Abstract
In the bipolar basidiomycete Pholiota microspora, a pair of homeodomain protein genes located at the A-mating-type locus regulates mating compatibility. In the present study, we used a DNA-mediated transformation system in P. microspora to investigate the homeodomain proteins that control the clamp formation. When a single homeodomain protein gene (A3-hox1 or A3-hox2) from the A3 monokaryon strain was transformed into the A4 monokaryon strain, the transformants produced many pseudoclamps but very few clamps. When two homeodomain protein genes (A3-hox1 and A3-hox2) were transformed either separately or together into the A4 monokaryon, the ratio of clamps to the clamplike cells in the transformants was significantly increased to ca. 50%. We therefore concluded that the gene dosage of homeodomain protein genes is important for clamp formation. When the sip promoter was connected to the coding region of A3-hox1 and A3-hox2 and the fused fragments were introduced into NGW19-6 (A4), the transformants achieved more than 85% clamp formation and exhibited two nuclei per cell, similar to the dikaryon (NGW12-163 × NGW19-6). The results of real-time reverse transcription-PCR confirmed that sip promoter activity is greater than that of the native promoter of homeodomain protein genes in P. microspora. Thus, we concluded that nearly 100% clamp formation requires high expression levels of homeodomain protein genes and that altered expression of the A-mating-type genes alone is sufficient to drive true clamp formation.
In basidiomycetous mushrooms, mating compatibility is controlled by one or two sets of multiple allelomorphic genes known as bipolar or tetrapolar mating systems, respectively (37). In tetrapolar mushrooms, such as Coprinopsis cinerea (5, 13), Laccaria bicolor (8, 11, 19), and Schizophyllum commune (10), the mating-type loci A and B, which are located on different chromosomes, regulate mating and clamp formation (9, 15, 27, 28). The A locus comprises multigenes encoding homeodomain proteins, and the B locus comprises multigenes encoding pheromones and pheromone receptor proteins (6, 13, 21, 25, 26, 30, 31, 32, 35, 36). On the basis of the homeodomain sequence, the mating-type proteins of the A locus are divided into two subgroups: HD1 and HD2 (20, 21). When an HD1 protein from one mate heterodimerizes with an HD2 protein from the other mate to form a functional regulatory protein, sexual compatibility is intracellularly recognized, and the A developmental pathway is initiated (3, 17, 23).
Few studies have examined the composition and function of mating-type loci in bipolar basidiomycetes. In a landmark study, Bakkeren and Kronstad (2) discovered that in bipolar fungus, Ustilago hordei, the A- and B-mating-type loci were fused into one nonrecombining mating-type region with two alleles. However, subsequent studies revealed that although both A- and B-mating-type homologs are found in bipolar mushrooms, they are present on different chromosomes, and only the A-mating-type homologs are related to mating compatibility (1, 16).
Although Pholiota microspora (P. nameko, Strophariaceae) has a very similar life cycle to other members of the order Agaricales, such as the tetrapolar mushroom C. cinerea, it has a bipolar A incompatibility factor and at least six different mating types (29). Ratanatragooldacha et al. (29) concluded that the bipolar A locus of P. microspora contains two functional subunits, Aα and Aβ, which appear to be located 0.3 centimorgan apart from each other on the same chromosome. Aimi et al. (1) sequenced and characterized the P. microspora genes encoding the homeodomain protein, hox1, and the pheromone receptor, rcb1, which are putative homologues of the HD1 protein and putative pheromone receptor protein genes in the tetrapolar basidiomycete C. cinerea, respectively. Restriction fragment length polymorphism and linkage analyses indicated that these two genes are present on different linkage groups and that only hox1 is involved in regulating mating incompatibility in P. microspora. A second homeodomain gene (A4-hox2) was discovered upstream of A4-hox1, and only two homeodomain protein genes exist in this Aα sublocus (33). Similarly, the bipolar mushroom Coprinellus disseminatus (16) contains two unlinked mating-type homologs (A and B), and only the homeodomain protein genes segregate with mating type. Moreover, the A factor of C. disseminatus encodes two tightly linked pairs of homeodomain transcription factors similar to the A-mating-type locus of C. cinerea. Due to the lack of a DNA-mediated transformation system in C. disseminatus, the C. disseminatus A and B homologues were transformed into C. cinerea, and sexual reactions similar to those of the homologous mating-type genes were elicited. Thus, the functions of the C. disseminatus mating type were studied in a tetrapolar mushroom, C. cinerea, instead of in a homologous bipolar species. In a previous study of P. microspora, we successfully constructed a DNA-mediated transformation system using a homologous selective marker (a carboxin resistance mutant gene of the succinate dehydrogenase iron-sulfur protein subunit) and a heterologous drug selective marker (hygromycin B phosphotransferase gene) (34). In the present study, we examined the functions of the P. microspora A-mating-type locus during clamp formation in vivo using our transformation system.
MATERIALS AND METHODS
Fungal strains.
Monokaryons of P. microspora were obtained by monospore isolation from the fruit bodies of various wild strains (24). The auxotrophic mutant monokaryons of P. microspora NGW19-6 (A4, pdx1) and NGW12-163 (A3, Arg4) were derived from the wild monokaryotic strains NGW19 (A4) and NGW12 (A3), respectively.
Mycelium preparation and DNA and RNA extraction.
To collect mycelium of auxotrophic mutant strain NGW19-6 and NGW12-163, five mycelial agar blocks (5 by 5 by 5 mm3) cut from an MYG plate (glucose [2%], malt extract [0.5%], yeast extract [0.5%], agar [1.5%]; pH 5.6) were transferred to 5 ml of liquid MYG medium (glucose [2%], malt extract [0.5%], yeast extract [0.5%]; pH 5.6) in a 100-ml Erlenmeyer flask. To collect the mycelia of cotransformants, the MYG plates and liquid medium contained 2.0 μg of carboxin/ml or 150 μg of hygromycin B/ml (in the case of two-step transformations, both drug reagents were mixed). The mycelium were grown at 25°C without shaking for 2 weeks and then harvested by filtration, frozen in liquid nitrogen, and ground to a fine powder using a mortar and pestle. Genomic DNA was extracted from the frozen mycelium according to the method described by Dellaporta et al. (7).
