Abstract
Edge Hill virus (EHV) is a mosquito-borne flavivirus isolated throughout Australia during mosquito surveillance programs. While not posing an immediate threat to the human population, EHV is a taxonomically interesting flavivirus since it remains the only member of the yellow fever virus (YFV) sub-group to be detected within Australia. Here we present both an antigenic and genetic investigation of collected isolates, and confirm taxonomic classification of the virus within the YFV-group. Isolates were not clustered based on geographical origin or time of isolation, suggesting that minimal genetic evolution of EHV has occurred over geographic distance or time within the EHV cluster. However, two isolates showed significant differences in antigenic reactivity patterns, and had a much larger divergence from the EHV prototype (19% nucleotide and 6% amino acid divergence), indicating a distinct subtype or variant within the EHV subgroup.
Keywords: Edge Hill virus, EHV, yellow fever virus, flavivirus
Edge Hill virus (EHV) is a mosquito-borne flavivirus that has been isolated during mosquito-surveillance programs conducted in the Northern Territory, Queensland, Western Australia, and New South Wales.1–3 While the Flavivirus genus is known to harbor serious global pathogens such as yellow fever virus (YFV), West Nile virus (WNV) and the dengue viruses (DENV),4 EHV has only once been implicated in human disease. In this instance, virus symptoms included myalgia, arthralgia and fatigue.5 Generally, EHV sero-conversion by humans is rare,6 indicating the virus is of low risk to the human population. Indeed EHV has mainly been associated with marsupial infections, since antibodies reactive to EHV have been detected in wallabies, kangaroos and bandicoots.6,7
Taxonomically, EHV is a unique flavivirus since it remains the only member of the YFV sub-group to be detected within Australia. Adding to the interest, historically the classification of EHV was briefly contentious. The virus was originally classified within the Uganda S antigenic complex via cross-neutralization tests using polyclonal antisera,8,9 but later RNA hybridization studies suggested EHV might share up to 70% sequence homology with DENV-2 virus,10 which belongs to a different antigenic clade.8,9 Moreover, monoclonal antibody 1B7, previously thought to be dengue specific, was found to cross-react with EHV in an ELISA,10 suggesting an antigenic similarity between these viruses. Antigenic homology to DENV-2 could have serious implications for DENV detection and subsequent dengue hemorrhagic fever epidemiology within Australia.11 However, subsequent partial nucleotide sequencing of EHV did not confirm a relationship with DENV-2.12,13 Rather, sequencing confirmed a homology with other members of the YFV group, and EHV is classified by the ICTV within this group.14 Geographically, the closest near-neighbour of EHV is Sepik virus (SEPV), which has been isolated in Papua New Guinea (PNG) but not in Australia.15,16 Reasons for the apparent geographic delineation of these viruses, and the notable absence of YFV in both countries, presumably involve availability of mosquito vectors and host organisms. Additionally, the prevalence of one strain could be exclusionary to other related strains.
Since the original isolation of EHV in a suburb of Cairns in 1961, we and others have isolated the virus in Western Australia, Queensland, New South Wales, and the Northern Territory (Table 1). Here we present both an antigenic and genetic investigation of these isolates. For antigenic analysis, monoclonal antibodies to EHV were raised using either the C281 or the PH235 isolate, and for historical purposes we also tested antibodies raised to DENV-2, along with some reference DENV monoclonals17 (Table 2). Monoclonal antibody (mAb) binding patterns of isolates were determined by inoculation of isolates onto confluent 96-well monolayers of C6/36 cells, incubation for up to 7 days, and subsequent assay by tissue culture enzyme immunoassay.18 The four antibodies raised to C281 recognized all EHV isolates: 3D11 was found to be EHV-specific, while 6A9 also recognized YFV, 7C6 cross-reacted with SEPV, and 8G2 reacted with both SEPV and Banzi virus (BANV). Of the antibodies produced to PH235, only 6F7 recognized all the EHV isolates. The remaining antibodies (3B11, 3G1, 5D3, and 7C3) failed to recognize isolates P1553 and V366. Interestingly, with the exception of 6F7, which also recognized SEPV, the monoclonal antibodies produced to PH235 did not react with other flaviviruses used in this study. Finally, monoclonal antibodies raised to DENV-2 were also tested against EHV and a panel of flaviviruses. These antibodies were found to recognize only viruses within the dengue group, except for 1D1, which recognized all flaviviruses tested. Five reference dengue monoclonal antibodies (15F3, 3H5, 5D4, 1H10 and 2H2) also failed to react with EHV.
