Abstract
Pyridine nucleotide transhydrogenase (PNT) catalyzes the direct transfer of a hydride-ion equivalent between NAD(H) and NADP(H) in bacteria and the mitochondria of eukaryotes. PNT was previously postulated to be localized to the highly divergent mitochondrion-related organelle, the mitosome, in the anaerobic/microaerophilic protozoan parasite Entamoeba histolytica based on the potential mitochondrion-targeting signal. However, our previous proteomic study of isolated phagosomes suggested that PNT is localized to organelles other than mitosomes. An immunofluorescence assay using anti-E. histolytica PNT (EhPNT) antibody raised against the NADH-binding domain showed a distribution to the membrane of numerous vesicles/vacuoles, including lysosomes and phagosomes. The domain(s) required for the trafficking of PNT to vesicles/vacuoles was examined by using amoeba transformants expressing a series of carboxyl-terminally truncated PNTs fused with green fluorescent protein or a hemagglutinin tag. All truncated PNTs failed to reach vesicles/vacuoles and were retained in the endoplasmic reticulum. These data indicate that the putative targeting signal is not sufficient for the trafficking of PNT to the vesicular/vacuolar compartments and that full-length PNT is necessary for correct transport. PNT displayed a smear of >120 kDa on SDS-PAGE gels. PNGase F and tunicamycin treatment, chemical degradation of carbohydrates, and heat treatment of PNT suggested that the apparent aberrant mobility of PNT is likely attributable to its hydrophobic nature. PNT that is compartmentalized to the acidic compartments is unprecedented in eukaryotes and may possess a unique physiological role in E. histolytica.
Pyridine nucleotide transhydrogenase (PNT) participates in the bioenergetic processes of the cell. PNT generally resides on the cytoplasmic membranes of bacteria and the inner membrane of mammalian mitochondria (3, 16) and utilizes the electrochemical proton gradient across the membrane to drive NADPH formation from NADH (14, 15, 39) according to the reaction H+out + NADH + NADP+↔H+in + NAD+ + NADPH, where “out” and “in” denote the cytosol and the matrix of the mitochondria, or the periplasmic space and the cytosol of bacteria, respectively.
PNT has been identified in several protozoan parasites, including Entamoeba histolytica (8, 51), Eimeria tenella (17, 47), Mastigamoeba balamuthi (11) Plasmodium falciparum (10), Plasmodium yoelii (6), and Plasmodium berghei (12). In general, PNT contains conserved structural units consisting of three domains, the NAD(H)-binding domain (domain I [dI]) and the NADP(H)-binding domain (domain III [dIII]), both of which face the matrix side of the eukaryotic mitochondria or the cytoplasmic side in bacteria, and the hydrophobic domain (domain II [dII]), containing 11 to 13 transmembrane regions. PNT from E. tenella and E. histolytica exists as a single polypeptide in an unusual configuration consisting of dIIb-dIII-dI-dIIa, with a 38-amino-acid-long linker region between dIII and dI (48).
E. histolytica, previously considered an “amitochondriate” protist, is currently considered to possess a mitochondrion-related organelle with reduced and divergent functions, the mitosome (1, 21, 23a, 26, 42). Our recent proteomic study of isolated mitosomes identified about 20 new constituents (26), together with four proteins previously demonstrated in E. histolytica mitosomes: Cpn60 (8, 19, 21, 42), Cpn10 (46), mitochondrial Hsp70 (2, 44), and mitochondrion carrier family (MCF) (ADP/ATP transporter) (7). Despite the early presumption of PNT being localized in mitosomes (8), based on the amino-terminal region rich in hydroxylated (five serines and threonines) and acidic (three glutamates) amino acids, which slightly resembles known mitochondrion- and hydrogenosome-targeting sequences (8, 35), PNT was not discovered in the mitosome proteome. We also doubted this premise because PNT was one of the major proteins identified in isolated phagosomes (32, 33). Thus, the intracellular localization and trafficking of PNT remain unknown.
In this report, we showed that E. histolytica PNT (EhPNT) is localized to various vesicles and vacuoles, including lysosomes and phagosomes, using wild-type amoebae and antiserum raised against recombinant EhPNT and an E. histolytica line expressing EhPNT with a carboxyl-terminal hemagglutinin (HA) epitope tag and anti-HA antibody. We also showed that all domains of EhPNT are required for its trafficking to the acidic compartment by using amoeba transformants expressing the HA tag or green fluorescent protein (GFP) fused with a region containing various domains of EhPNT.
MATERIALS AND METHODS
Cells, cultures, and reagents.
Trophozoites of E. histolytica strain HM-1:IMSS cl6 were maintained axenically in Diamond's BI-S-33 medium (9) at 35.5°C. Chinese hamster ovary (CHO) cells were maintained in F12 medium (Invitrogen, San Diego, CA) supplied with 10% fetal calf serum (Medical Biological Laboratory International, Woburn, MA) at 37°C with 5% CO2. Escherichia coli strains DH5α and BL21(DE3) were purchased from Life Technologies (Tokyo, Japan) and Novagen (Madison, WI), respectively. LysoTracker Red DND-99 and CellTracker Orange CMTMR [5-(and-6)-(((4-chloromethyl)benzoyl)amino)tetramethylrhodamine] were purchased from Molecular Probes (Eugene, OR). All other chemicals of analytical grade were purchased from Sigma-Aldrich unless otherwise stated.
Plasmid construction.
