Abstract
The genome sequences of the basidiomycete Agaricomycetes species Coprinopsis cinerea, Laccaria bicolor, Schizophyllum commune, Phanerochaete chrysosporium, and Postia placenta, as well as of Cryptococcus neoformans and Ustilago maydis, are now publicly available. Out of these fungi, C. cinerea, S. commune, and U. maydis, together with the budding yeast Saccharomyces cerevisiae, have been investigated for years genetically and molecularly for signaling in sexual reproduction. The comparison of the structure and organization of mating type genes in fungal genomes reveals an amazing conservation of genes regulating the sexual reproduction throughout the fungal kingdom. In agaricomycetes, two mating type loci, A, coding for homeodomain type transcription factors, and B, encoding a pheromone/receptor system, regulate the four typical mating interactions of tetrapolar species. Evidence for both A and B mating type genes can also be identified in basidiomycetes with bipolar systems, where only two mating interactions are seen. In some of these fungi, the B locus has lost its self/nonself discrimination ability and thus its specificity while retaining the other regulatory functions in development. In silico analyses now also permit the identification of putative components of the pheromone-dependent signaling pathways. Induction of these signaling cascades leads to development of dikaryotic mycelia, fruiting body formation, and meiotic spore production. In pheromone-dependent signaling, the role of heterotrimeric G proteins, components of a mitogen-activated protein kinase (MAPK) cascade, and cyclic AMP-dependent pathways can now be defined. Additionally, the pheromone-dependent signaling through monomeric, small GTPases potentially involved in creating the polarized cytoskeleton for reciprocal nuclear exchange and migration during mating is predicted.
THE MATING SYSTEMS IN BASIDIOMYCETES
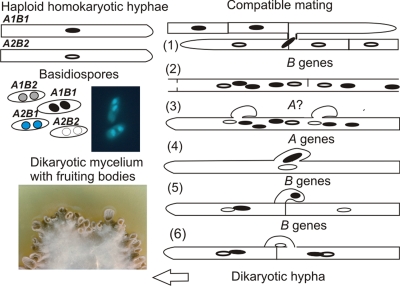
The typical life cycle of the higher basidiomycetes, presently classified as Agaricomycetes (Table 1) (34), consists of haploid, monokaryotic as well as dikaryotic stages, the latter of which prevails in nature. The monokaryotic mycelium contains nuclei of only one genetic type and is thus termed homokaryon. This mycelium, grown from haploid spores, in general contains one nucleus per cell (Fig. 1). The dikaryon is formed when genetically different homokaryons mate. In the saprotrophic agaricomycetes, the mushroom-forming species Schizophyllum commune and Coprinopsis cinerea (Table 1), mating is regulated by a tetrapolar system consisting of two unlinked genetic complexes, named A and B. The term tetrapolar reflects the fact that there are four possible mating interactions scored in matings between haploid strains derived from spores formed on the fruiting bodies of same parent dikaryon: a fully compatible interaction occurs when both A and B between the mates are of different specificities (Fig. 1), an incompatible interaction occurs when allelic specificities are all the same, and two semicompatible interactions occur when either A- or B-regulated development is turned on due to differences in either A or B genes.
Table 1.
Taxonomya and mating type systems of the fungal species discussed in the text
| Species | Mating system | Mating type loci | Reference(s) |
|---|---|---|---|
| Ascomycota | |||
| Saccharomyces cerevisiae | Bipolar | MATa and MATα | 42 |
| Schizosaccharomyces pombe | Bipolar | mat1P and mat1M | 72 |
| Neurospora crassa | Bipolar | mata and matA | 8 |
| Basidiomycota | |||
| Ustilaginomycotina | |||
| Ustilago hordei | Bipolar | Mat-1 and Mat-2, a and b linked | 5, 35 |
| Ustilago maydis | Tetrapolar | Biallelic a and multiallelic b | 5, 6 |
| Pucciniomycotina | |||
| Microbotryum violaceum | Bipolar | MAT A1 and MAT A2, a and b linked | 23 |
| Agaricomycotina | |||
| Tremellomycetes | |||
| Cryptococcus neoformans (teleomorph name: Filobasidiella neoformans) | Bipolar | Mata and Matα, sex chromosome | 35 |
| Agaricomycetes | |||
| Coprinellus disseminatus | Bipolar | Multiallelic, multiple subloci in A, B genes present | 41 |
| Coprinopsis cinerea | Tetrapolar | Multiallelic, multiple subloci in both A and B | 12 |
| Laccaria bicolor | Tetrapolar | Multiallelic A and B | 70 |
| Phanerochaete chrysosporium | Bipolar | Multiallelic A, B genes present | 63, 70 |
| Pholiota nameko | Bipolar | Multiallelic A, B genes present | 122 |
| Pleurotus djamor | Tetrapolar | Multiallelic A and B | 40 |
| Postia placenta | Bipolar? | A and B genes present | 62 |
| Schizophyllum commune | Tetrapolar | Multiallelic, multiple subloci in both A and B | 80 |
See reference 34.
Fig. 1.
Life cycle of the tetrapolar basidiomycete Schizophyllum commune. In the fruiting bodies, basidiospores with four different combinations of A and B mating type genes are produced. If both A and B genes are different in two developing, haploid mycelia, a fully compatible mating takes place. (1) B genes regulate the reciprocal nuclear migration, illustrated by movement of a nucleus via a hyphal fusion through broken septa from one haploid hypha into the other. (2) As a consequence of nuclear migration, multinucleate hyphae occur in the interaction zone of the mates. Unfused clamp connections (pseudoclamps) in the multinucleate hyphae are seen as a function of A-regulated development (3), which then controls the pairing of different nuclei followed by the hook cell formation and synchronous nuclear division (4). After separation of the sister nuclei, a cross wall is formed at the base of the hook and in the hypha (5). The growth of the hook toward and the subsequent fusion to the subapical cell are controlled by B genes and allow the movement of the nucleus from the hook into the subapical cell to complete clamp formation (6).
In S. commune, each mating type complex consists of two linked multiallelic loci: (i) Aα and Aβ and (ii) Bα and Bβ (80). In C. cinerea, the A complex also consists of two closely linked multiallelic loci, Aα and Aβ, while B consists of only one locus, as determined by recombination analyses (10, 12). For comparison, the plant pathogenic basidiomycetous smut fungus Ustilago maydis also contains two independent mating type loci (5), although the terminology is swapped, so that the multiallelic b locus corresponds to Aα or Aβ of the mushroom-forming agaricomycetes S. commune and C. cinerea, while the B counterpart in U. maydis is the a locus with only two specificities, a1 and a2 (5, 6, 43). On the basis of the phenotypes of the mating interactions between haploid strains with similar and different A and B loci, it is possible to determine the developmental pathway controlled by genes in each locus. In both S. commune and C. cinerea, the difference in one of the A or B subloci (α or β) is sufficient to activate the respective pathway of A- and B-controlled development. The B genes regulate reciprocal nuclear exchange and nuclear migration in both mates, while the A genes control the development of clamps involved in formation of dikaryotic hyphal compartments. This includes the initial pairing of the haploid nuclei with different A and B specificities and the synchronous division of the nuclear pair in association with the initial development of the clamp cell, the hook formation. Different B genes are then needed for the fusion of the hook cell to the subapical cell, which completes the clamp connection formation (4, 49, 80) (Fig. 1). In U. maydis, cell recognition and fusion, more reminiscent of the yeast system, is regulated by the a mating type locus, and only after fusion, the b mating type locus is called upon to regulate the formation of the dikaryotic hyphae with the ability to infect and grow in the host plant (6). Recently, the dikaryon of U. maydis also has been shown to be able to develop clamp-like structures (94).
In basidiomycetes, not all species exhibit a tetrapolar mating behavior with four potential mating interactions between spores from one fruiting body. In bipolar species (Table 1), only compatible or incompatible interactions are found, lacking the two semicompatible interactions described above. However, both A and B mating type genes can be identified, meaning that the bipolar state is likely a secondary development from tetrapolar progenitors. In the few well-studied bipolar species, such as the smut fungus Ustilago hordei and the rust Microbotryum violaceum (5, 23), the two mating type loci are tightly linked on the same chromosome, probably as a consequence of chromosomal rearrangements, including translocation (35). For the tremellomycete Cryptococcus neoformans, the development of a sex chromosome, linking not only the mating type genes but also genes involved in downstream signaling, was proposed (53, 57). However, bipolar mating type systems of agaricomycetes like Coprinellus disseminatus or Pholiota nameko appear to have followed a different evolutionary path, in which the B locus has lost its self/nonself discrimination ability and thus specificity, although not its other regulatory functions in development, perhaps as a consequence of a mutation or recombination event (41). This evolutionary route is substantiated by identification of self-recognizing B loci due to mutagenesis (80) as well as introduction of recombinant genes (28), leading to functionally bipolar mutant strains for S. commune and C. cinerea.
In addition to these functional and experimental analyses, the genomes of the agaricomycetes Phanerochaete chrysosporium, a lignin-degrading white rot fungus, the brown rot fungus Postia placenta, and ectomycorrhiza-forming Laccaria bicolor allow examination of putative A and B mating type and mating type-like loci (60, 62, 63). Early studies on the sexual reproduction by mating interactions suggested that Ph. chrysosporium has a primary homothallic, or self-fertile, mating system (1), while presently it is considered to be bipolar (70). Ph. chrysosporium has multinucleate hyphae without clamp connections, while the bipolar P. placenta (62) is known to form dikaryotic hyphae with rare clamp connections, and fruiting bodies can be found for both in nature. The mycorrhiza-forming L. bicolor more closely resembles S. commune and C. cinerea in that it has dikaryotic hyphae with clamp connections. It forms fruiting bodies in association with its host tree. The classical genetic studies, although performed originally by confrontation of a small number of homokaryons from the same fruiting body, suggested that the species is tetrapolar (24), which could be verified recently (61, 70).
MOLECULAR STRUCTURE OF A MATING TYPE GENES
Homeodomain protein-encoding genes.
With the development of transformation systems, creation and screening of genomic and cDNA libraries, and expression of the isolated genomic and cDNA clones, it was possible to reveal the complex organization and to understand the function of the A and B loci of basidiomycetes. The structures of the A mating type loci have been well characterized in C. cinerea, in which the predicted “archetype” A locus contains at least three pairs of genes, each pair encoding one member each of the homeodomain (HD) protein families HD1 and HD2. With sequencing data available, the terms Aα and Aß were replaced with a terminology of gene pairs a within the former Aα locus and b, c, and d in the former Aß locus. However, so far no functional c locus has been identified using transformation assays. The current understanding is that there are three subloci in C. cinerea: a, b, and d. No natural mating type locus containing all pairs has been found; instead, remnants of single genes, as well as pairs, were discovered, whose combinations provide the functional diversity for a high number of productive mating interactions in nature. The HD1 and HD2 proteins resemble the homedomain Matα2 and Mata1 mating type proteins of the budding yeast Saccharomyces cerevisiae, respectively (12). HD2 has a strong and HD1 a weak DNA binding activity, but HD1 carries nuclear localization signals and an activation domain. The N-terminal parts of HD1 and HD2 carry dimerization motifs. Heterodimerization of an HD1 with an HD2 protein is possible between allelic versions of different specificities present in one cell. The heterodimer creates an active transcription factor necessary for the activation of the target genes of the A mating type pathway (3, 50, 51, 77, 106).
