Abstract
African trypanosomes express a family of dually acylated, EF-hand calcium-binding proteins called the calflagins. These proteins associate with lipid raft microdomains in the flagellar membrane, where they putatively function as calcium signaling proteins. Here we show that these proteins bind calcium with high affinity and that their expression is regulated during the life cycle stage of the parasite, with protein levels approximately 10-fold higher in the mammalian bloodstream form than in the insect vector procyclic stage. We also demonstrate a role for the calflagins in mammalian infection, as inhibition of the entire calflagin family by RNA interference dramatically increased host survival and attenuated parasitemia in a mouse model of sleeping sickness. In contrast to infection with parental wild-type parasites, which demonstrated an unremitting parasitemia and death within 6 to 10 days, infection with calflagin-depleted parasites demonstrated prolonged survival associated with a sudden decrease in parasitemia at approximately 8 days postinfection. Subsequent relapsing and remitting waves of parasitemia thereafter were associated with alternate expression of the variant surface glycoprotein, suggesting that initial clearance was antigen specific. Interestingly, despite the notable in vivo phenotype and flagellar localization of the calflagins, in vitro analysis of the calflagin-deficient parasites demonstrated normal proliferation, flagellar motility, and morphology. Further analysis of the kinetics of surface antibody clearance also did not demonstrate a deficit in the calflagin-deficient parasites; thus, the molecular basis for the altered course of infection is independent of an effect on parasite cell cycle progression, motility, or degradation of surface-bound antibodies.
The protozoan parasite Trypanosoma brucei is the causative agent of African sleeping sickness, a fatal disease endemic to regions throughout sub-Saharan Africa. Incidence rates of human T. brucei infection have risen dramatically in the past 30 to 50 years, leading to renewed emphasis by the World Health Organization on disease surveillance and control among the millions of people at risk of infection. T. brucei also infects cattle to cause nagana, a disease which renders vast regions unsuitable for livestock and poses a major barrier to economic development in afflicted areas (30).
T. brucei is transmitted to its mammalian host via the bite of the infected tsetse fly, when parasites are introduced into the host circulation during a blood meal. To thrive in both the insect vector and the mammalian host, T. brucei has evolved digenetic life cycle stages; the two most commonly studied life cycle stages are the procyclic stage from the fly midgut and the bloodstream form found in the mammalian host. Programmed differentiation between these stages regulates broad aspects of parasite biology, enabling adaptation to either environment. In the mammalian host, avoiding clearance by the humoral immune response is particularly important, and T. brucei has evolved sophisticated mechanisms to this end. Bloodstream form parasites are covered by a thick monolayer of variant surface glycoprotein (VSG) that blocks the access of host antibodies to underlying invariant antigens (2). VSG is highly immunogenic, and VSG-specific antibodies facilitate the clearance of parasites from the blood. However, the parasite undergoes antigenic variation, a process whereby the parasite spontaneously switches from the expression of a single VSG type to that of another of the hundreds in its genomic repertoire. The new parasite clone is resistant to the existing antibodies and persists until antibodies to the new VSG are produced, thus selecting for another antigen variant and propagating the cycle. T. brucei has coevolved with primates for millions of years, and antigenic variation is not its sole means of immune evasion. Additional mechanisms such as host immunosuppression (20), motility-driven internalization and degradation of surface-bound antibodies (13, 22), and the shedding of VSG molecules (7) are each likely to contribute to the survival of T. brucei in the hostile environment of its mammalian host. However, the signaling and genetic pathways by which T. brucei regulates stage-specific adaptations to its environment remain poorly understood.
Calcium signaling plays critical roles in virtually every eukaryotic cell type, and trypanosomes are no exception. Regulated changes in intracellular calcium are important for trypanosome replication, differentiation, cell invasion, and virulence (24). The importance of calcium regulation in trypanosomes is further underscored by the presence of a specialized organelle, the acidocalcisome, which contains a major intracellular reservoir of calcium (9). Cellular responses to calcium fluctuations are mediated by calcium-binding proteins. In addition to calmodulin, T. brucei expresses a family of EF-hand proteins named the calflagins. They were discovered as the predominant T. brucei proteins to bind a hydrophobic resin in a calcium-dependent manner (35, 36). These proteins specifically localize to the flagellum, an organelle which, in addition to its obvious role in cellular motility, compartmentalizes signaling proteins, including adenylate cyclases and phosphodiesterases (26, 27). In the flagellum, calflagins associate with lipid raft microdomains, which in many cell types serve as recruitment platforms for signaling molecules (32). We have recently elucidated the role of acylation in calflagin trafficking and raft association and identified the calflagin-specific palmitoyl acyltransferase (11). However, the precise functions of these abundant proteins remain unknown. The calflagins show homology with Trypanosoma cruzi FCaBP, an immunodominant protein with similar flagellar enrichment yet an unidentified biologic function (12). Outside of the common EF-hand domains, the calflagins display only minimal homology to proteins in organisms outside the kinetoplastid lineage.
To elucidate the function of T. brucei calflagins, we assessed the consequences of calflagin inhibition to T. brucei in vivo during host infection and in vitro. We can now report for the first time that calflagin expression influences the outcome of parasite infection, as mice infected with calflagin-deficient cells demonstrate a sudden decrease in parasitemia approximately 1 week postinfection. This drop is associated with prolonged host survival and outgrowth of parasites expressing alternative VSG molecules, indicating selective pressure against the initial dominant VSG molecule and suggesting that the primary deficit of calflagin-deficient cells is one of enhanced sensitivity to the host adaptive immune response. We demonstrate that the molecular mechanisms underlying this effect are not due to altered parasite proliferation, motility, or clearance of surface-bound antibodies.
MATERIALS AND METHODS
Trypanosomes.
All of the T. brucei strains described in this study are derivatives of the procyclic 29-13 strain or the bloodstream form “single-marker” strain (34), both of which were originally derived from Lister strain 427 (8, 10) to coexpress the bacterial T7 polymerase and tetracycline repressor to enable drug-inducible transcription. Procyclic-stage parasites were cultured at 27°C with 3% CO2 in SDM-79 medium (5) supplemented with 10% fetal bovine serum (FBS; Gemini Bio-Products, West Sacramento, CA), 7.5 mg/liter hemin (Sigma, St. Louis, MO), 100 U/ml penicillin/streptomycin (Invitrogen, Carlsbad, CA), 50 μg/ml hygromycin (Invitrogen), and 15 μg/ml G418 (Invitrogen). Bloodstream form parasites were cultured at 37°C with 5% CO2 in HMI-9 medium (16) supplemented with 10% FBS, 10% serum plus medium complement (SAFC Biosciences),100 U/ml penicillin/streptomycin, and 15 μg/ml G418. Transfectants were cultured under continuous drug selection with 2.5 μg/ml phleomycin (Invivogen, San Diego, CA).