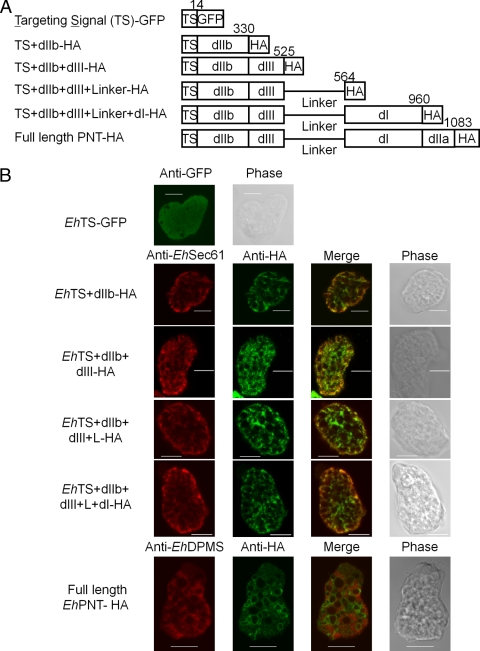
Standard techniques were used for routine DNA manipulation, subcloning, and plasmid construction as previously described (38). To produce E. coli recombinant proteins, a coding region corresponding to dI (amino acids [aa] 565 to 960) of EhPNT (EhPNTdI) was amplified from an E. histolytica cDNA library by using a pair of appropriate primers designed on the basis of the nucleotide sequences in the GenBank database (accession number L39933) (8), with BamHI and XhoI restriction enzyme sites. The sense and antisense primers were 5′-CGAGGATCCGATGTTATTTATTGGTATTCCAAAAG-3′ and 5′-CGTTCTCGAGTCATTCTTCTTCAGTTGAAAGA-3′, respectively, where boldface type indicates the BamHI or XhoI site. The PCR products were cloned into the BamHI- and XhoI-digested vector pET47b (Novagen, Madison, WI), and the resulting plasmid was designated pHisPNTdI. To generate vectors to express either full-length or truncated forms of EhPNT fused to HA in the amoeba, a protein-coding region corresponding to full-length EhPNT (GenBank accession number AAC41577) (aa 1 to 1083), the 14-aa amino-terminal region encompassing the putative targeting sequence (TS)+dIIb (aa 1 to 330), TS+dIIb+dIII (aa 1 to 525), TS+dIIb+dIII+linker (aa 1 to 564), and TS+dIIb+dIII+ linker+dI (aa 1 to 960) was amplified from an E. histolytica cDNA library by using a pair of appropriate primers and cloned into the BglII site of pEhExHA (29). The antisense primers were 5′-CGTGGATCCATGAAACATTTTCAACATTCTG-3′, 5′-CGTGGATCCGAATGATCTATTCATAGCTTTACAC-3′, 5′-CGTGGATCCTTCTTCATTTAATTCTTCAAATCCTTTC-3′, 5′-CGTGGATCCATCTTCTGCAAGAACTTTTGTTGGA-3′, and 5′-CGTGGATCCTTCTTCTTCAGTTGAAAGAGT-3′, respectively, where boldface type represents the BamHI site. The sense primer described above was used for these full-length or truncated forms of EhPNT to express in the amoeba. To generate a plasmid to express GFP fused to the EhPNT TS, a pair of oligonucleotides corresponding to the TS were generated, self-annealed, and cloned into the BglII site of pEhExGFP. The oligonucleotides were 5′-GATCTATGAGCACAAGTTCTAGTATTGAAGAAGAAGTGTTCAATTATA-3′ and 5′-GATCTATAATTGAACACTTCTTCTTCAATACTAGAACTTGTGCTCATA-3′, where boldface type represents the truncated BglII site. pEhExGFP was generated by the ligation of the GFP protein-coding region of pKT-MG (29) into the BglII-XhoI site of pEhEx (31). These constructs allowed the expression of PNT with the three tandem copies of the HA tag or GFP at the carboxyl terminus. The resulting plasmids were designated pPNTFL-HA, pTS/IIb-HA, pTS/IIb/III-HA, pTS/dIIb/dIII/L-HA, pTS/IIb/III/L/I-HA, and pPNTTS-GFP. The production of the Cpn60-HA transformant was previously described (26).
Amoeba transformation.
Plasmids generated as described above were introduced into amoeba trophozoites by lipofection as previously described (30). Geneticin (Invitrogen, San Diego, CA) was added at a concentration of 1 μg/ml at 24 h after transfection, and the Geneticin concentration was gradually increased for approximately 2 weeks until it reached 10 μg/ml.
Recombinant protein production.
pHisPNTdI was introduced into BL21(DE3) cells. The expression of the histidine-tagged EhPNTdI protein was induced with 1 mM isopropyl-β-thiogalactoside at 37°C for 3 h. After harvesting and washing three times with phosphate-buffered saline (PBS) (pH 7.4), the bacteria were lysed in B-PER reagent (Pierce, Rockford, IL) containing Complete Mini EDTA-free protease inhibitor cocktail (Roche Diagnostic, Mannheim, Germany) and mixed with 1.0 ml of a 50% slurry of Ni2+-nitrilotriacetic acid (NTA) His-Bind resin. The recombinant EhPNTdI-bound resin was washed in a column three times with 25 ml of buffer A (50 mM NaH2PO4, 300 mM NaCl [pH 8.0]) containing 20 mM imidazole. Bound proteins were eluted with buffer A containing 1 M imidazole and dialyzed against PBS.
Antibodies.
Anti-EhPNT antibody was raised against purified EhPNTdI in rabbit commercially (Operon, Tokyo, Japan). Anti-HA 11MO mouse monoclonal antibody was purchased from Berkeley Antibody (Berkeley, CA). Anti-EhSec61 α-subunit and anti-E. histolytica dolicol-P-mannose synthase (EhDPMS) antibodies were a gift from Rosana Sánchez-López (37). Anti-galactose/N-acetylgalactosamine inhibitable lectin (Hgl) monoclonal antibody (3F4) (23) was a gift from Barbara J. Mann and William A. Petri, Jr. Alexa Fluor 488- or 568-conjugated anti-mouse and anti-rabbit IgGs were purchased from Invitrogen. Alkaline phosphatase-conjugated goat anti-rabbit and goat anti-mouse IgGs were bought from Jackson ImmunoResearch Laboratories (Bar Harbor, ME). Anti-GFP rabbit antibody was purchased from Medical Biological Laboratory International.
Immunoprecipitation.
Approximately 3 × 106 cells of EhPNT-HA- and Cpn60-HA-expressing amoebae were lysed in lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton X-100, and 0.5 mg/ml E-64). The soluble lysate, after centrifugation at 15,000 × g, was incubated with protein G-Sepharose beads (30 μl of a 50% slurry) (Amersham Biosciences, Uppsala, Sweden) premixed with anti-HA antibody (0.5 μl) or anti-HA and anti-Hgl antibodies, respectively.
Immunoblot analysis.
Whole-cell lysate and immunoprecipitated samples were separated on either a 12% or 15% (wt/vol) SDS-polyacrylamide gel and subsequently electrotransferred onto nitrocellulose membranes (Hybond-C Extra; Amersham Biosciences, Little Chalfont, Bucks, United Kingdom) as described previously (41). The membranes were blocked by incubation in 5% nonfat dried milk in TBST (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, and 0.05% Tween 20) for 1.5 h at room temperature. The blots were reacted with primary anti-EhPNT rabbit or anti-HA mouse antibody at a dilution of 1:500 to 1:1,000. The membranes were washed with TBST and further reacted with alkaline phosphatase-conjugated anti-rabbit or anti-mouse IgG antibody (1:1,000) at room temperature for 1.5 h. After further washing with TBST, specific proteins were visualized with an alkaline phosphatase conjugate substrate kit (Bio-Rad, Hercules, CA).