From massive recombination analyses of S. commune, in the A complex nine allelic specificities in Aα and 32 in Aß have been calculated to exist in nature (80). Again, homeodomain proteins are encoded by A mating type genes, and the proteins named Y and Z have been analyzed from three allelic Aα specificities (105, 107). One Aß allele was partially sequenced, and there the respective homeodomain protein was called V (100). Activation of Aα occurs through the interaction of Y and Z proteins from different Aα specificities, in which the Y protein has the HD2 motif (100) but also a nuclear localization signal (90). Binding of Y to DNA, however, requires interaction with Z (2). The Y and Z interaction has been shown in protein affinity assays in vitro, and the nuclear localization signal of Y4 fused to GFP was transported into the nucleus when expressed in S. cerevisiae (90). Comparison of homeodomain protein sequences from S. commune with those from U. maydis, Cr. neoformans, C. cinerea, and Pleurotus djamor identifies two classes of homeodomain proteins and includes S. commune Aα Z proteins in HD1 and Aα Y proteins in HD2 homeodomain proteins (5). The structure of the A loci, however, is more complex and rather similar to the situation found in C. cinerea, as the genome sequence reveals additional genes with sequence similarities, which helps to explain the excessive number of allelic specificities. In U. maydis, the b alleles code for two proteins named bE and bW. Each contains a homeodomain-related motif and they, again, constitute an active heterodimeric transcription factor if the two subunits are derived from different allelic specificities (43, 44).
Homeodomain-encoding genes are conserved in both tetrapolar and bipolar basidiomycetes.
The screening of the L. bicolor genome (60) with known HD1 and HD2 sequences from C. cinerea revealed the gene pair Lba1 and Lba2, encoding HD1 and HD2 proteins, respectively (70), without the redundancy observed in the C. cinerea A locus (12). The HD1 and HD2 proteins of L. bicolor have the same conserved motifs identified in corresponding proteins of C. cinerea, suggesting functional similarity. Only one pair of homeodomain genes was also detected in the tetrapolar oyster mushroom, Pleurotus djamor (40), and in the A mating type locus of bipolar Pholiota nameko (122). Another bipolar species, Coprinellus disseminatus, contains multiallelic Aα and Aβ loci, each locus with an HD1 and an HD2 protein-encoding gene (41).
In P. placenta, the genes mip1 and β-fg, known from previous work to flank the A mating type loci in other basidiomycetes (39, 41), were used to identify one pair of HD1- and HD2-encoding genes in both of the two putative A loci located on different scaffolds in the sequenced dikaryotic genome (62). The HD protein sequences show 40 to 50% similarity to other basidiomycete mating type proteins. This represents the level of similarity typical of the A mating type proteins, both between species and between allelic sequences within species. The Aα mating type locus with two homeodomain-encoding genes similar to those in other agaricomycetes and located next to the mip1 gene has also been identified in Ph. chrysosporium (63).
The structure and function of HD-protein pairs of tetrapolar C. cinerea, S. commune, and Pleurotus djamor, as well as those of bipolar Coprinellus disseminatus and Pholiota nameko, are comparable to the organization and function of bE and bW proteins in the tetrapolar basidiomycete U. maydis (44). In all these cases, a high number of allelic variants of HD proteins exist. The multiple pairs of genes encoding HD proteins in the C. cinerea A locus are probably duplications of one original gene pair (45, 51). In S. commune, the evolution of multiple specificities in the A genes awaits detailed analyses, since so far mainly Aα alleles were investigated and very little information is available from genes in the Aβ locus (100). In contrast to the tetrapolar and bipolar species with a high number of allelic variants of HD proteins, the bipolar basidiomycete Cr. neoformans contains only two specificities, MATa and MATα, and the number of homeodomain genes is also lower. The SXI1α gene in MATα encodes an HD1 protein and SXI2a in MATa codes for an HD2 protein (5, 35, 53).
MOLECULAR STRUCTURE OF B MATING TYPE GENES
Pheromones and receptors.
The B mating type genes in S. commune and C. cinerea were shown to code for multiallelic pheromone systems and for receptor systems. The first pheromone- and receptor-containing locus had been identified in the basidiomycete U. maydis, where the two alleles a1 and a2 at the a locus contain the pheromone genes mfa1 and mfa2 and the pheromone receptor genes pra1 and pra2 (9). Both a1 and a2 were shown to be comparable to the genes encoding the lipopeptide a factor, while both receptors share similatity with the Ste3 pheromone receptor in the yeast S. cerevisiae. Comparison of the structure of the cloned alleles of Bα and Bβ loci from S. commune (104, 112, 117), and also that of the B locus from C. cinerea (30, 74), led to detection of pheromone- and receptor-encoding genes also in these multiallelic loci. Here, a more redundant manner is seen than in U. maydis. The S. commune Bα1 and Bβ1 alleles contain both a receptor gene of the S. cerevisiae Ste3 type and several lipopeptide pheromone precursor-encoding genes, respectively. The most complex structure so far found is for the Bβ2 allele, which codes for one receptor and eight pheromones, with three more pheromones probable from the genomic sequence (22).
The Bβ2 allele was of special interest due to the numerous primary and secondary mutations that had been induced earlier to show that reversion of tetrapolar to bipolar mating systems is possible and indicating multiple genes responsible for B mating type function (76, 81, 82). The primary mutations were screened for induction of B-regulated development, thus rendering the mutant functionally bipolar and indicating constitutive signaling without mating (76). Secondary mutations were then induced, suppressing the constitutive phenotype and returning phenotypically to a nonconstitutive situation not exactly the same as that of the wild type but associated with loss of some specificity in tetrapolar mating interactions (81, 82). It was expected that the primary mutations should be in the receptor gene, rendering it constitutively active or responsive to (some) self-pheromones. The respective secondary mutations would then correspond to the loss of (some) pheromone genes or their respective functions (47). Both predictions were proven, since the molecular analysis of known Bβ2 mutants could show corresponding mutations in pheromone- and receptor-encoding genes.
The pheromones contain farnesylation signals making them comparable to the a factor of S. cerevisiae. The small-lipopeptide pheromones result from the posttranslation modification of both N and C termini of the translated amino acid sequence. The pheromone-encoding amino acid sequences are deduced on the basis of the carboxyl terminus sequence CAAX, where C is cysteine, A is aliphatic, and X is any residue. The receptors in basidiomycetes are G-protein-coupled receptors (GPCRs) with seven membrane-passing domains (73, 112, 117, 119). The subclass of fungal, Ste3-like GPCRs is too divergent from rhodopsin-like and other serpentine receptors to allow modeling based on a known X-ray structure (7, 109). Recently, the yeast Ste3p was purified from transfected 293E cells (101), which might in the future allow structural and biochemical analyses of yeast as well as basidiomycete receptors.
Expression of pheromones and receptors from basidiomycetes in yeast.
S. cerevisiae has been used as a heterologous expression system for basidiomycete receptors and pheromones (19, 31). The a mating type cells of S. cerevisiae have been used to express S. commune pheromones, and α mating type cells have been used to express compatible receptors. Culture supernatant of S. commune (31) or cell culture supernatant from yeast cells expressing one of the S. commune pheromone genes (19) has been used to activate a compatible S. commune receptor expressed in yeast. The strong reporter signals obtained by the pheromone receptor interaction in yeast suggested that the activation of the downstream signaling pathway took place via the yeast heterotrimeric G protein and a mitogen-activated protein kinase (MAPK) cascade. This was further confirmed by deletion of the gene STE4 encoding the heterotrimeric G protein β subunit from the yeast host, which lowered dramatically the induction of the reporter system by S. commune pheromones. The deletion of the yeast SST2 gene, involved in desensitizing of the Gα subunit, increased the reporter activity (19). The interaction with the yeast signaling cascade was also improved when the third cytoplasmic loop and proximal C terminus of the heterologous receptor protein known to be involved in heterotrimeric G-protein interaction were mutated for better homology to the yeast receptor protein (31).
The use of the heterologous expression system also made it possible to compare S. commune pheromone modification (19) with yeast a factor biosynthesis (37). The tests with yeast strains carrying either a wild-type or mutated member of the genes belonging to the a factor biosynthesis pathway (37) indicated that S. commune pheromone precursors can be farnesylated by Ram1/Ramp2, and for the removal of the C-terminal signal AAX, endoproteolysis by Rce1 or Ste24 is essential. Ste14 could also play an active role in carboxymethylation of S. commune pheromones. On the other hand, the yeast N-terminal proteases Ste23 and Ax1 seemed not to be required for production of an active S. commune pheromone. The deletion of a factor transporter Ste6p did not prevent the secretion of S. commune pheromones, suggesting an independent export of the heterologous pheromone from yeast (19).
In C. cinerea, the yeast heterologous expression of pheromones and receptors was used to analyze the activity of a mutant constitutive receptor gene by coexpressing a wild-type or a mutant receptor with a compatible pheromone (73). It was also shown that a synthetic pheromone whose structure was deduced on the basis of the C. cinerea pheromone sequence data (12) was able to activate the C. cinerea receptor expressed in yeast (73). Unfortunately, the synthetic pheromone had no effect when applied to growing C. cinerea hyphae, leaving the question about the site of pheromone/receptor interaction in filamentous fungi unanswered.
S. commune Bnull strain.
In addition to heterologous expression in yeast, for S. commune an elegant system is available to screen the receptor/pheromone interactions by expression in a Bnull strain in which a large deletion removes all functions of the B complex (21, 22, 27, 28). The S. commune Bnull strain has been transformed independently with all functionally characterized Bα and Bβ pheromone- and receptor-encoding genes (21). The transformants were then mated with wild-type tester strains representing all Bα and Bβ mating types.
When a receptor is activated by a pheromone of different specificity, a dikaryon can be formed at the side of the induced receptor, providing that the transformant and tester strain had compatible A mating types (21, 27). This assay allowed grouping of the mature pheromones on the basis of the similarity in their 11- to 15-amino-acid sequences into five clusters, each activating a set of Bα or Bβ receptors. Surprisingly, in three pheromone groups, two pheromones interacted with receptors from both Bα and Bβ, which had not been observed before. The closer examination of one of these cases suggested that the Bα8 and Bβ1 receptor proteins are highly similar in structure and that the mature pheromones Bbp2(4) and Bbp2(5) differ by only one amino acid. In the other two pheromone groups activating both Bα and Bβ receptors, the pheromone sequences differed by two amino acids, while the sequence information about identity/similarity of the respective Bα and Bβ receptors is still missing. These observations are extremely intriguing when the evolutionary origin of the multiallelic, multispecific B genes is considered (21, 28, 48, 87).
Mutational analysis of pheromone receptor specificity domains.
The Bnull strain has also been used to functionally characterize chimeric receptors. To identify specificity domains within the receptor proteins, N-terminal, central, and C-terminal parts of Bar1 and Bar2, encoded by Bα1 and Bα2, were interchanged (27). The swapping changed dramatically the properties of the receptors in recognizing different pheromones, either expressed by transformation in the Bnull strain or from a wild-type tester strain for all Bα specificities, and led to identification of new phenotypes for receptors: constitutive, nonspecific (promiscuous), and highly selective (28). The use of one single pheromone at a time from the set of pheromones encoded in the wild-type locus showed that each pheromone has its own profile for induction of different receptors, while at the same time each receptor has a profile of pheromones accepted as ligands for activation of sexual development (22, 28). This was even true for the promiscuous receptors, which are responsive to all nine specificities if mated with wild-type tester strains but which still excluded single pheromones when these were tested one at a time. This did show that the interaction of each single pheromone with a given receptor is dependent on recognition sites within the receptor molecule. Single point mutations at selected sites in the chimeric receptors could prove this theory, since altered specificity or reversion of a constitutive receptor to pheromone dependency was observed upon introduction of a single point mutation (28). This model is in accordance with the hypothesized multistate interaction model of GPCRs with their ligands (13, 25).
