Abstract
Arginine methylation is a widespread posttranslational modification of proteins catalyzed by a family of protein arginine methyltransferases (PRMTs). In Saccharomyces cerevisiae and mammals, this modification affects multiple cellular processes, such as chromatin remodeling leading to transcriptional regulation, RNA processing, DNA repair, and cell signaling. The protozoan parasite Trypanosoma brucei possesses five putative PRMTs in its genome. This is a large number of PRMTs relative to other unicellular eukaryotes, suggesting an important role for arginine methylation in trypanosomes. Here, we present the in vitro and in vivo characterization of a T. brucei enzyme homologous to human PRMT6, which we term TbPRMT6. Like human PRMT6, TbPRMT6 is a type I PRMT, catalyzing the production of monomethylarginine and asymmetric dimethylarginine residues. In in vitro methylation assays, TbPRMT6 utilizes bovine histones as a substrate, but it does not methylate several T. brucei glycine/arginine-rich proteins. As such, it exhibits a relatively narrow substrate specificity compared to other T. brucei PRMTs. Knockdown of TbPRMT6 in both procyclic form and bloodstream form T. brucei leads to a modest but reproducible effect on parasite growth in culture. Moreover, upon TbPRMT6 depletion, both PF and BF exhibit aberrant morphologies indicating defects in cell division, and these defects differ in the two life cycle stages. Mass spectrometry of TbPRMT6-associated proteins reveals histones, components of the nuclear pore complex, and flagellar proteins that may represent TbPRMT6 substrates contributing to the observed growth and morphological defects.
Posttranslational methylation of proteins on arginine residues has multiple roles in a wide array of cellular functions, such as chromatin remodeling leading to transcription activation or repression, RNA processing, DNA repair, and various forms of cell signaling (5, 6, 8, 9, 52, 70, 98). The process of arginine methylation involves the transfer of methyl groups from S-adenosyl-methionine (AdoMet) to arginine residues of proteins and is catalyzed by a group of enzymes known as protein arginine methyltransferases (PRMTs). PRMTs themselves are further divided into four classes, depending on the type of methylated arginine generated. The largest PRMT class comprises the type I enzymes, as characterized by the first discovered PRMT, PRMT1. Type I PRMTs initially catalyze the formation of monomethylated arginine (MMA) on the terminal ω-nitrogen, followed by addition of a second methyl group on the same ω-nitrogen, which yields asymmetric dimethylarginine (ADMA). The type II PRMTs are a smaller group, presently consisting only of PRMT5 and its homologues. These enzymes also catalyze the synthesis of MMA on the terminal ω-nitrogen, but in contrast to the type I enzymes, type II PRMTs add a second methyl group to the adjacent terminal ω-nitrogen, resulting in symmetric dimethylarginine (SDMA). Almost all known eukaryotic cells possess at least one type I PRMT and one type II PRMT in the form of PRMT1 and PRMT5 homologues, respectively (3). Less is understood about the type III and type IV PRMTs. Type III PRMTs catalyze the production of only MMA. Thus, far, the Trypanosoma brucei homologue of human PRMT7, TbPRMT7, is the only enzyme thought to be exclusively type III, and the specificity of the mammalian homologue PRMT7 is controversial (27, 54, 64). Finally, the type IV PRMTs catalyze MMA on the δ-nitrogen of arginine but to date have been described to occur only in fungi (63, 69).
PRMT substrates are varied and include chromatin-associated proteins, signaling proteins, and a large number of RNA binding proteins (RBPs) (5). RBPs are usually methylated within glycine/arginine-rich (GAR) regions (68), often within canonical RGG motifs. However, methylation of arginine residues in noncanonical regions is becoming more apparent, suggesting a more complex specificity than initially thought (6, 97). Thus, a large number of PRMT substrates cannot be identified based on their sequences and so must be empirically defined.
The homologues of PRMT6 in humans and other higher eukaryotes comprise a family of type I PRMTs involved in transcription and DNA repair (28, 53). PRMT6 exhibits a relatively narrow substrate specificity, with the currently known substrates being HMG1A (66, 87, 106), histone subunits (32, 37, 38), DNA polymerase beta (20), and several components of the HIV virus (10, 39, 40) as well as PRMT6 itself (28). The human enzyme is reported to display an exclusively nuclear localization pattern (28), consistent with its known roles in nuclear processes. Detailed in vitro studies showed that human PRMT6 catalyzes methyl transfers in a distributive manner, depositing the first methyl group and creating MMA, dissociating from the substrate, and then rebinding to the methyl mark and forming ADMA (53). Homologues of PRMT6 are apparently absent from the genomes of most single-celled eukaryotes, with the exception of Trypanosoma brucei and, possibly, Dictyostelium (3).
The kinetoplastid protozoan T. brucei is the causative agent of African sleeping sickness. Kinetoplastid parasites, including T. brucei, Trypanosoma cruzi, and Leishmania spp., exhibit several unique features, one of the most striking of which is the absence of gene regulation at the level of transcription (13, 14). Instead, these parasites regulate several posttranscriptional processes, including RNA stability, translation, and RNA editing, to control gene expression. This unusual mode of gene regulation necessitates the involvement of a large number of RBPs, a few of which have been identified (26, 35, 48, 61, 83, 91, 92). Correspondingly, the T. brucei genome encodes a large number of RBPs. Because many of these RBPs contain GAR motifs, they are in turn proposed targets of regulation by arginine methylation (17; L. K. Read, unpublished results).
Previously, we identified five putative PRMTs in the T. brucei genome, which is, to our knowledge, the highest number in a single-celled eukaryote (3, 73). In this study, we present an in vitro and in vivo characterization of the T. brucei homologue of the human PRMT6 protein, which we term TbPRMT6. TbPRMT6 is a type I PRMT with a relatively narrow substrate specificity compared to those of other T. brucei PRMTs. Knockdown of TbPRMT6 in both procyclic form (PF) and bloodstream form (BF) T. brucei leads to a modest but reproducible effect on parasite growth in culture as well as differential defects in cell division. Mass spectrometry of TbPRMT6-associated proteins reveals several potential substrates that may contribute to these growth and morphological defects.
MATERIALS AND METHODS
Cloning and expression of TbPRMT6.
The gene carrying the TbPRMT6 open reading frame (Tb927.5.3960) was PCR amplified from oligo(dT)-primed cDNA extracted from procyclic form (PF) T. brucei (strain 927 Eatro 1.1) RNA using the primers PRMT6-5′-BamHI (5′-GAGGATCCATGGAGTCCGGAGGGTTTG-3′) and PRMT6-3′-HindIII (5′-GGAAGCTTTTTTAACTCGAGCTCAATG-3′) (the restriction enzyme-cut sites are underlined). The resultant product was cloned into pJET (CloneJet cloning kit; Fermentas). TbPRMT6 was excised from pJET-TbPRMT6 and ligated into the BamHI and HindIII sites of both the pET21a and the pET42a vectors (Novagen). The resultant plasmids were then transformed into Rosetta strain Escherichia coli cells (Novagen) for expression. Glutathione S-transferase (GST)-tagged TbPRMT6 (from pET42a-TbPRMT6) was purified using single-step glutathione-agarose (Invitrogen) and a standard GST purification protocol. TbPRMT6 with a C-terminal six-histidine tag was produced from pET21a-TbPRMT6 and was purified by a standard purification protocol using TALON resin (Clontech).
Antibodies.
Polyclonal antibodies against TbPRMT6 and TbPRMT7 were raised by injecting rabbits with full-length GST-TbPRMT6 or GST-TbPRMT7 (27) (Proteintech, Inc.). Affinity-purified polyclonal antipeptide antibodies against TbPRMT1 were purchased from Bethyl Laboratories. Monoclonal antibodies against protein C were purchased from Sigma. Antibodies against Hsp70 and the C-terminal domain (CTD) of RNA polymerase II were generously provided by James Bangs (University of Wisconsin) and Vivian Bellafatto (University of Medicine and Dentistry of New Jersey), respectively. Antibodies against MRP2 were previously described (1).
Cell culture and RNAi.
PF T. brucei strain 29-13 (from George A. M. Cross, Rockefeller University), which contains integrated genes for the T7 RNA polymerase and the tetracycline repressor, were grown in SDM-79 medium supplemented with 15% fetal bovine serum (FBS), as indicated previously (75), unless otherwise noted. Bloodstream form (BF) single-marker T. brucei cells (also provided by George A. M. Cross) were cultured in HMI-9 medium supplemented with 10% FBS and 10% Serum Plus (SAFC) (75). For creation of cells expressing double-stranded RNA (dsRNA) targeting TbPRMT6 for RNA interference (RNAi), the full-length TbPRMT6 open reading frame was excised from pET42a-TbPRMT6 using BamHI and HindIII and ligated into the BamHI-HindIII sites of the tetracycline-inducible RNAi vector p2T7-177 (95), creating p2T7-177-TbPRMT6. NotI-linearized p2T7-177-TbPRMT6 was transfected into PF and BF cells, and cells harboring this construct were selected with 2.5 μg/ml phleomycin. Clones were obtained by serial dilution and grown over the indicated time periods in the absence or presence of 2.5 μg/ml tetracycline. For exogenous expression of TbPRMT6, we engineered the vector pLEW79-MH-TAP to express TbPRMT6 with a C-terminal myc–six-histidine TAP tag (43). Full-length TbPRMT6 was amplified using the primers PRMT6 5′ HindIII (5′-GGAAGCTTATGGAGTCCGGAGGGTTTG-3′) and TbPRMT6 3′ BamHI (5′-GAGGATCCTTTTAACTCGAGCTCAATG-3′) (restriction sites are underlined), cloned into the cloning vector pJET, and finally cloned into the HindIII-BamHI sites of pLEW79-MH-TAP. The resultant vector, pLEW79-TbPRMT6-MH-TAP, was transfected into 29-13 cells, and clones were selected by limiting dilution. Expression of tetracycline-inducible TbPRMT6-MH-TAP was verified by Western blotting against both TbPRMT6 and the myc tag, which confirmed the expression of the native 41-kDa TbPRMT6 protein as well as the approximately 69-kDa tagged TbPRMT6-MH-TAP protein.