Table 1.
Details of Edge Hill virus isolates used in this study.1
| Strain | Place of isolation | Date of isolation | Source |
|---|---|---|---|
| C281* | Edge Hill (Cairns) QLD | 1961 | Cx. annulirostris |
| P1553 | Marble Bar (Pilbara region-WA) | 16/3/94 | Cx. ENM#92 |
| GU0068 | Normanton, QLD | 13/4/2000 | Ae. normanensis |
| PH235 | Newman (Pilbara region-WA) | 18–21/3/79 | Ae. bancroftianus |
| SW42148 | Leschenault Inlet (South-West Region-WA) | 15/1/96 | Ae. camptorhynchus |
| K22005 | Broome (Kimberley region-WA) | 28/3/96 | Ae. vigilax |
| V366 | Darwin, NT | 5/1/83 | Ae. vigilax |
| 19542 | unknown | before 1976 | unknown |
| 22144 | Batemans Bay (NSW) | 17/1/95 | Ae. vigilax |
| 22441 | Batemans Bay (NSW) | 17/1/95 | Ae. vigilax |
| 22897 | Batemans Bay (NSW) | 23/1/95 | Ae. vigilax |
| 22905 | Batemans Bay (NSW) | 23/1/95 | Ae. vigilax |
| 22969 | Batemans Bay (NSW) | 23/1/95 | Ae. vigilax |
| 23056 | Batemans Bay (NSW) | 6/2/95 | Ae. vigilax |
| 23060 | Batemans Bay (NSW) | 6/2/95 | Ae. vigilax |
| 23072 | Batemans Bay (NSW) | 6/2/95 | Ae. vigilax |
| 23462 | Batemans Bay (NSW) | 14/2/95 | Ae. vigilax |
| 23543 | Batemans Bay (NSW) | 20/2/95 | Ae. vigilax |
| 23562 | Batemans Bay (NSW) | 20/2/95 | Ae. vigilax |
| 23703 | Batemans Bay (NSW) | 14/2/95 | Ae. vigilax |
| 24045 | Tathra (NSW) | 7/3/95 | Ae. vigilax |
| 24880 | Ballina (NSW) | 10/4/95 | Ae. procax |
| 25716 | Maclean (NSW) | 5/4/95 | Ae. vigilax |
| 26314 | Boggabilla (NSW) | 11/3/96 | Cx. annulirostris |
| 33029 | Port Stephens (NSW) | 19/2/96 | Ae. vigilax |
Prototype strain.
Table 2.
Binding patterns of monoclonal antibodies as determined by ELISA when tested with EHV isolates and a selection of flaviviruses.