Here, a natural discriminatory system has been found that is perfectly explained by interaction of the ligand with different, distant parts of a receptor to determine mating specificity. The correct three-dimensional receptor folding pattern and the ligand-induced conformational change leading to pheromone signaling thus are dependent on multiple interactions of amino acids to allow stabilization of the induced receptor state in the case of a compatible interaction and avoid interacting with nonactivating pheromones. This also might explain why very delicate differences that needed to be balanced in S. commune could not be investigated in yeast, where the expression of S. commune receptor genes generally allowed the investigation of downstream signaling, but the differentiation between specificities was in the end different from the respective answers received when using homologous transformation assays.
Comprehensive structure of B locus alleles.
In S. commune, the sequencing of all pheromones and receptors from nine Bα and nine Bβ alleles is not yet complete. However, the arrangement of the entire B mating type complexes in two strains (Bα3; Bβ2 and Bα3; Bβ3) has been resolved at sequence level, giving interesting material to interpret functional and evolutionary implications. Among these facts are the sequence homologies determining recombination rates between Bα and Bβ (21). The loci have been determined to be linked at 0.6 to 3.5 centimorgans (80), which already indicated a high level of diversity among natural isolates in the structure of respective B loci. This unique complexity can now be revisited using the genome sequence of S. commune strain H4-8.
In C. cinerea, more extensive information on the structure of the B locus is available. The B locus arrangement in C. cinerea consists of three tandemly arranged groups of genes, each group consisting of one receptor-encoding gene and one to three pheromone-encoding genes. The sequencing of 13 known, allelic B mating type specificities of different geographic origin indicated that group 1 is represented by two, group 2 by five, and group 3 by seven alleles (89), allowing the generation of 70 different B mating specificities by recombination between the gene groups. The phylogenetic analysis of C. cinerea receptors divided the proteins into two main clusters, indicating an early duplication of an ancestral gene. Each cluster had members from the three gene groups, suggesting that the receptors in groups 1, 2, and 3 had not developed independently as separate lines but that, instead, genetic material was exchanged followed by diversification. Recently, the C. cinerea Okayama strain 7 genome sequence revealed the receptors of all three groups of genes (89) but also a duplication of the receptor gene from group 2 and two new pheromone genes in group 1 (70). In addition, three putative nonmating type, receptor-like protein-encoding genes were identified, two of them linked to the B locus and the third gene elsewhere in the genome. The receptor genes belonging to the groups 1, 2, and 3 were named CcSTE3.1, CcSTE3.2a (and CcSTE3.2b for the duplicated one), and CcSTE3.3. The nonmating type receptor genes linked to the B locus were named CcSTE3-2151 and CcSTE3-2153 based on the protein identifications (70). It is of interest to speculate on the function of the proteins encoded by the additional receptor genes, which so far have never been detected in any functional test. The genome sequence of S. commune allowed the detection of a similar situation with four new pheromone receptor-like genes, indicating a common trait for mushroom-forming basidiomycetes.
Exploring B mating type genes in newly sequenced agaricomycete genomes.
The analysis of the region containing a cluster of STE3-like sequences in the L. bicolor genome led to identification of three genes encoding Ste3-like receptors, each with a pheromone-encoding gene in close proximity, and two nonmating type receptor genes with no pheromone genes in proximity. The arrangement of the B mating type gene cluster in L. bicolor and C. cinerea turned out to be highly conserved (70). A high number of pheromone-like genes with CAAX motifs at the C terminus occur throughout the L. bicolor genome, and several of them have multiple homologs in the C. cinerea genome. The function of these sequences has not yet been established. Oligoarray analysis in L. bicolor indicated that all the receptor-like genes were expressed but only the three with pheromone genes in close proximity were strongly expressed together with the pheromones in vegetative mycelium, ectomycorrhiza, and fruiting bodies (70).
In the bipolar Ph. chrysosporium and tetrapolar Pleurotus djamor (40), the putative B mating type locus seems to be represented by receptors homologous to CcSTE3.1 and CcSTE3.2, respectively. A pheromone is encoded in close proximity to the Pleurotus djamor receptor, while no pheromone genes were linked to the receptor genes in bipolar Ph. chrysosporium (70). In addition, a gene encoding a pheromone receptor-like protein was also identified in the Ph. chrysosporium genome (70). In P. placenta, the organization of the putative B mating type locus appears to be more complex (62). The B locus is represented by two allelic multimember clusters of receptor- and pheromone-encoding genes in two scaffolds of the dikaryotic genome. The phylogenetic analysis of all Ste3-type GPCRs of P. placenta revealed members of the three subfamilies defined for C. cinerea in addition to one receptor of each haploid genome, which did not group with the others, suggesting a new subtype or function other than mating. A high number of genes encoding putative pheromones occur between the receptors and in the flanking regions in each cluster. Both Ph. chrysosporium and P. placenta are assumed to represent bipolar mating type systems (62, 70), and therefore the role of the B mating type genes in these fungi is still unclear. Perhaps they share the reason for bipolar behavior with Coprinellus disseminatus, where the B factor receptors Ste3.1 and Ste3.2 are not linked to mating type determination but still to function in clamp formation and fruiting (41). On the other hand, in the bipolar basidiomycete Cr. neoformans, the pheromone receptors, Ste3α and Ste3a in MATα and MATa alleles, respectively, are responsible for pheromone signaling and critical for mating specificity (35, 119). Here, the clustering of mating-associated genes led to linkage of pheromone/receptor and homeodomain transcription factor loci.
SIGNALING PATHWAYS ASSOCIATED WITH B MATING TYPE GENES
In agaricomycetes, the downstream components in the signaling pathways regulated by A and B mating type genes have yet to be analyzed in detail. In order to induce sexual development, mates have to respond to the ligand binding of the pheromone receptor via a downstream signaling cascade leading to dikaryotization (Fig. 1 and 2). Therefore, the last part of this review will concentrate on pheromone signaling associated with pheromone response.
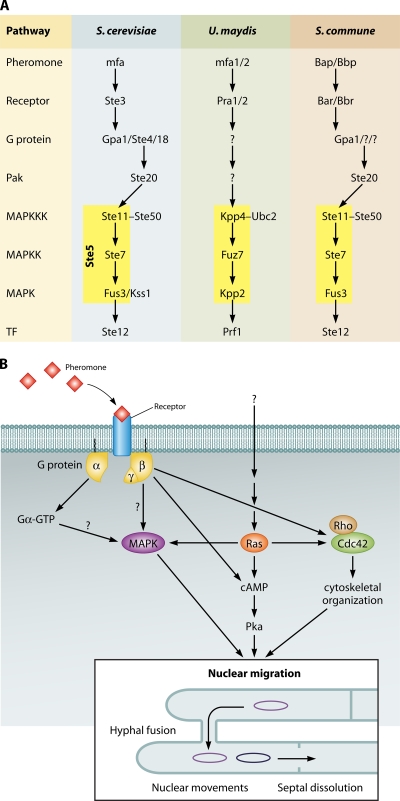
Fig. 2.
Putative signaling components in fungal mating interactions. (A) The well-known components of the signaling pathway associated with mating in the ascomycete S. cerevisiae (78) and in the ustilaginomycete U. maydis (126), as well as the known and putative signaling in the agaricomycete S. commune, are shown. The arrows illustrate the direction of signaling from pheromone/receptor interaction via G protein and MAPK cascade to the transcription factor (TF). Note that in S. cerevisiae the Ste4/18 (βγ) complex interacts with Ste20 (a PAK-like kinase), which then signals further to the MAPK cascade containing the scaffolding protein Ste5. No Ste5 has been found in the S. commune genome, while an ortholog to Ste20 is detected. A homolog to Ste50, involved in mating response, invasive/filamentous growth, and osmotolerance, and acting as an adaptor that links the G protein-associated Cdc42-Ste20 complex to Ste11 in S. cerevisiae, has been identified in U. maydis (Ubc2 [46]) and in S. commune. (B) A hypothetical model of signaling components in the agaricomycete S. commune after G protein activation by pheromone/receptor interaction. The released G protein subunits activate a homologous MAPK cascade. Perceived functions include induction of enzymes necessary for hyphal fusions and septal dissolution, which might also respond to the cAMP-dependent Pka pathway. In addition to the MAPK cascade, Gβγ has been shown to activate Cdc42, thus controlling cytoskeletal rearrangement necessary for nuclear exchange and migration.
The main function of the pheromone/receptor system in S. cerevisiae and U. maydis is to induce the expression of genes that encode proteins involved in attracting and directing the growth of mates toward each other. These proteins also play a role in the fusion of the conjugating cells and establishing an infectious plant pathogenic dikaryon in U. maydis. In agaricomycetes like S. commune and C. cinerea, hyphal fusions, or anastomoses, can occur between all vegetative hyphae and thus are thought to be independent of mating types. Therefore, the pheromone/receptor interaction is assumed to be relevant for events following fusion, specifically in the reciprocal exchange and migration of nuclei between the mates (84).
In the filamentous ascomycete Neurospora crassa, the differences between vegetative and sexual hyphal fusions have been investigated by screening the N. crassa genome for proteins known to play a central role in cell fusion during mating in S. cerevisiae (18, 26). No orthologs for Fus1, Fus2, Fig1, and Fig2 proteins were found (26). In S. cerevisiae, the expression of these proteins located at the tips of shmoos, the conjugating cells necessary for efficient mating, is regulated upon pheromone response. Instead, a pheromone-regulated membrane protein from S. cerevisiae, Prm1 (32), was shown to be part of the general fusion machinery in N. crassa (26). Deletion of the prm1-like gene led to 50% reduction of vegetative cell fusions and caused a complete block of fusion needed for sexual reproduction in the hook cells after fertilization, suggesting that vegetative and sexual hyphal fusions may share functions of the same proteins (17). A preliminary screening of the S. commune genome suggests a comparable situation in this basidiomycete; no orthologs are detected for FUS1, FUS2, FIG1, and FIG2 proteins, while a putative ortholog for Prm1 is found.
In N. crassa, mutations in the heterotrimeric G protein that were necessary for sexual development had no effect on vegetative fusions while effectively blocking sexual development (18). These observations point at the possibility of distinguishing two types of hyphal fusions. In agaricomycetes, this would lead to the question of whether it is possible to identify these two fusion events. One type would represent vegetative anastomoses independent of heterotrimeric G protein, whereas the other type would be hyphal fusions controlled by pheromone and receptor interaction and signaling through heterotrimeric G proteins. For sexually controlled fusion events in agaricomycetes, the fusion of a clamp cell with the peg formed at the surface of the subterminal cell also represents an example.
FUNCTION OF G PROTEINS DURING MATING IN BASIDIOMYCETES
Upon pheromone binding, the heterotrimeric G protein located at the plasma membrane, by virtue of the lipid tails in its Gα and Gγ subunits, will be activated by the ligand-bound GPCR that functions as a guanine exchange factor (GEF) for the Gα subunit. The conformational change associated with GPD to GTP exchange in Gα leads to release of Gα from the receptor and Gβγ subunit from Gα. Both the released Gα-GTP and Gβγ may activate downstream processes, out of which the Gβγ-activated signaling pathway of S. cerevisiae mating is known in detail (78, 102). From this point of view, the activation of downstream signaling in S. cerevisiae with heterologous expression of pheromone receptor and pheromone-encoding genes from S. commune (19, 31) and C. cinerea (73) strongly supports the idea that the respective heterotrimeric G protein is involved in downstream signaling in mating interactions of these fungi.