In vitro methylation assays.
S-Adenosyl-[methyl-3H]methionine ([3H]AdoMet) (65 to 82 Ci/mmol) was purchased from Amersham or Perkin Elmer. Methylation assays were performed essentially as described in references 73 and 74, with the following modifications. Purified TbPRMT6-His (3 μg) was incubated in 50 μl of phosphate-buffered saline (PBS) with specified substrate (typically 3 μg; 10 μg for histones) and 2 μCi [3H]AdoMet for 14 h at room temperature. The substrates used in this study are as follows: the synthetic peptide H-CGRGRGRGRGRGRGRG-NH2 (12), myelin basic protein (MBP) (Sigma), bovine mixed histones (IIAS; Sigma), His-RBP16 (35), GST-TbRGG1 (74), and GST-TbRGG2 (26).
High-resolution amino acid analysis of acid hydrolysates of TbPRMT6 products.
Reactions were carried out overnight as indicated above using either 3 μg of GST-TbPRMT6 alone or GST-TbPRMT6 with 10 μg bovine histones. The reactions were precipitated with 50% trichloroacetic acid (TCA) and washed with ice-cold 100% acetone, and the resultant TCA pellet was acid hydrolyzed to component amino acids under a vacuum for 20 h at 110°C. The samples were dried and resuspended in 50 μl of water, followed by addition of 500 μl of sodium citrate buffer (pH 2.2) and 1.0 μmol each of unlabeled standards of ω-NG-monomethylarginine (acetate salt) (MMA), ω-NG, NG-dimethylarginine (hydrochloride) (ADMA), and ω-NG, NG′-dimethylarginine (di(p-hydroxyazobenzene-p′-sulfonate salt) (SDMA) (all purchased from Sigma) in a total of 1 ml. The reaction was then analyzed by cation exchange chromatography as detailed previously (82).
Subcellular fractionation of TbPRMT6-PTP cells.
PF 29-13 cells were fractionated into cytoplasmic and nuclear fractions using methods described previously (102). Mitochondria were purified by the method of Harris et al. (33). To establish the efficiency of fractionation, Western blot analyses were performed with antibodies specific for cytoplasmic Hsp70, nuclear RNA polymerase II CTD, and mitochondrial TbRGG2 (26).
Microscopy of TbPRMT6 RNAi cells.
TbPRMT6 RNAi cells (1 × 106) in PBS were spread on poly-l-lysine-coated slides (Electron Microscope Sciences) and adhered at room temperature for 20 min. Cells were then fixed in 4% paraformaldehyde for 15 min. Fixation was stopped by washing the slides twice for 5 min each in PBS with 0.1 M glycine (pH 8.5). Cells were permeabilized for 5 min by addition of PBS with 0.025% (wt/vol) Triton X-100 and then washed with PBS for 5 min. Slides were then immersed in methanol at −20°C overnight. Cells were then rehydrated by three washes of PBST (PBS with 0.1% Tween 20) for 5 min each, followed by staining with 0.1 μg/ml DAPI (4′,6-diamidino-2-phenylindole) in PBS for 5 min, and two final washes with PBS. Slides were mounted using a coverslip and Vectashield (Vector Laboratories) as an antifade. Cells were visualized using a Zeiss Axio Observer inverted microscope and Zeiss Axiovision software.
RESULTS
Identification and sequence analysis of TbPRMT6.
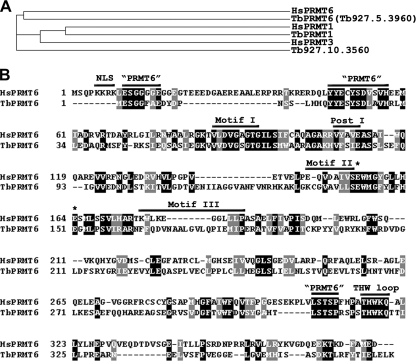
Of the five putative PRMTs in the annotated T. brucei genome database, three have previously been characterized in our laboratory. This includes canonical homologues of the human type I PRMT1 and type II PRMT5 proteins (73, 74) as well as a unique type III enzyme known as TbPRMT7 (27). A fourth enzyme (Tb927.10.3560), as yet uncharacterized, appears to be the homologue of human PRMT3. The fifth putative PRMT (Tb927.5.3960) shares overall amino acid sequence identities with human PRMT1 but appears to be most similar to human PRMT6 (HsPRMT6) when sequences outside the catalytic core are analyzed. To determine the relationship of Tb927.5.3960 to human PRMTs, we used CLUSTALW (http://www.ebi.ac.uk/Tools/clustalw2/index.html) to align the amino acid sequences of three human type I PRMTs, HsPRMT1, HsPRMT3, and HsPRMT6, with putative type I PRMTs from T. brucei. A cladogram of this alignment (Fig. 1 A) demonstrates that Tb927.5.3960 is most similar to HsPRMT6. On the basis of this alignment and analyses described below, we coined this protein TbPRMT6. All active PRMTs share several common motifs required for binding to AdoMet and substrate proteins and catalysis of the methyl transfer (3, 6, 49). In Fig. 1B, we align the amino acid sequences of HsPRMT6 and TbPRMT6, demonstrating that TbPRMT6 possesses all of these motifs (motifs I, post I, II, and III and the double E and THW loops) (Fig. 1), although motif III of TbPRMT6 diverges from the canonical motif. Across the regions of motif 1 through the THW loop, the human and T. brucei enzymes share 31% identity and 45.7% similarity. While the proteins are generally less conserved outside this region, we identified several shared motifs between TbPRMT6 and HsPRMT6 that are absent in other type I PRMTs (Fig. 1B, “PRMT 6”) and which presumably contribute to the homology of the two proteins as shown in Fig. 1A. TbPRMT6 appears to lack the N-terminal nuclear localization signal (NLS) present in HsPRMT6. Whereas human PRMT6 is exclusively nuclear (28), the absence of an evident NLS in TbPRMT6 suggested that its localization may differ from that of its human counterpart (see below). The homologous proteins in the related kinetoplastid parasites T. cruzi (two distinct contigs, Tc00.1047053507057.30 and Tc00.1047053506947.80) and Leishmania major (LmjF16.0030), which are 57.1% and 47.3% identical to TbPRMT6 at the amino acid level, respectively, also lack an evident N-terminal NLS. On the basis of the primary sequence analysis, the TbPRMT6 gene appears to encode an active PRMT enzyme.
Fig. 1.
Sequence analysis of TbPRMT6. (A) CLUSTALW-generated cladogram of the human PRMT1, PRMT3, and PRMT6 proteins with the trypanosomes TbPRMT1, TbPRMT6 (Tb927.5.3960), and Tb927.10.3560. (B) Sequence comparison of human PRMT6 (HsPRMT6) and TbPRMT6. Horizontal lines indicate conserved PRMT domains required for AdoMet binding and enzymatic activity (see text) as well as the nuclear localization signal (NLS) present only in the human enzyme. Regions typically found in mammalian PRMT6 enzymes are also labeled as “PRMT6” regions. The asterisks indicate the position of the conserved “double E” loop amino acids. A black background with white text indicates identical amino acids; a gray background with white text indicates conserved amino acids.
Characterization of TbPRMT6 activity.
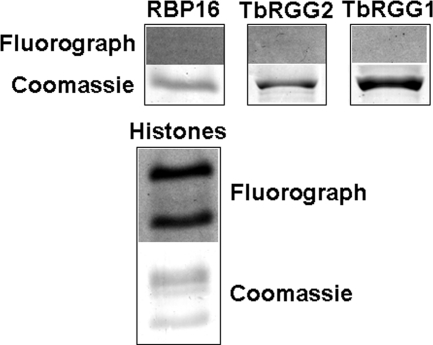
To determine whether TbPRMT6 possesses PRMT activity, we conducted in vitro methyltransferase assays using methyl-[H3]-labeled AdoMet, recombinant six-histidine-tagged TbPRMT6, and several potential substrate proteins. Because HsPRMT6 methylates the tail of histone H3 (37, 38), we tested a mixture of bovine histones as an in vitro substrate for TbPRMT6. Additionally, we tested three T. brucei proteins involved in RNA metabolism, RBP16 (75), TbRGG1 (34), and TbRGG2 (26). All three of these proteins have glycine/arginine-rich (GAR) regions that are common sites of protein arginine methylation, and all three are in vitro substrates for other trypanosome PRMTs (27, 73, 74). Surprisingly, as shown in Fig. 2, we observed no methylation of RBP16, TbRGG1, or TbRGG2 by TbPRMT6, even after a 1-month exposure to film. Other common PRMT substrates, such as an RG-containing peptide and myelin basic protein, were also not methylated by TbPRMT6 in these assays (data not shown). In contrast, TbPRMT6 did demonstrate PRMT activity toward proteins in the bovine mixed-histone preparation, with the specific substrates appearing to be histone H3 and H4 according to their migration on SDS-PAGE gel (Fig. 2). These data demonstrate that TbPRMT6 is an active PRMT with a narrow substrate range, similar to HsPRMT6. TbPRMT6 differs from the other trypanosome PRMTs characterized to date in this regard, since those enzymes all methylate a wide range of substrates (27, 73, 74).
Fig. 2.