Calcium-binding analysis.
Recombinant proteins were purified from BL21 cells expressing 6×His-tagged fusions from the pET23b vector (Novagen, San Diego, CA) and analyzed by a calcium overlay assay (21). Equimolar amounts of protein were boiled for 7 min in 1× Laemmli sample buffer, fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to nitrocellulose membranes. The membranes were washed three times for 20 min each in overlay buffer (5 mM MgCl2, 60 mM KCl, 10 mM imidazole-HCl, pH 6.8). Membranes were then either stained with Ponceau S or soaked in overlay buffer plus 1 mCi/liter 45CaCl2 (Perkin-Elmer, Wellesley, MA) for 10 min, rinsed with 50% ethanol for 5 min, dried, and exposed to autoradiography film for 14 h.
Generation of calflagin RNA interference (RNAi) lines.
The RNAi target sequence was selected on the basis of its conservation among the calflagin genes. A lack of similarity to off-target genes was verified using the RNAit software (29). The target sequence was amplified from genomic DNA isolated as previously described (23), using primers 5′-CTCGAGCATTGTCCAGCGTGCGTTTGA-3′ and 5′-AAGCTTGGATCGCCACATACCTGCAAC-3′. The 345-bp PCR product was ligated into the pCRII-TOPO vector (Invitrogen) and subcloned into pZJM (33) with XhoI and HindIII to create the pZJM-calflagin RNAi construct. pZJM-calflagin was linearized by digestion with NotI and ethanol precipitated. For transfection of procyclic-form T. brucei, 100 μg linearized pZJM-calflagin in 100 μl sterile water were mixed with 2.25 × 107 parasites in 450 μl of electroporation medium (120 mM KCl, 0.15 mM CaCl2, 9.2 mM K2HPO4, 25 mM HEPES, 2 mM EDTA, 4.75 mM MgCl2, 69 mM sucrose, pH 7.6), transferred to a 0.4 cm gap cuvette, electroporated twice (1.4 kV, 25 μF), and collected in 5 ml of fresh SDM-79. For transfection of bloodstream form T. brucei, 100 μg linearized pZJM-calflagin in 100 μl sterile water was mixed with 2.4 × 107 parasites in 400 μl Cytomix (120 mM KCl, 0.15 mM CaCl2, 10 mM K2HPO4/KH2PO4, 25 mM HEPES, 2 mM EGTA, 5 mM MgCl2, 0.5% glucose, 100 μg/ml bovine serum albumin [BSA], 1 mM hypoxanthine, pH 7.6), transferred to a 0.2-cm gap cuvette, electroporated once (1.4 kV, 25 μF), and collected in 36 ml HMI-9. Drug selection was initiated 6 to 16 h posttransfection with 2.5 μg/ml phleomycin.
Western blotting.
Antiserum specific for calflagin Tb24, which reacts with Tb44 and Tb17, was generated in our laboratory (32). For protein detection, equal cell numbers of parasites were lysed in 2× SDS-PAGE sample buffer, separated by SDS-PAGE, and transferred to nitrocellulose membranes for Western blotting and detection by enhanced chemiluminescence (Amersham) as previously described (32). For Western blotting, anti-Tb24 antibody was used at 1:2,000 and anti-tubulin antibody (E7 ascites; Iowa Hybridoma Bank) was used at 1:1,000. A horseradish peroxidase-conjugated anti-mouse secondary antibody was used at 1:5,000. The resulting signal was compared across a range of serial dilutions to determine the linear range of detection, and values were normalized against the tubulin signal. Image acquisition and analysis were performed with Kodak 1D 3.5.4.
Immunofluorescence microscopy.
Parasites were washed twice in phosphate-buffered saline (PBS), resuspended in 4% paraformaldehyde, and settled onto poly-l-lysine-coated slides for 30 min. Fixative was quenched with 50 mM glycine for 15 min. Cells were permeabilized with 0.2% Triton X-100 in PBS for 15 min, incubated in blocking buffer (2% normal goat serum, 1% BSA in PBS) for 30 min, probed with calflagin antiserum (32) diluted 1:1,000 in blocking buffer for 3 h, washed for 30 min with PBS, incubated in Alexa Fluor 488-conjugated goat anti-mouse secondary antibody (Invitrogen) diluted 1:200 in blocking buffer, and washed again as after the primary antibody incubation. Cells were then incubated with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) for 10 to 20 s, washed for 2 min with ultrapure water, and mounted in Gelvatol containing 100 mg/ml 1,4-diazabicyclo[2.2.2]octane (DABCO). Immunofluorescence microscopy was performed with a Leica DM IRE2 microscope with a 100× objective under oil. Image acquisition was performed with OpenLab (Leica, Wetzlar, Germany), and image analysis, including deconvolution, was performed using Volocity software (Improvision, Seattle, WA).
In vitro parasite growth.
Wild-type single-marker T. brucei blood forms and calflagin RNAi cells were seeded at an initial density of 105/ml with and without 1 μg/ml tetracycline. Daily measurements of cell density were taken with a Z1 series Coulter Counter (Beckman-Coulter, Kansas City, MO) with a lower diameter threshold set to 3 μm and an upper threshold set to 10 μm. Each day, the parasites were passaged to a density of less than 105 cells/ml to maintain the parasites in the logarithmic phase of growth.
Mouse infections.
All animal experiments were approved by the Animal Care and Use Committee of Northwestern University. Infection of 4- to 6-week-old male BALB/c mice (The Jackson Laboratory, Bar Harbor, ME) was initiated by intravenous injection of 100 bloodstream form T. brucei parasites in 100 μl of PBS supplemented with 5% d-glucose and prechilled to 4°C. Mice were watered with 1 mg/ml doxycycline (Sigma) in 5% sucrose beginning 2 days prior to infection. RNAi of parasite cultures was also induced 2 days prior to infection by the addition of 1 μg/ml tetracycline to the culture medium. Following an initial 3-day period before the parasites reached the threshold of detection, daily parasitemias were measured by diluting tail snip blood into red blood cell (RBC) lysis buffer and manually counting the parasites on a Neubauer hemacytometer. Moribund mice or those with parasitemias above 109 cells/ml were euthanized in accordance with our animal care protocol. Survival was monitored daily throughout the course of infection. At selected time points, peripheral blood was smeared onto glass slides, prepared with Wright-Giemsa stain, and visualized on a microscope with a 100× objective. VSG switching was analyzed by diluting peripheral blood into HMI-9, culturing it for 24 to 48 h to expand the parasites over the background host blood cells, and conducting immunofluorescence microscopy as described above with an antibody specific for VSG Lister 427-2 (kind gift of George Cross) at a dilution of 1:10,000. To determine the precise VSG expressed, RNA was extracted with TriZol reagent (Invitrogen), converted to cDNA with the Superscript III First Strand Synthesis kit for reverse transcription-PCR (Invitrogen), amplified with a 5′ VSG-nonspecific primer and a 3′ poly(A) primer, ligated into pCR-Blunt-II-TOPO (Invitrogen), and sequenced with plasmid-derived primers.