Structure and function of Gα subunits in mating interactions.
In yeasts and filamentous ascomycetes, two and three Gα subunit-encoding genes are known, respectively (56). Three Gα subunit-encoding genes are also reported to be present in the basidiomycete S. commune (85, 120). Screening of the S. commune genome suggested the existence of an additional Gα gene not reported before. Previously, four Gα subunits had been reported only for U. maydis (88). All these proteins contain the domains typical for Gαs, the GTP-binding and GTPase domains (96). Additionally, the four S. commune Gα proteins share an N-terminal consensus sequence (MGXCXS/MGCXXS) with methionine, glycine, and cysteine separated by one amino acid for myristoylation or directly adjacent for palmitoylation and hence for plasma membrane localization. In C. cinerea, this motif is, however, recognized in only one of the three putative Gαs in GenBank.
For functional analysis, three Gα-protein-encoding genes, GP-A, GP-B, and GP-C, have been mutated in S. commune. The constitutively active Gα proteins were then ectopically expressed in a haploid strain under the promoter of the hydrophobin sc3 gene (120). Unfortunately, the promoter is monokaryon specific, and probably for this reason, no alteration of mating behavior and only a slight reduction of aerial hyphal formation were seen in the homokaryotic transformants with constitutively active ScGP-A and ScGP-C, while the mutated genes blocked fruiting in a dikaryon (120). A corresponding mutation in gpb had no effect on dikaryosis or fruiting. In later studies, it was described that the active gpa and gpc genes elevated intracellular cyclic AMP (cAMP) concentration (121). Interestingly, both proteins share 72% identical amino acids, whereas they show only 46% identity to GP-B.
The S. commune thn1 gene encodes an ortholog to regulators of G protein signaling (RGS), known as Sst2 in S. cerevisiae and FlbA in Aspergillus nidulans (20). The products of these genes regulate the rate of conversion of GTP-bound to GDP-bound Gα, thus controlling the intensity and duration of G protein signaling by working as GTPase activating proteins (GAPs) on the Gα. The thin mutation, as in the constitutively active gpa and gpc genes, reduces the growth of aerial hyphae and, when homoallelic in a dikaryon, inhibits fruiting body formation (20).
In basidiomycetes, it is not yet known which of the known Gαs interacts with pheromone receptors. In S. cerevisiae, Gpa1 is necessary for sexual development and interacts with both S. cerevisiae ligand-bound pheromone receptors, Ste2 and Ste3 (42). Deduced from positive results in heterologous pheromone and receptor expression studies of yeast, pheromone receptors Bbr1 and Bbr2 of S. commune interacted with S. cerevisiae Gpa1 (19, 31). In the heterologous expression of C. cinerea pheromone receptors, unmodified Gpa1p responded only weakly, while Gα chimeras containing the five last amino acids from mammalian Gα16 or from U. maydis Gpa3 performed better (73).
In the basidiomycete Cr. neoformans, two Gα proteins, Gpa2 and Gpa3, are involved in regulating pheromone signaling by interacting with the pheromone receptor (36). Gpa2 promotes mating, and Gpa3 inhibits the interaction. Genetic regulation is also different in that gpa2 expression is induced by pheromone signaling and gpa3 via the nutrient pathway (36). It has been suggested that involvement of two Gα subunits in sexual development could be evolutionary conserved among fungi that express several Gα proteins (36).
Screening of the S. commune genome with known Gα proteins leads to detection of at least 11 Gα-like proteins in addition to the four Gαs with conserved GTP-binding and GTPase domains, further complicating the picture. Similar screening of C. cinerea and L. bicolor genomes also reveals several members of Gα-like proteins. A high number of Gα-like proteins has been reported for the genome of P. placenta (62), The function of these proteins is unknown, and the closer examination of the sequences for GTP binding and GTPase domains shows only weak similarity. The phylogenetic analysis of all putative P. placenta Gαs separated the Gα-like proteins clearly from the Gαs with proper GTP binding and GTPase domains (62).
G protein-associated downstream pathways.
The structure and function of Gβ and Gγ subunits of heterotrimeric G proteins have obtained less attention in agaricomycetes than the Gα subunits, due to the fact that the Gα subunit is thought to be involved in dissociation of the heterotrimeric G protein and in releasing Gβγ for MAPK induction upon pheromone binding (19, 73). However, experimental proof for either Gα or Gβγ signaling is still pending. In the sequenced genomes of C. cinerea, S. commune, and P. placenta, as well as in Lentinula edodes (56), there appears to be only one highly identical (90%) Gβ protein which also is conserved to Gβ subunits of ascomycetes (56). In addition, the genomes of the sequenced agaricomycetes contain several Gβ-like proteins with the typical WD-40 domains. The number of true Gγ-encoding sequences is probably two for agaricomycetes (56), and their amino acid sequence identity is close to 70%. In the S. commune genome, the two Gγs are located close to each other on the same scaffold and share 68% identity.
The mating of S. commune compatible strains leads to strong induction of the expression of pheromone- and receptor-encoding genes (112). A comparable phenomenon is seen in U. maydis, where the pheromone mfa1 transcripts increased when the cells were treated with a compatible a2 pheromone (126). This signaling could be attributed to MAPK- and cAMP-dependent signaling (16, 65, 126). While a MAPK cascade leads to activation of the transcription factor Prf1—which does not show high similarity to Ste12 of S. cerevisiae—cAMP signaling occurs via protein kinase A (Pka). In the ascomycete N. crassa, mating and fruiting body formation also involve two pathways: the MAPK cascade and cAMP signaling with Pka activation (56).
For agaricomycetes, Gα and Gβγ activation of MAPK signaling is expected to lead to phosphorylation of a transcription factor with similarity to S. cerevisiae Ste12 or U. maydis Prf1. The activated transcription factor then will repress/activate target genes necessary for changing the haploid hyphal growth pattern to B-regulated development and, ultimately, to dikaryotic growth and fruiting body development. As for the MAPK pathway, orthologs to the members of the S. cerevisiae MAPK cascade, including Ste20, Ste11, Ste7, Fus3, and Ste12, can be identified in S. commune (Fig. 2A). The complexity of the pheromone response pathway and cross talk with cAMP signaling in the basidiomycete U. maydis (44, 46, 126) prompted a search for other genes potentially involved in sexual reproduction. For cAMP signaling, the genes for Ras, adenylate cyclase, and the regulatory and catalytic subunits of Pka are also recognized in the S. commune genome.
MONOMERIC GTPases Ras AND Cdc42
A high number of small GTPases are suggested to be involved in regulating hyphal growth and morphogenesis (78). Among these proteins, two monomeric GTPases, Ras and Cdc42, have gained special attention. Ras is known to be involved in the regulation of cAMP/Pka signaling (52), which, in turn, has been shown to be involved in fruiting body development in C. cinerea and S. commune (99, 110, 111, 123). Cdc42 functions in pathways linking extracellular growth signals to the actin cytoskeleton, a response often initiated by binding of a signal molecule to a GPCR (95). The Ras and Cdc42 GTPases are conserved molecular switches, which are normally anchored to the plasma membrane by a C-terminal farnesyl or geranyl-geranyl tail. The ability to bind and hydrolyze GTP allows the GTPases to exist either in an active, GTP-bound form or in an inactive, GDP-bound form. Upstream regulatory proteins are involved in the cycling of Ras and Cdc42 between GTP and GPD binding forms and the GTPase-activating proteins (GAPs) facilitate efficient GTP hydrolysis, while guanine nucleotide exchange factors (GEFs) catalyze the exchange of bound GDP for GTP. Nucleotide dissociation inhibitors (GDIs) help to keep the small GTPases in soluble form (15).
Ras proteins.
The function of Ras proteins encoded by two different genes, Ras1 and Ras2, has been extensively analyzed in S. cerevisiae (108), Cr. neoformans (68, 69, 114, 115, 118), and U. maydis (52, 64). Several aspects complicate the understanding of the function of Ras proteins, as Ras gene expression is often controlled by nutritional and environmental factors. Through activation of adenylate cyclase and increased cAMP concentrations, the Pka pathway is induced. However, this pathway is also controlled by a Gα subunit, and thus cross talk has to be considered (113). In Cr. neoformans, Ras1 is involved in regulation of early stages of mating, and it is necessary for pheromone production (115), while the cAMP production and Pka pathway are controlled by Gpa1 (118). In U. maydis, dominant/constitutively active Ras2 changed the growth pattern from yeast-like to filamentous, while dominant active Ras1 elevated pheromone expression, probably via increased cAMP levels (52, 64). Also, the expression of both Ras proteins varies. In Cr. neoformans, Ras1 expression is higher than Ras2 and the protein functions mainly overlap (114). Interestingly, in Cr. neoformans Ras1 is also an effector to Cdc24, the GEF of Cdc42, and in this way is involved in the regulation of actin organization at high temperatures (68, 69). In Schizosaccharomyces pombe with only one Ras gene, Ras protein is activated by two different GEFs, Efc25 and Ste6. Depending on which Ras GEF is binding, different effectors are activated: one couples Ras regulation to the MAPK pathway and the other to Cdc42 signaling (75). In U. maydis, the Ras2 GEF, sql2, with similarity to S. cerevisiae Cdc25, has been isolated as a suppressor of the morphology induced by a dominant/constitutively active gpa3 mutation, which again shows the interaction between Gα and Ras signaling (64).
In S. commune, two ras genes with 45% amino acid identity can be found. Ras1 is about 60% and Ras2 over 80% identical with C. cinerea, L. bicolor (79), and Suillus bovinus Ras1 and Ras2 proteins in GenBank, respectively. While Ras2 awaits functional analysis, constitutively active Ras1 in S. commune (121), as well as the effect of deletion of a Ras-specific Gap gene, gap1 (97), has been investigated. These two mutations of the Ras signaling pathway would be expected to have a similar phenotype, since both mutants lead to accumulation of active, GTP-bound Ras. Indeed, the phenotypes are largely overlapping but not identical. Both mutations led to decreased hyphal growth and a disoriented, curving growth pattern. Homozygous crosses of Δgap1 strains produced a strong phenotype in dikaryons, with a failure in clamp formation. The trapped nucleus in the nonfused clamp-like structure subsequently was rescued by side branch formation originating from the mother cell (97). The growth of the branch and the outgrowing clamp cell seemed attracted toward each other, indicating involvement of pheromone recognition between the two monokaryotic cells. This view is supported by the fact that in unfused clamp cells, pheromone receptor expression is strongly upregulated (E. Kothe, unpublished data).
A third phenotype of gap1 deletions was seen in poorly developed pseudolamellae, altered hymenium formation, and lack of spore production during fruiting body development in homozygous crosses (97). Together, these results indicate that Ras1 has an effect on hyphal growth, clamp connection, and fruiting body development. The existence of the second ras gene in S. commune requires reinvestigation of the effect of Ras proteins on cAMP-, MAPK-, and Rho-dependent signaling and morphogenesis in order to reveal whether the two Ras proteins have different functions, as seen in U. maydis (64), or are more or less redundant in function, as is the case for the two Ras proteins in S. cerevisiae and Cr. neoformans. The characterization of Ras GEFs would also be informative for understanding the specificity of Ras signaling.