To prepare total RNA from NGW19-6, NGW12-163, and the transformants, the mycelium was grown on PDA (potato extract with 2% [wt/vol] glucose and 1.5% agar]) at 25°C for 2 weeks, after which the mycelium, along with three square agar blocks (5 by 5 by 5 mm3), was transferred to a piece of sterilized cellophane (40 by 40 mm2) on an MYG plate and grown at 25°C for a week. To isolate total RNA, the mycelium was scraped from the cellophane, frozen in liquid nitrogen, and ground to a fine powder with a mortar and pestle. An RNeasy minikit (Qiagen, Tokyo, Japan) was used to extract RNA from the powder, and the integrity of total RNA was examined by separation on a 1.0% agarose gel. A 1:10 dilution of stock total RNA was used for real-time reverse transcription-PCR (RT-PCR).
Amplification of A3-hox1 and A3-hox2 genes.
To introduce homeodomain protein genes to the NGW19-6 strain, A3-hox1 or A3-hox2 DNA fragments from the NGW12-163 strain were amplified. The A3-hox1 gene was amplified with MipF and 163mipR6 (Fig. 1), and the A3-hox2 gene was amplified with Hox2-A3-R1 and 163mipF6. The A3-hox1 gene amplification conditions consisted of an initial denaturation at 94°C for 3 min, followed by 35 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 3.5 min, and then a final extension at 72°C for 10 min. The amplification conditions for A3-hox2 consisted of an initial denaturation at 94°C for 3 min, followed by 35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 2.5 min, and then a final extension at 72°C for 10 min.
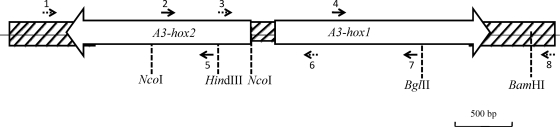
Fig. 1.
Map of A3-hox1 and A3-hox2. The positions of primers used for DNA amplification, the Southern hybridization probe, and the cutting sites of restriction enzymes are indicated. The dashed arrows show the primer positions used for the amplification of DNA fragments, and the solid arrows indicate the primer position used for making the Southern hybridization probe. Primers: 1, Hox2-A3-R1; 2, 163mipF-d5; 3, 163mipR6; 4, Hox1-A3-3RACE1; 5, 163mipR-d7; 6, 163mipF6; 7, 163mipF4; 8, MipF.
To introduce the DNA fragment containing A3-hox1 and A3-hox2 into NGW19-6, the genomic DNA fragments of both A3-hox1 and A3-hox2 from P. microspora NGW12-163 were amplified by using primers MipF and Hox2-A3-R1. PCR was performed with an initial denaturation at 94°C for 5 min, followed by 30 cycles of 30 s at 94°C, 30 s at 58°C, and 5 min at 72°C. The PCR product was subcloned into the pT7Blue(R) T-vector (Novagen, Darmstadt, Germany) to create pMBhox12.
Cotransformation method.
The DNA-mediated transformation method was performed with pMBsip2 or pMBhph1, as described in our previous study (34). pMBsip2 carries a carboxin resistance gene, and pMBhph1 carries a hygromycin B resistance gene. The homeodomain protein gene and the selective plasmid were introduced together into NGW19-6. For each transformation, 5 × 106 protoplasts, 5 to 10 μg of plasmid DNA, and 10 to 15 μg of amplified DNA containing the homeodomain protein gene were used. After the colonies appeared on the regeneration plate, they were individually subcultured onto fresh MYG plates containing 2 μg of carboxin/ml and/or 200 μg of hygromycin B/ml, as appropriate. After a 7- to 10-day incubation at 25°C, the mycelium edges of the colonies were microscopically examined for clamp-cell formations.
To introduce two separate homeodomain protein genes, A3-hox1 and A3-hox2, into NGW19-6, a two-step transformation was performed. In the first step, the A3-hox2 gene and pMBsip2 were transformed into the NGW19-6 strain. Then, carboxin-resistant transformants expressing A3-hox2 were identified (Hox2-1, Hox2-2), and one strain (Hox2-1) was used as the host strain for the second cotransformation with A3-hox1 and pMBhph1.
DAPI and Fluorescent Brightener 28 staining and microscopic observation.
Autoclaved slide glass was dipped into 1.0% agar medium and then placed in a sterilized plate. The mycelium was put on the glass-containing agar, incubated for 5 to 7 days, and then stained for 20 min with a solution of 50 μg of DAPI (4′,6′-diamidino-2-phenylindole; Merck, Darmstadt, Germany)/ml, which stains nuclei, and 20 μg of Fluorescent Brightener 28 (Sigma-Aldrich, St. Louis, MO)/ml, which stains the cell wall. The stained slides were studied with a Nikon Eclipse 50i microscope (Nikon, Tokyo, Japan).
Construction of two plasmid vectors for overexpression of the A3-hox1 and A3-hox2 genes.
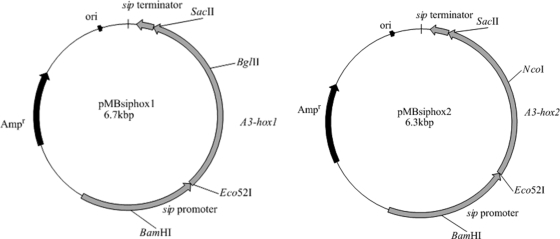
To determine whether the high expression level of homeodomain protein genes may increase the ratio of clamps in P. microspora, we connected the sip (iron-sulfur protein subunit of succinate dehydrogenase) promoter (34), which is expressed continually in the citrate cycle (tricarboxylic acid), to A3-hox1 and A3-hox2. First, the A3-hox1 and A3-hox2 gene fragments were amplified using A3-hox1-Eco52I/A3-hox1-SacII and A3-hox2-Eco52I/A3-hox2-SacII, respectively, and NGW12-163 mycelium DNA as the template. Thermal cycling parameters were as follows: initial denaturation at 94°C for 4 min; followed by 30 cycles of 94°C for 30 s, 57°C for 30 s, 72°C for 2 min; and a final extension at 72°C for 10 min. The amplified fragments were digested with EcoR52I and SacII. Second, the fragment containing pT7Blue(R) T-vector (Novagen), sip promoter and terminator, was amplified using Ip-pro-Eco52I and Ip-ter-SacII primers and pMBsip1 (34) as a template. The amplification conditions were as follows: initial denaturation at 94°C for 4 min; followed by 30 cycles of 94°C for 30 s, 58°C for 30 s, 72°C for 5 min; and a final 10-min extension at 72°C. The PCR product was also digested with EcoR52I and SacII. Then, the two kinds of digested fragments were ligated to form plasmids named pMBsiphox1 and pMBsiphox2 (Fig. 2). The identity of these plasmids was confirmed by sequencing.