| Viruses |
mAbs prepared to EHV strain C281 |
mAbs prepared to EHV strain PH235 |
|||||||
| 3D11a | 6A9a | 7C6a | 8G2b | 3B11b | 3G1c | 5D3c | 7C3c | 6F7b | |
| EHV C281* | + | + | + | + | + | + | + | + | + |
| EHV PH235* | + | + | + | + | + | + | + | + | + |
| EHV 26314 | + | + | + | + | w | w | − | w | + |
| EHV P1553 | + | + | + | + | − | − | − | − | + |
| EHV V366 | + | + | + | + | − | − | − | − | + |
| BANV | − | − | − | + | − | − | − | − | − |
| YFV | − | + | − | − | − | − | − | − | − |
| SEPV | − | − | + | + | − | − | − | − | + |
| DENV 1 | − | − | − | − | − | − | − | − | − |
| DENV 2 | − | − | − | − | − | − | − | − | − |
| DENV 3 | − | − | − | − | − | − | − | − | − |
| DENV 4 | − | − | − | − | − | − | − | − | − |
| MVEV | − | − | NT | − | − | − | − | − | − |
| KOKV** | − | − | NT | − | − | − | − | − | − |
|
mAbs prepared to DENV 2 |
mAbs prepared to DENV 1, 2, 3 or 417 |
||||||||
| 1D1b | 2C5-2b | 3A9b | 3B2-1b | 15F3c | 3H5b | 5D4c | 1H10c | 2H2a | |
| EHV C281* | + | − | − | − | − | − | − | − | − |
| EHV PH235* | + | − | − | − | − | − | − | − | − |
| EHV 26314 | + | − | − | − | − | − | − | − | − |
| EHV P1553 | + | − | − | − | − | − | − | − | − |
| EHV V366 | + | − | − | − | − | − | − | − | − |
| BANV | + | − | − | − | − | − | − | − | − |
| YFV | + | − | − | − | − | − | − | − | − |
| DENV 1 | + | − | + | + | + | − | − | − | + |
| DENV 2 | + | + | + | + | − | + | − | − | + |
| DENV 3 | + | − | + | − | − | − | + | − | + |
| DENV 4 | + | − | − | − | − | − | − | + | + |
| MVEV | + | − | − | − | NT | − | − | NT | NT |
| KOKV** | + | − | − | − | NT | − | − | NT | NT |
Legend: +, positive; −, negative; w, weak reaction; NT, not tested.
C281, PH235 and other EHV isolates (except 26314, P1553 and V366) had identical reaction patterns.
Viral protein reactivity of some mAbs was determined by Western blot under non-reducing conditions:
Antibodies 3B11, 6A9, 7C6, and 2H2 bind to prM.
Antibodies 8G2, 6F7, 1D1, 2C5-2, 3A9, 3B2-1 and 3H5 bind to E.
Viral protein recognized by 3G1, 5D3, 7C3, 15F3, 5D4 and 1H10 could not be determined by Western blot.
Kokobera virus (KOKV).
Our monoclonal antibody analysis confirmed that EHV is antigenically similar to YFV, BANV and SEPV, and lacks antigenic similarity to DENV-2. Interestingly, the monoclonal antibodies created using the PH235-based immunogen had a different reaction pattern compared to the antibodies induced by a C281-based immunogen, despite antigenic similarity between both isolates. This difference in antibody panel reactivity is probably due to differences in mouse immunization procedures, since the PH235 antibodies were created from immunizations with extracts of infected suckling mouse brain (see Supplementary Material, SM), whereas the C281-derived immunogens comprised purified but inactivated virus cultured in Vero cells (D. Phillips, personal communication).
The most interesting observation from the antigenic studies was the distinct reaction pattern of isolates P1553 and V366, from Western Australia and the Northern Territory respectively. Testing against the panel of C281-derived antibodies clearly indicated these isolates were antigenically more similar to the EHV prototype than our representative YFV, BANV and SEPV isolates, however they showed a distinct antigenic pattern from the EHV prototype when tested against the PH235-derived antibody panel. These data indicate that P1553 and V366 represent a second antigenic subtype of EHV.