Cdc42 and actin.
In the yeast S. cerevisiae, membrane-localized Cdc42 is required for proper bud site selection and for the development of mating projections where Cdc42 links pheromone-dependent Gβγ activation to polarized growth. Cdc42-specific GEF Cdc24 is involved in interaction with several proteins like Far1, Ste20, Bem1, and Bni1 (78). Through these interactions, GTP-bound Cdc42 activates weakly the MAPK cascade, but more importantly, it influences the proper distribution and orientation of the actin cytoskeleton for the tip growth of the conjugating cells. In yeast mating interactions, Far1p appears to play two central roles: it is necessary for cell cycle arrest but also for the polarized growth of the mates.
In S. commune, the ectopically expressed, constitutively active mutant allele of cdc42 regulates branch site selection and subsequent branch development of monokaryotic hyphae (116). In mating interactions, most of the hyphal fusions involve side branches that either grow toward each other or grow toward the hypha of the mate (84). These side branches show a strong actin cytoskeleton (M. Raudaskoski, unpublished data). The tip of the hook at clamp formation also shows actin accumulation during growth and fusion to the subapical cell (92), a process regulated by B mating type genes and Ras (80, 97). A role for Ccdc42 in B-regulated mating interactions can be inferred from poor dikaryotization, along with irregular clamp connections in mating of strains both expressing a constitutively active cdc42 allele (116). Protein(s) should be identified that is needed to link the signal from pheromone/receptor interaction, and potentially also Ras, through Cdc42 to actin. Specifically, the respective GEF that can be postulated to play a central role leading to polarization of the actin cytoskeleton would be of interest.
PHEROMONE RESPONSE AND NUCLEAR MIGRATION
Mating in yeast cells and in U. maydis necessitates synchronization of the mitotic phase prior to the fusion of the conjugating cells. The cell cycle arrest is relevant to allow the nuclear fusion for becoming diploid in S. cerevisiae or to allow the establishment of a dikaryotic hypha waiting for signals to start the pathogenic growth in planta in U. maydis. In S. cerevisiae, the gene responsible for cell cycle arrest at the G1 phase of the mitotic cell cycle is Far1 (78). An ortholog to this gene is not found in the U. maydis genome (126) nor in the S. commune genomes. Instead, in U. maydis, several cell-cycle-associated genes were repressed in a microarray analysis of pheromone response, indicating cell cycle arrest in spite of the absence of Far1. In S. commune, a function in cell cycle arrest might also be envisioned from phenotypic analyses. When intercellular nuclear movements have been followed in living hyphae, or when such movements in fixed hyphae have been labeled for visualization of nuclei and microtubules, synchronous nuclear divisions were always observed in close connection with hyphal fusions (83, 84).
An interesting feature specifically well investigated in S. commune and C. cinerea is the nuclear exchange following hyphal anastomosis of cells with different B mating types (Fig. 1 and 2B). This B-specific nuclear migration was followed by looking for dikaryotization of distal hyphal parts in compatible matings. The intercellular nuclear migration in S. commune was characterized with a speed of 1 to 5 mm min−1 (103), while speeds from 1 to 3 mm h−1 for C. cinerea (49) and as fast as 40 mm h−1 for Coprinellus congregatus (91) were reported. The reports based on cytological observations on nuclear migration indicated that the nuclei move with a speed 10 to 20 times their hyphal growth rate (71). The extensive microtubule tracks seen in migration hyphae around nuclei (84) suggest that microtubule tracks, together with microtubule-associated motors, could be used in intercellular nuclear exchange and migration. Genes for kinesins, performing mainly plus-end-directed transport, and dynein, for minus-end-directed transport, can be identified in the basidiomycete genomes. In U. maydis, 10 kinesin genes and 2 genes encoding the N-terminal and C-terminal parts of one dynein heavy chain have been identified (98). A similar situation with a bipartite dynein gene and several kinesins can also be found in S. commune (E. Kothe and M. Raudaskoski, unpublished data).
In S. cerevisiae, it is reported that a mating-specific Gα, Gpa1, interacts with kinesin-14 (Kar-3), a minus-end-directed microtubule-associated motor, in a pheromone-responsive way (125). The interaction regulates pheromone-induced nuclear migration to the shmoo tip. Gpa1 was immunoprecipitated with Kar3, showing protein-protein interaction, and Gpa1 and Kar3 were visualized at the shmoo tip after pheromone treatment. The use of Gpa1 mutants indicated that Gpa1 affects positively the association of Kar3 to the shmoo tip as well as the dynamics and orientation of microtubules. These results suggest that Gpa1 serves as an externally regulated position determinant anchoring Kar3, a role not previously known for Gα proteins (125).
In S. commune hyphal fusions, not only an actin cytoskeleton but also prominent microtubules extending from the nuclear spindle pole body toward the hyphal tip (84) are seen, reminescent of the nuclear position at the yeast shmoo tip. In S. commune, this nuclear position might thus also be Gα dependent, separating sexual from vegetative hyphal fusions.
At the time of clamp fusion, in addition to actin cytoskeleton (92), strong microtubule tracks are involved in nuclear positioning at the fusion point (84, 86). Thus, there are two B-regulated processes in S. commune with similarities to the Gα-controlled processes in conjugating yeast cells that warrant further investigation of nuclear positioning during hyphal fusion and clamp formation. In animal cells, Gαs activated by ligand binding to a GPCR move from the membrane to the cytosol and bind to tubulin and microtubules, making the microtubule cytoskeleton more dynamic (124). Gα proteins are also suggested to link extracellular signals to microtubule dynamics via Rho GEF and actin-associated formin activity (29). In view of these recent results, coupling the B-regulated gene expression with cytoskeleton rearrangement during mating in basidiomycetes seems likely.
DOWNSTREAM TARGETS FOR A AND B MATING TYPE GENES
Downstream of the major regulators of sexual differentiation, the A and B mating type genes, several target genes have been identified. In C. cinerea, num1 was isolated by REMI mutagenesis and screening for altered nuclear migration. The mutation led to unilateral mating, where the mutant acts as a nuclear donor. The gene num1 encodes a 217-amino-acid protein with leucine zippers at the N and C termini and a coiled-coil domain in the C-terminal half. Its transcription is downregulated by activation of the B-regulated pathway. On the basis of its structure, Num1 is thought to be a part of a larger protein complex (58). In S. commune, differential RNA display identified brt1, which is repressed during B-regulated development, and mat1, upregulated in fully compatible matings. The gene brt1 appears to encode a translation initiation inhibitor (54), while the product of mat1 shows similarity to a peptide transporter (55). These genes are detected in the genome of other agaricomycetes, but no further functional characterization is available.
In C. cinerea, two genes have been shown to be involved in regulating clamp connection formation: pcc1 and clp1 (38, 66). Orthologs to the pcc1 gene are found in other fungal and eukaryotic genomes, while clp1 appears to be restricted to basidiomycetes (94). The gene pcc1 encodes a transcription factor with a HMG box and is suggested to repress A-regulated genes in the homokaryon, while the A pathway activates the expression of clp1 in the dikaryon. The clp1 gene product is thought to repress the expression of pcc1 in the dikaryon, which in turn would allow the transcription of genes necessary for clamp formation (45, 94).
FUTURE PROSPECTS
Present knowledge on the structure and function of mating type genes in fungi has already shown that homeodomain transcription factors are involved in the regulation of mating throughout the fungal kingdom (11). Although the pheromone/receptor system seems to be basically conserved, differences in its regulation, the newly in silico discovered receptor-like proteins, and cross talk of several intracellular signal transduction pathways make the understanding of the origin and function of the pheromone receptor system a challenge, especially in the highly complex systems of agaricomycetes.
In S. cerevisiae (42), Schizosaccharomyces pombe (72), and N. crassa (8), the genes of MAT loci regulate the transcription of pheromone- and receptor-encoding genes in haploid cells. In agaricomycetes, the receptor- and pheromone-encoding genes of the B mating type loci themselves regulate the specificity of the reciprocal exchange and migration of nuclei at the beginning of mating interactions (11), and they are involved in morphogenesis at the stages of clamp formation and fruiting body development. Noteworthy is that during clamp formation the nuclei with different A and B mating type genes become separated from each other, one into the forming clamp cell and the other into the new subapical cell, thus leading to a temporarily monokaryotic stage (Fig. 1), reminiscent of the situation at the beginning of a yeast mating in which pheromone/receptor signaling plays a central role. This allows future investigation of the output of pheromone signaling with a new direction, potentially even with regard to the still completely unanswered question of how the different nuclei might be recognizing each other, organizing themselves into pairs at the beginning of mating, and then maintaining the pairing in the dikaryon (Fig. 1). This leads to another interesting question about the function of both A and B mating type genes in agaricomycetes without clamp connections on dikaryotic hyphae, like Amanita regalis, Suillus bovinus (93), and Tricholoma species (59).
In the pheromone response pathway of agaricomycetes, the deletion or silencing (14, 33, 67) of the heterotrimeric G-protein α subunits would clarify which one is or which ones are interacting with the pheromone receptor. It would also allow functional analyses which could finally show whether G protein activation is regulating hyphal fusions. In contrast, G protein signaling limited to the time after hyphal fusions would then be responsible only for the induction of nuclear movements and breakdown of hyphal cross walls (84) necessary for nuclear exchange and migration between the mates. In addition to gene silencing, it seems necessary and possible now to identify target genes by transcriptome analyses. This will allow a more complete view of the regulatory pathways involved in sexual differentiation in basidiomycetes. The answers that can now be obtained in addition to in silico analyses of the fungal genomes not only will allow an understanding of the tetrapolar versus bipolar mating systems in these fungi but will also link up to general signal transduction pathways common to all fungi and even to all eukaryotes.
ACKNOWLEDGMENTS
This work was supported by a grant from Niemi Foundation (M.R.).
The help with image processing of Fig. 1 given by Mika Keränen to M.R. is highly appreciated. U. Kües is acknowledged for helpful comments on the manuscript. The S. commune genome sequence has been made available by the U.S. Department of Energy Joint Genome Institute supported by the Office of Science (contract no. DE-AC02-05CH11231).
Biographies
Marjatta Raudaskoski received her Ph.D. degree from Turku University in 1973. Her thesis, entitled On the Origin of New Incompatibility Alleles and the Nuclear Migration in the Basidiomycete Schizophyllum commune, included genetics work started with J. R. Raper at Harvard University. Research stays at Tel Aviv University, Israel, and at the University of Guelph, Canada, preceded her appointment as professor in plant physiology and anatomy at Helsinki University in 1982. She learned the tools of molecular biology at J. G. H. Wessels' laboratory, University of Groningen, Netherlands, and at Carlene Raper's laboratory at the University of Vermont. Her research interests aside from fungal genetics and molecular cell biology include ectomycorrhizae and cytoskeletal functions in fungi and pollen tube growth. Now retired, she has moved back to Turku University, where she is integrating fungal genomics with research on signaling pathways and visualization tools for nuclear migration in S. commune.