Motility analysis.
For single-cell tracking, glass slides and coverslips were treated with 1 M HCl for 10 min, rinsed thoroughly with ultrapure water, coated with poly-l-glutamic acid sodium salt (Sigma) for 10 min, rinsed again with ultrapure water, and allowed to dry. The coverslip was placed upon two pieces of double-sided tape spaced approximately 2 cm apart. The sides of the coverslip were then sealed with nail polish, leaving a small opening for the insertion of a pipette tip, and allowed to dry. Approximately 10 μl of trypanosomes at 106 cells/ml in medium was introduced into the chamber. Differential interference contrast (DIC) images were obtained at 7 frames/s over a 30-s time span with a 20× objective on a Zeiss Axioimager Z1 microscope (Zeiss, Thornwood, NY), and point-to-point tracking was performed with Volocity software (Improvision). Cell classification was performed in accordance with the definition of Baron et al. (1). For population analysis of motility by the sedimentation assay, T. brucei procyclic forms at 5 × 106 cells/ml in SDM79 were dispensed into separate optical cuvettes for each time point. Every 2 h over a 12-h time span, the optical density at 600 nm of the culture was measured first while it was still settled and again following vigorous resuspension by repeated pipetting. The difference between these measurements was plotted against the incubation time.
Flow cytometry.
To determine the clearance of surface-bound antibody, we employed a temperature-dependent pulse-chase labeling method similar to previously published methods (13) by using 5 ×106 blood form parasites labeled with monoclonal anti-VSG Lister 427-2 antibody. Briefly, cells were preincubated with propidium iodide (PI; BD Pharmingen, San Diego, CA) at 1 μg/ml, enabling live/dead exclusion, and then labeled at 4°C with 1:4,000 primary antibody for 15 min. Cells were incubated at 37°C for various amounts of time (0 to 600 s), permitting antibody internalization, and then washed with 10 volumes of ice-cold PBS, labeled with 1:400 goat anti-mouse 647 (Molecular Probes) for 15 min, and fixed in 1% paraformaldehyde. DNA was labeled with DAPI (2 μg/ml), and cells were filtered through a 40-μm mesh. Acquisition of relative cell size, dead cell exclusion, DNA content, and surface VSG expression were accomplished on a Becton Dickinson LSR II with 355-nm UV and 488-nm and 633-nm argon ion lasers and standard filter sets. Forward-angle light scatter and side light scatter plots were used as indicators of cell size. DAPI, PI, and allophycocyanin signals were collected for at least 10,000 cells gated for PI exclusion and mononuclear DNA content on DAPI-A/W bivariate analysis. In all experiments, the positive gate was determined by analyzing unstained controls. Relative mean fluorescence intensity (MFI) determination, compensation, and postacquisition analysis were accomplished using FlowJo 8.8.4 (Treestar Inc., Ashland, OR) on a Macintosh platform.
Statistical analysis.
For calculation of significance for continuous variables (parasite motility run velocity and meandering index, fluorescence intensity, an unpaired Student t test was employed. For calculation of significance for categorical data (motility classification analysis), a Fisher two-tailed exact test was performed. For calculation of significance for survival curves, a log-rank test using the WinSTAT add-in for Microsoft Excel was used.
RESULTS
Calflagin gene organization and expression.
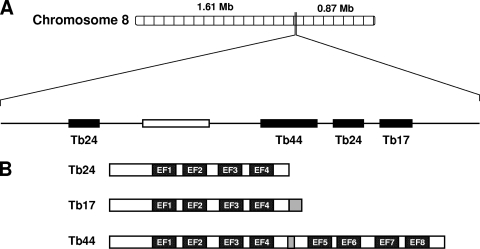
The genes encoding the calflagin family are located in a 7.6-kb region of chromosome 8 (Fig. 1A). Calflagin Tb24, the gene with the greatest sequence homology to T. cruzi FCaBP, is currently annotated in the genome as existing in two copies. One of these was previously identified as Tb1.7g (35). These genes show >98% identity at both the DNA (647/654) and protein (214/218) levels. Since we were unable to resolve a difference between these proteins in our experiments, we have adopted the nomenclature described above and henceforth refer to both copies as Tb24. Another calflagin gene, Tb17, shows >98% identity with both Tb24 genes but also contains a C-terminal extension encoding a 15-amino-acid peptide not found in either Tb24 or FCaBP. The protein encoded by Tb44 contains a duplication of a 186-amino-acid Tb24 core sequence separated by a small 8-amino-acid spacer peptide (Fig. 1B). The gene cluster expansion of this family is unique to T. brucei, since only a Tb24 homolog is found in other kinetoplastid parasites.
Fig. 1.
Genome organization and domain structure of the calflagins. (A) The calflagin genes are organized in a tandem array (black rectangles) in a 7.6-kb region of chromosome 8. Calflagins Tb44, Tb24, and Tb17 are offset from an additional copy of Tb24 by another gene encoding a putative amino acid transporter (open rectangle). (B) Calflagins Tb24 and Tb17 each have EF-hand domains (closed rectangles). Calflagin Tb17 also contains a 15-amino-acid extension at the C terminus relative to Tb24 (gray). Calflagin Tb44 has a duplication of a 186-amino-acid Tb24/Tb17 core sequence, separated by a short 8-amino-acid hinge peptide (gray).
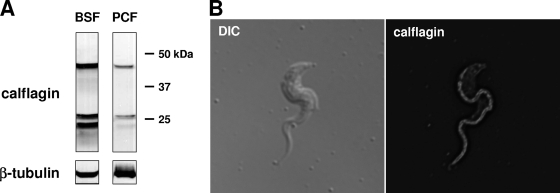
To investigate the biochemical and functional properties of the calflagin gene family, we expressed and purified recombinant Tb24 in Escherichia coli and then used it to generate polyclonal antisera in mice. Consistent with the high degree of sequence similarity among the calflagins, antiserum raised against Tb24 reacted with three distinct bands in parasite lysates, corresponding to the predicted molecular masses of Tb44, Tb17, and Tb24 (Fig. 2A). While these proteins were expressed in both life cycle stages of the parasite, expression was markedly higher in bloodstream form parasites. Densitometric analysis of parasite lysate serial dilutions, using equal numbers of parasites and normalizing against tubulin, estimated this difference to be approximately 10-fold greater in bloodstream forms than in procyclic cells. Immunofluorescence microscopy with calflagin-specific antiserum demonstrated the flagellar localization of these proteins (Fig. 2B).