Enzymatic activity of recombinant TbPRMT6. Three micrograms of TbPRMT6-His was incubated in the presence of [3H]AdoMet and the indicated substrate in PBS for 14 h at 22°C. Products were resolved by SDS-PAGE and Coomassie blue stained (bottom panels). Gels were then treated with EnH3ance, dried, and exposed to film for 1 month (top panels) (Fluorograph).
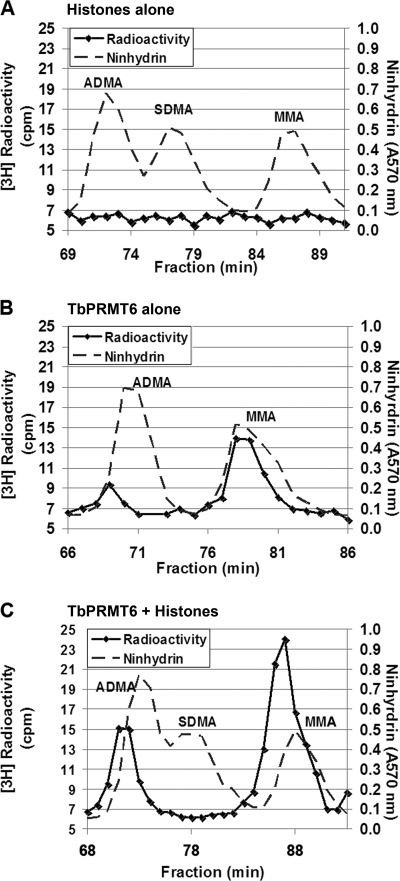
HsPRMT6 is a type I PRMT that synthesizes MMA followed by ADMA in a distributive enzymatic manner (28, 53). To determine whether TbPRMT6 similarly exhibits type I methyl transfer, we performed in vitro methyltransferase reactions in the presence or absence of bovine histones under the same conditions as those shown in Fig. 2. Following the reactions, proteins were TCA precipitated and acid hydrolyzed to single amino acids. The products were separated by high-resolution chromatography and compared to known standards for MMA, ADMA, and SDMA. As shown in Fig. 3 C, TbPRMT6 catalyzed the formation of both MMA and ADMA reactions on the histone substrates, typical of a type I PRMT. There was no evidence of SDMA production by TbPRMT6. It is important to note that, in these experiments, the peaks of radioactivity eluted approximately 1 min earlier than the MMA and ADMA standards detected by ninhydrin analysis. Because of their mass and pKa differences, tritiated amines and amino acids elute slightly earlier on high-resolution chromatography than their hydrogen counterparts (30, 44, 46, 99). The elution of radioactivity here agrees with that observed previously for tritiated MMA and ADMA (55, 58, 65). Thus, we conclude that MMA and ADMA are the major methyl species detected. The absence of any methylated amino acids in a reaction without the enzyme (Fig. 3A) demonstrates that TbPRMT6 catalyzes the production of these methylarginine derivatives on bovine histones. Notably, when TbPRMT6 was analyzed in the absence of substrate, we also observed MMA and ADMA production (Fig. 3B). This indicates that, like human PRMT6 (28), TbPRMT6 is also able to catalyze automethylation. In summary, these data demonstrate that TbPRMT6 is a type I PRMT.
Fig. 3.
Amino acid analysis of methylarginine derivatives formed by TbPRMT6. (A) Ten micrograms of the bovine histone substrate alone was incubated in the presence of [3H]AdoMet in PBS for 14 h at 22°C. The proteins were precipitated with 50% trichloroacetic acid and digested into amino acids by acid hydrolysis. Amino acids were analyzed by cation exchange chromatography in the presence of unlabeled ADMA, SDMA, and MMA standards (dotted lines). Two hundred microliters of each fraction (one-fifth of the total fraction) was removed for radioactivity analysis, 100 μl was removed for ninhydrin analysis, and the fractions were counted three times for 3 min each (solid lines). (B) Three micrograms of TbPRMT6-His in the absence of additional substrate was incubated and analyzed as described for panel A. (C) Ten micrograms of bovine histones was incubated with three micrograms of TbPRMT6-His and incubated and analyzed as described for panel A.
TbPRMT6 subcellular localization and expression through the life cycle.
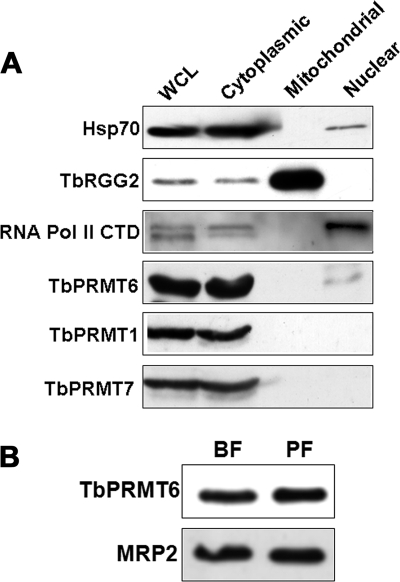
Having shown that TbPRMT6 is an active PRMT, we next wanted to characterize the in vivo properties of the enzyme. In humans, HsPRMT6 is the only PRMT that displays an exclusively nuclear localization. Because the HsPRMT6 NLS is not conserved in TbPRMT6 (Fig. 1), it was of interest to determine the subcellular localization of the trypanosome enzyme. To this end, we analyzed equal protein amounts (20 μg) of nuclear, cytoplasmic, and mitochondrially enriched fractions from PF T. brucei by immunoblotting with anti-TbPRMT6 antibodies. Antibodies against Hsp70, TbRGG2, and the RNA polymerase II CTD were used as markers for cytoplasm, mitochondria, and nuclei, respectively (26, 31, 45). As seen in Fig. 4 A, TbPRMT6 appears almost exclusively cytoplasmic. No signal was detected in mitochondrially enriched fractions. We did, however, observe a faint signal for TbPRMT6 in nuclear extract. The TbPRMT6 signal in the nuclear fraction may result from a small amount of cytoplasmic contamination, as suggested by the faint nuclear signal observed for the cytoplasmic Hsp70. However, when we analyzed the localization of TbPRMT1 and TbPRMT7 in the same fractions, these enzymes were clearly absent from the nucleus and appeared entirely cytoplasmic, as shown previously (27; M. Pelletier, submitted for publication). Thus, TbPRMT6 is primarily cytoplasmic, although we cannot rule out that a small amount of enzyme is also present in the nucleus. The predominant cytoplasmic localization of TbPRMT6 is in direct contrast to the exclusively nuclear localization of its human counterpart.
Fig. 4.
Subcellular localization and life cycle-dependent expression of TbPRMT6. (A) PF cells were separated into cytoplasmic and nuclear fractions. Mitochondrial extracts were also prepared in a separate fractionation scheme. Equal amounts of protein (20 μg) from whole-cell lysates (WCL), cytoplasm, mitochondria, and nuclei were fractionated on SDS-PAGE gel. Antibodies against Hsp70 (cytoplasmic), TbRGG2 (mitochondrial), and the RNA polymerase II CTD (nuclear) were used to monitor fractionation efficiency. TbPRMT1 and TbPRMT7 are two cytoplasmic PRMTs that were also used as controls. (B) Determination of TbPRMT6 levels in the procyclic form (PF) and bloodstream form (BF) life cycle stages of T. brucei. Total cell extract (equivalent of 5 × 106 cells) from 29-13 PF cells and single-marker BF cells were resolved by SDS-PAGE and immunoblotted using an antibody against TbPRMT6. As a loading control, levels of MRP2 (a protein known to be equivalent between PF and BF life cycles [92]) were also analyzed by Western blotting.
We next asked whether TbPRMT6 is expressed in both insect and human infective stages of the T. brucei life cycle. Equal protein amounts of PF and BF whole-cell extracts were analyzed by immunoblotting with anti-TbPRMT6 antibodies. Loading was normalized using antibodies against MRP2, a protein that is equally expressed in both life cycle stages (92). Figure 4B shows that TbPRMT6 is expressed at essentially equal levels in the PF and BF life cycle stages, suggesting that there is no life cycle-dependent regulation of TbPRMT6 levels.
Effects of RNAi-mediated knockdown of TbPRMT6.
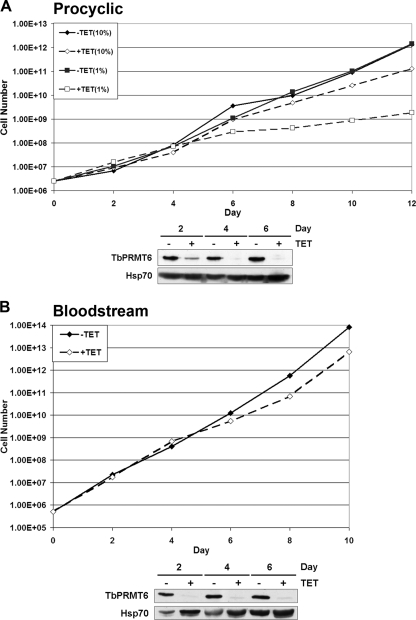
Previous studies from our laboratory demonstrated that targeted depletion of TbPRMT1, TbPRMT5, and TbPRMT7 individually has no effect on the growth of PF cells in vitro (27, 73, 74). Whether this is due to redundancy between PRMTs or the fact that RNAi does not completely eliminate the protein of interest is currently unknown. Here, we analyzed the effect of RNAi-mediated depletion of TbPRMT6 on the growth of PF and BF T. brucei. We utilized the p2T7-177 RNAi vector (96) to generate clonal PF and BF lines expressing TbPRMT6 RNAi. We then induced TbPRMT6 RNAi using tetracycline and monitored cell growth for 12 days (PF) (Fig. 5 A) or 10 days (BF) (Fig. 5B). Depletion of TbPRMT6 protein was confirmed on several days during the time course by immunoblotting with anti-TbPRMT6 antibodies (Fig. 5A and B). In both PF (Fig. 5A) and BF (Fig. 5B) cells, we observed a modest, but highly reproducible, slow-growth phenotype beginning on day 6 upon induction of TbPRMT6 RNAi under normal culture conditions. This growth phenotype was also evident in multiple analyzed clones of both PF and BF lineages (data not shown). To further investigate the effect of TbPRMT6 downregulation on T. brucei growth, we cultured PF cells under nutrient deprivation (1% FBS) and induced the RNAi under these “stressed” conditions (Fig. 5A). Uninduced cells grew at similar rates in 10% and 1% sera. However, when TbPRMT6 RNAi was induced, cells grown in 1% FBS displayed a significantly lower growth rate than did uninduced cells or induced cells in 10% FBS (Fig. 5A). Thus, TbPRMT6 is essential for optimal growth of both PF and BF T. brucei. These data suggest that TbPRMT6 has a role in cellular growth that is not shared by the other trypanosome PRMTs investigated to date.