We subsequently performed a genetic investigation of EHV isolates via nucleotide sequencing. Several consensus primers were tested for RT-PCR amplification of RNA extracted from EHV infected cell culture supernatant. Only one primer pair (FG1 and FG2)19 successfully amplified any of the EHV isolates—yielding an approximately 1kb portion within the viral NS5 gene. The lack of amplification from the other primer pairs tested suggests significant divergence of EHV from other more characterized flaviviruses. Indeed, the recent report of the full genomic sequence of a single strain of EHV, published after the completion of the present study, has yielded additional information on the evolution of EHV and other members of the YFV group, and will be useful for the design of improved flavivirus-group consensus primers throughout the genome.20
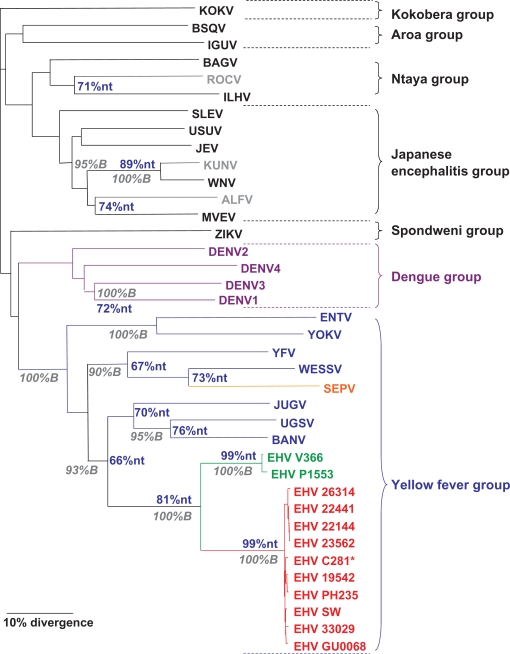
Nucleotide (Fig. 1) and amino acid (SM) sequence analysis of the amplified products indicated homology within the YFV group for all EHV isolates as expected, supported by 100% of bootstrap replicates. Moreover, despite geographic disparity, most EHV isolates had a very high degree of genetic similarity, with less than 1% nucleotide (nt) divergence. Clustering based on geographic origin or time of isolation was not observed, indicating that genetic evolution within the EHV cluster has been minimal over geographic distance and time. However, isolates P1553 and V366 showed a much larger divergence from the EHV C281 prototype (19% nt and 6% amino acid divergence). This degree of divergence is larger than the genetic distance between Kunjin and West Nile viruses (11% nt and 1% amino acid divergence), which are currently classified as separate subtypes within the WNV virus species.14 However the divergence is distinctly smaller than the divergence between DENV-1 and DENV-3 (72% nt), Murray valley encephalitis virus (MVEV) and Alfuy virus (ALFV) (74% nt), and BANV and Uganda S virus (UGSV) (76% nt), indicating classification as a separate virus is not warranted. Combined with our antigenic data, these results clearly indicate that isolates P1553 and V366 represent a distinct subtype or variant of EHV.
Figure 1.
Phylogenic analysis of EHV nucleotide sequences. EHV and other mosquito-borne flavivirus nucleotide sequences (660 nucleotides) of the NS5 gene were aligned using Clustal W.23 Phylogenetic analysis was performed with the PHYLIP analysis package24 using the following parameters: DNA distances were obtained from the Kimura 2-parameter algorithm, phylogenetic relationships determined with the neighbour-joining algorithm, and statistical significance of clusters calculated by bootstrapping 100 replicates and subsequent determination of consensus trees. Bootstrap (B) values are shown in the grey italics, and percent nucleotide similarity (nt) is shown in blue. EHV strains are shown in red, whereas EHV variants are shown in green. Scale bar indicates the distance length of 10 percent divergence.
With only 2 representative strains, the origin and divergence of the EHV variant cannot be determined, but full-length sequence analysis could provide valuable information. Current data appears to preclude a recombination event, and the geographic location of the variant isolates indicates that they most likely co-exist with the prototype strain. Such a coexistence of two EHV subtypes within the YFV group suggests that strain exclusion is perhaps an unlikely scenario for the absence of other YFV group viruses in Australia. Recently Australia has seen the emergence of both JEV21 and a new KOKV-like virus,22 both of which are suspected to have been introduced from PNG. The determination of a second subtype of EHV, and detections of Sepik virus in PNG,15 indicate a serious possibility of the introduction of further YFV-group viruses into Australia. We recommend that virulence studies in mouse models of both Sepik and the EHV variant be performed to assess risks of disease in man.