Erika Kothe studied biology at Philipps-Universität Marburg, Germany. In 1988, she earned her Ph.D. with her thesis entitled The F420-Reducing Hydrogenase from Methanococcus voltae: Purification and Characterization of the Enzyme and Genetic Investigations. After postdoctoral studies in Burlington, Vermont, with R. C. Ullrich and in Groningen, Netherlands, with J. G. H. Wessels, she earned a habilitation in genetics and mycology at Philipps-Universität Marburg for her thesis entitled The Sexual Development of Schizophyllum commune: Molecular Genetics of Mating Specificity. Since 1997, she has been professor of microbial phytopathology at the Friedrich-Schiller-Universität in Jena, Germany. Her research interests are mating type genes and sexual development in basidiomycetes, specifically the white rot fungus S. commune (since 1989); mutually beneficial symbiotic interactions of basidiomycetes in ectomycorrhizae (since 1996); and heavy-metal resistance in fungi and streptomycetes, with application to the improvement of bioremediation (since 2000).
Footnotes
Published ahead of print on 26 February 2010.
REFERENCES
- 1..Alic M., Letzring C., Gold M. H. 1987. Mating system and basidiospore formation in the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl. Environ. Microbiol. 53:1464–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asada Y., Yue C., Wu J., Shen G. P., Novotny C. P., Ullrich R. C. 1997. Schizophyllum commune Aα mating-type proteins, Y and Z, form complexes in all combinations in vitro. Genetics 147:117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asante-Owusu R. N., Banham A. H., Böhnert H. U., Mellor E. J., Casselton L. A. 1996. Heterodimerization between two classes of homeodomain proteins in the mushroom Coprinus cinereus brings together potential DNA-binding and activation domains. Gene 175:25–31 [DOI] [PubMed] [Google Scholar]
- 4.Badalyan S. M., Polak E., Hermann R., Aebi M., Kües U. 2004. Role of peg formation in clamp cell fusion of homobasidiomycete fungi. J. Basic Microbiol. 44:167–177 [DOI] [PubMed] [Google Scholar]
- 5.Bakkeren G., Kämper J., Schirawski J. 2008. Sex in smut fungi: structure, function and evolution of mating-type complexes. Fungal Genet. Biol. 45S:15–21 [DOI] [PubMed] [Google Scholar]
- 6.Banuett F. 1995. Genetics of Ustilago maydis, a fungal pathogen that induces tumors in maize. Annu. Rev. Genet. 29:179–208 [DOI] [PubMed] [Google Scholar]
- 7.Bjarnadóttir T. K., Gloriam D. E., Hellstrand S. H., Kristiansson H., Fredriksson R., Schiöth H. B. 2006. Comprehensive repertoire and phylogenetic analysis of the G protein-coupled receptors in human and mouse. Genomics 88:263–273 [DOI] [PubMed] [Google Scholar]
- 8.Bobrowicz P., Pawlak R., Correa A., Bell-Pedersen D., Ebbole D. J. 2002. The Neurospora crassa pheromone precursor genes are regulated by the mating type locus and the circadian clock. Mol. Microbiol. 45:795–804 [DOI] [PubMed] [Google Scholar]
- 9.Bölker M., Urban M., Kahmann R. 1992. The a mating type locus of U. maydis specifies cell signaling components. Cell 68:441–450 [DOI] [PubMed] [Google Scholar]
- 10.Brown A. J., Casselton L. A. 2001. Mating in mushrooms: increasing the chances but prolonging the affair. Trends Genet. 17:393–400 [DOI] [PubMed] [Google Scholar]
- 11.Casselton L. A. 2008. Fungal sex genes—searching for the ancestors. Bioessays 30:711–714 [DOI] [PubMed] [Google Scholar]
- 12.Casselton L. A., Olesnicky N. S. 1998. Molecular genetics of mating recognition in basidiomycete fungi. Microbiol. Mol. Biol. Rev. 62:55–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christopoulus A., Kenakin T. 2002. G-protein-coupled receptor allosterism and complexing. Pharmacol. Rev. 54:323–374 [DOI] [PubMed] [Google Scholar]
- 14.de Jong J. F., Deelstra H. J., Wösten H. A., Lugones L. G. 2006. RNA-mediated gene silencing in monokaryons and dikaryons of Schizophyllum commune. Appl. Environ. Microbiol. 72:1267–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Etienne-Manneville S., Hall A. 2002. Rho GTPases in cell biology. Nature 420:629–635 [DOI] [PubMed] [Google Scholar]
- 16.Feldbrügge M., Kämper J., Steinberg G., Kahmann R. 2004. Regulation of mating and pathogenic development in Ustilago maydis. Curr. Opin. Microbiol. 7:666–672 [DOI] [PubMed] [Google Scholar]
- 17.Fleissner A., Diamond S., Glass N. L. 2009. The Saccharomyces cerevisiae PRM1 homolog in Neurospora crassa is involved in vegetative and sexual cell fusion events but also has postfertilization functions. Genetics 181:497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleissner A., Simonin A. R., Glass N. L. 2008. Cell fusion in the filamentous fungus, Neurospora crassa. Methods Mol. Biol. 475:21–38 [DOI] [PubMed] [Google Scholar]
- 19.Fowler T. J., DeSimone S. M., Mitton M. F., Kurjan J., Raper C. A. 1999. Multiple sex pheromones and receptors of a mushroom-producing fungus elicit mating in yeast. Mol. Biol. Cell 10:2559–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fowler T. J., Mitton M. F. 2000. Scooter, a new active transposon in Schizophyllum commune, has disrupted two genes regulating signal transduction. Genetics 156:1585–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fowler T. J., Mitton M. F., Rees E. I., Raper C. A. 2004. Crossing the boundary between the Bα and Bß mating-type loci in Schizophyllum commune. Fungal Genet. Biol. 41:89–101 [DOI] [PubMed] [Google Scholar]
- 22.Fowler T. J., Mitton M. F., Vaillancourt L. J., Raper C. A. 2001. Changes in mate recognition through alterations of pheromones and receptors in the multisexual mushroom fungus Schizophyllum commune. Genetics 158:1491–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fraser J. A., Hsueh Y.-P., Findley K., Heitman J. 2008. Evolution of the mating-type locus: the basidiomycetes, p. 19–34 InHeitman J., Kronstad J. W., Taylor J. W., Casselton L. A., (ed.), Sex in fungi ASM Press, Washington, DC [Google Scholar]
- 24.Fries N., Mueller G. M. 1984. Incompatibility systems, cultural features and species circumscriptions in the ectomycorrhizal genus Laccaria (Agaricales). Mycologia 76:633–642 [Google Scholar]
- 25.Gether U. 2000. Uncovering molecular mechanisms involved in activation of G protein-coupled receptors. Endocrinol. Rev. 21:90–113 [DOI] [PubMed] [Google Scholar]
- 26.Glass N. L., Rasmussen C., Roca M. G., Read N. D. 2004. Hyphal homing, fusion and mycelial interconnectedness. Trends Microbiol. 12:135–141 [DOI] [PubMed] [Google Scholar]
- 27.Gola S., Hegner J., Kothe E. 2000. Chimeric pheromone receptors in the basidiomycete Schizophyllum commune. Fungal Genet. Biol. 30:191–196 [DOI] [PubMed] [Google Scholar]
- 28.Gola S., Kothe E. 2003. The little difference: in vivo analysis of pheromone discrimination in Schizophyllum commune. Curr. Genet. 42:276–283 [DOI] [PubMed] [Google Scholar]
- 29.Goulimari P., Knieling H., Engel U., Grosse R. 2008. LARG and mDia1 link Galpha12/13 to cell polarity and microtubule dynamics. Mol. Biol. Cell 19:30–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halsall J. R., Milner M. J., Casselton L. A. 2000. Three subfamilies of pheromone and receptor genes generate multiple B mating specificities in the mushroom Coprinus cinereus. Genetics 154:1115–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hegner J., Siebert-Bartholmei C., Kothe E. 1999. Ligand recognition in multiallelic pheromone receptors from the basidiomycete Schizophyllum commune studied in yeast. Fungal Genet. Biol. 26:190–197 [DOI] [PubMed] [Google Scholar]
- 32.Heiman M. G., Walter P. 2000. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J. Cell Biol. 151:719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heneghan M. N., Costa A. M., Challen M. P., Mills P. R., Bailey A., Foster G. D. 2007. A comparison of methods for successful triggering of gene silencing in Coprinus cinereus. Mol. Biotechnol. 35:283–296 [DOI] [PubMed] [Google Scholar]
- 34.Hibbett D. S., Binder M., Bischoff J. F., Blackwell M., Cannon P. F., Eriksson O. E., Huhndorf S., James T., Kirk P. M., Lücking R., Thorsten Lumbsch H., Lutzoni F., Matheny P. B., McLaughlin D. J., Powell M. J., Redhead S., Schoch C. L., Spatafora J. W., Stalpers J. A., Vilgalys R., Aime M. C., Aptroot A., Bauer R., Begerow D., Benny G. L., Castlebury L. A., Crous P. W., Dai Y. C., Gams W., Geiser D. M., Griffith G. W., Gueidan C., Hawksworth D. L., Hestmark G., Hosaka K., Humber R. A., Hyde K. D., Ironside J. F., Kõljalg U., Kurtzman C. P., Larsson K. H., Lichtwardt R., Longcore J., Miadlikowska J., Miller A., Moncalvo J. M., Mozley-Standridge S., Oberwinkler F., Parmasto E., Reeb V., Rogers J. D., Roux C., Ryvarden L., Sampaio J. P., Schüssler A., Sugiyama J., Thorn R. G., Tibell L., Untereiner W. A., Walker C., Wang Z., Weir A., Weiss M., White M. M., Winka K., Yao Y. J., Zhang N. 2007. A higher-level phylogenetic classification of the fungi. Mycol. Res. 111:509–547 [DOI] [PubMed] [Google Scholar]
- 35.Hsueh Y. P., Heitman J. 2008. Orchestration of sexual reproduction and virulence by the fungal mating-type locus. Curr. Opin. Microbiol. 11:517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsueh Y. P., Xue C., Heitman J. 2007. G protein signaling governing cell fate decisions involves opposing Gα subunits in Cryptococcus neoformans. Mol. Biol. Cell 18:3237–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huyer G., Kistler A., Nouvet F. J., George C. M., Boyle M. L., Michaelis S. 2006. Saccharomyces cerevisiae a-factor mutants reveal residues critical for processing, activity, and export. Eukaryot. Cell 5:1560–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inada K., Morimoto Y., Arima T., Murata Y., Kamada T. 2001. The clp1 gene of the mushroom Coprinus cinereus is essential for A-regulated sexual development. Genetics 157:133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Isaya G., Sakati W. R., Rollins R. A., Shen G. P., Hanson L. C., Ullrich R. C., Novotny C. P. 1995. Mammalian mitochondrial intermediate peptidase: structure/function analysis of a new homologue from Schizophyllum commune and relationship to thimet oligopeptidases. Genomics 28:450–461 [DOI] [PubMed] [Google Scholar]
- 40.James T. Y., Liou S. R., Vilgalys R. 2004. The genetic structure and diversity of the A and B mating-type genes from the tropical oyster mushroom, Pleurotus djamor. Fungal Genet. Biol. 41:813–825 [DOI] [PubMed] [Google Scholar]