Fig. 2.
Physical maps of plasmids pMBsiphox1 and pMBsiphox2. The NcoI, BamHI, Eco52I, SacII, and BglII recognition sites are indicated. Arrows indicate the direction of transcription.
Southern hybridization.
Southern hybridization analysis of the transformants was performed to analyze the integration of the transforming DNA. Genomic DNA (0.3 to 0.5 μg) from NGW19-6, NGW12-163, and the cotransformants was digested for 5 h at 37°C in a 500-μl reaction mixture containing 20 U of restriction enzymes in the buffer supplied by the manufacturer (Toyobo, Osaka, Japan). The digested fragments were concentrated by ethanol precipitation and then electrophoretically separated in a 1.0% agarose gel and blotted onto a nylon membrane (Hybond-N+; Amersham Biosciences, London, United Kingdom). DNA hybridization probes were labeled and detected by using DIG-High Prime DNA labeling and detection kits (Roche Diagnostics, Tokyo, Japan). We used nested PCR to label the probe. To detect the A3-hox1 gene in the transformants, we amplified a partial A3-hox1 sequence using primers Hox1-A3-3RACE1 and 163mipF4. To detect the A3-hox2 in the transformants, we amplified a partial A3-hox2 sequence with primers 163mipF-d5 and 163mip-d7 (Table 1). Also, these two probes were used for transformants from the pMBhox12 transformation.
Table 1.
Primers used in the present study
| Primer | Sequence (5′→3′) | Description |
|---|---|---|
| A3-hox1FNcoI | CCATGGACGCACGAGTAACAGAAA | A3-hox1 from NGW12-163 strain |
| A3-hox1RBamHI | GGATCCAAAATTTTCAATCAAGGTC | A3-hox1 from NGW12-163 strain |
| A3hox2FEcoRI | GAATTCGCCATGGTATCCGATCTG | A3-hox2 from NGW12-163 strain |
| A3hox2RBamHI | GGATCCAGCGACGAAAAGCATTAT | A3-hox2 from NGW12-163 strain |
| A4hox1FNdeI | CATATGGCCTCCGCCGTGGACCTCAGA | A4-hox1 from NGW19-6 strain |
| A4hox1RBamHI | GGATCCAGAAGATGGCAGATCAAT | A4-hox1 from NGW19-6 strain |
| A4hox2FNcoI | ATTACAACCATGGTGTCGACCGCA | A4-hox2 from NGW19-6 strain |
| A4hox2RSmaI | CCCGGGAATAGCAACAGAAAAGCAT | A4-hox2 from NGW19-6 strain |
| MipF | GCAGAGCTAGCCAAATTACACGAA | Used for amplification of the fragment containing A3-hox1 |
| 163mipR6 | TTGCTGGGACTGAACG | Used for amplification of the fragment containing A3-hox1 |
| Hox2-A3-R1 | CGCAGGGGTAGGATGTTATGGATT | Used for amplification of the fragment containing A3-hox2 |
| 163mipF6 | CATATGCTATTCCGGACA | Used for amplification of the fragment containing A3-hox2 |
| 163mipF-d5 | AAGGCTCAGGAAGAAGGGGAG | Used for amplification of the partial A3-hox2 |
| 163mipR-d7 | TACCTCTGCACATCTTACCAATC | Used for amplification of the partial A3-hox2 |
| Hox1-A3-3RACE1 | CCGGGCTAACTGATTACTCCATG | Used for amplification of the partial A3-hox1 |
| 163mipF4 | ATTTGATATGGGTAGCGG | Used for amplification of the partial A3-hox1 |
| A3-hox1 forward | CGGAATGCTTGAACTTGAAGTAGAG | Used for real-time RT-PCR of A3-hox1 |
| A3-hox1 reverse | ACTGGGATGGAATCTAGAACTTTGC | Used for real-time RT-PCR of A3-hox1 |
| A3-hox2 forward | GCTCAGGAAGAAGGGGAGAAATAG | Used for real-time RT-PCR of A3-hox2 |
| A3-hox2 reverse | CAATCGGTCTAAGAAAGAGGGAATAC | Used for real-time RT-PCR of A3-hox2 |
| A4-hox1 forward | ATTCCAGAAGCCACCTCTAACG | Used for real-time RT-PCR of A4-hox1 |
| A4-hox1 reverse | GCGGGTTGATGAATGTATGATTG | Used for real-time RT-PCR of A4-hox1 |
| A4-hox2 forward | CGCAAAAGCGTATCAGGCAG | Used for real-time RT-PCR of A4-hox2 |
| A4-hox2 reverse | GCTGAAGGAGTGACTTTACCCAAT | Used for real-time RT-PCR of A4-hox2 |
| Actin forward | TCGGTCTTGAGGCTGCTGGT | Used for real-time RT-PCR of actin |
| Actin reverse | AGTCAACTCCTTCTGCATACGGTC | Used for real-time RT-PCR of actin |
| ActindpF | CRGGTGTCMTGGTYGGWATGG | Used for partial actin gene amplification |
| ActindpR | CRRGVGGVGCRACGATCTTGAC | Used for partial actin gene amplification |
| Ip-d1R | TCGACGCAGATGGCACT | |
| Actin up F2 | CTTCAATGTCAGGATACCACGCTTC | Used for partial actin gene amplification |
| Actin down R2 | CACACCTTCCACAAAAAAAAACC | Used for partial actin gene amplification |
| Hox1-A3-R1 | GGAACAGAGAGGCATAGTGATAGA | Used for amplification of the DNA fragment containing A3-hox1 and A3-hox2 |
| A3-hox1-Eco52I | GGCCACGGCCGACATGGACGCACGAGTAACAG | A3-hox1 from NGW12-163 strain |
| A3-hox1-SacII | GGGAGCCGCGGATGTAAAATTTTCAATCAAG | A3-hox1 from NGW12-163 strain |
| A3-hox2-Eco52I | AACATCGGCCGACATGGTATCCGATCTGTCCT | A3-hox2 from NGW12-163 strain |
| A3-hox2-SacII | ATAGACCGCGGAAAAGCATTATGCAGTAGCA | A3-hox2 from NGW12-163 strain |
Real-time PCR assay.