Fig. 2.
Expression and localization of the calflagins. (A) The calflagins are predominantly expressed in the bloodstream form. Western blotting with calflagin-specific antiserum on lysates from 4 × 105 cell equivalents of bloodstream form (BSF) and procyclic-form (PCF) cells demonstrates an ∼10-fold increase in bloodstream form T. brucei. (B) The calflagins are restricted to the flagellum. Immunofluorescence microscopy of bloodstream form T. brucei with calflagin antiserum, shown next to DIC light microscopy, demonstrates flagellar concentration.
Calflagins bind calcium with high affinity.
The elution of the calflagins from a Phenyl Sepharose column upon calcium chelation (36) suggests that they undergo calcium-induced conformational changes that expose hydrophobic domains. Consistent with this model, the calflagins contain multiple EF-hand calcium-binding sites in which two orthogonal alpha helices flank an ∼12-residue calcium-binding loop (25). Calflagins Tb24 and Tb17 each contain four potential calcium-binding sites, of which one is degenerate, with mismatches in the critical −Y and −Z positions (25). Calflagin Tb44 has eight potential sites, two of which are degenerate. The presence of multiple EF-hand domains within a single protein is thought to promote cooperativity of calcium binding and subsequent conformational changes.
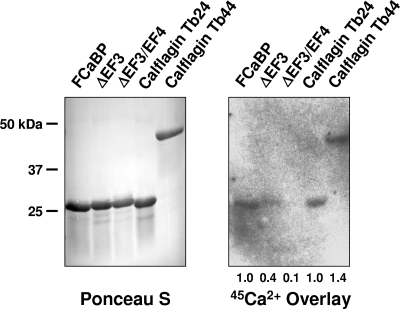
We engineered and purified recombinant calflagins Tb24 and Tb44 and assessed their ability to bind calcium by 45Ca2+ overlay analysis. In this technique, proteins are separated by SDS-PAGE, transferred to a nitrocellulose membrane, and incubated with buffer containing 45Ca2+. Autoradiography of the membrane then allows qualitative assessment of calcium binding by the refolded protein (21). The control proteins used in our assay were T. cruzi FCaBP, which contains two calcium-binding sites (Kmhigh = 9 μM, Kmlow = 120 μM), the FCaBP ΔEF3 mutant protein, in which the high-affinity site is abolished but the low-affinity site remains intact, and the FCaBP ΔEF3/EF4 double-mutant protein, in which binding at both sites has been abolished (6, 19). Calflagins Tb24 and Tb44 gave signals that were 1.0 and 1.4 times that of FCaBP, while the control ΔEF3 and ΔEF3EF4 proteins gave signals that were 0.4 and 0.1 times that of FCaBP and comparable to that of the wild-type FCaBP control (Fig. 3). Therefore, we conclude that the calflagins directly bind to calcium with a binding capacity and affinity similar to those of FCaBP. Furthermore, since E. coli lacks myristoylation and palmitoylation, calcium binding of calflagin does not require N-terminal acylation, as is true of FCaBP (6, 19).
Fig. 3.
Recombinant calflagins bind calcium with high affinity. Purified recombinant calflagins and control proteins were fractionated by SDS-PAGE, transferred to nitrocellulose, and either stained with Ponceau S to analyze total protein or overlaid with a buffer containing 45Ca2+ to assess calcium binding. The total CaCl2 concentration in this assay was 9 μM. Control proteins are T. cruzi FCaBP, which has a high-affinity Ca2+ binding site (Km = 9 μM) and a low-affinity site (Km = 120 μM), and point mutant proteins that abolish the high-affinity site (ΔEF3) or both sites (ΔEF3EF4). The relative band intensities were compared by scanning densitometry, and the values listed below the autoradiogram are relative to those obtained with FCaBP.
Calflagin-deficient parasites demonstrate normal in vitro proliferation.
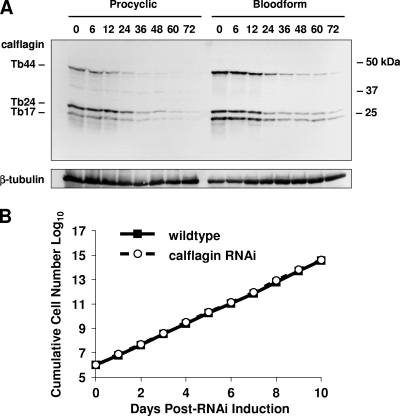
To test the necessity of calflagins for trypanosome viability, we utilized an RNAi approach that allows for drug-inducible inhibition of gene expression. The high degree of sequence identity among the calflagin genes enabled simultaneous inhibition of the entire family with a single targeting construct. Unlike mammalian cells, which require individual short hairpin RNA transcripts for effective RNAi, T. brucei processes a larger double-stranded RNA (dsRNA) into many potential effectors, resulting in an efficient and straightforward method to degrade mRNA in a specific manner. The 345-bp calflagin target sequence was subcloned into the pZJM plasmid, which provides flanking opposing T7 promoters for dsRNA synthesis and sequences for integration into the tubulin intergenic region (34). Transfection of this construct into T. brucei strains engineered to coexpress T7 RNA polymerase and tetracycline repressor permitted tetracycline-inducible expression of dsRNA that rapidly and specifically led to a depletion of protein (Fig. 4A). RNAi induction caused calflagin protein levels to drop to less than 10% within 48 h after induction.
Fig. 4.
Depletion of the calflagins does not affect parasite growth in culture. (A) Calflagin RNA was induced in both procyclic form (PCF) and bloodstream form (BSF) cells by the addition of 1 μg/ml tetracycline and monitored for calflagin expression. At the indicated time points, parasites were removed and 2.5 × 106 cell equivalents of lysate were analyzed by Western blotting for calflagin and β-tubulin (for normalization). (B) Bloodstream form calflagin RNAi cells and parental wild-type T. brucei single-marker parasites were cultured at a starting density of 1 × 105 cells/ml and monitored daily for growth in culture. At each measurement, parasite cultures were diluted to prevent cells from entering stationary phase. The cumulative cell number at each time point represents cell density × cumulative dilution factor.