Fig. 5.
Effect of TbPRMT6 depletion on procyclic form (PF) and bloodstream form (BF) T. brucei cell growth. (A) PF 29-13 cells harboring the tetracycline (TET)-regulatable RNAi vector p2T7-177-TbPRMT6 were treated in the absence of tetracycline (solid lines) or in the presence of 5 μg/ml tetracycline (dotted lines) and counted every 2 days. RNAi induction and cell growth analyses were performed with both 10% fetal bovine serum (FBS; diamonds) and 1% FBS (squares). The lower panel shows Western blotting of TbPRMT6 on days 2, 4, and 6 following tetracycline induction of RNAi. Hsp70 was used as a loading control. (B) BF single-marker cells transfected with the RNAi vector p2T7-177-TbPRMT6 were uninduced (solid line) or induced as described for panel A (dotted line). The lower panel shows the Western blotting of TbPRMT6 and Hsp70 on days 2, 4, and 6 following tetracycline induction.
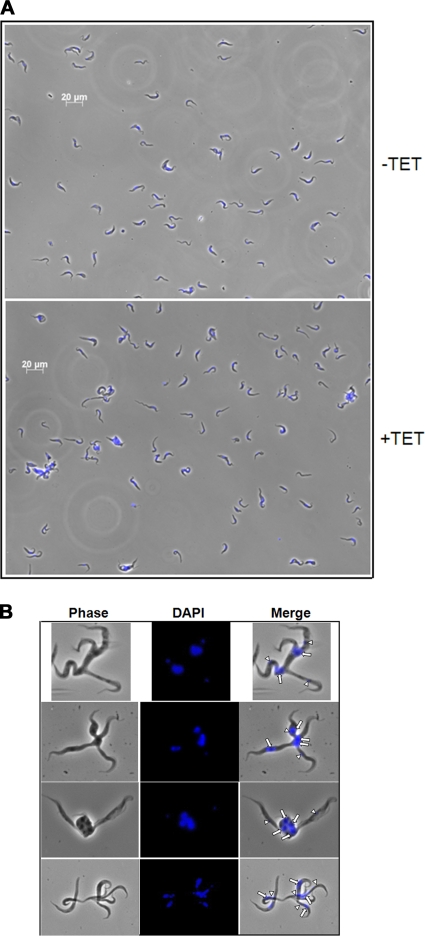
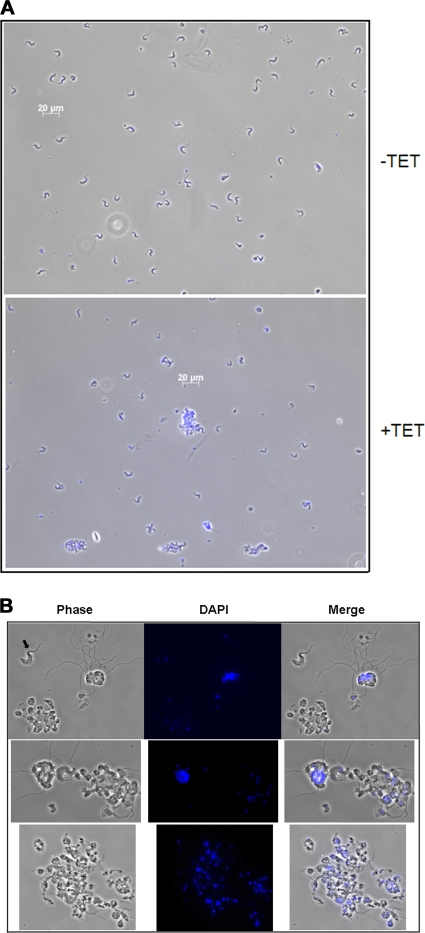
In addition to the evident growth effect following loss of TbPRMT6, we also observed some striking morphological effects in the induced PF TbPRMT6 RNAi cells, even when the cells were grown under normal conditions (10% FBS) (Fig. 6 A). Starting on about day 4 following induction of RNAi and carrying into days 6 and 8, cells began displaying an unusual morphology, which we termed “hydra.” These cells display a single body with multiple half-formed cell “heads” that protrude from the central, often nucleus-containing body. The heads of the hydra occasionally contained DAPI-staining kinetoplast DNA or nuclear DNA but not normal numbers of both, and they were frequently devoid of DNA (Fig. 6B, white arrows [indicating nuclei] and white arrowheads [indicating kinetoplasts]). PF cells with the hydra morphology were most abundant on day 4, where they comprised almost 5% of the induced cells counted, but they also persisted on days 6 and 8 following tetracycline induction. We next analyzed the morphology of BF TbPRMT6 RNAi cells on day 6 following tetracycline addition (Fig. 7 A). While some hydra-like cells were observed in TbPRMT6-depleted BF cells, these cells generally exhibited a different morphology that was characterized by giant rounded cells containing large numbers of both nuclei and kinetoplasts (Fig. 7B). Apparent detachment of the flagella was also evident in several of these giant cells. Giant cells comprised approximately 10% of the BF cells fixed to the microscopy slide. In contrast, no PF or BF cells with these distinct morphologies were ever observed in the uninduced TbPRMT6 RNAi cell cultures. In an attempt to further define a defect in cell division, we counted nuclei and kinetoplasts in at least 200 PF cells for days 4 and 6 or at least 300 BF cells for day 6, (i.e., the approximate time frame at which the growth effect reproducibly manifested) following induction in uninduced and induced cultures. However, we did not detect any significant deviance from the normal 1N1K-to-2N2K distribution of nuclei and kinetoplasts in the cells exhibiting normal morphologies (data not shown). Together, these results suggest that the PF hydra morphology and the BF giant cells are the result of a defect in cell division in a population of cells depleted of TbPRMT6 and that this in turn contributes to the slowing of growth observed on day 6 of the RNAi induction.
Fig. 6.
Microscopy of procyclic form cells exhibiting aberrant morphologies upon TbPRMT6 depletion. (A) Morphological changes evident in PF TbPRMT6 RNAi cells. Cells grown in the presence (+TET) or absence (−TET) of tetracycline for 6 days were fixed to slides, stained with DAPI, and analyzed by microscopy. (B) Images of PF “hydra” cells, characterized by a central body containing nuclei (white arrows) with multiple heads with or without kinetoplasts (white arrowheads).
Fig. 7.
Microscopy of bloodstream form cells exhibiting aberrant morphologies upon TbPRMT6 depletion. (A) Morphological changes evident in BF TbPRMT6 RNAi cells. Cells grown in the presence (+TET) or absence (−TET) of tetracycline for 6 days were fixed to slides, stained with DAPI, and analyzed by microscopy. (B) Images of the giant cells with detached flagella evident in BF TbPRMT6 RNAi cells. Note the BF cell with normal morphology (black arrow) for scale reference.
Identification of TbPRMT6-associated proteins.