We used the actin gene as the housekeeping gene. A partial actin gene in P. microspora was cloned by the degenerate PCR primers ActindpF and ActindpR. Primers for A3-hox1, A3-hox2, A4-hox1, A4-hox2, and actin were designed according to their cDNA sequences by using GENETYX 9.0 (Genetyx, Tokyo, Japan). The primers were designed according to the principles of primer design, and 3 to 6 bp of the 3′ site were designed to cross the intron in the primer spanning the intron. All primers were tested to ensure amplification of single bands with no primer-dimers. Plasmid extraction was performed according to the method modified by Birnboim (4). Four 10-fold dilutions of plasmid were performed to construct standard curves. Real-time PCR was conducted using the RNA-direct SYBR green Real-Time PCR master mix (Toyobo, Osaka, Japan) and Linegene (BioFlux, Hangzhou, China). Each reaction was run twice. The cycling parameters were 90°C for 30 s to activate thermostable DNA polymerase, 61°C for 20 min to reverse transcription, 95°C for 30 s predenaturation, and then 35 cycles of 95°C for 15 s, 60°C for 15 s, and 74°C for 30 s. Melting curves were determined according to the manufacturer's instructions. After real-time RT-PCR, samples were also run on a 1.5% agarose gel to confirm amplification specificity. The data analysis was performed according to the manipulation's instructions.
Plasmid construction for yeast two hybrids.
To synthesize the cDNA of A3-hox1, A3-hox2, A4-hox1, and A4-hox2, we isolated total RNA from a dikaryon strain (NGW19-6 × NGW12-163) by using an RNeasy minikit (Qiagen, Tokyo, Japan). We used 100 ng of total RNA as a template for first-strand cDNA synthesis in the presence of oligo(dT) (First-Strand cDNA synthesis kit; Takara Shigama, Japan). The primer pairs A3-hox1FNcoI and A3-hox1RBamHI, A3-hox2FEcoRI and A3-hox2RBamHI, A4-hox1FNdeI and A4hox1RBamHI, and A4-hox2FNcoI and A4-hox2RSmaI were used to amplify the nearly full-length A3-hox1, A3-hox2, A4-hox1, and A4-hox2 cDNAs, respectively. The amplified cDNA fragments of A3-hox1, A3-hox2, A4-hox1, and A4-hox2 were digested with NcoI and BamHI, EcoRI and BamHI, NdeI and BamHI, and NcoI and SmaI, respectively. Then, each fragment was cloned into plasmid pGADT7 (Clontech, Madison, WI) to generate an in-frame fusion with the C terminus of the GAL4 DNA-binding domain and the fragments of A3-hox2 and A4-hox2 were cloned into plasmid pGBKT7 (Clontech) to generate an in-frame fusion with the C terminus of the GAL4 activation domain. Each construction was sequenced across the ligation junction, and no deviations from the expected sequences were found.
Yeast transformation and α-galactosidase assay.
The Matchmaker two-hybrid system 3 (Clontech) and the lithium acetate-mediated method was used for the yeast transformation (14). The transformation was performed according to the manufacturer's manual (Clontech). We performed a two-step transformation. First, we introduced pGBKT7-A4-hox2 or pGBKT7-A3-hox2 into the Y187 strain (Trp− Leu−) (12) and selected the transformants on SD agar plates (0.67% yeast nitrogen base without amino acid, 2% agar, appropriate dropout supplement, and 2% glucose) lacking l-tryptophan. Then, each plasmid (pGADT7-A4-hox1, pGADT7-A4-hox2, pGADT7-A3-hox1, and pGADT7-A3-hox2) was introduced into the transformant with pGBKT7-A4-hox2 or pGBKT7-A3-hox2, as appropriate. The transformants were selected on SD agar plates lacking l-tryptophan and l-leucine. pGADT7-T and pGBKT7-Lam were used as negative controls. The α-galactosidase assay was performed according to the manufacturer's instructions (Clontech). One unit of α-galactosidase was defined as the amount of enzyme that hydrolyzes 1 μmol of p-nitrophenyl-α-d-galactoside to p-nitrophenol and d-galactose in 1 min at 30°C in acetate buffer (pH 4.5) (22).
RESULTS
A single introduced hox gene is insufficient to induce true clamps in high frequency.
To confirm that the introduction of a single compatible homeodomain protein gene is sufficient for clamp formation, A3-hox1 DNA fragments or A3-hox2 DNA fragments were cotransformed into the A4 strain NGW19-6, using pMBsip2 as a carboxin-resistant selective marker. The A3-hox1 DNA fragments contained an ∼260-bp partial A3-hox2 DNA fragment, the 206-bp spacing fragment between A3-hox1 and A3-hox2, and the A3-hox1 coding and terminator region. The A3-hox2 DNA fragments contained an ∼500-bp partial A3-hox1 DNA fragment, the 206-bp spacing fragment between A3-hox1 and A3-hox2, and the A3-hox2 coding and terminator region (Fig. 1).
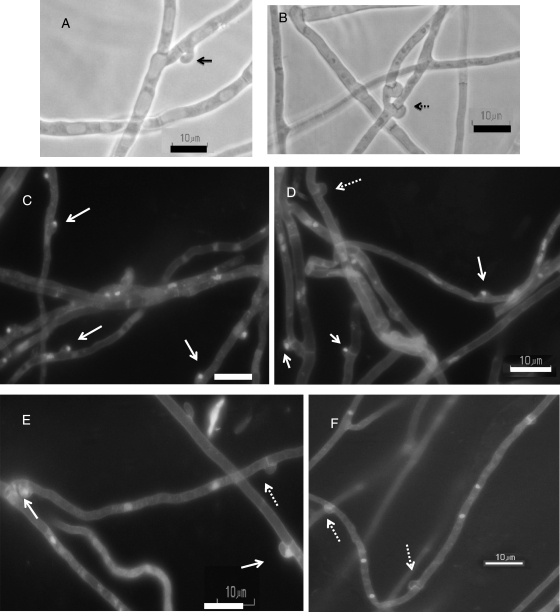
We collected carboxin-resistant regenerated colonies and microscopically examined their hook cell fusion after growth on MYG plates. Clamplike cells were present in 7 of 146 carboxin-resistant transformants from the transformation with A3-hox1 and in 16 of 111 carboxin-resistant transformants from the transformation with A3-hox2. However, all of the transformants with the clamplike cells contained mostly pseudoclamps with only rare clamps (see Fig. 3 A and B). Partial pseudoclamp data are shown in Table 2. The Hox1-1 and Hox1-2 strains containing introduced A3-hox1 had a ratio of clamps to total clamplike cells of <1%. The Hox2-1 and Hox2-1 strains containing introduced A3-hox2 had a ratio of fused hook cell to total clamplike cells of 4%. DAPI and Fluorescent Brightener 28 staining showed that the nuclei were trapped within the hook cell (Fig. 3C).