Deglycosylation of PNT.
The EhPNT-HA-expressing transformant was cultured with 3 μg/ml of tunicamycin (Sigma, St. Louis, MO) for 24 h to inhibit asparagine-linked glycosylation according to a protocol described previously (22). For the chemical deglycosylation of EhPNT, EhPNT and fetuin were dried in a Speed Vac. Ice-cold trifluoromethanesulfonic acid (TFMS)-anisol (3:2, vol/vol [100 μl]) was added, and the samples were incubated for 4 h at 4°C under N2 according to a method described previously (13). The reaction was stopped by slowly adding 200 μl ice-cold H2O-pyridine (73:10, vol/vol) containing 0.1% SDS to the mixture. Anisol was extracted three times with 250 μl ethyl ether. Dialysis was performed against 2 mM pyridine acetate buffer. After dialysis, the samples were subjected to SDS-PAGE and silver staining. Immunoprecipitated EhPNT was also digested with PNGase F (New England Biolabs, Ipswich, MA), an amidase that cleaves between the innermost N-acetylglucosamine and asparagine residues of asparagine-linked glycoproteins, according to the manufacturer's instructions.
Immunofluorescence assay and organelle staining.
Amoeba transformant or wild-type amoebae in a logarithmic growth phase were harvested, transferred into 8-mm round wells on glass slides, and incubated for 30 min at 35°C to let trophozoites attach to the glass surface. An indirect immunofluorescence assay was performed as previously described (36). Briefly, amoebae were fixed with 3.7% paraformaldehyde in PBS for 10 min at room temperature. The cells were then permeabilized with 0.05% Triton X-100 in PBS for 5 min. The samples were reacted with anti-EhPNT (1:100), anti-EhSec61 α-subunit (1:30), anti-EhDPMS antibody (1:30), anti-GFP (1:1,000) rabbit antibody, anti-HA (1:1,000) mouse antibody, or preimmune rabbit serum (1:100). The samples were then reacted with Alexa Fluor 488- or 568-conjugated anti-mouse or anti-rabbit IgG (1:1,000).
For the staining of endosomes or late endosomes/lysosomes, amoebae were incubated with BI-S-33 medium containing 2 mg/ml rhodamine isothiocyanate (RITC)-dextran for 10 to 60 min (for endosomes) or LysoTracker Red DND-99 (Molecular Probes, Eugene, OR) (1:500) for 12 h (for late endosomes/lysosomes), respectively. To visualize phagosomes, CHO cells prestained with 10 μM CellTracker Orange or 20 μM CellTracker Blue were added to E. histolytica trophozoites in 8-mm wells on a glass slide and incubated for 10 to 60 min. The samples were examined on an LSM 510 Meta confocal laser scanning microscope (Carl Zeiss, Thornwood, NY). Images were further analyzed by using LSM510 software.
RESULTS
Identification of two isotypes of PNT in E. histolytica.
Two isozymes of PNT have been identified in the E. histolytica genome database at Pathema (http://pathema.tigr.org/tigr-scripts/pathema/) (EHI_055400 and EHI_014030, corresponding to GenBank accession numbers XP_001914099 and AAC41577, respectively). They showed 90% mutual amino acid identity. The latter EhPNT isotype (EH_014030) is 1,083 aa long with a predicted molecular mass of 117.0 kDa and a pI of 5.39. This predicted protein is identical to the PNT protein previously reported (GenBank accession number AAC41577) (8). The dIII, dI, and dIIa regions of EHI_055400 (1,098 aa) are conserved except for a single amino acid substitution in dIII. EHI_055400 also contains a 5-aa extension at the amino terminus (MSLLL) and 9 aa substitutions in the putative TS (aa 1 to 14 of EHI_014030) (data not shown). In addition, dIIb of EHI_055400 contained 65 aa substitutions and two (4- and 6-aa-long) block insertions; the linker region also contains 24 aa substitutions. Since the domains involved in catalysis in EHI_055400 and EHI_014030 are totally conserved, and the two genes are expressed at comparable levels as steady-state mRNA by quantitative reverse transcriptase PCR (data not shown), we further studied only EHI_014030 in the present work. EHI_001930 (GenBank accession number XP_653216), which is annotated as the PNT β-subunit, showed no significant homology to either of the two EhPNTs described above, while this sequence showed similarity with the β-subunit from other organisms and, thus, was excluded from this study.
Expression of PNT in E. histolytica trophozoites.
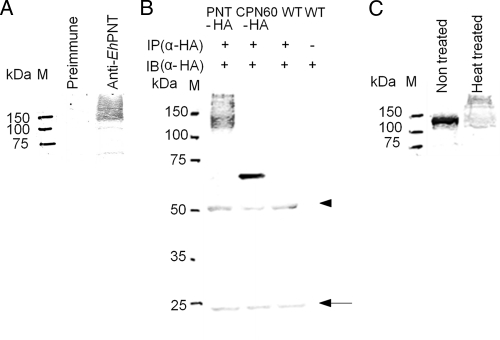
Immunoblot analysis of the trophozoite lysate using anti-EhPNT antibody showed a smear of >120 kDa (Fig. 1A). The size of the smear was unexpected because the predicted molecular masses of EHI_055400 and EHI_014030 were 119.0 and 117.0 kDa, respectively. To verify that this was not due to the cross-reactivity of anti-EhPNT antibody, we immunoprecipitated EhPNT from the EhPNT-HA-expressing transformant with anti-HA antibody, followed by immunoblotting with anti-HA (Fig. 1B) or anti-EhPNT (data not shown) antibody. Immunoprecipitated EhPNT-HA was also recognized as a smear of >120 kDa, similar to that of endogenous EhPNT.
Fig. 1.
Expression of PNT in E. histolytica. (A) Immunoblot analysis of native EhPNT. Approximately 10 μg of total lysate was electrophoresed on a 12% SDS-polyacrylamide gel and subjected to an immunoblot assay with anti-EhPNT antibody or preimmune serum. M, molecular mass marker. (B) Immunoprecipitation of EhPNT. The lysates derived from the transformant expressing either EhPNT-HA (“PNT-HA”) or EhCpn60-HA (“CPN60-HA”) and the wild-type strain (“WT”) were subjected to immunoprecipitation with anti-HA antibody, followed by immunoblot analysis with anti-HA antibody. Lysate derived from the wild-type amoebae was also used directly for immunoblot analysis as a control. An arrowhead and an arrow indicate heavy and light chains of anti-HA antibody, respectively. IP, immunoprecipitation; IB, immunoblot. (C) Effect of heat treatment on the mobility of EhPNT on an SDS-PAGE gel. Approximately 10 μg of total lysate was electrophoresed on a 12% SDS-polyacrylamide gel and subjected to an immunoblot assay with anti-EhPNT antibody. M, molecular mass marker.