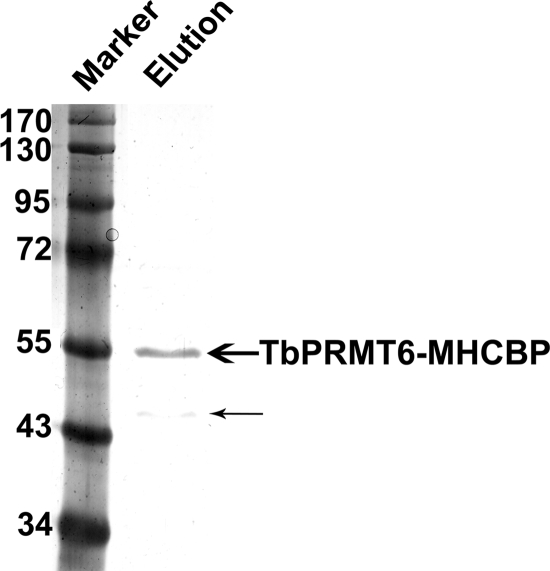
Because the associations between PRMTs and their substrates are often relatively stable, some PRMT substrates, as well as upstream activators, have been identified using various protein-protein interaction strategies (29, 89, 90). To begin to understand the molecular basis of the growth and morphological defects in TbPRMT6 RNAi cells, we identified TbPRMT6-interacting proteins using affinity purification followed by mass spectrometry. To this end, we generated a PF T. brucei cell line expressing TbPRMT6 with a C-terminal Myc-His-TAP tag (TbPRMT6-MHT) in a tetracycline-inducible manner, a strategy that often yields relevant in vivo complexes (34, 72, 84). We purified TbPRMT6-MHT over sequential IgG and calmodulin columns, analyzed the resultant eluted proteins by silver staining (Fig. 8), and subjected the proteins to liquid chromatography-tandem mass spectrometry (LC–MS-MS) analysis. By silver staining, the primary protein in the eluate was the expected size of TbPRMT6-MHCBP (TbPRMT6 with the Myc-His-calmodulin binding protein tag), and only one other substoichiometric protein at approximately 45 kDa was apparent (Fig. 8). However, with the use of mass spectrometry, 43 proteins, in addition to TbPRMT6, were identified by two or more peptides. Of these, almost half were presumed to be common contaminants present in TAP-based purifications, based on mass spectrometry studies from other laboratories (85, 101, 104, 105) and our previous experience (see Table S1 in the supplemental material). It was striking that the remaining TbPRMT6-associated proteins could be readily classified into three major groups (nucleocytoplasmic transport proteins, histones, and flagellar proteins) (Table 1). Of the seven proteins involved in nucleocytoplasmic transport, six were identified in a recent proteomic study of nuclear pore complexes (NPCs) (18). TbPRMT6-associated NPC proteins were diverse, including members of the B-propeller, a-solenoid, B-sandwich, and FG-repeat classes. Together, these proteins are predicted either to be present on the outer rings of the NPCs on both the nuclear and cytoplasmic faces (TbSec13, TbNup82, TbNup96, and TbNup158) or to line the NPC channel (TbNup98 and TbNup149) (18). The seventh protein listed in Table 1 under nucleocytoplasmic transport, NUP-1, is thought to be a major component of the trypanosome nuclear lamina (81). The second major group of proteins that copurified with TbPRMT6 were histones (H2A, H2B, an H2B variant, H3, an H3 variant, and two alleles of H4) (Table 1). This finding, together with the ability of TbPRMT6 to methylate bovine histones and the established function of PRMT6-catalyzed histone methylation in humans (32, 37, 38), suggests that trypanosome histones may be bona fide substrates of TbPRMT6. The third class of TbPRMT-interacting proteins comprised proteins involved in flagellar function and structure (Table 1). Interestingly, several flagellar proteins have been reported to play a role in cell division in T. brucei (11, 50, 78, 79), including the TbPRMT6-associated KMP-11 protein, depletion of which was shown to cause inappropriate nuclear division (57). Finally, TbPRMT6 also copurified with two proteins reported or suggested to be involved in RNA metabolism (the RNA binding protein TbRBD3 [21] and the putative RNA helicase TbDed1, which also associated with TbPRMT5 [73]). The low stoichiometric levels of these proteins relative to the level for TbPRMT6 itself (Fig. 8) are consistent with the idea that these proteins act as substrates for TbPRMT6 rather than as stable components of a TbPRMT6-containing complex. Overall, these data suggest multiple mechanisms by which TbPRMT6-catalyzed arginine methylation could affect cell growth and morphology, including modulation of nucleocytoplasmic transport of macromolecules, chromatin modification, and modulation of RNA processing.
Fig. 8.
Silver staining of TAP-tagged purified TbPRMT6 complexes. TbPRMT6–Myc-His-TAP was expressed in PF 29-13 cells and purified using standard tandem affinity purification. The eluate was resolved using SDS-PAGE and silver stained. In addition to the major band representing TbPRMT6-MHCBP (TbPRMT6–Myc-His-calmodulin binding protein) (bold arrow), a protein of approximately 45 kDa was observed (thin arrow).
Table 1.
Proteins identified in TbPRMT6-MHT eluatesa
| Protein group and designation in GeneDB | No. of unique peptides | Protein description |
|---|---|---|
| Nucleocytoplasmic transport proteins | ||
| Tb09.211.4780 | 2 | TbNup82, putative |
| Tb10.61.2630 | 3 | Protein transport protein Sec13, putative |
| Tb10.6k15.3670 | 3 | TbNup96, putative |
| Tb11.03.0140 | 5 | TbNup158, putative |
| Tb927.2.4230 | 5 | NUP-1 protein, putative |
| Tb927.3.3180 | 4 | TbNup98, putative |
| Tb927.4.2880 | 4 | TbNup149, putative |
| Histones | ||
| Tb10.61.1090 | 2 | Histone H3 variant, putative |
| Tb10.496.0330 | 6 | Histone H2B, putative |
| Tb11.02.5250 | 2 | Histone H2B variant, putative |
| Tb927.1.2430 | 2 | Histone H3, putative |
| Tb927.2.2670 | 4 | Histone H4, putative |
| Tb927.5.4170 | 6 | Histone H4, putative |
| Tb927.7.2820 | 2 | Histone H2A, putative |
| Flagellar proteins | ||
| Tb09.211.4511 | 3 | Kinetoplastid membrane protein KMP-11 |
| Tb927.3.4290 | 8 | PFR1 |
| Tb927.8.4970 | 4 | PFR2 |
| Tb927.10.9570 | 2 | PFC14 |
| Other | ||
| Tb10.61.2130 | 2 | ATP-dependent DEAD/H RNA helicase, putative (TbDed1p) |
| Tb927.6.4440 | 3 | TbDRBD3 |
| Tb927.8.4580 | 2 | Hypothetical protein, conserved |
| Tb927.8.4780 | 2 | Hypothetical protein, conserved |
TbPRMT6-MHT was purified by tandem affinity chromatography, and resulting eluates were analyzed by mass spectrometry. The sequences for the peptides identified were compared to known sequences in the annotated trypanosome database GeneDB (http://www.genedb.org/). The identified proteins were further categorized based on their known or putative functions. The number of unique peptides identified for each protein is shown. Proteins for which only one peptide was identified were excluded from the analysis.
DISCUSSION
In this article, we report the in vitro and in vivo characterization of a PRMT from the protozoan parasite T. brucei. This enzyme, which we term TbPRMT6, is most similar to PRMT6 in the family of mammalian protein arginine methyltransferases. The mammalian PRMT6 enzymes have been shown to function in DNA repair and transcriptional control (20, 37, 38). However, PRMT6 homologues appear to be absent from Saccharomyces cerevisiae and most other protozoa (3). Despite the evolutionary distance between humans and trypanosomes, HsPRMT6 and TbPRMT6 are similar in that they exhibit type 1 PRMT activity and catalyze automethylation. The consequences of this automethylation are currently unknown. TbPRMT6 also resembles human PRMT6 in its apparently narrow substrate range, and it differs from the other, more promiscuous T. brucei PRMTs in this regard (27, 73, 74). In vitro, TbPRMT6 methylated a mixture of bovine histones but did not methylate several other substrates, including three trypanosome GAR proteins. TbPRMT6 histone methylation may be of functional significance since histones are an in vivo substrate of human PRMT6, and we found that TAP-tagged TbPRMT6 expressed in PF T. brucei copurified with several histones (see below). In contrast to the properties listed above, HsPRMT6 and TbPRMT6 differ in their subcellular localization, with the human enzyme exhibiting an exclusively nuclear localization, while TbPRMT6 is predominantly cytoplasmic. It is important to note, however, that the cytoplasmic steady-state localization of TbPRMT6 does not preclude the possibility that this protein has nuclear functions. Several human PRMTs shuttle between the cytoplasm and nucleus, and human PRMT5 appears predominantly cytoplasmic at steady state despite several well-documented roles in transcription and cell cycle progression (23, 41, 51, 80, 86, 94, 103). While experiments using leptomycin B, an inhibitor of crm1/exportin1-dependent nuclear export, did not provide evidence for nucleocytoplasmic shuttling of TbPRMT6 (data not shown), we cannot rule out that TbPRMT6 shuttles through a different pathway or that a small amount of nuclear enzyme is sufficient for this protein's nuclear functions.
Of the four T. brucei PRMTs that have been characterized to date, TbPRMT6 is the only enzyme whose downregulation leads to a growth defect. While this defect is modest under normal culture conditions, it becomes pronounced when PF cells are nutrient starved. Thus, TbPRMT6 performs a nonredundant function that is critical for cell growth. TbPRMT6 depletion also leads to morphological defects in a subset of both PF and BF cells. Aberrant PF cells appeared to have initiated, but not completed, cell division. In addition, there often appeared to be multiple nuclei at the junctions of partially divided cells, suggesting that nuclear segregation was impaired in cells depleted of TbPRMT6. In BF, giant rounded cells with large numbers of nuclei and kinetoplasts accumulated, often with detached flagella, again indicating defects in cytokinesis in cells with decreased TbPRMT6 levels.
Mass spectrometry analysis of TbPRMT6-associated proteins highlighted several cellular pathways that could contribute to an essential function of the enzyme. Notably, histones were a major class of proteins that were associated with TbPRMT6-MHT in PF cells. We obtained multiple peptide hits for seven different trypanosome histone proteins. This finding, coupled with the ability of HsPRMT6 to methylate histones in vivo and our demonstration that TbPRMT6 methylates bovine histones in vitro (Fig. 2), suggests that histones might be an in vivo substrate for TbPRMT6. The identification of histones as putative TbPRMT substrates was somewhat surprising, as analyses of histone modifications in T. brucei to date have identified acetylation, phosphorylation, and lysine methylation but no methylation of arginine residues (25). Previous analyses of histone modifications were performed using Edman degradation (42, 62). Recently developed mass spectrophotometric techniques for detection of methylarginines (93) may be useful for identification of previously overlooked methylarginine residues in histones of T. brucei. If such modifications are detected in wild-type cells, it will be of interest to analyze histone modifications in TbPRMT6-depleted cells to reveal whether this enzyme catalyzes histone arginine methylation and whether it, in turn, affects chromatin structure and gene regulation.
A second family of proteins that was highly represented in TbPRMT6-MHT purification was that comprising components of the nuclear pore complex (NPC). There are two different possibilities to account for the association of TbPRMT6 with the NPC, and these scenarios are not mutually exclusive. First, as suggested above, TbPRMT6 may itself be a shuttling protein. As such, NPC components may have been isolated in combination with TbPRMT6 that was itself undergoing nucleocytoplasmic transport at the time of cell lysis. Alternatively, some NPC components may be substrates for TbPRMT6. Arginine methylation is well known to affect the subcellular localization of many proteins and RNAs (2, 8, 60, 76, 88). In some instances, methylation-induced alterations in nucleocytoplasmic transport have been directly attributed to modification of specific shuttling proteins, but a role for methylation of NPC components can easily be envisioned.