Fig. 3.
Configuration of clamps and pseudoclamps and DAPI and Fluorescent Brightener 28 staining of nuclei and cell walls in the cotransformants. (A) Pseudoclamps in Hox2-1. (B) Clamps in Hox2-1. (C) Pseudoclamps with staining of nuclei and cell walls in Hox2-1. (D) Pseudoclamps and clamps in Hox2-hox1-1. (E) Pseudoclamps and clamps in Hox1,2-1. (F) Clamps in Shox1-1. The solid and dashed arrows indicate the pseudoclamps and fused hook cells, respectively. Bars, 10 μm.
Table 2.
Ratio of clamps among total clamplike cells
| Strain | No. of clamps | Total no. of clamplike cells | Clamp ratio (%) | Description |
|---|---|---|---|---|
| NGW19-6 × NGW12-163 | 141 | 165 | 85.4 | Wild-type dikaryon |
| Hox1-1 | 0 | 146 | 0 | A3-hox1 transformants |
| Hox1-2 | 1 | 127 | 0.8 | A3-hox1 transformants |
| Hox2-1 | 8 | 221 | 3.6 | A3-hox2 transformants |
| Hox2-2 | 3 | 153 | 2.0 | A3-hox2 transformants |
| Hox2-hox1-1 | 59 | 112 | 52.7 | Transformants introduced with A3-hox1 to Hox2-1 |
| Hox2-hox1-2 | 64 | 150 | 42.7 | Transformants introduced with A3-hox1 to Hox2-1 |
| Hox2-hox1-3 | 43 | 108 | 39.8 | Transformants introduced with A3-hox1 to Hox2-1 |
| Hox12-1 | 69 | 133 | 51.9 | Transformants introduced with pMBhox12 |
| Hox12-2 | 52 | 99 | 52.5 | Transformants introduced with pMBhox12 |
| Hox12-3 | 69 | 205 | 33.6 | Transformants introduced with pMBhox12 |
| Shox1-1 | 120 | 138 | 89.1 | Transformants introduced with pMBsiphox1 |
| Shox1-2 | 107 | 123 | 89.1 | Transformants introduced with pMBsiphox1 |
| Shox2-1 | 120 | 140 | 85.7 | Transformants introduced with pMBsiphox2 |
| Shox2-2 | 73 | 84 | 86.9 | Transformants introduced with pMBsiphox2 |
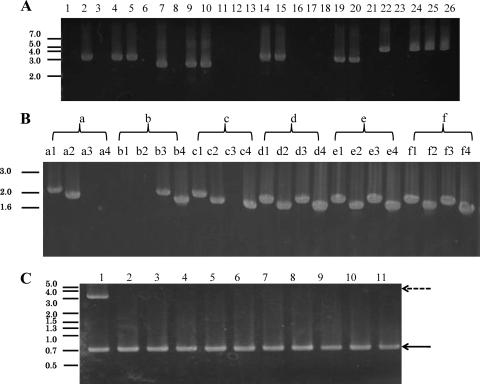
Using PCR amplification, we detected the band of the entire A3-hox1 DNA fragment in all of the A3-hox1 transformants with mostly pseudoclamps. The partial data is shown in Fig. 4 A (Hox1-1 and Hox1-2 strains, lanes 4 and 5). In Southern hybridization with a partial A3-hox1 DNA fragment as the probe, hybridization bands were detected in all of the A3-hox1 transformants with mostly pseudoclamps (data not shown). No entire fragments were detected, but a partial A3-hox1 DNA fragment was detected in most of the transformants with no clamps (see Fig. 4C). All of the A3-hox2 transformants with mostly pseudoclamps shared the similar results for PCR (Fig. 4A, Hox2-1 and Hox2-2 strains, lanes 9 and 10) and Southern hybridization (data not shown). These results confirm that the A3-hox1 or A3-hox2 gene was ectopically integrated into the chromosomes of transformants with clamplike cells.
Fig. 4.
PCR of the host strains, A3-hox1 and A3-hox2 transformants. The position and size in kilobase pair are indicated on the left. (A) PCR amplification of the host strains and transformants. Lanes 1 to 5 show PCR results of host strains and A3-hox1 transformants using the primers MipF and 163mipR6. Lane 1, NGW19-6 (A4); lane 2, NGW12-163 (A3); lane 3, control (transformants with no clamps); lane 4, Hox1-1 (transformants with pseudoclamps); lane 5, Hox1-2 (transformants with pseudoclamps). Lanes 6 to 10 show PCR results of host strains and A3-hox2 transformants using the primers Hox2-A3-R1 and 163mipF6. Lane 6, NGW19-6 (A4); lane 7, NGW12-163 (A3); lane 8, control (transformants with no clamps); lane 9, Hox2-1 (transformants with pseudoclamps); lane 10, Hox2-2 (transformants with pseudoclamps). Lanes 11 to 15 show PCR results of wild-type strains and transformants with pMBsiphox1 using the primers Ip-d1R and A3-hox1-SacII. Lane 11, NGW19-6 (A4); lane 12, NGW12-163 (A3); lane 13, control (transformants with no clamps); lane 14, Shox1-1 (transformants with clamps); lane 15, Shox1-2 (transformants with clamps). Lanes 16 to 20 show PCR results of the host strain and transformants introduced with pMBsiphox2 using the primers Ip-d1R and A3-hox2-SacII. Lane 16, NGW19-6 (A4); lane 17, NGW12-163 (A3); lane 18, control (transformants with no clamps); lane 19, Shox2-1 (transformants with clamps); lane 20, Shox2-2 (transformants with clamps). Lanes 21 to 26 show the amplification of DNA fragments containing A3-hox1 and A3-hox2 in the wild-type strains and the transformants using the primers Hox1-A3-R1 and Hox2-A3-R1. Lane 21, NGW19-6 (A4); lane 22 NGW12-163 (A3); lane 23, control (transformants with no clamps); lanes 24 to 26, Hox1,2-1, Hox1,2-2, and Hox1,2-3 (transformants with increased fusion hook cell). (B) PCR results of four homeodomain protein genes (A4-hox1, A4-hox2, A3-hox1, and A3-hox2) in host strains and transformants. The amplification order of each strain is A4-hox1 (using the primers A4-hox1FNdeI and A4-hox1RBamHI), A4-hox2 (using the primers A4-hox1FNcoI and A4-hox1RBamHI), A3-hox1 (using the primers A3-hox1FNcoI and A3-hox1RBamHI), and A3-hox2 (using the primers A3-hox2FEcoRI and A3-hox2RBamHI). Lanes a, NGW19-6 (A4); lanes b NGW12-163 (A3); lanes c Hox2-1 (the host of second transformation); lanes d to f, Hox2-hox1-1, Hox2-hox1-2, and Hox2-hox1-3 (transformants with increased hook cell fusion). (C) PCR results of A3-hox1 and partial actin gene in Hox1-1 and A3-hox1 transformants without clamps. The primers MipF and 163mipR6, actin2 up F2, and actin2 down R2 were used for amplification of A3-hox1 and the partial actin gene, respectively. The dashed arrow indicates the PCR amplification band of A3-hox1, and the solid line arrow shows the PCR amplification band of partial actin gene. Lane 1, Hox1-1; lanes 2 to 11, A3-hox1 transformants without clamps.