We tested for the necessity of calflagin expression for cell proliferation in T. brucei by culturing calflagin RNAi lines in the presence or absence of tetracycline. Cell density was measured daily, and cultures were diluted as necessary to prevent parasites from entering stationary phase. Tetracycline itself had no effect on the growth of wild-type cells (data not shown). Over 10 days of drug induction, calflagin RNAi cells continued to proliferate at the same rate as untransfected cells (Fig. 4B). These results demonstrated successful calflagin depletion by ∼90% with preserved in vitro viability and cell proliferation.
Parasitemia is attenuated and survival prolonged in mice infected with calflagin-deficient T. brucei.
Given the increased expression of calflagins in the infectious life cycle stage of T. brucei, we hypothesized that calflagin depletion might have an effect specific to the environment of the mammalian bloodstream. Mice were infected intravenously with either calflagin RNAi or parental wild-type parasites. RNAi induction was initiated 2 days prior to infection by the addition of tetracycline to parasite culture medium and was maintained during infection by the addition of doxycycline to the drinking water of the mice. Consistent with the high susceptibility of BALB/c mice to T. brucei infection (14), we found that inoculation with 100 wild-type parasites was lethal to 100% of infected BALB/c mice within 10 days (Fig. 5A, n = 7). For both wild-type cells and calflagin RNAi lines, host mortality was closely correlated with parasitemia. Death was not observed in mice whose parasitemia was below 5 × 108 cells/ml the day before, and no mouse with parasitemia above this threshold survived for more than 48 h. Wild-type parasitemias first rose above our threshold of detection (1 × 105 cells/ml) beginning on day 4 postinfection and increased approximately 10-fold per day thereafter (Fig. 5B). For no single mouse infected with wild-type parasites did parasitemia drop between successive measurements. These mice demonstrated an unremitting parasitemia, consistent with the lack of effective parasite clearance, resulting in 0% survival by day 10 (n = 7).
Fig. 5.
Mice infected with calflagin-deficient parasites exhibit prolonged survival and suppressed parasitemias. (A) Mice were infected intravenously with 100 calflagin RNAi (dashed line, open circles) or parental wild-type (WT; solid line, closed circles) bloodstream form T. brucei cells. Parasite cultures were preinduced with 1 μg/ml tetracycline for 2 days. To maintain induction in the mammalian host, 1 mg/ml doxycycline (dox) was added to the drinking water of mice beginning 2 days before infection and the drinking water was changed every other day. Survival was monitored daily and is shown by a Kaplan-Meier curve. At 5 days postinfection, peripheral blood from a single mouse in each group containing 2.5 × 105 parasites was mixed with SDS-PAGE sample buffer and analyzed by anti-calflagin Western blotting (inset). (B) At the indicated days postinfection, 2.5 μl of peripheral blood was obtained by tail snip and diluted 1:10 into RBC lysis buffer and parasites were enumerated on a hemacytometer.
Infection with calflagin-deficient parasites, by contrast, resulted in markedly pronged survival (Fig. 5A). By the time all of the mice infected with wild-type parasites succumbed at day 10, 67% of the mice infected with calflagin-depleted parasites remained alive. Of these, half (33% of the original cohort) survived with undetected parasitemias (<105) until the conclusion of the experiment at day 21 (n = 9). This outcome could not be explained by a nonspecific protective effect of doxycycline, since administration of this drug had no protective effect for mice infected with wild-type parasites (data not shown). Corresponding to their prolonged survival, mice infected with calflagin-deficient cells exhibited a dramatically different parasitemia profile (Fig. 5B). In the first 7 days postinfection, these cells proliferated in the host at a normal rate. At day 8, however, the parasitemia in surviving animals infected with calflagin-deficient parasites dropped below the threshold of detection. These mice demonstrated a second and even a third wave of parasitemia at 4- to 5-day intervals. This situation is very similar to the pattern observed with the naturally occurring infection of polyclonal parasites where relapsing/remitting waves of parasitemia occur as the natural consequence of the interplay between host adaptive immunity and parasite antigenic variation.
Relapsing parasitemia in calflagin RNAi infection is associated with antigenic variation.
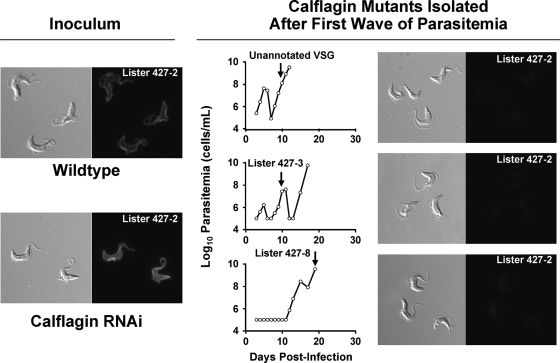
Given the sudden decrease in parasitemia during infection with calflagin RNAi cells, we next tested whether subsequent rises in parasitemia were associated with antigenic variation (VSG expression switching). Immunofluorescence microscopy was conducted with antiserum specific for VSG MITat 1.2 (VSG 221), the dominant VSG expressed by both wild-type and calflagin RNAi populations at the time of inoculation (Fig. 6). Parasites isolated from the bloodstreams of calflagin RNAi-infected mice at various time points during the second or third wave of parasitemia demonstrated a loss of reactivity with the MITat 1.2-specific serum compared to the input parasites, supporting the notion that sudden drops in parasitemia represented antigen-specific clearance. To confirm this, we isolated cDNA from harvested parasites and determined the identities of their expressed VSGs by sequencing. From these later peaks, we identified expressed MITat 1.3, MITat 1.8, and two previously unidentified VSGs, yet not the parental MITat 1.2 VSG.
Fig. 6.
Secondary peaks of calflagin RNAi parasitemia are associated with alternative VSG expression. Mice were inoculated with wild-type or calflagin RNAi blood form parasites expressing the MITat 1.2 VSG (inoculum, left). Parasitemias were followed, and the VSG expressed at the first or second relapse (arrows) was determined by cDNA cloning and sequencing. MITat 1.2 immunofluorescence was negative in all cases (right). The specific VSG expressed in each of these animals is indicated in the parasitemia plot.
Calflagin-deficient parasites have normal motility.