We also identified four flagellar proteins in association with TbPRMT6, although no defects in cell motility were evident in those TbPRMT6 RNAi cells with normal morphology. However, several flagellar proteins of T. brucei are reported to affect both the cell cycle and cellular division. While we did not detect specific cell cycle defects in the majority TbRMT6-depleted cells, up to 5% of PF cells exhibited the dramatically altered morphology described above that we termed “hydra.” A similar morphology was observed when the paraflagellar rod protein pfr2 was depleted in PF cells (79). As stated above, the aberrant forms present in cultures of TbPRMT6-depleted BF cells were even more striking. These altered morphologies indicate that cells depleted for TbPRMT6 exhibit defects in cytokinesis, and these defects are more pronounced in the BF stage. Interestingly, depletion of various flagellar components or flagellar interacting proteins leads to aberrant cytokinesis and/or abnormal numbers of nuclei/kinetoplasts, and these defects are often more severe in BF cells than in PF cells (11, 47, 50, 78). Loss of another TbPRMT6-interacting protein, KMP-11, leads to altered cytokinesis, attachment of flagella, and multiple nuclei (57). Together, these data suggest a model whereby TbPRMT6-catalyzed methylation of a subset of flagellar proteins is essential for proper cell division in T. brucei.
Finally, two proteins that have been shown or are presumed to function in RNA metabolism, TbRBD3 and TbDed1, were isolated with TbPRMT6-MHT. In general, RBPs constitute a major class of arginine methyl proteins in both mammals and yeast (4, 36, 59, 70, 89). Methylation of RBPs generally leads to dramatic alterations in protein-protein interactions (7, 15, 19, 24, 56, 67, 100) and, less often, in protein-RNA interactions. Estevez (21) recently reported that TbRBD3 binds to a subset of developmentally regulated transcripts and promotes their stabilization. Because TbRBD3 is equally expressed in PF and BF T. brucei, it was proposed that its role in developmentally modulated RNA stabilization may result from life cycle-specific posttranslational modifications. Thus, it will be interesting to determine the methylation status of TbRBD3 in PF and BF. TbDed1 is a DEAD box protein and a putative RNA helicase that has been reported in TAP pulldowns of RNA polymerase II and a noncanonical nuclear poly(A) polymerase in T. brucei (16, 22), although its functions have not been elucidated. Notably, we also identified TbDed1 in association with the SDMA-producing TbPRMT5 protein from T. brucei cytoplasm (73). In addition, the human homologue of TbDed1, DDX3, associates with the type I PRMT PRMT8 (71). Recent analyses in our laboratory suggest that TbDed1 contains both ADMA and SDMA modifications (S. G. Menon, J. C. Fisk, and L. K. Read, unpublished observations). Future studies will be focused on the roles of arginine methylation in modulating the actions of this apparently multifunctional DEAD box protein in trypanosome gene regulation.
Collectively, the studies presented here identify TbPRMT6 as a type I PRMT with a key function in both PF and BF T. brucei. The enzyme exhibits a narrow substrate range, which must include proteins that are critical for cell growth. Analysis of the methylation status and function of the TbPRMT6-associated proteins reported here will provide insight into the essential roles this enzyme and its methylarginine function in trypanosomes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jay Bangs and Vivian Bellofatto for antibodies, George Cross for cell lines, and David Campbell for leptomycin B. Thanks are due to Yuko Ogata at SBRI for mass spectrophotometric analysis of TbPRMT6 complexes.
L.K.R. was supported by NIH grant no. R01 AI060260, and S.G.C. was supported by NIH grant no. R37 GM026020. C.Z.-L. was supported by a Ruth L. Kirschstein National Research Service Award (NRSA) from the NIH (GM078761). J.C.F. was supported in part by NIH NRSA postdoctoral fellowship no. F32 AI07718501.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 23 April 2010.
REFERENCES
- 1.Ammerman M. L., Fisk J. C., Read L. K. 2008. gRNA/pre-mRNA annealing and RNA chaperone activities of RBP16. RNA 14:1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki K., Ishii Y., Matsumoto K., Tsujimoto M. 2002. Methylation of Xenopus CIRP2 regulates its arginine- and glycine-rich region-mediated nucleocytoplasmic distribution. Nucleic Acids Res. 30:5182–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachand F. 2007. Protein arginine methyltransferases: from unicellular eukaryotes to humans. Eukaryot. Cell 6:889–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachand F., Silver P. A. 2004. PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits. EMBO J. 23:2641–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bedford M. T. 2007. Arginine methylation at a glance. J. Cell Sci. 120:4243–4246 [DOI] [PubMed] [Google Scholar]
- 6.Bedford M. T., Clarke S. G. 2009. Protein arginine methylation in mammals: who, what, and why. Mol. Cell 33:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bedford M. T., Frankel A., Yaffe M. B., Clarke S., Leder P., Richard S. 2000. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J. Biol. Chem. 275:16030–16036 [DOI] [PubMed] [Google Scholar]
- 8.Bedford M. T., Richard S. 2005. Arginine methylation an emerging regulator of protein function. Mol. Cell 18:263–272 [DOI] [PubMed] [Google Scholar]
- 9.Boisvert F.-M., Chenard C. A., Richard S. 2005. Protein interfaces in signaling regulated by arginine methylation. Sci. STKE 2005:re2. [DOI] [PubMed] [Google Scholar]
- 10.Boulanger M.-C., Liang C., Russell R. S., Lin R., Bedford M. T., Wainberg M. A., Richard S. 2005. Methylation of Tat by PRMT6 regulates human immunodeficiency virus type 1 gene expression. J. Virol. 79:124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broadhead R., Dawe H. R., Farr H., Griffiths S., Hart S. R., Portman N., Shaw M. K., Ginger M. L., Gaskell S. J., McKean P. G., Gull K. 2006. Flagellar motility is required for the viability of the bloodstream trypanosome. Nature 440:224–227 [DOI] [PubMed] [Google Scholar]
- 12.Cimato T. R., Tang J., Xu Y., Guarnaccia C., Herschman H. R., Pongor S., Aletta J. M. 2002. Nerve growth factor-mediated increases in protein methylation occur predominantly at type I arginine methylation sites and involve protein arginine methyltransferase 1. J. Neurosci. Res. 67:435–442 [DOI] [PubMed] [Google Scholar]
- 13.Clayton C., Shapira M. 2007. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol. Biochem. Parasitol. 156:93–101 [DOI] [PubMed] [Google Scholar]
- 14.Clayton C. E. 2002. Life without transcriptional control? From fly to man and back again. EMBO J. 21:1881–1888(Erratum, 21:3917.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cote J., Richard S. 2005. Tudor domains bind symmetrical dimethylated arginines. J. Biol. Chem. 280:28476–28483 [DOI] [PubMed] [Google Scholar]
- 16.Das A., Li H., Liu T., Bellofatto V. 2006. Biochemical characterization of Trypanosoma brucei RNA polymerase II. Mol. Biochem. Parasitol. 150:201–210 [DOI] [PubMed] [Google Scholar]
- 17.De Gaudenzi J., Frasch A. C., Clayton C. 2005. RNA-binding domain proteins in kinetoplastids: a comparative analysis. Eukaryot. Cell 4:2106–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeGrasse J., DuBois K., Devos D., Siegel T., Sali A., Field M., Rout M., Chait B. 2009. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol. Cell. Proteomics 8:2119–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El-Andaloussi N., Valovka T., Toueille M., Hassa P. O., Gehrig P., Covic M., Hubscher U., Hottiger M. O. 2007. Methylation of DNA polymerase beta by protein arginine methyltransferase 1 regulates its binding to proliferating cell nuclear antigen. FASEB J. 21:26–34 [DOI] [PubMed] [Google Scholar]
- 20.El-Andaloussi N., Valovka T., Toueille M., Steinacher R., Focke F., Gehrig P., Covic M., Hassa P. O., Schar P., Hubscher U., Hottiger M. O. 2006. Arginine methylation regulates DNA polymerase beta. Mol. Cell 22:51–62 [DOI] [PubMed] [Google Scholar]
- 21.Estevez A. M. 2008. The RNA-binding protein TbDRBD3 regulates the stability of a specific subset of mRNAs in trypanosomes. Nucleic Acids Res. 36:4573–4586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Etheridge R. D., Clemens D. M., Gershon P. D., Aphasizhev R. 2009. Identification and characterization of nuclear non-canonical poly(A) polymerases from Trypanosoma brucei. Mol. Biochem. Parasitol. 164:66–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fabbrizio E., El Messaoudi S., Polanowska J., Paul C., Cook J. R., Lee J.-H., Negre V., Rousset M., Pestka S., Le Cam A., Sardet C. 2002. Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep. 3:641–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng Q., Yi P., Wong J., O'Malley B. W. 2006. Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol. Cell. Biol. 26:7846–7857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Figueiredo L. M., Cross G. A. M., Janzen C. J. 2009. Epigenetic regulation in African trypanosomes: a new kid on the block. Nat. Rev. Microbiol. 7:504–513 [DOI] [PubMed] [Google Scholar]
- 26.Fisk J. C., Ammerman M. L., Presnyak V., Read L. K. 2008. TbRGG2, an essential RNA editing accessory factor in two Trypanosoma brucei life cycle stages. J. Biol. Chem. 283:23016–23025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fisk J. C., Sayegh J., Zurita-Lopez C., Menon S., Presnyak V., Clarke S. G., Read L. K. 2009. A type III protein arginine methyltransferase from the protozoan parasite Trypanosoma brucei. J. Biol. Chem. 284:11590–11600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frankel A., Yadav N., Lee J., Branscombe T. L., Clarke S., Bedford M. T. 2002. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J. Biol. Chem. 277:3537–3543 [DOI] [PubMed] [Google Scholar]
- 29.Friesen W. J., Paushkin S., Wyce A., Massenet S., Pesiridis G. S., Van Duyne G., Rappsilber J., Mann M., Dreyfuss G. 2001. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol. Cell. Biol. 21:8289–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gottschling H., Freese E. 1962. A tritium isotope effect on ion exchange chromatography. Nature 196:829–831 [DOI] [PubMed] [Google Scholar]