- 41.James T. Y., Srivilai P., Kües U., Vilgalys R. 2006. Evolution of the bipolar mating system of the mushroom Coprinellus disseminatus from its tetrapolar ancestors involves loss of mating-type-specific pheromone receptor function. Genetics 172:1877–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson A. D. 1995. Molecular mechanisms of cell-type determination in budding yeast. Curr. Opin. Genet. Dev. 5:552–558 [DOI] [PubMed] [Google Scholar]
- 43.Kahmann R., Romeis T., Bölker M., Kämper J. 1995. Control of mating and development in Ustilago maydis. Curr. Opin. Genet. Dev. 5:559–564 [DOI] [PubMed] [Google Scholar]
- 44.Kahmann R., Schirawski J. 2007. Mating in the smut fungi: from a to b to the downstream cascades, p. 377–387 InHeitman J., Kronstad J. W., Taylor J. W., Casselton L. A. (ed.), Sex in fungi ASM Press, Washington, DC [Google Scholar]
- 45.Kamada T. 2002. Molecular genetics of sexual development in the mushroom Coprinus cinereus. Bioessays 24:449–459 [DOI] [PubMed] [Google Scholar]
- 46.Klosterman S. J., Martinez-Espinoza A. D., Andrews D. L., Seay J. R., Gold S. E. 2008. Ubc2, an ortholog of the yeast Ste50p adaptor, possesses a basidiomycete-specific carboxy terminal extension essential for pathogenicity independent of pheromone response. Mol. Plant Microbe Interact. 21:110–121 [DOI] [PubMed] [Google Scholar]
- 47.Kothe E. 1999. Mating types and pheromone recognition in the homobasidiomycete Schizophyllum commune. Fungal Genet. Biol. 27:146–152 [DOI] [PubMed] [Google Scholar]
- 48.Kothe E., Gola S., Wendland J. 2003. Evolution of multispecific mating-type alleles for pheromone perception in the homobasidiomycete fungi. Curr. Genet. 42:268–275 [DOI] [PubMed] [Google Scholar]
- 49.Kües U. 2000. Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol. Mol. Biol. Rev. 64:316–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kües U., Asante-Owusu R. N., Mutasa E. S., Tymon A. M., Pardo E. H., O'Shea S. H., Göttgens B., Casselton L. A. 1994. Two classes of homeodomain proteins specify the multiple a mating types of the mushroom Coprinus cinereus. Plant Cell 6:1467–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kües U., Richardson W. V., Tymon A. M., Mutasa E. S., Göttgens B., Gaubatz S., Gregoriades A., Casselton L. A. 1992. The combination of dissimilar alleles of the Aα and Aβ gene complexes, whose proteins contain homeo domain motifs, determines sexual development in the mushroom Coprinus cinereus. Genes Dev. 6:568–577 [DOI] [PubMed] [Google Scholar]
- 52.Lee N., Kronstad J. W. 2002. Ras2 controls morphogenesis, pheromone response, and pathogenicity in the fungal pathogen Ustilago maydis. Eukaryot. Cell 1:954–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lengeler K. B., Fox D. S., Fraser J. A., Allen A., Forrester K., Dietrich F. S., Heitman J. 2002. Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot. Cell 1:704–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lengeler K. B., Kothe E. 1999a. Identification and characterization of brt1, a gene down-regulated during B-regulated development in Schizophyllum commune. Curr. Genet. 35:551–556 [DOI] [PubMed] [Google Scholar]
- 55.Lengeler K. B., Kothe E. 1999b. Mated: a putative peptide transporter of Schizophyllum commune expressed in dikaryons. Curr. Genet. 36:159–164 [DOI] [PubMed] [Google Scholar]
- 56.Li L., Wright S. J., Krystofova S., Park G., Borkovich K. A. 2007. Heterotrimeric G protein signaling in filamentous fungi. Annu. Rev. Microbiol. 61:423–452 [DOI] [PubMed] [Google Scholar]
- 57.Loftus B. J., Fung E., Roncaglia P., Rowley D., Amedeo P., Bruno D., Vamathevan J., Miranda M., Anderson I. J., Fraser J. A., Allen J. E., Bosdet I. E., Brent M. R., Chiu R., Doering T. L., Donlin M. J., D'Souza C. A., Fox D. S., Grinberg V., Fu J., Fukushima M., Haas B. J., Huang J. C., Janbon G., Jones S. J., Koo H. L., Krzywinski M. I., Kwon-Chung J. K., Lengeler K. B., Maiti R., Marra M. A., Marra R. E., Mathewson C. A., Mitchell T. G., Pertea M., Riggs F. R., Salzberg S. L., Schein J. E., Shvartsbeyn A., Shin H., Shumway M., Specht C. A., Suh B. B., Tenney A., Utterback T. R., Wickes B. L., Wortman J. R., Wye N. H., Kronstad J. W., Lodge J. K., Heitman J., Davis R. W., Fraser C. M., Hyman R. W. 2005. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307:1321–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Makino R., Kamada T. 2004. Isolation and characterization of mutations that affect nuclear migration for dikaryosis in Coprinus cinereus. Curr. Genet. 45:149–156 [DOI] [PubMed] [Google Scholar]
- 59.Mankel A., Krause K., Kothe E. 2002. Identification of a hydrophobin gene that is developmentally regulated in the ectomycorrhizal fungus Tricholoma terreum. Appl. Environ. Microbiol. 68:1408–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martin F., Aerts A., Ahrén D., Brun A., Danchin E. G., Duchaussoy F., Gibon J., Kohler A., Lindquist E., Pereda V., Salamov A., Shapiro H. J., Wuyts J., Blaudez D., Buée M., Brokstein P., Canbäck B., Cohen D., Courty P. E., Coutinho P. M., Delaruelle C., Detter J. C., Deveau A., DiFazio S., Duplessis S., Fraissinet-Tachet L., Lucic E., Frey-Klett P., Fourrey C., Feussner I., Gay G., Grimwood J., Hoegger P. J., Jain P., Kilaru S., Labbé J., Lin Y. C., Legué V., Le Tacon F., Marmeisse R., Melayah D., Montanini B., Muratet M., Nehls U., Niculita-Hirzel H., Oudot-Le Secq M. P., Peter M., Quesneville H., Rajashekar B., Reich M., Rouhier N., Schmutz J., Yin T., Chalot M., Henrissat B., Kües U., Lucas S., Van de Peer Y., Podila G. K., Polle A., Pukkila P. J., Richardson P. M., Rouzé P., Sanders I. R., Stajich J. E., Tunlid A., Tuskan G., Grigoriev I. V. 2008. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature 452:88–9257. [DOI] [PubMed] [Google Scholar]
- 61.Martin F., Selosse M. A. 2008. The Laccaria genome: a symbiont blueprint decoded. New Phytol. 180:296–310 [DOI] [PubMed] [Google Scholar]
- 62.Martinez D., Challacombe J., Morgenstern I., Hibbett D., Schmoll M., Kubicek C. P., Ferreira P., Ruiz-Duenas F. J., Martinez A. T., Kersten P., Hammel K. E., Vanden Wymelenberg A., Gaskell J., Lindquist E., Sabat G., Bondurant S. S., Larrondo L. F., Canessa P., Vicuna R., Yadav J., Doddapaneni H., Subramanian V., Pisabarro A. G., Lavín J. L., Oguiza J. A., Master E., Henrissat B., Coutinho P. M., Harris P., Magnuson J. K., Baker S. E., Bruno K., Kenealy W., Hoegger P. J., Kües U., Ramaiya P., Lucas S., Salamov A., Shapiro H., Tu H., Chee C. L., Misra M., Xie G., Teter S., Yaver D., James T., Mokrejs M., Pospisek M., Grigoriev I. V., Brettin T., Rokhsar D., Berka R., Cullen D. 2009. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanism of lignocellulose conversion. Proc. Natl. Acad. Sci. U. S. A. 106:1954–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martinez D., Larrondo L. F., Putnam N., Gelpke M. D., Huang K., Chapman J., Helfenbein K. G., Ramaiya P., Detter J. C., Larimer F., Coutinho P. M., Henrissat B., Berka R., Cullen D., Rokhsar D. 2004. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat. Biotechnol. 22:695–700 [DOI] [PubMed] [Google Scholar]
- 64.Müller P., Katzenberger J. D., Loubradou G., Kahmann R. 2003. Guanyl nucleotide exchange factor Sql2 and Ras2 regulate filamentous growth in Ustilago maydis. Eukaryot. Cell 2:609–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Müller P., Leibbrandt A., Teunissen H., Cubasch S., Aichinger C., Kahmann R. 2004. The Gß-subunit-encoding gene bpp1 controls cyclic-AMP signaling in Ustilago maydis. Eukaryot. Cell 3:806–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murata Y., Fujii M., Zolan M. E., Kamada T. 1998. Molecular analysis of pcc1, a gene that leads to A-regulated sexual morphogenesis in Coprinus cinereus. Genetics 149:1753–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Namekawa S. H., Iwabata K., Sugawara H., Hamada F. N., Koshiyama A., Chiku H., Kamada T., Sakaguchi K. 2005. Knockdown of LIM15/DMC1 in the mushroom Coprinus cinereus by double-stranded RNA-mediated gene silencing. Microbiology 151:3669–3678 [DOI] [PubMed] [Google Scholar]
- 68.Nichols C. B., Ferreyra J., Ballou E. R., Alspaugh J. A. 2009. Subcellular localization directs signaling specificity of the Cryptococcus neoformans Ras1 protein. Eukaryot. Cell 8:181–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nichols C. B., Perfect Z. H., Alspaugh J. A. 2007. A Ras1-Cdc24 signal transduction pathway mediates thermotolerance in the fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 63:118–1130 [DOI] [PubMed] [Google Scholar]
- 70.Niculita-Hirzel H., Labbé J., Kohler A., le Tacon F., Martin F., Sanders I. R., Kües U. 2008. Gene organization of the mating type regions in the ectomycorrhizal fungus Laccaria bicolor reveals distinct evolution between the two mating type loci. New Phytol. 180:329–342 [DOI] [PubMed] [Google Scholar]
- 71.Niederpruem D. J. 1980. Direct studies of dikaryotization in Schizophyllum commune. I. Live inter-cellular nuclear migration patterns. Arch. Microbiol. 128:172–178 [DOI] [PubMed] [Google Scholar]
- 72.Nielsen O., Davey J. 1995. Pheromone communication in the fission yeast Schizosaccharomyces pombe. Semin. Cell Biol. 6:95–104 [DOI] [PubMed] [Google Scholar]
- 73.Olesnicky N. S., Brown A. J., Dowell S. J., Casselton L. A. 1999. A constitutively active G-protein-coupled receptor causes mating self-compatibility in the mushroom Coprinus. EMBO J. 18:2756–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.O'Shea S. F., Chaure P. T., Halsall J. R., Olesnicky N. S., Leibbrandt A., Connerton I. F., Casselton L. A. 1998. A large pheromone and receptor gene complex determines multiple B mating type specificities in Coprinus cinereus. Genetics 148:1081–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Papadaki P., Pizon V., Onken B., Chang E. C. 2002. Two ras pathways in fission yeast are differentially regulated by two ras guanine nucleotide exchange factors. Mol. Cell. Biol. 22:4598–4606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Parag Y. 1962. Mutations in the B incompatibility factor of Schizophyllum commune. Proc. Natl. Acad. Sci. U. S. A. 48:743–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pardo E. H., O'Shea S. F., Casselton L. A. 1996. Multiple versions of the A mating type locus of Coprinus cinereus are generated by three paralogous pairs of multiallelic homeobox genes. Genetics 144:87–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Park H. O., Bi E. 2007. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 71:48–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rajashekar B., Kohler A., Johansson T., Martin F., Tunlid A., Ahrén D. 2009. Expansion of signal pathways in the ectomycorrhizal fungus Laccaria bicolor—evolution of nucleotide sequences and expression patterns in families of protein kinases and RAS small GTPases. New Phytol. 183:365–379 [DOI] [PubMed] [Google Scholar]