Supplementary Material
Acknowledgments
We thank Dr. Natalie Prow for antigenic analysis of Sepik virus. We also acknowledge Andrew Falconar’s contributions to the original generation of some of the dengue virus monoclonal antibodies used in this study. The study was partially funded by the National Health and Medical Research Council of Australia.
Current Addresses
JM—Division of Experimental Therapeutics, Department of Medicine, Columbia University, 630W 168th St, Box 84, New York, NY 10032 USA.
MP—Integrative Computational Sciences, Lilly Singapore Centre for Drug Discovery, 8A Biomedical Grove, Singapore 138648.
JSM—Australian Biosecurity CRC, Curtin University of Technology, Perth, WA6845 Australia.
DP—Inverness Medical Innovations Australia Pty Ltd, 532 Seventeen Mile Rocks Road, Sinnamon Park QLD 4073 Australia.
SM—VIDO University of Saskatchewan 120 Veterinary Rd, Saskatoon, SK S7 N5E3, Canada.
GG—CSIRO Livestock Industries, Australian Animal Health Laboratory, Geelong, Victoria, Australia.
DG—Australian Biosecurity CRC, Curtin University of Technology, Perth, WA6845 Australia.
Disclosures
This manuscript has been read and approved by all authors. This paper is unique and is not under consideration by any other publication and has not been published elsewhere. The authors and peer reviewers of this paper report no conflicts of interest. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.Doherty RL, Carley JG, Mackerras MJ, Marks EN. Studies of arthropod-borne virus infections in Queensland. III. Isolation and characterization of virus strains from wild-caught mosquitoes in north Queensland. Australian Journal of Experimental Biology. 1963;41:17–40. doi: 10.1038/icb.1963.2. [DOI] [PubMed] [Google Scholar]
- 2.van den Hurk AF, Nisbet DJ, Foley PN, Ritchie SA, Mackenzie JS, Beebe NW. Isolation of arboviruses from mosquitoes (Diptera: Culicidae) collected from the Gulf Plains region of northwest Queensland, Australia. J Med Entomol. 2002;39:786–92. doi: 10.1603/0022-2585-39.5.786. [DOI] [PubMed] [Google Scholar]
- 3.Russell RC. Arboviruses and their vectors in Australia: an update on the ecology and epidemiology of some mosquito-borne arboviruses. Reviews of Medical and Veterinary Entomology. 1995;83:141–58. [Google Scholar]
- 4.Lindenbach BD, Rice CM. Flaviviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Fourth Edition. Vol. 1. Lippincott Williams & Wilkins; Philadelphia: 2001. pp. 991–1041. [Google Scholar]
- 5.Aaskov JG, Phillips DA, Wiemers MA. Possible clinical infection with Edge Hill virus. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1993;87:452–3. doi: 10.1016/0035-9203(93)90032-l. [DOI] [PubMed] [Google Scholar]
- 6.Hawkes RA, Boughton CR, Naim HM, Wild J, Chapman B. Arbovirus infections of humans in New South Wales. Seroepidemiology of the flavivirus group of togaviruses. The Medical Journal of Australia. 1985;143:555–61. [PubMed] [Google Scholar]
- 7.Doherty RL, Carley JG, Gorman BM. Studies of arthropod-borne virus infections in Queensland. IV. Further serological investigations of antibodies to group B arboviruses in man and animals. Australian Journal of Experimental Biology. 1964;42:149–64. doi: 10.1038/icb.1964.16. [DOI] [PubMed] [Google Scholar]
- 8.Calisher CH, Karabatsos N, Dalrymple JM, et al. Antigenic relationships between flaviviruses as determined by cross-neutralisation tests with polyclonal antisera. Journal of General Virology. 1989;70:37–43. doi: 10.1099/0022-1317-70-1-37. [DOI] [PubMed] [Google Scholar]