- 31.Grondal E. J., Evers R., Kosubek K., Cornelissen A. W. 1989. Characterization of the RNA polymerases of Trypanosoma brucei: trypanosomal mRNAs are composed of transcripts derived from both RNA polymerase II and III. EMBO J. 8:3383–3389(Erratum, 8:4359.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guccione E., Bassi C., Casadio F., Martinato F., Cesaroni M., Schuchlautz H., Luscher B., Amati B. 2007. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 449:933–937 [DOI] [PubMed] [Google Scholar]
- 33.Harris M. E., Moore D. R., Hajduk S. L. 1990. Addition of uridines to edited RNAs in trypanosome mitochondria occurs independently of transcription. J. Biol. Chem. 265:11368–11376 [PubMed] [Google Scholar]
- 34.Hashimi H., Zikova A., Panigrahi A. K., Stuart K. D., Lukes J. 2008. TbRGG1, an essential protein involved in kinetoplastid RNA metabolism that is associated with a novel multiprotein complex. RNA 14:970–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayman M. L., Read L. K. 1999. Trypanosoma brucei RBP16 is a mitochondrial Y-box family protein with guide RNA binding activity. J. Biol. Chem. 274:12067–12074 [DOI] [PubMed] [Google Scholar]
- 36.Herrmann F., Bossert M., Schwander A., Akgun E., Fackelmayer F. O. 2004. Arginine methylation of scaffold attachment factor A by heterogeneous nuclear ribonucleoprotein particle-associated PRMT1. J. Biol. Chem. 279:48774–48779 [DOI] [PubMed] [Google Scholar]
- 37.Hyllus D., Stein C., Schnabel K., Schiltz E., Imhof A., Dou Y., Hsieh J., Bauer U.-M. 2007. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 21:3369–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iberg A. N., Espejo A., Cheng D., Kim D., Michaud-Levesque J., Richard S., Bedford M. T. 2008. Arginine methylation of the histone H3 tail impedes effector binding. J. Biol. Chem. 283:3006–3010 [DOI] [PubMed] [Google Scholar]
- 39.Invernizzi C. F., Xie B., Frankel F. A., Feldhammer M., Roy B. B., Richard S., Wainberg M. A. 2007. Arginine methylation of the HIV-1 nucleocapsid protein results in its diminished function. AIDS 21:795–805 [DOI] [PubMed] [Google Scholar]
- 40.Invernizzi C. F., Xie B., Richard S., Wainberg M. A. 2006. PRMT6 diminishes HIV-1 Rev binding to and export of viral RNA. Retrovirology 3:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jansson M., Durant S. T., Cho E.-C., Sheahan S., Edelmann M., Kessler B., La Thangue N. B. 2008. Arginine methylation regulates the p53 response. Nat. Cell Biol. 10:1431–1439 [DOI] [PubMed] [Google Scholar]
- 42.Janzen C. J., Fernandez J. P., Deng H., Diaz R., Hake S. B., Cross G. A. M. 2006. Unusual histone modifications in Trypanosoma brucei. FEBS Lett. 580:2306–2310 [DOI] [PubMed] [Google Scholar]
- 43.Jensen B. C., Kifer C. T., Brekken D. L., Randall A. C., Wang Q., Drees B. L., Parsons M. 2007. Characterization of protein kinase CK2 from Trypanosoma brucei. Mol. Biochem. Parasitol. 151:28–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kleene S. J., Toews M. L., Adler J. 1977. Isolation of glutamic acid methyl ester from an Escherichia coli membrane protein involved in chemotaxis. J. Biol. Chem. 252:3214–3218 [PubMed] [Google Scholar]
- 45.Klein K. G., Olson C. L., Donelson J. E., Engman D. M. 1995. Molecular comparison of the mitochondrial and cytoplasmic hsp70 of Trypanosoma cruzi, Trypanosoma brucei and Leishmania major. J. Eukaryot. Microbiol. 42:473–476 [DOI] [PubMed] [Google Scholar]
- 46.Klein P. D., Szczepanik P. A. 1967. Fine structure of isotopically labeled amino acids determined by ion exchange chromatography. Anal. Chem. 39:1276–1281 [DOI] [PubMed] [Google Scholar]
- 47.Kohl L., Robinson D., Bastin P. 2003. Novel roles for the flagellum in cell morphogenesis and cytokinesis of trypanosomes. EMBO J. 22:5336–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koller J., Muller U. F., Schmid B., Missel A., Kruft V., Stuart K., Goringer H. U. 1997. Trypanosoma brucei gBP21. An arginine-rich mitochondrial protein that binds to guide RNA with high affinity. J. Biol. Chem. 272:3749–3757 [DOI] [PubMed] [Google Scholar]
- 49.Krause C. D., Yang Z.-H., Kim Y.-S., Lee J.-H., Cook J. R., Pestka S. 2007. Protein arginine methyltransferases: evolution and assessment of their pharmacological and therapeutic potential. Pharmacol. Ther. 113:50–87 [DOI] [PubMed] [Google Scholar]
- 50.LaCount D. J., Barrett B., Donelson J. E. 2002. Trypanosoma brucei FLA1 is required for flagellum attachment and cytokinesis. J. Biol. Chem. 277:17580–17588 [DOI] [PubMed] [Google Scholar]
- 51.Lacroix M., Messaoudi S. E., Rodier G., Le Cam A., Sardet C., Fabbrizio E. 2008. The histone-binding protein COPR5 is required for nuclear functions of the protein arginine methyltransferase PRMT5. EMBO Rep. 9:452–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lake A. N., Bedford M. T. 2007. Protein methylation and DNA repair. Mutat. Res. 618:91–101 [DOI] [PubMed] [Google Scholar]
- 53.Lakowski T. M., Frankel A. 2008. A kinetic study of human protein arginine N-methyltransferase 6 reveals a distributive mechanism. J. Biol. Chem. 283:10015–10025 [DOI] [PubMed] [Google Scholar]
- 54.Lee J.-H., Cook J. R., Yang Z.-H., Mirochnitchenko O., Gunderson S. I., Felix A. M., Herth N., Hoffmann R., Pestka S. 2005. PRMT7, a new protein arginine methyltransferase that synthesizes symmetric dimethylarginine. J. Biol. Chem. 280:3656–3664 [DOI] [PubMed] [Google Scholar]
- 55.Lee J., Sayegh J., Daniel J., Clarke S., Bedford M. T. 2005. PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J. Biol. Chem. 280:32890–32896 [DOI] [PubMed] [Google Scholar]
- 56.Lee Y.-H., Coonrod S. A., Kraus W. L., Jelinek M. A., Stallcup M. R. 2005. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc. Natl. Acad. Sci. U. S. A. 102:3611–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Z., Wang C. C. 2008. KMP-11, a basal body and flagellar protein, is required for cell division in Trypanosoma brucei. Eukaryot. Cell 7:1941–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin W. J., Gary J. D., Yang M. C., Clarke S., Herschman H. R. 1996. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J. Biol. Chem. 271:15034–15044 [DOI] [PubMed] [Google Scholar]
- 59.Liu Q., Dreyfuss G. 1995. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol. Cell. Biol. 15:2800–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lukong K. E., Richard S. 2004. Arginine methylation signals mRNA export. Nat. Struct. Mol. Biol. 11:914–915 [DOI] [PubMed] [Google Scholar]
- 61.Madison-Antenucci S., Hajduk S. L. 2001. RNA editing-associated protein 1 is an RNA binding protein with specificity for preedited mRNA. Mol. Cell 7:879–886 [DOI] [PubMed] [Google Scholar]
- 62.Mandava V., Fernandez J. P., Deng H., Janzen C. J., Hake S. B., Cross G. A. M. 2007. Histone modifications in Trypanosoma brucei. Mol. Biochem. Parasitol. 156:41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McBride A. E., Zurita-Lopez C., Regis A., Blum E., Conboy A., Elf S., Clarke S. 2007. Protein arginine methylation in Candida albicans: role in nuclear transport. Eukaryot. Cell 6:1119–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miranda T. B., Miranda M., Frankel A., Clarke S. 2004. PRMT7 is a member of the protein arginine methyltransferase family with a distinct substrate specificity. J. Biol. Chem. 279:22902–22907 [DOI] [PubMed] [Google Scholar]
- 65.Miranda T. B., Sayegh J., Frankel A., Katz J. E., Miranda M., Clarke S. 2006. Yeast Hsl7 (histone synthetic lethal 7) catalyses the in vitro formation of omega-N(G)-monomethylarginine in calf thymus histone H2A. Biochem. J. 395:563–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miranda T. B., Webb K. J., Edberg D. D., Reeves R., Clarke S. 2005. Protein arginine methyltransferase 6 specifically methylates the nonhistone chromatin protein HMGA1a. Biochem. Biophys. Res. Commun. 336:831–835 [DOI] [PubMed] [Google Scholar]
- 67.Naeem H., Cheng D., Zhao Q., Underhill C., Tini M., Bedford M. T., Torchia J. 2007. The activity and stability of the transcriptional coactivator p/CIP/SRC-3 are regulated by CARM1-dependent methylation. Mol. Cell. Biol. 27:120–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Najbauer J., Johnson B. A., Young A. L., Aswad D. W. 1993. Peptides with sequences similar to glycine, arginine-rich motifs in proteins interacting with RNA are efficiently recognized by methyltransferase(s) modifying arginine in numerous proteins. J. Biol. Chem. 268:10501–10509 [PubMed] [Google Scholar]
- 69.Niewmierzycka A., Clarke S. 1999. S-Adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. Identification of a novel protein arginine methyltransferase. J. Biol. Chem. 274:814–824 [DOI] [PubMed] [Google Scholar]
- 70.Pahlich S., Zakaryan R. P., Gehring H. 2006. Protein arginine methylation: Cellular functions and methods of analysis. Biochim. Biophys. Acta 1764:1890–1903 [DOI] [PubMed] [Google Scholar]
- 71.Pahlich S., Zakaryan R. P., Gehring H. 2008. Identification of proteins interacting with protein arginine methyltransferase 8: the Ewing sarcoma (EWS) protein binds independent of its methylation state. Proteins 72:1125–1137 [DOI] [PubMed] [Google Scholar]
- 72.Panigrahi A. K., Zíková A., Dalley R. A., Acestor N., Ogata Y., Anupama A., Myler P. J., Stuart K. 2008. Mitochondrial complexes in trypanosoma brucei: a novel complex and a unique oxidoreductase complex. Mol. Cell. Proteomics 7:534–545 [DOI] [PubMed] [Google Scholar]
- 73.Pasternack D. A., Sayegh J., Clarke S., Read L. K. 2007. Evolutionarily divergent type II protein arginine methyltransferase in Trypanosoma brucei. Eukaryot. Cell 6:1665–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pelletier M., Pasternack D. A., Read L. K. 2005. In vitro and in vivo analysis of the major type I protein arginine methyltransferase from Trypanosoma brucei. Mol. Biochem. Parasitol. 144:206–217 [DOI] [PubMed] [Google Scholar]
- 75.Pelletier M., Read L. K. 2003. RBP16 is a multifunctional gene regulatory protein involved in editing and stabilization of specific mitochondrial mRNAs in Trypanosoma brucei. RNA 9:457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pintucci G., Quarto N., Rifkin D. B. 1996. Methylation of high molecular weight fibroblast growth factor-2 determines post-translational increases in molecular weight and affects its intracellular distribution. Mol. Biol. Cell 7:1249–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reference deleted.