To examine whether the smear was due to posttranslational modifications such as glycosylation, we treated immunoprecipitated EhPNT-HA with TFMS or PNGase F. The pattern of immunoblots with anti-HA antibody was not affected, while the apparent molecular mass of control fetuin decreased (Fig. 2 A and B). In addition, the treatment of the trophozoites with tunicamycin did not affect the mobility of EhPNT, while the mobility of control Hgl increased (Fig. 2C). These data are consistent with the notion that EhPNT is not glycosylated. We next examined whether aberrant mobility is due to unusual tertiary structures. We compared the patterns of the amoebic lysates, mixed with a one-third volume of 4× SDS-PAGE sample buffer (0.25 M Tris-HCl [pH 6.8], 8% SDS, and 8% 2-mercaptoethanol) and either incubated at 95°C for 5 min or left unheated, on SDS-PAGE gels. When the sample was electrophoresed without heating, EhPNT was observed as a polypeptide of the predicted size (Fig. 1C). The exclusion of 2-mercaptoethanol did not affect mobility (data not shown). A similar observation was previously reported for membrane proteins, including serotonin transporter (24) and severe acute respiratory syndrome (SARS)-associated coronavirus membrane protein (18). In the latter case, three hydrophobic regions of 10 to 35 aa were shown to be responsible for the heat-induced aggregation of the membrane protein (18), suggesting that the heat-induced change of mobility of EhPNT on SDS-PAGE is likely due to the hydrophobic nature of the protein.
Fig. 2.
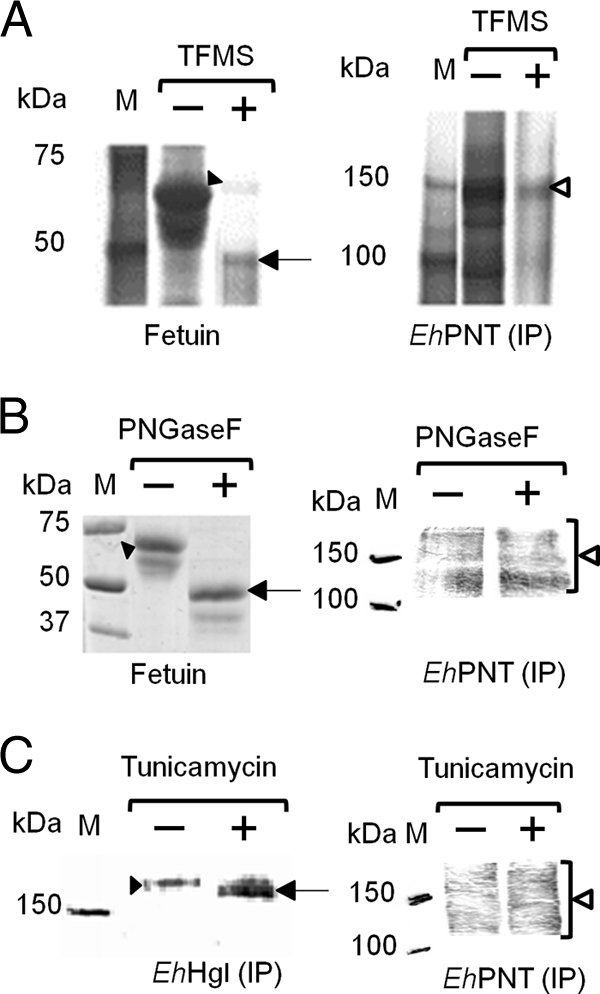
Deglycosylation of EhPNT. (A and B) Deglycosylation by TFMS or PNGase F. The lysates obtained from the transformant expressing EhPNT-HA were subjected to immunoprecipitation (IP) with anti-HA antibody, followed by TFMS or PNGase F treatment and immunoblot detection with anti-HA antibody (right). Fetuin was used as a control, and gels were stained with silver (A) or Coomassie brilliant blue (B) (left). Filled arrowheads indicate untreated fetuin. Arrows indicate fetuin deglycosylated by TFMS (A) or PNGase F (B). Open arrowheads indicate EhPNT. (C) Deglycosylation by tunicamycin. Lysates from the transformant expressing EhPNT-HA cultured with tunicamycin were subjected to immunoprecipitation with either anti-EhHgl or anti-HA antibody, followed by immunoblot analysis with either anti-EhHgl or anti-HA antibody, respectively. A filled arrowhead or an arrow indicates untreated or deglycosylated EhHgl, respectively. An open arrowhead indicates EhPNT.
Subcellular distribution of EhPNT.
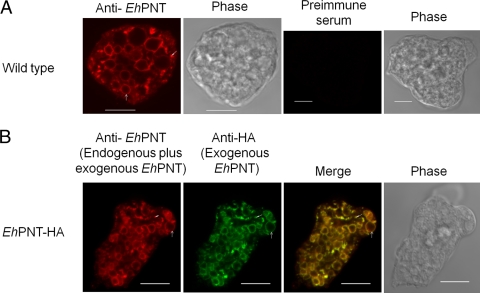
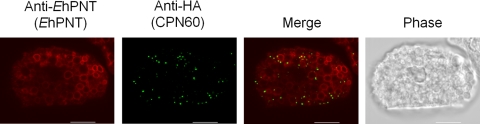
An immunofluorescence assay using anti-EhPNT antibody showed that EhPNT is associated with the membranes of vesicles and vacuoles varying in size, or sometimes dot-like structures, scattered throughout the cytosol (Fig. 3A). We also examined the intracellular distribution of EhPNT using the amoebic transformant that expressed EhPNT (EHI_014030) with the carboxyl-terminal HA tag. The pattern of exogenous EhPNT-HA (EHI_014030) and that of endogenous EhPNT (a sum of EHI_055400 and EHI_014030) plus exogenous EhPNT (EHI_014030) were indistinguishable (Fig. 3B). Since EhPNT was previously postulated to be localized to mitosomes (8), we next examined the localization of EhPNT and EhCpn60, the authentic marker of mitosomes, in the amoebic transformant expressing EhCpn60-HA (26) using anti-EhPNT and anti-HA antibodies. No colocalization of EhPNT and EhCpn60 was observed (Fig. 4).
Fig. 3.