- 9.Madrid ATD, Porterfield JS. The flaviviruses (group B arboviruses): a cross-neutralisation study. Journal of General Virology. 1974;23:91–6. doi: 10.1099/0022-1317-23-1-91. [DOI] [PubMed] [Google Scholar]
- 10.Blok J, Henchal EA, Gorman BM. Comparison of dengue viruses and some other flaviviruses by cDNA-RNA hybridisation analysis and detection of a close relationshipo between dengue virus serotype 2 and Edge Hill virus. Journal of General Virology. 1984;65:2173–81. doi: 10.1099/0022-1317-65-12-2173. [DOI] [PubMed] [Google Scholar]
- 11.Hanna JN, Ritchie SA, Richards AR, et al. Multiple outbreaks of dengue serotype 2 in north Queensland, 2003/04. Aust NZJ Public Health. 2006;30:220–5. doi: 10.1111/j.1467-842x.2006.tb00861.x. [DOI] [PubMed] [Google Scholar]
- 12.Kuno G, Chang GJ, Tsuchiya KR, Karabatsos N, Cropp CB. Phylogeny of the genus Flavivirus. J Virol. 1998;72:73–83. doi: 10.1128/jvi.72.1.73-83.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maher-Sturgess SL, Forrester NL, Wayper PJ, et al. Universal primers that amplify RNA from all three flavivirus subgroups. Virol J. 2008;5:16. doi: 10.1186/1743-422X-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fauquet C, Mayo MA, Maniloff J, Desselberger U, Ball LA. Virus Taxonomy VIIIth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press; San Diego, CA: 2005. [Google Scholar]
- 15.Johansen CA, van den Hurk AF, Ritchie SA, et al. Isolation of Japanese encephalitis virus from mosquitoes (Diptera: Culicidae) collected in the Western Province of Papua New Guinea, 1997–1998. Am J Trop Med Hyg. 2000;62:631–8. doi: 10.4269/ajtmh.2000.62.631. [DOI] [PubMed] [Google Scholar]
- 16.Kuno G, Chang GJ. Characterization of Sepik and Entebbe bat viruses closely related to yellow fever virus. Am J Trop Med Hyg. 2006;75:1165–70. [PubMed] [Google Scholar]
- 17.Henchal EA, Gentry MK, McCown JM, Brandt WE. Dengue virus-specific and flavivirus group determinants indentified with monoclonal antibodies by indirect immunoflourescence. American Journal of Tropical Medicine and Hygiene. 1982;31:830–6. doi: 10.4269/ajtmh.1982.31.830. [DOI] [PubMed] [Google Scholar]
- 18.Adams SC, Broom AK, Sammels LM, et al. Glycosylation and Antigenic variation among Kunjin virus isolates. Virology. 1995;206:49–56. doi: 10.1016/s0042-6822(95)80018-2. [DOI] [PubMed] [Google Scholar]
- 19.Fulop L, Barrett ADT, Phillpotts R, Martin K, Leslie D, Titball RW. Rapid identification of flaviviruses based on conserved NS5 gene sequences. Journal of Virological Methods. 1993;44:179–88. doi: 10.1016/0166-0934(93)90053-t. [DOI] [PubMed] [Google Scholar]
- 20.Grard G, Moureau G, Charrel RN, Holmes EC, Gould EA, de Lamballerie X. Genomics and evolution of Aedes-borne flaviviruses. J Gen Virol. 2010;91:87–94. doi: 10.1099/vir.0.014506-0. [DOI] [PubMed] [Google Scholar]
- 21.Van Den Hurk AF, Montgomery BL, Northill JA, et al. Short report: the first isolation of Japanese encephalitis virus from mosquitoes collected from mainland Australia. Am J Trop Med Hyg. 2006;75:21–5. [PubMed] [Google Scholar]
- 22.Nisbet DJ, Lee KJ, van den Hurk AF, et al. Identification of new flaviviruses in the Kokobera virus complex. J Gen Virol. 2005;86:121–4. doi: 10.1099/vir.0.80381-0. [DOI] [PubMed] [Google Scholar]
- 23.Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 24.Felsenstein J. PHYLIP—Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–6. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.