- 78.Ralston K. S., Hill K. L. 2006. Trypanin, a component of the flagellar Dynein regulatory complex, is essential in bloodstream form African trypanosomes. PLoS Pathog. 2:e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ralston K. S., Lerner A. G., Diener D. R., Hill K. L. 2006. Flagellar motility contributes to cytokinesis in Trypanosoma brucei and is modulated by an evolutionarily conserved dynein regulatory system. Eukaryot. Cell 5:696–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rho J., Choi S., Seong Y. R., Cho W. K., Kim S. H., Im D. S. 2001. Prmt5, which forms distinct homo-oligomers, is a member of the protein-arginine methyltransferase family. J. Biol. Chem. 276:11393–11401 [DOI] [PubMed] [Google Scholar]
- 81.Rout M. P., Field M. C. 2001. Isolation and characterization of subnuclear compartments from Trypanosoma brucei. Identification of a major repetitive nuclear lamina component. J. Biol. Chem. 276:38261–38271 [DOI] [PubMed] [Google Scholar]
- 82.Sayegh J., Webb K., Cheng D., Bedford M. T., Clarke S. G. 2007. Regulation of protein arginine methyltransferase 8 (PRMT8) activity by its N-terminal domain. J. Biol. Chem. 282:36444–36453 [DOI] [PubMed] [Google Scholar]
- 83.Sbicego S., Alfonzo J. D., Estevez A. M., Rubio M. A. T., Kang X., Turck C. W., Peris M., Simpson L. 2003. RBP38, a novel RNA-binding protein from trypanosomatid mitochondria, modulates RNA stability. Eukaryot. Cell 2:560–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schimanski B., Nguyen T. N., Gunzl A. 2005. Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryot. Cell 4:1942–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schwede A., Ellis L., Luther J., Carrington M., Stoecklin G., Clayton C. 2008. A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucleic Acids Res. 36:3374–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Scoumanne A., Zhang J., Chen X. 2009. PRMT5 is required for cell-cycle progression and p53 tumor suppressor function. Nucleic Acids Res. 37:4965–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sgarra R., Lee J., Tessari M. A., Altamura S., Spolaore B., Giancotti V., Bedford M. T., Manfioletti G. 2006. The AT-hook of the chromatin architectural transcription factor high mobility group A1a is arginine-methylated by protein arginine methyltransferase 6. J. Biol. Chem. 281:3764–3772 [DOI] [PubMed] [Google Scholar]
- 88.Shen E. C., Henry M. F., Weiss V. H., Valentini S. R., Silver P. A., Lee M. S. 1998. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 12:679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Swiercz R., Person M. D., Bedford M. T. 2005. Ribosomal protein S2 is a substrate for mammalian PRMT3 (protein arginine methyltransferase 3). Biochem. J. 386:85–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tang J., Kao P. N., Herschman H. R. 2000. Protein-arginine methyltransferase I, the predominant protein-arginine methyltransferase in cells, interacts with and is regulated by interleukin enhancer-binding factor 3. J. Biol. Chem. 275:19866–19876 [DOI] [PubMed] [Google Scholar]
- 91.Vanhamme L., Perez-Morga D., Marchal C., Speijer D., Lambert L., Geuskens M., Alexandre S., Ismaili N., Goringer U., Benne R., Pays E. 1998. Trypanosoma brucei TBRGG1, a mitochondrial oligo(U)-binding protein that co-localizes with an in vitro RNA editing activity. J. Biol. Chem. 273:21825–21833 [DOI] [PubMed] [Google Scholar]
- 92.Vondruskova E., van den Burg J., Zikova A., Ernst N. L., Stuart K., Benne R., Lukes J. 2005. RNA interference analyses suggest a transcript-specific regulatory role for mitochondrial RNA-binding proteins MRP1 and MRP2 in RNA editing and other RNA processing in Trypanosoma brucei. J. Biol. Chem. 280:2429–2438 [DOI] [PubMed] [Google Scholar]
- 93.Wang H., Straubinger R. M., Aletta J. M., Cao J., Duan X., Yu H., Qu J. 2009. Accurate localization and relative quantification of arginine methylation using nanoflow liquid chromatography coupled to electron transfer dissociation and orbitrap mass spectrometry. J. Am. Soc. Mass Spectrom. 20:507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang L., Pal S., Sif S. 2008. Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Mol. Cell. Biol. 28:6262–6277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wickstead B., Ersfeld K., Gull K. 2002. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol. Biochem. Parasitol. 125:211–216 [DOI] [PubMed] [Google Scholar]
- 96.Wirtz E., Leal S., Ochatt C., Cross G. A. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89–101 [DOI] [PubMed] [Google Scholar]
- 97.Wooderchak W. L., Zang T., Zhou Z. S., Acuna M., Tahara S. M., Hevel J. M. 2008. Substrate profiling of PRMT1 reveals amino acid sequences that extend beyond the “RGG” paradigm. Biochemistry 47:9456–9466 [DOI] [PubMed] [Google Scholar]
- 98.Wysocka J., Allis C. D., Coonrod S. 2006. Histone arginine methylation and its dynamic regulation. Front. Biosci. 11:344–355 [DOI] [PubMed] [Google Scholar]
- 99.Xie H., Clarke S. 1993. Methyl esterification of C-terminal leucine residues in cytosolic 36-kDa polypeptides of bovine brain. A novel eucaryotic protein carboxyl methylation reaction. J. Biol. Chem. 268:13364–13371 [PubMed] [Google Scholar]
- 100.Xu W., Chen H., Du K., Asahara H., Tini M., Emerson B. M., Montminy M., Evans R. M. 2001. A transcriptional switch mediated by cofactor methylation. Science 294:2507–2511 [DOI] [PubMed] [Google Scholar]
- 101.Zamudio J. R., Mittra B., Chattopadhyay A., Wohlschlegel J. A., Sturm N. R., Campbell D. A. 2009. Trypanosoma brucei spliced leader RNA maturation by the cap 1 2′-O-ribose methyltransferase and SLA1 H/ACA snoRNA pseudouridine synthase complex. Mol. Cell. Biol. 29:1202–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zeiner G. M., Sturm N. R., Campbell D. A. 2003. Exportin 1 mediates nuclear export of the kinetoplastid spliced leader RNA. Eukaryot. Cell 2:222–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhao Q., Rank G., Tan Y. T., Li H., Moritz R. L., Simpson R. J., Cerruti L., Curtis D. J., Patel D. J., Allis C. D., Cunningham J. M., Jane S. M. 2009. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat. Struct. Mol. Biol. 16:304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zikova A., Panigrahi A. K., Dalley R. A., Acestor N., Anupama A., Ogata Y., Myler P. J., Stuart K. 2008. Trypanosoma brucei mitochondrial ribosomes: affinity purification and component identification by mass spectrometry. Mol. Cell. Proteomics 7:1286–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ziková A., Panigrahi A. K., Uboldi A. D., Dalley R. A., Handman E., Stuart K. 2008. Structural and functional association of Trypanosoma brucei MIX protein with cytochrome c oxidase complex. Eukaryot. Cell 7:1994–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zou Y., Webb K., Perna A. D., Zhang Q., Clarke S., Wang Y. 2007. A mass spectrometric study on the in vitro methylation of HMGA1a and HMGA1b proteins by PRMTs: methylation specificity, the effect of binding to AT-rich duplex DNA, and the effect of C-terminal phosphorylation. Biochemistry 46:7896–7906 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.