- 80.Raper J. 1966. Sexuality of higher fungi, p. 1–283 The Roland Press, New York, NY [Google Scholar]
- 81.Raper C. A., Raper J. R. 1973. Mutational analysis of a regulatory gene for morphogenesis in Schizophyllum. Proc. Natl. Acad. Sci. U. S. A. 70:1427–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Raper J. R., Raudaskoski M. 1968. Secondary mutations at the Bβ incompatibility locus of Schizophyllum commune. Heredity 23:109–117 [Google Scholar]
- 83.Raudaskoski M. 1973. Light and electron microscope study of unilateral mating between a secondary mutant and a wild-type strain of Schizophyllum commune. Protoplasma 76:35–48 [DOI] [PubMed] [Google Scholar]
- 84.Raudaskoski M. 1998. The relationship between B mating type genes and nuclear migration in Schizophyllum commune. Fungal Genet. Biol. 24:207–227 [DOI] [PubMed] [Google Scholar]
- 85.Raudaskoski M., Pardo A. G., Tarkka M. T., Gorfer M., Hanif M., Laitiainen E. 2001. Small GTPases, cytoskeleton and signal transduction in filamentous homobasidiomycetes, p. 123–136 InGeitman A., Cresti M., Heath I. B. (ed.), Cell biology of plant and fungal tip growth. NATO Science Series I: life and behavioural sciences, vol. 328 IOS Press, Washington, DC [Google Scholar]
- 86.Raudaskoski M., Rupeš I., Timonen S. 1991. Immunofluorescence microscopy of the cytoskeleton in filamentous fungi after quick-freezing and low-temperature fixation. Exp. Mycol. 15:167–173 [Google Scholar]
- 87.Raudaskoski M., Stamberg J., Bawnik N., Koltin Y. 1976. Mutational analysis of natural alleles at the B incompatibility factor of Schizophyllum commune: α2 and ß6. Genetics 83:507–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Regenfelder E., Spellig T., Hartmann A., Lauenstein S., Bölker M., Kahmann R. 1997. G proteins in Ustilago maydis: transmission of multiple signals? EMBO J. 16:1934–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Riquelme M., Challen M. P., Casselton L. A., Brown A. J. 2005. The origin of multiple B mating specificities in Coprinus cinereus. Genetics 170:1105–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Robertson C. I., McMahon Kende A., Toenjes K., Novotny C. P., Ullrich R. C. 2002. Evidence for interaction of Schizophyllum commune Y mating-type proteins in vivo. Genetics 160:1461–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ross I. 1976. Nuclear migration rates in Coprinus congregatus: a new record? Mycologia 68:418–422 [Google Scholar]
- 92.Runeberg P., Raudaskoski M., Virtanen I. 1986. Cytoskeletal elements in the hyphae of the homobasidiomycete Schizophyllum commune visualized with indirect immunofluorescence and NBD-phallacidin. Eur. J. Cell Biol. 41:25–32 [Google Scholar]
- 93.Salo V., Niini S. S., Virtanen I. I., Raudaskoski M. 1989. Comparative immunocytochemistry of the cytoskeleton in filamentous fungi with dikaryotic and multinucleate hyphae. J. Cell Sci. 94:11–24 [Google Scholar]
- 94.Scherer M., Heimel K., Starke V., Kämper J. 2006. The Clp1 protein is required for clamp formation and pathogenic development of Ustilago maydis. Plant Cell 18:2388–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schmidt A., Hall M. N. 1998. Signaling to the actin cytoskeleton. Annu. Rev. Cell Dev. Biol. 14:305–338 [DOI] [PubMed] [Google Scholar]
- 96.Schmoll M. 2008. The information highways of a biotechnological workhorse-signal transduction in Hypocrea jecorina. BMC Genomics 9:1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schubert D., Raudaskoski M., Knabe N., Kothe E. 2006. Ras GTPase-activating protein Gap1 of the homobasidiomycete Schizophyllum commune regulates hyphal growth orientation and sexual development. Eukaryot. Cell 5:683–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schuchardt I., Assmann D., Thines E., Schuberth C., Steinberg G. 2005. Myosin-V, kinesin-1, and kinesin-3 cooperate in hyphal growth of the fungus Ustilago maydis. Mol. Biol. Cell 16:5191–5201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schwalb M. N. 1974. Effect of adenosine 3′,5′-cyclic monophosphate on the morphogenesis of fruit bodies of Schizophyllum commune. Arch. Mikrobiol. 96:17–20 [DOI] [PubMed] [Google Scholar]
- 100.Shen G. P., Park D. C., Ullrich R. C., Novotny C. P. 1996. Cloning and characterization of a Schizophyllum gene with Aβ6 mating-type activity. Curr. Genet. 29:136–142 [DOI] [PubMed] [Google Scholar]
- 101.Shi C., Kaminskyj S., Caldwell S., Loewen M. C. 2007. A role for a complex between activated G protein-coupled receptors in yeast cellular mating. Proc. Natl. Acad. Sci. U. S. A. 104:5395–5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Slessareva J. E., Dohlman H. G. 2006. G protein signaling in yeast: new components, new connections, new compartments. Science 314:1412–1413 [DOI] [PubMed] [Google Scholar]
- 103.Snider P. J. 1968. Nuclear movements in Schizophyllum. Symp. Soc. Exp. Biol. 22:261–383 [PubMed] [Google Scholar]
- 104.Specht C. A. 1995. Isolation of the Bα and Bβ mating-type loci of Schizophyllum commune. Curr. Genet. 28:374–379 [DOI] [PubMed] [Google Scholar]
- 105.Specht C. A., Stankis M. M., Giasson L., Novotny C. P., Ullrich R. C. 1992. Functional analysis of the homeodomain-related proteins of the Aα locus of Schizophyllum commune. Proc. Natl. Acad. Sci. U. S. A. 89:7174–7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Spit A., Hyland R. H., Mellor E. J., Casselton L. A. 1998. A role for heterodimerization in nuclear localization of a homeodomain protein. Proc. Natl. Acad. Sci. U. S. A. 95:6228–6233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stankis M. M., Specht C. A., Yang H., Giasson L., Ullrich R. C., Novotny C. P. 1992. The Aα mating locus of Schizophyllum commune encodes two dissimilar multiallelic homeodomain proteins. Proc. Natl. Acad. Sci. U. S. A. 89:7169–7173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Toda T., Uno I., Ishikawa T., Powers S., Kataoka T., Broek D., Cameron S., Broach J., Matsumoto K., Wigler M. 1985. In yeast, Ras proteins are controlling elements of adenylate cyclase. Cell 40:27–36 [DOI] [PubMed] [Google Scholar]
- 109.Topiol S., Sabio M. 2009. X-ray structure breakthroughs in the GPCR transmembrane region. Biochem. Pharmacol. 78:11–20 [DOI] [PubMed] [Google Scholar]
- 110.Uno I., Ishikawa T. 1976. Effect of cyclic AMP on glycogen phosphorylase in Coprinus macrorhizus. Biochim. Biophys. Acta 452:112–120 [DOI] [PubMed] [Google Scholar]
- 111.Uno I., Yamaguchi M., Ishikawa T. 1974. The effect of light on fruiting body formation and adenosine 3′:5′-cyclic monophosphate metabolism in Coprinus macrorhizus. Proc. Natl. Acad. Sci. U. S. A. 71:479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vaillancourt L. J., Raudaskoski M., Specht C. A., Raper C. A. 1997. Multiple genes encoding pheromones and a pheromone receptor define the Bβ1 mating-type specificity in Schizophyllum commune. Genetics 146:541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang Y., Pierce M., Schneper L., Güldal C. G., Zhang A., Tavazoie S., Broach J. R. 2004. Ras and Gpa2 mediate one branch of a redundant glucose signaling pathway in yeast. PLoS Biol. 2:610–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Waugh M. S., Nichols C. B., DeCesare C. M., Cox G. M., Heitman J., Alspaugh J. A. 2002. Ras1 and Ras2 contribute shared and unique roles in physiology and virulence of Cryptococcus neoformans. Microbiology 148:191–201 [DOI] [PubMed] [Google Scholar]
- 115.Waugh M. S., Vallim M. A., Heitman J., Alspaugh J. A. 2003. Ras1 controls pheromone expression and response during mating in Cryptococcus neoformans. Fungal Genet. Biol. 38:110–121 [DOI] [PubMed] [Google Scholar]
- 116.Weber M., Salo V., Uuskallio M., Raudaskoski M. 2005. Ectopic expression of a constitutively active Cdc42 small GTPase alters the morphology of haploid and dikaryotic hyphae in the filamentous homobasidiomycete Schizophyllum commune. Fungal Genet. Biol. 42:624–637 [DOI] [PubMed] [Google Scholar]
- 117.Wendland J., Vaillancourt L. J., Hegner J., Lengeler K. B., Laddison K. J., Specht C. A., Raper C. A., Kothe E. 1995. The mating-type locus Bα1 of Schizophyllum commune contains a pheromone receptor gene and putative pheromone genes. EMBO J. 14:5271–5278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xue C., Bahn Y. S., Cox G. M., Heitman J. 2006. G protein-coupled receptor Gpr4 senses amino acids and activates the cAMP-PKA pathway in Cryptococcus neoformans. Mol. Biol. Cell 17:667–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xue C., Hsueh Y. P., Heitman J. 2008. Magnificent seven: roles of G protein-coupled receptors in extracellular sensing in fungi. FEMS Microbiol. Rev. 32:1010–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yamagishi K., Kimura T., Suzuki M., Shinmoto H. 2002. Suppression of fruit-body formation by constitutively active G-protein α-subunits ScGP-A and ScGP-C in the homobasidiomycete Schizophyllum commune. Microbiology 148:2797–2809 [DOI] [PubMed] [Google Scholar]
- 121.Yamagishi K., Kimura T., Suzuki M., Shinmoto H., Yamaki K. J. 2004. Elevation of intracellular cAMP levels by dominant active heterotrimeric G protein alpha subunits ScGP-A and ScGP-C in homobasidiomycete, Schizophyllum commune. Biosci. Biotechnol. Biochem. 68:1017–1026 [DOI] [PubMed] [Google Scholar]
- 122.Yi R., Tachikawa T., Ishikawa M., Mukaiyama H., Bao D., Aimi T. 2008. Genomic structure of the A mating-type locus in a bipolar basidiomycete, Pholiota nameko. Mycol. Res. 113:240–248 [DOI] [PubMed] [Google Scholar]
- 123.Yli-Mattila T. 1987. The effect of UV-light on cAMP level in the basidiomycete Schizophyllum commune. Physiol. Plant. 69:451–455 [Google Scholar]
- 124.Yu J. Z., Dave R. H., Allen J. A., Sarma T., Rasenick M. M. 2009. Cytosolic Gαs acts as an intracellular messenger to increase microtubule dynamics and promote neurite outgrowth. J. Biol. Chem. 284:10462–10472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zaichick S. V., Metodiev M. V., Nelson S. A., Durbrovskyi O., Draper E., Cooper J. A., Stone D. E. 2009. The mating-specific Gα interacts with a kinesin-14 and regulates pheromone-induced nuclear migration in budding yeast. Mol. Biol. Cell 20:2820–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zarnack K., Eichhorn H., Kahmann R., Feldbrügge M. 2008. Pheromone-regulated target genes respond differentially to MAPK phosphorylation of transcription factor Prf1. Mol. Microbiol. 69:1041–1053 [DOI] [PubMed] [Google Scholar]