Two separated introduced hox genes increase the frequency of clamps.
Because transformation with a single compatible homeodomain protein gene was not sufficient for clamp formation, we examined whether a pair of homeodomain protein genes was needed for clamp formation. We selected Hox2-1 as the host strain for the second transformation and introduced A3-hox1 into it by using pMBhph1. The Hox2-1 strain is a single homeodomain protein gene transformant expressing A3-hox2. About 200 colonies that were resistant to carboxin and hygromycin B were collected and grown on new MYG plates containing both antibiotics, and the fusion of hook cells were assessed by microscopy. Among these 200 colonies, 21 colonies seem to increase with clamps, implying that they might receive a copy of A3-hox1 gene. The ratio of clamps to total clamplike cells was calculated in these colonies. These colonies contained increased ratios of clamps (ca. 50%), and partial clamp data are shown in Table. 2. DAPI and Fluorescent Brightener 28 staining confirmed that some nuclei trapped in the hook cells and that some hooks were fused without nuclei (Fig. 3D).
Using PCR amplification, we detected A4-hox1, A4-hox2, A3-hox1, and A3-hox2 in the genomes of all the cotransformants with increased ratios of clamps, and partial data are shown in Fig. 4B (Hox2-hox1-1, Hox2-hox1-2, and Hox2-hox1-3 strain, lanes d, e, and f). Southern hybridization analysis also confirmed that the A3-hox2 gene was still present in the chromosomes of these transformants with the same detective band as the host strain Hox2-1 and that A3-hox1 was ectopically integrated into the chromosomes of the cotransformants (data not shown).
Two introduced combined hox genes also increase the frequency of clamps.
By successively introducing a pair of homeodomain protein genes to A4 strain NGW19-6, a significant increase in true clamps was found in the transformants. So, we wondered whether the same phenomenon occurs after transformation with A3-hox1 and A3-hox2 gene fragments that are linked together like the native genes. Using pMBsip2, we cotransformed pMBhox12, which was obtained by connecting the fragment of A3-hox1 and A3-hox2 gene to pT7Blue(R) T-vector, into the A4 strain NGW19-6, along with marker plasmid pMBsip2. We collected ∼120 regeneration colonies and placed them on new MYG plates that contained 2 μg of carboxin/ml. Eight transformants with clamplike cells were found among these carboxin-resistant colonies, and partial clamp formation data are shown in Table. 2. The ratio of clamps to total clamplike cells (ca. 50%) and the mycelium configuration in these transformants (Hox1,2-1, Hox1,2-2, and Hox1,2-3 strains) are similar to those of transformants that were successively transformed with A3-hox1 and A3-hox2 (see Table 2 and Fig. 3E).
PCR amplification indicated that DNA fragments containing A3-hox1 and A3-hox2 exist in almost transformants with clamps, and partial data are shown in Fig. 4A (Hox1,2-1, Hox1,2-2, and Hox1,2-3 strains, lanes 24, 25, and 26). Southern hybridization confirmed that both A3-hox1 and A3-hox2 were ectopically integrated into the chromosomal DNA (data not shown).
Greater expression of the hox genes drives the real clamp formation.
When A3-hox1 or A3-hox2 was introduced into A4 strain NGW19-6, clamps were only rarely detected in the cotransformants. When A3-hox1 and A3-hox2, either separately or together, were introduced into NGW19-6, ca. 50% clamp formation was detected in the transformants expressing two hox genes. Thus, the following experiments were conducted to determine the effect of greater expression of the hox genes on true clamp formation. We connected the code regions of A3-hox1 and A3-hox2 to the sip promoter and constructed pMBsiphox1 and pMBsiphox2, respectively (see Fig. 2). Using a carboxin-resistant selective marker, we introduced pMBsiphox1 or pMBsiphox2 into A4 strain NGW19-6. In each transformation, ∼150 regenerated colonies were collected and grown on MYG plates containing 2.0 μg of carboxin/ml, and then the clamplike cell formation was examined microscopically. In the transformation of pMBsiphox1, there were 23 colonies containing clamplike cells. The ratios of clamps to clamplike cells in these cotransformants were calculated, and representative data are shown in Table 2. The representative Shox1-1 and Shox1-2 colonies with introduced pMBsiphox1 exhibited >85% real clamps among the clamplike cells (see Table 2). Nuclei and cell wall staining of the mycelium of these two transformants confirmed that the majority of clamplike cells were not pseudoclamps and that most cells contained two nuclei (see Fig. 3F and Table 3). The transformation of pMBsiphox2 yielded 30 carboxin-resistant transformants with clamplike cells. In these cotransformants, the ratios of real clamps among the clamplike cells and the numbers of nuclei per cell are similar to those of the cotransformants with pMBsiphox1 (see Table 2).
Table 3.
Nuclei per cell in transformants with pMBsiphox1 and pMBsiphox2
| Strain | No. of cells with: |
Total no. of cells counted | Ratio (%)a | |||
|---|---|---|---|---|---|---|
| No nucleus | One nucleus | Two nuclei | Three nuclei | |||
| Shox1-7 | 13 | 9 | 99 | 7 | 128 | 77.3 |
| Shox1-51 | 6 | 3 | 87 | 1 | 97 | 89.7 |
| Shox2-4 | 1 | 8 | 94 | 1 | 104 | 90.3 |
| Shox2-15 | 5 | 7 | 88 | 3 | 103 | 85.4 |
That is, the ratio of two nuclei in the total counted cells.