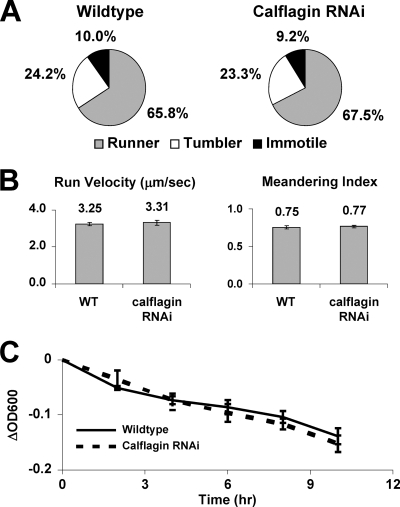
The markedly prolonged host survival and the flagellar localization of these proteins led us to surmise that perhaps they are essential for parasite motility. We hypothesized that our calflagin knockdown would have defective motility, since the putative function of this protein is calcium-dependent signaling in flagellar motility or sensation. We therefore evaluated the motility of calflagin-deficient cells by tracing >100 individual cells of both wild-type and calflagin RNAi cells and quantifying several parameters of motility (Fig. 7). Trypanosome directional movement occurs by alternating periods of “tumbles,” during which the flagellum assumes a bent-hook shape as the parasite rotates in three dimensions, and “runs,” during which tip-to-base flagellar beating propels the parasite forward in a corkscrew-like manner (15). We adopted a published classification scheme whereby individual cells were classified as “runners” if they sustained directional movement for at least 5 s over a 30-s span, “tumblers” if flagellar beating was apparent but directional movement was not maintained, or “immotile” if no movement was seen (1). The proportions of these classes were similar for wild-type and calflagin RNAi cells. Similarly, the average velocities of parasites during runs were comparable (3.25 ± 0.09 μm/s for wild-type cells and 3.31 ± 0.14 μm/s for calflagin RNAi cells [mean ± standard error of the mean; P = 0.72]). The ability of the parasites to maintain a straight path during runs was ascertained by calculating the “meandering index” of cells, equal to the net displacement divided by the point-to-point distance traveled by each cell. Thus, a value of 1.0 would represent a path along a perfectly straight line, while values closer to 0 would represent more circuitous paths. Again, no significant difference was seen (0.75 ± 0.04 for wild-type cells, 0.77 ± 0.03 for calflagin RNAi cells [mean ± standard error of the mean; P = 0.49]). Motility defects in trypanosomes can also be quantified at the population level by the sedimentation assay. In this approach, cells with impaired motility are found to settle to the bottom of an optical cuvette more rapidly that wild-type parasites, as measured by the difference between the optical densities before and after vigorous resuspension (3, 28). Calflagin RNAi parasites had no measurable motility deficit by this assay, staying in suspension as readily as wild-type cells throughout a 10-h incubation period. Therefore, we conclude that parasites depleted of calflagin do not demonstrate altered in vitro motility characteristics.
Fig. 7.
Calflagin depletion does not alter normal flagellar motility in vitro. (A) A chamber was created by affixing a glass coverslip on a single piece of tape mounted on a glass slide. Movies 30 s long were recorded, and >100 individual cells of each strain were tracked using Volocity software. The observer was blinded to the sample identities, and individual cells were classified as “runners” if they sustained directional motility for at least 5 s, “tumblers” if they failed to sustain directional movement but still exhibited net displacement, and “immotile” if they did not move from the point of origin. (B) For cells that did undergo runs during the movie, the average velocity during the run was calculated. The linearity of the run was measured by calculating the “meandering index,” defined as the net displacement divided by the point-to-point displacement. (C) Motility of procyclic cells was also measured by the sedimentation assay, in which parasites in suspension are allowed to settle for various lengths of time. The ability of cells to stay in suspension, which serves as a measurement of flagellar motility, was recorded by calculating the difference between the optical density (ΔOD600) of the settled culture and that of the culture following vigorous resuspension. WT, wild type.
Calflagin-deficient parasites clear surface-bound antibody normally.
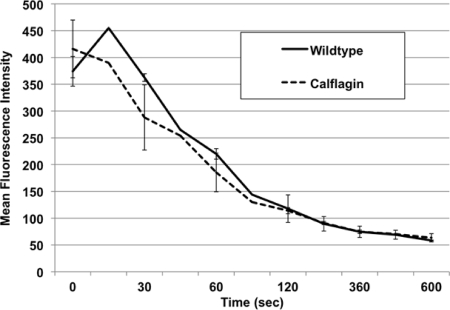
One mechanism of T. brucei immune evasion is rapid clearance of surface-bound antibodies of the bloodstream parasite to prevent complement-mediated lysis (13, 22). We hypothesized that parasites deficient in the calflagins may clear surface-bound antibodies at a slower rate than wild-type parasites, thus rendering them more susceptible to the early host immune response during infection. We therefore incubated blood form parasites with antisera specific for their expressed VSG, mimicking the first wave of adaptive humoral immunity during infection (Fig. 6). We monitored surface clearance by the reduction in MFI as labeled cells were warmed to and incubated at 37°C over time. The data supported the null hypothesis, as we found both the magnitude of initial labeling and rate of MFI decay were equal between wild-type and calflagin-deficient parasites (Fig. 8). We propose that the principal mechanisms of prolonged animal survival, while temporally related to the principal events surrounding the host adaptive immune response, are unlikely to be a result of impaired clearance of surface-bound antibody.
Fig. 8.
Surface antibody clearance is unaffected by calflagin depletion. Live bloodstream form parasites were labeled at 4°C with anti-VSG antibody and then incubated at 37°C for various lengths of time (0 to 600 s) to allow surface clearance of antibody prior to secondary labeling and fixation. Flow cytometric analysis was performed, and the resulting MFIs of the wild-type and calflagin-depleted cells in the positive population were compared. Neither the initial staining nor the rate of clearance differed significantly between groups. More than 10,000 live nucleated cells were analyzed (n = 3).
DISCUSSION
Dissecting the mechanisms underlying parasite survival in the mammalian host can facilitate the development of novel specific therapies for trypanosomiasis. The observations described here point to a previously unrecognized role for the calflagins in T. brucei mammalian infection. These results also have implications for the related kinetoplastid pathogens T. cruzi and Leishmania spp., since calflagin homologues are present in these organisms. Calcium handling and calcium-regulated signaling are critical processes. The calflagins, with their flagellar localization, acylation-dependent lipid raft association, and multiple EF-hand domains, likely contribute to calcium-mediated signaling. They have previously been shown to interact with a hydrophobic resin in a calcium-dependent manner (36). However, it remained possible that the interaction of calflagins with the resin occurred via association with other proteins in the cell lysate used in the assay. Indeed, the calflagin homologue of T. cruzi, FCaBP, interacts with another protein which itself binds calcium (6). 45Ca2+ overlay analysis indicates that recombinant calflagins, denatured and refolded on a nitrocellulose membrane, directly bind calcium ions (Fig. 3). The intensity of the 45Ca2+ signal for both calflagins Tb24 and Tb44 indicates an affinity similar to that of FCaBP (Km, ∼9 μM) and clearly higher than that of the FCaBP EF3 mutant protein (Km, ∼120 μM).