(A) Subcellular localization of EhPNT in wild-type amoebae. Wild-type amoebae were fixed, and an immunofluorescence assay was performed by using anti-EhPNT (red) and preimmune sera. Bar, 10 μm. Arrows and arrowheads indicate representative vacuolar and dot-like structures of EhPNT, respectively. (B) Colocalization of endogenous and exogenous epitope-tagged PNT in E. histolytica. EhPNT-HA-overexpressing amoebae were fixed, and an immunofluorescence assay was performed by using anti-EhPNT (red) and anti-HA (green) antibodies.
Fig. 4.
Lack of association of EhPNT with mitosomes. The amoebic transformant expressing EhCpn60-HA was stained with anti-HA (green) and anti-EhPNT (red) antibodies to visualize EhCpn60 and EhPNT, respectively. Bar, 10 μm.
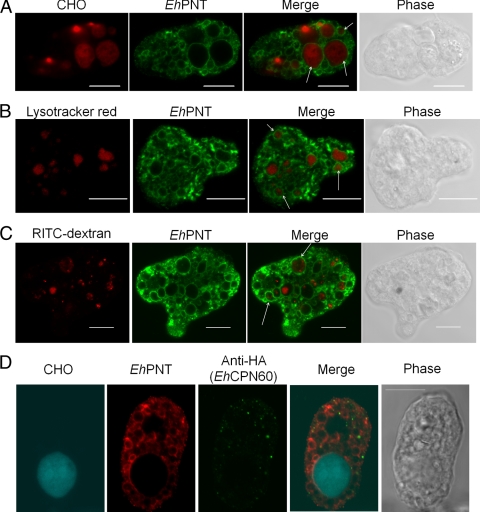
Since EhPNT was previously detected in isolated phagosomes (32, 33), we examined the localization of EhPNT during the phagocytosis of CHO cells. Wild-type amoebae were incubated with CellTracker Orange-loaded CHO cells for 10 to 60 min to allow the ingestion of CHO cells. An immunofluorescence assay using anti-EhPNT antibody (Fig. 5A) showed that the phagocytosed CHO cells were associated with EhPNT at all time points (10, 20, and 60 min; only the images at 60 min are shown). The percentage of association gradually increased during the course of phagocytosis (65% ± 6%, 79% ± 9%, and 80% ± 8% at 10, 30, and 60 min, respectively). We then examined whether EhPNT is localized to lysosomes using LysoTracker Red, a membrane-diffusible probe accumulated in acidic organelles (5). We found that the LysoTracker-labeled acidic compartment, the size and number of which were consistent with previous findings (28, 36), was associated with EhPNT under steady-state conditions (79% ± 6% association) (Fig. 5B). To see whether EhPNT is also associated with endosomes, we examined the localization of an endocytosed fluid-phage marker, RITC-dextran. EhPNT was only partially associated with RITC-dextran-containing endosomes, which were observed as tiny dot-like structures or a multivesicular body as previously shown (29), at each time point (10, 30, or 60 min; only the images at 60 min are shown) (Fig. 5C). We also examined whether EhPNT is associated with mitosomes during the phagocytosis of CHO cells. An immunofluorescence assay using the amoebic transformant expressing EhCpn60-HA, CellTracker Blue-loaded CHO cells, anti-EhPNT, and anti-HA antibody showed no colocalization of mitosomes and EhPNT (Fig. 5D).
Fig. 5.
Localization of EhPNT to phagosomes, lysosomes, and endosomes. (A) Association of EhPNT with phagosomes. Amoebae were incubated with CellTracker Orange-loaded CHO cells (red) for 60 min, fixed, and reacted with anti-EhPNT antibody (green). Arrows indicate representative phagocytosed CHO cells associated with EhPNT. (B) Association of EhPNT with lysosomes. Amoebae were labeled with LysoTracker (red) and subjected to an immunofluorescence assay with anti-EhPNT antibody (green). Arrows indicate representative lysosomes associated with EhPNT. (C) Association of EhPNT with the fluid-phase marker. Amoebae were incubated with medium containing RITC-dextran (red) for 1 h. The cells were fixed and reacted with anti-EhPNT antibody (green). Arrows indicate representative endocytosed RITC-dextran associated with EhPNT. (D) Subcellular localization of phagosomes, mitosomes, and EhPNT. The amoebic transformant expressing EhCpn60-HA was incubated with CellTracker Blue-loaded CHO cells (blue) for 60 min, fixed, and reacted with anti-EhPNT (red) and anti-EhCpn60 (green) antibodies. Bar, 10 μm.
All domains are essential for the vesicular/vacuolar distribution of EhPNT.
To define the domain necessary for vesicular/vacuolar targeting of EhPNT, we created amoeba transformants expressing the HA-tagged or GFP-fused protein containing various domains of PNT (Fig. 6A). The amoebic transformant expressing GFP fused with the 14-aa amino-terminal putative TS showed a cytoplasmic distribution (Fig. 6B), although the sequence MSTSSSIEEEVFNY appeared to contain the elements implicated for the TS (rich in hydroxylated and hydrophobic amino acids). The transformants expressing TS+dIIb-HA, TS+dIIb+dIII-HA, TS+dIIb+dIII+L-HA, or TS+dIIb+dIII+L+dI-HA showed a distribution that overlapped that of the endoplasmic reticulum (ER), visualized with anti-EhSec61 α-subunit antibody (27). The ER pattern was also confirmed with anti-EhDPMS antibody (Fig. 6B). Full-length EhPNT-HA did not overlap the ER visualized with either anti-EhSec61 α-subunit or anti- EhDPMS antibody. Fractionation of the amoeba lysate followed by immunoblot analysis with anti-HA or anti-GFP antibodies showed that all truncated forms of EhPNT except for TS-GFP were partitioned into the 5,000 × g and 100,000 × g pellet fractions, while TS-GFP was fractionated into the 100,000 × g supernatant fraction (data not shown).
Fig. 6.
Localization of a series of truncated EhPNT proteins. (A) Schematic representation of HA epitope-tagged or GFP-fused recombinant EhPNT used in the study. Domains, epitope (or GFP), and amino acid numbers are shown. (B) Localization of epitope-tagged or GFP-fused carboxyl-terminally truncated EhPNT recombinant proteins. The transformants expressing EhTS-GFP, EhTS+dIIb-HA, EhTS+dIIb+dIII-HA, EhTS+dIIb+dIII+L-HA, and EhTS+dIIb+dIII+L+dI-HA and EhPNT-HA were subjected to an immunofluorescence assay using anti-HA (or anti-GFP for EhTS-GFP) and anti-EhSec61 α-subunit (or anti-EhDPMS for the EhPNT-HA transformant) antibodies. Bar, 10 μm.