Amplification with the primers the Ip-d1R and A3-hox1-SacII, which correspond to the near 5′ end of the sip promoter and the 3′ end of the A3-hox1 gene, respectively, yielded a band of the expected size (∼3.5 kbp) in almost cotransformants, and partial strains Shox1-1 and Shox1-2 were shown in Fig. 4A (lanes 14 and 15). No bands were amplified from the genomic DNA of host strains NGW19-6 and NGW12-163 (see Fig. 4A, lanes 11 and 12). In Southern hybridization, the partial A3-hox1 gene sequence was used as the probe. BamHI and BglII, located at the sip promoter and the A3-hox1 gene, respectively, were used to cut the genomic DNA. A band of the expected sized (∼2.3 kbp), including the partial sip promoter and the A3-hox1 gene, was detected in Shox1-1 and Shox1-2 strain, and a band with a different size (∼4.7 kbp) was detected in the A3 strain NGW12-163 (data not shown). Also, other bands exist in Shox1-2 strain, which may result from a different type of ectopic integration (data not shown). Similar results were obtained for the Shox2-1 and Shox2-2 strains introduced with pMBsiphox2 (see Fig. 4A, lanes 19 and 20). These results suggest that the fused DNA fragment containing the sip promoter and A3-hox1 or A3-hox2 was ectopically integrated into the chromosome of NGW19-6.
Different growth conditions were observed in different kinds of transformants.
In the transformation introduced with single A3-hox1, the cotransformants had a different mycelium configuration than other carboxin-resistant transformants, with no clamplike cells. The A3-hox1 cotransformants had procumbent mycelium, uneven colony borders, slower mycelium growth (∼0.13 cm/day in the MYG plate without carboxin) than NGW19-6 (∼0.26 cm/day), and brown deposits around the inoculum. The A3-hox2 cotransformants had relatively abundant aerial mycelia, uneven colony borders, and slower mycelium growth (∼0.14 cm/day). The carboxin-resistant transformants with no clamplike cells demonstrated abundant mycelia, smooth colony borders, and faster growth, similar to that of the host strain, NGW19-6.
Cotransformants with two combined or separately introduced hox genes, when grown on MYG plates without carboxin and hygromycin B, showed abundant aerial mycelium, faster growth (∼0.17 cm/day) than transformants containing a single introduced homeodomain protein gene, and a radiating mycelium configuration similar to that of the wild-type dikaryon (NGW19-6 × NGW12-163).
In the transformants with greater expression of the introduced hox gene, most of the colonies also had abundant aerial mycelia, a moderate growth rate (∼0.18 cm/day on MYG plates without carboxin), and a radiating mycelium configuration similar to that of the wild-type dikaryon (NGW19-6 × NGW12-163).
Different expression amounts of four hox genes in different kinds of transformants.
When A3-hox1 or A3-hox2 was introduced into A4 strain NGW19-6, clamps were only rarely detected in the cotransformants (representative strains, Hox1-1, Hox1-2, Hox2-1, and Hox2-2). When A3-hox1 and A3-hox2, either separately or together, were introduced into NGW19-6, ca. 50% clamp formation was detected in all of the cotransformants expressing two hox genes (representative strains, Hox2-hox1-1, Hox2-hox1-2, Hox1-hox1-3, Hox1,2-2, and Hox1,2-3). When A3-hox1 or A3-hox2 connected to the sip promoter was used for transformation, there was >85% real clamp among clamplike cells in the cotransformants (representative strains, Shox1-1, Shox1-2, Shox2-1, and Shox2-2). Thus, we considered the possibility that hook cell fusion is affected by the expression level of homeodomain protein genes. Therefore, the rationale for the following experiments of real-time RT-PCR is to measure the expression amount of the hox gene, which may directly affect the clamp formation, in these different kinds of transformants. The level of transcription was determined in triplicate for all transformants.
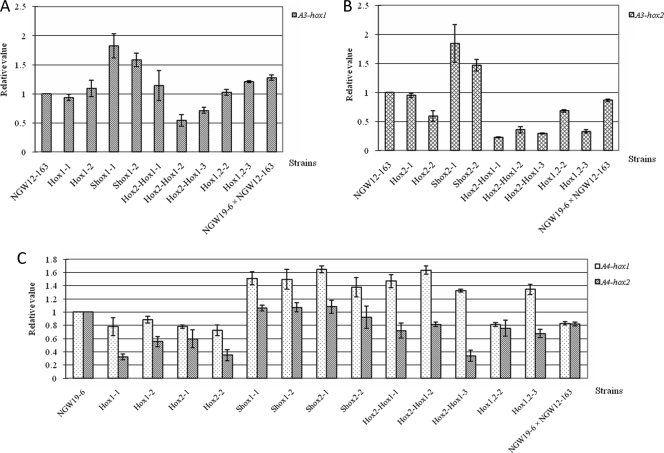
The quantities of A3-hox1 and A3-hox2 transcription in host strains NGW12-163 and NGW19-6 were used as reference values and set at 100%. In Hox1-1 and Hox1-2 strains, which contain only A3-hox1, the relative values of A3-hox1 (0.94 and 1.09) were close to that of NGW12-163 (see Fig. 5). In the Hox2-1 and Hox2-2 strains, which contain only A3-hox2, the relative values of A3-hox2 were 0.95 and 0.60 (mean). In the Hox1-1, Hox1-2, Hox2-1, and Hox2-2 strains, the relative values of A4-hox1 were ∼0.80, and the values of A4-hox2 ranged from 0.3 to 0.6. In the transformants with successively introduced A3-hox2 and A3-hox1, the Hox2-hox1-1, Hox2-hox1-2, and Hox2-hox1-3 strains, the transcription levels of A3-hox1 were different (range, 0.5 to 1.2). Their relative values of A3-hox2 were <0.4, which is different than the value in the Hox2-1 strain. This phenomenon is very interesting. It seems that after the introduction of A3-hox1 into the Hox2-1 strain, the expression of A3-hox2 was suppressed from 0.95 to <0.4. In the Hox1,2-2 and Hox1,2-3 strains, which contain both A3-hox2 and A3-hox1, the mean of the relative value of A3-hox1 was near that of A3 strain NGW12-163, and the relative values of A3-hox2 were 0.68 and 0.33, respectively. Compared to the transformants that contain only a single homeodomain protein gene (Hox1-1, Hox1-2, Hox2-1, and Hox2-2), the levels of A4-hox1 transcription in the Hox2-hox1-1, Hox2-hox1-2, Hox2-hox1-3, Hox1,2-2, and Hox1,2-3 strains are increased (range, 0.8 to 1.6), while their transcription of A4-hox2, with the exception of Hox2-hox1-3, was also increased (range, 0.7 to 1.0). These results suggest that the gene dosage of homeodomain protein genes also affects the expression amount of four homeodomain protein genes (A4-hox1, A4-hox2, A3-hox1, and A3-hox2). In the wild-type dikaryon (NGW19-6 × NGW12-163), except for the relative value of A3-hox1 (>1.2), the transcription level of the other three homeodomain protein genes was ∼0.9. Compared to the transformants containing both A3-hox1 and A3-hox2, the wild-type dikaryon (NGW19-6 × NGW12-163) had a greater expression level of A3-hox2 and a lower expression level of A4-hox1.
