, Sittler T, Ideker T. The plasmodium protein network diverges from those of other eukaryotes. Nature. 2005;438:108–12. doi: 10.1038/nature04135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Forst CV, Schulten K. Phylogenetic analysis of metabolic pathways. J Mol Evol. 2001;52:471–89. doi: 10.1007/s002390010178. [DOI] [PubMed] [Google Scholar]
- 62.Heymans M, Singh A. Deriving phylogenetic trees from the similarity analysis of metabolic pathways. Bioinformatics. 2003;19(Suppl 1):i138–46. doi: 10.1093/bioinformatics/btg1018. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y, Li S, Skogerb G, et al. Phylophenetic properties of metabolic pathway topologies as revealed by global analysis. BMC Bioinformatics. 2006;7:252. doi: 10.1186/1471-2105-7-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Agrafioti I, Swire J, Abbott J, Huntley D, Butcher S, Stumpf MPH. Comparative analysis of the saccharomyces cerevisiae and caenorhabdits elegans protein interaction networks. BMC Evolutionary Biology. 2005;5(23) doi: 10.1186/1471-2148-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pennisi E. Modernizing the tree of life. Science. 2003;300 doi: 10.1126/science.300.5626.1692. [DOI] [PubMed] [Google Scholar]
- 66.Hillis DM, Zwickl D, Guttel R. Tree of life. University of Texas; [Google Scholar]
- 67.Keeling PJ, Luker MA, Palmer JD. Evidence from beta-tubulin phylogeny that microsporidia evolved from within the fungi. Mol Biol Evol. 2000;17(1):23–31. doi: 10.1093/oxfordjournals.molbev.a026235. [DOI] [PubMed] [Google Scholar]
- 68.Tanriverdi S, Widmer G. Differential evolution of repetitive sequences in cryptosporidium parvum and cryptosporidium hominis. Infect Genet Evol. 2006;6(2):113–22. doi: 10.1016/j.meegid.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 69.Xu P, et al. The genome of cryptosporidium hominis. Nature. 2004;431(7012):1107–12. doi: 10.1038/nature02977. [DOI] [PubMed] [Google Scholar]
- 70.Hashim A, Mulcahy G, Bourke B, Clyne M. Interaction of cryptosporidium hominis and cryptosporidium parvum with primary human and bovine intestinal cells. Infect Immun. 2006;74(1):99–107. doi: 10.1128/IAI.74.1.99-107.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Out H, Sayood K. A new sequence distance measure for phylogenetic tree construction. Bioinformatics. 2003;19(16):2122–30. doi: 10.1093/bioinformatics/btg295. [DOI] [PubMed] [Google Scholar]