DISCUSSION
Mitosomes have been identified in several parasitic protozoan lineages such as E. histolytica, Giardia intestinalis, Trachipleistophora hominis, and Cryptosporidium parvum (21, 34, 42, 43, 50). E. histolytica was previously considered to be an early-branching “amitochondriate,” as it lacks conventional mitochondria as well as other organelles typically found in most eukaryotes, such as peroxisomes, the rough ER, and the Golgi apparatus. However, the discovery of genes encoding the mitochondrial proteins Cpn60, PNT, mt-hsp70, ADP/ATP transporter, and Cpn10 (2, 7, 8, 21, 46) indicated that E. histolytica is the secondary “amitochondriate.” While only a half-dozen proteins were shown to be localized to mitosomes (1, 2, 7, 8, 19, 21, 42, 44, 46), we recently discovered by proteomic analysis of isolated mitosomes that sulfate activation is the major pathway compartmentalized in mitosomes. Three enzymes consisting of the pathway and additional proteins required for the pathway, including sodium/sulfate symporter, mitochondrion carrier family protein, and chaperons, were identified in the mitosomal proteome. Although PNT was not discovered in the proteome, it was previously postulated to be mitosomal based on the resemblance of the amino-terminal region of EhPNT to the potential mitochondrion-targeting peptide. However, it is important that the putative TS of EhPNT is certainly not a canonical mitochondrion-targeting peptide because it lacks basic residues such as arginine or lysine.
Despite the premise, we showed, in this study, that EhPNT was distributed to the membrane of vesicles and vacuoles, including lysosomes, phagosomes, and endosomes, but not to mitosomes. In addition, we showed that GFP fused with the amino-terminal putative TS of EhPNT was distributed to the cytosol, which disproved the premise that the amino-terminal domain of EhPNT functions as a putative organelle-targeting sequence. The 15-aa-long amino terminus of Cpn60 was not sufficient for the targeting of either luciferase or GFP to mitosomes (1; our unpublished data), although the removal of the first 15 aa of Cpn60 caused a mislocalization of the protein in the cytoplasm (42). Therefore, a role of the amino-terminal transit peptide for mitosomal transport remains obscure.
The association of PNT with the acidified compartments (lysosomes and phagosomes) (Fig. 5) is unprecedented in eukaryotes, where PNT is usually localized to the inner membrane of mitochondria (48). The dependence of transhydrogenase activity of EhPNT on pH was demonstrated; the rate of transhydrogenation was higher at an acidic pH (5.5) than at a neutral pH (7.0 to 8.0) (49). The generation of NADPH by membrane-associated PNT (49) depends upon a proton-motive force, which is likely generated by V-ATPase localized to the acidified compartment and also the nonacidified compartment. While the colocalization of V-ATPase and EhPNT has not been directly demonstrated, phagosomes contained major components of V-ATPase, as shown by the proteomic analysis of purified phagosomes (32, 33). Therefore, it is conceivable that amebic PNT localized in the acidic environment possesses enzymological properties suitable for acidic environments. Since our attempt to repress EhPNT expression by gene silencing failed (our unpublished data), the physiological role of EhPNT has not been elucidated. However, EhPNT may be involved in the detoxification of reactive oxygen and nitrogen species by supplying NADPH as a reducing power by using a proton gradient across the lysosomal and phagosomal membranes. During tissue invasion, E. histolytica adapts to changing oxygen tensions as it goes from the anaerobic colonic lumen to an oxygen-rich environment in the tissue (40). Additionally, the parasite must cope with cytotoxic reactive oxygen and nitrogen species that are produced and released by activated phagocytes that are attracted to the site of infection (4, 20, 40).
Although we cannot exclude the possibility that truncated EhPNT was misfolded and aggregated in the cell, our immunofluorescence and cellular fractionation data are consistent with the premise that all truncated EhPNT was retained in the ER, which is suggestive of the default mechanisms of retention of multiple transmembrane proteins in the ER. The possibility that truncated EhPNT is retained in the heavy microsomal fraction was previously suggested (1), where the majority (93%) of luciferase activity of the recombinant protein consisted of the 67-aa-long amino-terminal portion of PNT fused to firefly luciferase was associated with the mixed membrane fraction and was as susceptible to trypsin degradation as cytosolic luciferase from control parasites. Our present data also support the interpretation of the study that the fusion protein is embedded in the ER membrane and not targeted to mitosomes. We also showed that the aberrant mobility of PNT on SDS-PAGE gels (Fig. 1) was not likely due to either N-linked or O-linked glycosylation but was due to the hydrophobic nature of EhPNT. Altogether, E. histolytica PNT represents a novel class of PNT localized to lysosomes and appears to have evolved uniquely in this organism.
ACKNOWLEDGMENTS
We thank Rosana Sánchez-López, Universidad Nacional Autónoma de México, for anti-EhSec61 α-subunit and anti-EhDPMS antibodies and Barbara J. Mann and William A. Petri, Jr., University of Virginia Health System, for anti-Hgl antibody (3F4). We also thank Takashi Makiuchi for helpful discussions.
This work was supported by a grant-in-aid for creative scientific research (grant 18GS0314) and a grant-in-aid for scientific research (grants 18GS0314, 18050006, and 18073001) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to T.N.; a grant for Research on Emerging and Re-Emerging Infectious Diseases from the Ministry of Health, Labor, and Welfare of Japan (grant H20-Shinkosaiko-Ippan-016); and a grant for research to promote the development of anti-AIDS pharmaceuticals from the Japan Health Sciences Foundation to T.N. (grant KAA1551).
Footnotes
Published ahead of print on 9 April 2010.