Investigations of calflagin expression indicated an upregulation in bloodstream form parasites which, in turn, suggested a possible role in infection. Indeed, we found that calflagin-deficient parasites were attenuated in a mouse model of sleeping sickness. The fact that calflagin-deficient cells exhibit a normal growth rate in vitro and in vivo during days 1 to 6 postinfection suggests that their deficit is not simply one of impaired proliferation. Rather, parasitemias involving calflagin-deficient parasites deviate from those of wild-type infections when they undergo a sudden decrease to undetectable levels approximately 1 week postinfection (Fig. 5). That this drop is followed by the emergence of a parasite population expressing alternative VSGs (Fig. 6) implies that initial clearance is antigen specific and due to host adaptive immunity. The exact mechanism for this effect has not been delineated by this work. We propose that the most parsimonious explanation for these findings is that calflagins promote parasite resistance to early host immunity. Thus, calflagin-deficient cells behave like wild-type cells in the absence of immune selection, with a difference only manifest when the host mounts an adaptive immune response around day 7 postinfection.
It should be noted that the parasite strain analyzed in this work has different properties than the pleomorphic T. brucei strains found in nature. As pleomorphic strains reach high levels of parasitemia in the mammalian host, they differentiate from a long slender proliferative form into a short stumpy cell cycle-arrested form. This transition both prolongs the survival of the host and preadapts the parasite for transmission back into the tsetse fly. The strain analyzed in these studies, instead, is a laboratory-passaged monomorphic strain which, over time, has lost this differentiation behavior in the host but retains the capacity to do so in response to a membrane-permeable cyclic AMP analog (18). The waxing-and-waning parasitemia profile of calflagin-deficient parasites is similar to that observed during the natural course of pleomorphic parasite infections. Peripheral blood smears, however, revealed no difference in the morphology of the calflagin-deficient cells in the bloodstream (data not shown). Therefore, we believe that immune susceptibility, rather than cellular differentiation, underlies the phenotype of calflagin deficiency.
Given the flagellar localization of the calflagins, we assessed the motility of calflagin RNAi lines. Although it is not yet established whether motility itself is critical during parasite infection, flagellar proteins are clearly important to the viability of bloodstream form T. brucei (4). Motility mediates the sorting of surface-bound antibodies to the flagellar pocket for endocytosis and degradation (13, 22), a property likely to promote immune evasion through resistance against complement-mediated lysis and/or opsonization in vivo. Perhaps surprisingly, depletion of the calflagins had no demonstrable effect on T. brucei motility in vitro (Fig. 7). These assays have been previously used to classify motility mutants with various degrees of impairment and are sensitive enough to detect even subtle deficits (1). The RNAi effect was significant, reducing protein by >90% and eliciting a phenotype during infection, yet we cannot exclude the possibility that only a small amount of calflagin is necessary to support an essential function. Also, our functional assays tested baseline T. brucei motility in enriched medium and so might miss a change in flagellar motility triggered by calcium-dependent signaling, such as that which occurs in Ca2+-dependent hyperactivation of sperm cell motility in the female reproductive tract (17). Calcium regulation has likewise been implicated in flagellar wave reversal in the trypanosomatid Crithidia oncopelti (31). Finally, we cannot exclude the possibility that a genetic deletion (knockout) would reveal an in vitro phenotype and thus conclude only that trypanosomes expressing a few percent of normal levels of calflagin do not have detectable defects in proliferation, morphology, or motility.
To explore the possibility that calflagin-deficient cells might be deficient in clearing surface-bound antibodies, thereby being rendered more susceptible to host immunity, we examined the rate of anti-VSG antibody binding and clearance by flow cytometry. We did not find a difference between calflagin-deficient and wild-type cells. These results imply that neither in vitro motility nor defective antibody clearance likely underlies the prolonged host survival seen after infection with calflagin-deficient parasites.
Taken together, these results indicate an important role for the calflagins during T. brucei infection. These effects are independent of any demonstrable effect on normal in vitro growth, morphology, motility, or clearance of surface-bound antibody. Future elucidation of the mechanism underlying this phenotype will be important for understanding the molecular basis of the host-parasite relationship during mammalian infection.
ACKNOWLEDGMENTS
We thank Kent Hill for advice on motility analysis, Markus Engstler for advice on the antibody clearance studies, and George Cross for the MITat 1.2 (VSG 221)-specific antiserum.
This work was supported by National Institutes of Health grants AI46781 and HL75822 to D.M.E. and HD47349 to C.L.E. and an award from the Children's Research Foundation of Chicago to C.L.E. B.T.E. was supported by predoctoral fellowships HL94026 from the NIH and 0515554Z from the American Heart Association.
Footnotes
Published ahead of print on 23 April 2010.
REFERENCES
- 1.Baron D. M., Ralston K. S., Kabututu Z. P., Hill K. L. 2007. Functional genomics in Trypanosoma brucei identifies evolutionarily conserved components of motile flagella. J. Cell Sci. 120:478–491 [DOI] [PubMed] [Google Scholar]
- 2.Barry J. D., McCulloch R. 2001. Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv. Parasitol. 49:1–70 [DOI] [PubMed] [Google Scholar]
- 3.Bastin P., Pullen T. J., Sherwin T., Gull K. 1999. Protein transport and flagellum assembly dynamics revealed by analysis of the paralysed trypanosome mutant snl-1. J. Cell Sci. 112:3769–3777 [DOI] [PubMed] [Google Scholar]
- 4.Broadhead R., Dawe H. R., Farr H., Griffiths S., Hart S. R., Portman N., Shaw M. K., Ginger M. L., Gaskell S. J., McKean P. G., Gull K. 2006. Flagellar motility is required for the viability of the bloodstream trypanosome. Nature 440:224–227 [DOI] [PubMed] [Google Scholar]