REFERENCES
- 1.Aguilera P., Barry T., Tovar J. 2008. Entamoeba histolytica mitosomes: organelles in search of a function. Exp. Parasitol. 118:10–16 [DOI] [PubMed] [Google Scholar]
- 2.Bakatselou C., Kidgell C., Clark C. C. 2000. A mitochondrial-type hsp70 gene of Entamoeba histolytica. Mol. Biochem. Parasitol. 110:177–182 [DOI] [PubMed] [Google Scholar]
- 3.Bizouarn T., Fjellstrom O., Meuller J., Axelsson M., Bergkvist A., Johansson C., Karlsson G., Rydstrom J. 2000. Proton translocating nicotinamide nucleotide transhydrogenase from E. coli. Mechanism of action deduced from its structural and catalytic properties. Biochim. Biophys. Acta 1457:211–228 [DOI] [PubMed] [Google Scholar]
- 4.Bogdan C., Rollinghoff M., Diefenbach A. 2000. Reactive oxygen and reactive nitrogen intermediates in innate and specific immunity. Curr. Opin. Immunol. 12:64–76 [DOI] [PubMed] [Google Scholar]
- 5.Bucci C., Thomsen P., Nicoziani P., McCarthy J., van Deurs B. 2000. Rab7: a key to lysosome biogenesis. Mol. Biol. Cell 11:467–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlton J. M., Angiuoli S. V., Carucci D. J. 2002. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii. Nature 419:512–519 [DOI] [PubMed] [Google Scholar]
- 7.Chan K. W., Slotboom D. J., Cox S., Embley T. M., Fabre O., van der Giezen M., Harding M., Horner D. S., Kunji E. R., Leon-Avila G., Tovar J. 2005. A novel ADP/ATP transporter in the mitosome of the microaerophilic human parasite Entamoeba histolytica. Curr. Biol. 15:737–742 [DOI] [PubMed] [Google Scholar]
- 8.Clark C. G., Roger A. J. 1995. Direct evidence for secondary loss of mitochondria in Entamoeba histolytica. Proc. Natl. Acad. Sci. U. S. A. 92:6518–6521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diamond L. S., Harlow D. R., Cunnick C. C. 1978. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 72:431–432 [DOI] [PubMed] [Google Scholar]
- 10.Gardner M. J., Barrell B. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gill E. E., Diaz-Triviño S., Barberà M. J., Silberman J. D., Stechmann A., Gaston D., Tamas I., Roger A. J. 2007. Novel mitochondrion-related organelles in the anaerobic amoeba Mastigamoeba balamuthi. Mol. Microbiol. 66:1306–1320 [DOI] [PubMed] [Google Scholar]
- 12.Hall N., Karras M., Sinden R. E. 2005. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science 307:82–86 [DOI] [PubMed] [Google Scholar]
- 13.Hiltpold A., Frey M., Hulsmeier A., Kohler P. 2000. Glycosylation and palmitoylation are common modifications of Giardia variant surface proteins. Mol. Biochem. Parasitol. 109:61–65 [DOI] [PubMed] [Google Scholar]
- 14.Hoek J. B., Rydstrom J. 1988. Physiological roles of nicotinamide nucleotide transhydrogenase. Biochem. J. 254:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson J. B. 1991. The proton-translocating nicotinamide adenine dinucleotide transhydrogenase. J. Bioenerg. Biomembr. 23:715–741 [DOI] [PubMed] [Google Scholar]
- 16.Jackson J. B., Peake S. J., White S. A. 1999. Structure and mechanism of proton-translocating transhydrogenase. FEBS Lett. 464:1–8 [DOI] [PubMed] [Google Scholar]
- 17.Kramer R. A., Tomchak L. A., McAndrew S. J., Becker K., Hug D., Pasamontes L., Humbelin M. 1993. An Eimeria tenella gene encoding a protein with homology to the nucleotide transhydrogenases of Escherichia coli and bovine mitochondria. Mol. Biochem. Parasitol. 60:327–331 [DOI] [PubMed] [Google Scholar]
- 18.Lee Y.-N., Chen L.-K., Ma H.-C., Yang H.-H., Li H.-P., Lo S.-Y. 2005. Thermal aggregation of SARS-CoV membrane protein. J. Virol. Methods 129:152–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leon-Avila G., Tovar J. 2004. Mitosomes of Entamoeba histolytica are abundant mitochondrion-related remnant organelles that lack a detectable organeller genome. Microbiology 150:1245–1250 [DOI] [PubMed] [Google Scholar]
- 20.MacMicking J., Xie Q. W., Nathan C. 1997. Nitric oxide and macrophage function. Annu. Rev. Immunol. 15:323–350 [DOI] [PubMed] [Google Scholar]
- 21.Mai Z., Ghosh S., Frisardi M., Rosenthal B., Rogers R., Samuelson J. 1999. Hsp60 is targeted to a cryptic mitochondrion-derived organelle (“crypton”) in the microaerophilic protozoan parasite Entamoeba histolytica. Mol. Cell. Biol. 19:2198–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mann B. J., Torian B. E., Vedvick T. S., Petri W. A., Jr 1991. Sequence of a cysteine-rich galactose-specific lectin of Entamoeba histolytica. Proc. Natl. Acad. Sci. U. S. A. 88:3248–3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mann B. J., Chung C. Y., Dodson J. M., Ashley L. S., Braga L. L., Snodgrass T. L. 1993. Neutralizing monoclonal antibody epitopes of the Entamoeba histolytica galactose adhesin map to the cysteine-rich extracellular domain of the 170-kilodalton subunit. Infect. Immun. 61:1772–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Maralikova B., Ali V., Nakada-Tsukui K., Nozaki T., van der Giezen M., Henze K., Tovar J. 2010. Bacterial-type oxygen detoxification and iron-sulfur cluster assembly in amoebal relict mitochondria. Cell. Microbiol. 12:331–342 [DOI] [PubMed] [Google Scholar]
- 24.McLane M. W., Hatzidimitriou G., Yuan J., McCann U., Ricaurte G. 2007. Heating induces aggregation and decreases detection of serotonin transporter protein on Western blots. Synapse 61:875–876 [DOI] [PubMed] [Google Scholar]
- 25.Meza I. 1992. Entamoeba histolytica: phylogenetic consideration. Arch. Med. Res. 23:1–5 [PubMed] [Google Scholar]
- 26.Mi-ichi F., Yousuf M. A., Nakada-Tsukui K., Nozaki T. 2009. Mitosomes in Entamoeba histolytica contain a sulphate activation pathway. Proc. Natl. Acad. Sci. U. S. A. 106:21731–21736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitra B. N., Saito-Nakano Y., Nakada-Tsukui K., Sato D., Nozaki T. 2007. Rab11B small GTPase regulates secretion of cysteine proteases in the enteric protozoan parasite Entamoeba histolytica. Cell. Microbiol. 9:2112–2125 [DOI] [PubMed] [Google Scholar]