- 5.Brun R., Schonenberger 1979. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 36:289–292 [PubMed] [Google Scholar]
- 6.Buchanan K. T., Ames J. B., Asfaw S. H., Wingard J. N., Olson C. L., Campana P. T., Araujo A. P., Engman D. M. 2005. A flagellum-specific calcium sensor. J. Biol. Chem. 280:40104–40111 [DOI] [PubMed] [Google Scholar]
- 7.Carrington M., Carnall N., Crow M. S., Gaud A., Redpath M. B., Wasunna C. L., Webb H. 1998. The properties and function of the glycosylphosphatidylinositol-phospholipase C in Trypanosoma brucei. Mol. Biochem. Parasitol. 91:153–164 [DOI] [PubMed] [Google Scholar]
- 8.Cross G. A. 1975. Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology 71:393–417 [DOI] [PubMed] [Google Scholar]
- 9.Docampo R., Moreno S. N. 2001. The acidocalcisome. Mol. Biochem. Parasitol. 114:151–159 [DOI] [PubMed] [Google Scholar]
- 10.Doyle J. J., Hirumi H., Hirumi K., Lupton E. N., Cross G. A. 1980. Antigenic variation in clones of animal-infective Trypanosoma brucei derived and maintained in vitro. Parasitology 80:359–369 [DOI] [PubMed] [Google Scholar]
- 11.Emmer B. T., Souther C., Toriello K. M., Olson C. L., Epting C. L., Engman D. M. 2009. Identification of a palmitoyl acyltransferase required for protein sorting to the flagellar membrane. J. Cell Sci. 122:867–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engman D. M., Krause K. H., Blumin J. H., Kim K. S., Kirchhoff L. V., Donelson J. E. 1989. A novel flagellar Ca2+-binding protein in trypanosomes. J. Biol. Chem. 264:18627–18631 [PubMed] [Google Scholar]
- 13.Engstler M., Pfohl T., Herminghaus S., Boshart M., Wiegertjes G., Heddergott N., Overath P. 2007. Hydrodynamic flow-mediated protein sorting on the cell surface of trypanosomes. Cell 131:505–515 [DOI] [PubMed] [Google Scholar]
- 14.Greenblatt H. C., Diggs C. L., Rosenstreich D. L. 1984. Trypanosoma rhodesiense: analysis of the genetic control of resistance among mice. Infect. Immun. 44:107–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill K. L. 2003. Biology and mechanism of trypanosome cell motility. Eukaryot. Cell 2:200–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirumi H., Hirumi K. 1994. Axenic culture of African trypanosome bloodstream forms. Parasitol. Today 10:80–84 [DOI] [PubMed] [Google Scholar]
- 17.Ho H. C., Suarez S. S. 2001. Hyperactivation of mammalian spermatozoa: function and regulation. Reproduction 122:519–526 [DOI] [PubMed] [Google Scholar]
- 18.Laxman S., Riechers A., Sadilek M., Schwede F., Beavo J. A. 2006. Hydrolysis products of cAMP analogs cause transformation of Trypanosoma brucei from slender to stumpy-like forms. Proc. Natl. Acad. Sci. U. S. A. 103:19194–19199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maldonado R. A., Mirzoeva S., Godsel L. M., Lukas T. J., Goldenberg S., Watterson D. M., Engman D. M. 1999. Identification of calcium binding sites in the trypanosome flagellar calcium-acyl switch protein. Mol. Biochem. Parasitol. 101:61–70 [DOI] [PubMed] [Google Scholar]
- 20.Mansfield J. M., Paulnock D. M. 2005. Regulation of innate and acquired immunity in African trypanosomiasis. Parasite Immunol. 27:361–371 [DOI] [PubMed] [Google Scholar]
- 21.Maruyama K., Mikawa T., Ebashi S. 1984. Detection of calcium binding proteins by 45Ca autoradiography on nitrocellulose membrane after sodium dodecyl sulfate gel electrophoresis. J. Biochem. 95:511–519 [DOI] [PubMed] [Google Scholar]
- 22.McLintock L. M., Turner C. M., Vickerman K. 1993. Comparison of the effects of immune killing mechanisms on Trypanosoma brucei parasites of slender and stumpy morphology. Parasite Immunol. 15:475–480 [DOI] [PubMed] [Google Scholar]
- 23.Medina-Acosta E., Cross G. A. 1993. Rapid isolation of DNA from trypanosomatid protozoa using a simple ‘mini-prep’ procedure. Mol. Biochem. Parasitol. 59:327–329 [DOI] [PubMed] [Google Scholar]
- 24.Moreno S. N., Docampo R. 2003. Calcium regulation in protozoan parasites. Curr. Opin. Microbiol. 6:359–364 [DOI] [PubMed] [Google Scholar]
- 25.Nakayama S., Kretsinger R. H. 1994. Evolution of the EF-hand family of proteins. Annu. Rev. Biophys. Biomol. Struct. 23:473–507 [DOI] [PubMed] [Google Scholar]
- 26.Oberholzer M., Marti G., Baresic M., Kunz S., Hemphill A., Seebeck T. 2007. The Trypanosoma brucei cAMP phosphodiesterases TbrPDEB1 and TbrPDEB2: flagellar enzymes that are essential for parasite virulence. FASEB J. 21:720–731 [DOI] [PubMed] [Google Scholar]
- 27.Paindavoine P., Rolin S., Van Assel S., Geuskens M., Jauniaux J. C., Dinsart C., Huet G., Pays E. 1992. A gene from the variant surface glycoprotein expression site encodes one of several transmembrane adenylate cyclases located on the flagellum of Trypanosoma brucei. Mol. Cell. Biol. 12:1218–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ralston K. S., Lerner A. G., Diener D. R., Hill K. L. 2006. Flagellar motility contributes to cytokinesis in Trypanosoma brucei and is modulated by an evolutionarily conserved dynein regulatory system. Eukaryot. Cell 5:696–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Redmond S., Vadivelu J., Field M. C. 2003. RNAit: an automated web-based tool for the selection of RNAi targets in Trypanosoma brucei. Mol. Biochem. Parasitol. 128:115–118 [DOI] [PubMed] [Google Scholar]
- 30.Simarro P. P., Jannin J., Cattand P. 2008. Eliminating human African trypanosomiasis: where do we stand and what comes next? PLoS Med. 5:e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugrue P., Hirons M. R., Adam J. U., Holwill M. E. 1988. Flagellar wave reversal in the kinetoplastid flagellate Crithidia oncopelti. Biol. Cell 63:127–131 [DOI] [PubMed] [Google Scholar]
- 32.Tyler K. M., Fridberg A., Toriello K. M., Olson C. L., Cieslak J. A., Hazlett T. L., Engman D. M. 2009. Flagellar membrane localization via association with lipid rafts. J. Cell Sci. 122:859–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z., Morris J. C., Drew M. E., Englund P. T. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275:40174–40179 [DOI] [PubMed] [Google Scholar]
- 34.Wirtz E., Leal S., Ochatt C., Cross G. A. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89–101 [DOI] [PubMed] [Google Scholar]
- 35.Wu Y., Deford J., Benjamin R., Lee M. G., Ruben L. 1994. The gene family of EF-hand calcium-binding proteins from the flagellum of Trypanosoma brucei. Biochem. J. 304:833–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu Y., Haghighat N. G., Ruben L. 1992. The predominant calcimedins from Trypanosoma brucei comprise a family of flagellar EF-hand calcium-binding proteins. Biochem. J. 287:187–193 [DOI] [PMC free article] [PubMed] [Google Scholar]