- 28.Nakada-Tsukui K., Saito-Nakano Y., Ali V., Nozaki T. 2005. A retromerlike complex is a novel Rab7 effector that is involved in the transport of the virulence factor cysteine protease in the enteric protozoan parasite Entamoeba histolytica. Mol. Biol. Cell 16:5294–5303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakada-Tsukui K., Okada H., Mitra B. N., Nozaki T. 2009. Phosphatidylinositol-phosphates mediate cytoskeletal reorganization during phagocytosis via a unique modular protein consisting of RhoGEF/DH and FYVE domains in the parasitic protozoon Entamoeba histolytica. Cell. Microbiol. 11:1471–1491 [DOI] [PubMed] [Google Scholar]
- 30.Nozaki T., Asai T., Sanchez L. B., Kobayashi S., Nakazawa M. 1999. Characterization of the gene encoding serine acetyltransferase, a regulated enzyme of cysteine biosynthesis from the protist parasites Entamoeba histolytica and Entamoeba dispar. Regulation and possible function of the cysteine biosynthetic pathway in Entamoeba. J. Biol. Chem. 274:32445–32452 [DOI] [PubMed] [Google Scholar]
- 31.Nozaki T., Asai T., Kobayashi S., Ikegami F., Noji M., Saito K., Takeuchi T. 1998. Molecular cloning and characterization of the genes encoding two isoforms of cysteine synthase in the enteric protozoan parasite Entamoeba histolytica. Mol. Biochem. Parasitol. 97:33–44 [DOI] [PubMed] [Google Scholar]
- 32.Okada M., Huston C. D., Mann B. J., Petri W. A., Jr., Kita K., Nozaki T. 2005. Proteomic analysis of phagocytosis in the enteric protozoan parasite Entamoeba histolytica. Eukaryot. Cell 4:827–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okada M., Huston C. D., Oue M., Mann B. J., Petri W. A., Jr., Kita K., Nozaki T. 2006. Kinetics and strain variation of phagosome proteins of Entamoeba histolytica by proteomic analysis. Mol. Biochem. Parasitol. 145:171–183 [DOI] [PubMed] [Google Scholar]
- 34.Riordan C. E., Ault J. G., Langreth S. G., Keithly J. S. 2003. Cryptosporidium parvum Cpn60 targets a relict organelle. Curr. Genet. 44:138–147 [DOI] [PubMed] [Google Scholar]
- 35.Roger A. J., Svard S. G., Tovar J., Clark C. G., Smith M. W., Gillin F. D., Sogin M. L. 1998. A mitochondrial-like chaperonin 60 gene in Giardia lamblia: evidence that diplomonads once harboured an endosymbiont related to the progenitor of mitochondria. Proc. Natl. Acad. Sci. U. S. A. 95:229–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saito-Nakano Y., Yasuda T., Nakada-Tsukui K., Leippe M., Nozaki T. 2004. Rab5-associated vacuoles play a unique role in phagocytosis of the enteric protozoan parasite Entamoeba histolytica. J. Biol. Chem. 279:49497–49507 [DOI] [PubMed] [Google Scholar]
- 37.Salgado M., Villagómez-Castro J. C., Rocha-Rodríguez R., Sabanero-López M., Ramos M. A., Alagón A., López-Romero E., Sánchez-López R. 2005. Entamoeba histolytica: biochemical and molecular insights into the activities within microsomal fractions. Exp. Parasitol. 110:363–373 [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed.Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 39.Sazanov L. A., Jackson J. B. 1994. Proton-translocating transhydrogenase and NAD- and NADP-linked isocitrate dehydrogenases operate in a substrate cycle which contributes to fine regulation of the tricarboxylic acid cycle activity in mitochondria. FEBS Lett. 344:109–116 [DOI] [PubMed] [Google Scholar]
- 40.Stanley S. L., Jr 2003. Amoebiasis. Lancet 361:1025–1034 [DOI] [PubMed] [Google Scholar]
- 41.Tokoro M., Asai T., Kobayashi S., Takeuchi T., Nozaki T. 2003. Identification and characterization of two isoenzymes of methionine γ-lyase from Entamoeba histolytica: a key enzyme of sulfur-amino acid degradation in an anaerobic parasitic protist that lacks forward and reverse trans-sulfuration pathways. J. Biol. Chem. 278:42717–42727 [DOI] [PubMed] [Google Scholar]
- 42.Tovar J., Fischer A., Clark C. G. 1999. The mitosome, a novel organelle related to mitochondria in the amitochondrial parasite Entamoeba histolytica. Mol. Microbiol. 32:1013–1021 [DOI] [PubMed] [Google Scholar]
- 43.Tovar J., Leon-Avila G., Sanchez L. B., Sutak R., Tachezy J., van der Giezen M. 2003. Mitochondrial remnant organelles of Giardia function in iron-sulphur protein maturation. Nature 426:172–176 [DOI] [PubMed] [Google Scholar]
- 44.Tovar J., Cox S. S. E., van der Giezen M. 2007. A mitosome purification protocol based on Percoll density gradients and its use in validating the mitosomal nature of Entamoeba histolytica mitochondrial Hsp70. Methods Mol. Biol. 390:167–177 [DOI] [PubMed] [Google Scholar]
- 45.van der Giezen M. 2009. Hydrogenosomes and mitosomes: conservation and evolution of functions. J. Eukaryot. Microbiol. 56:221–231 [DOI] [PubMed] [Google Scholar]
- 46.van der Giezen M., Leon-Avila G., Tovar J. 2005. Characterization of chaperonin 10 (Cpn10) from the intestinal human pathogen Entamoeba histolytica. Microbiology 151:3107–3115 [DOI] [PubMed] [Google Scholar]
- 47.Vermeulen A. N., Kok J. J., Van Den Boogart P., Dijkema R., Claessens J. A. J. 1993. Eimeria refractile body proteins contain two potentially functional characteristics: transhydrogenase and carbohydrate transport. FEMS Microbiol. Lett. 110:223–229 [DOI] [PubMed] [Google Scholar]
- 48.Weston C. J., Venning J. D., Jackson J. B. 2002. The membrane-peripheral subunits of transhydrogenase from Entamoeba histolytica are functional only when dimerized. J. Biol. Chem. 277:26163–26170 [DOI] [PubMed] [Google Scholar]
- 49.Weston C. J., White S. A., Jackson J. B. 2001. The unusual transhydrogenase of Entamoeba histolytica. FEBS Lett. 488:51–54 [DOI] [PubMed] [Google Scholar]
- 50.Williams B. A., Hirt R. P., Lucocq J. M., Embley T. M. 2002. A mitochondrial remnant in the microsporidian Trachipleistophora hominis. Nature 418:865–869 [DOI] [PubMed] [Google Scholar]
- 51.Yu Y., Samuelson J. 1994. Primary structure of an Entamoeba histolytica nicotinamide nucleotide transhydrogenase. Mol. Biochem. Parasitol. 68:323–328 [DOI] [PubMed] [Google Scholar]