Fig. 5.
Transcription levels of four homeodomain protein genes (A3-hox1, A3-hox2, A4-hox1, and A4-hox2) in the host strain, dikaryon, and transformants. (A) Level of transcription of A3-hox1. The transcription of A3-hox1 in the A3 strain NGW12-163 was used as a reference value and set at 100%. (B) The quantity of transcription of A3-hox2. The transcription of A3-hox2 in the A3 strain NGW12-163 was used as reference values and set at 100%. (C) Level of transcription of A4-hox1 and A4-hox2. The transcription of A4-hox1 and A4-hox2 in A4 strain NGW19-6 was used as a reference value for A4-hox1 and A4-hox2 in other transformants and the dikaryon, respectively, and set at 100%. The error bars indicate standard deviations (n = 3). The sources of strains and cotransformants were as follows: NGW12-163 (A3 strain); NGW19-6 (A4 strain); Hox1-1 and Hox1-2 (introduced with A3-hox1); Hox2-1 and Hox2-2 (introduced with A3-hox2); Shox1-1 and Shox1-2 (introduced with pMBsiphox1); Shox2-1 and Shox2-2 (introduced with pMBsiphox2); Hox2-hox1-1, Hox2-hox1-2, and Hox2-hox1-3 (separately introduced with A3-hox1 and A3-hox2); Hox1,2-2 and Hox1,2-3 (introduced with combined A3-hox1 and A3-hox2); and NGW19-6 × NGW12-163 (wild dikaryon).
In the transformants Shox1-1 and Shox1-2 with introduced pMBsiphox1, the relative values of A3-hox1 (1.5) were greater than those of the Hox1-1 and Hox1-2 strains. In the transformants Shox2-1 and Shox2-2 with introduced pMBsiphox2, the transcription quantity of A3-hox2 (1.4) was greater than that of the Hox2-1 and Hox2-2 strains. From these results, we conclude that the promoter activity of sip is higher than the activity of the native promoter of homeodomain protein genes in P. microspora. Meanwhile, in the Shox1-1, Shox1-2, Shox2-1, and Shox2-2 strains, the transcription amount of A4-hox1 is increased (1.4), and the relative values of A4-hox2 were near 1.0.
Weak protein interaction was detected between Hox proteins from different A mating types.
When A3-hox1 or A3-hox2 was introduced into A4 strain NGW19-6, clamps were only rarely detected in the cotransformants. The reason for this may be relate to the interaction intensity of Hox proteins from different A mating type in P. microspora. Thus, we examined the Hox proteins interaction in this species by using the yeast two-hybrid system. We examined the α-galactosidase activity in yeast transformants with different plasmids containing the hox gene (Table 4). No α-galactosidase activity was detected in negative controls (GADT7-T/GBKT7-Lam). GAD and GBK fusions with the same homeodomain gene (GAD-A4-hox2/GBK-A4-hox2, GAD-A3-hox2/GBK-A3-hox2) did not activate transcription. This result indicates that homeodomain protein genes from the same A mating type do not undergo a protein-protein interaction. Non-self-combinations (GAD-A4-hox2/GBK-A3-hox1, GAD-A3-hox2/GBK-A4-hox1) had activated MEL1, indicating that A4-Hox2, A3-Hox1, A3-Hox2, and A4-Hox1 interact. However, the values were very low, ca. 0.033 and 0.023 mU/ml per cell, respectively, suggesting that their interaction intensity is very weak. The GAD-A4-hox2/GBK-A4-hox1 combination also had no α-galactosidase activity. A very low level of α-galactosidase activity, which may be only background activity, was detected between GAD-A3-hox2 and GAD-A3-hox1.
Table 4.
Two-hybrid analysis of A4-Hox1, A4-Hox2, A3-Hox1, and A3-Hox2 interaction
| GAL4 domaina |
α-Galactosidase activity (mU/ml per cell) | |
|---|---|---|
| DNA binding | Activation | |
| GAD-T | GBK-lam | – |
| GAD-A4-hox2 | GBK-A3-hox1 | + |
| GBK-A3-hox2 | – | |
| GBK-A4-hox1 | – | |
| GBK-A4-hox2 | – | |
| GAD-A3-hox2 | GBK-A3-hox1 | – |
| GBK-A3-hox2 | – | |
| GBK-A4-hox1 | + | |
| GBK-A4-hox2 | – | |
GAD and GBK denote the Clontech cloning vector with the GAL4 DNA binding (GADT7) and activation (GBKT7) domains, respectively. GAD-A4-hox2 and GAD-A3-hox2 denote vectors containing the A4-hox2 and A3-hox2 cDNAs described in Materials and Methods.
DISCUSSION
In the bipolar mushroom C. disseminatus, the functions of mating type were studied in a tetrapolar mushroom, C. cinerea, instead of in a homologous bipolar species (16). In this research, we used a homologous transformation system to determine the functions of the A mating type in bipolar mushroom P. microspora. This approach identifies individual proteins, and the functions of HD proteins we verified are truly the mating-type determinants.
In a previous study, it was found that pheromone receptor protein genes in the P. microspora are not part of the MAT locus and only homeodomain protein genes are involved in the mating incompatibility (1). However, how do the homeodomain proteins in these species determine the mating identity? Can it be confirmed that homeodomain proteins control dikaryosis and clamp formation through transformation studies in this bipolar species? To address these questions, a single homeodomain protein gene (A3-hox1 or A3-hox2) from A3 strain was introduced into an A4 strain. Unfortunately, few fusion hooks were detected in the cotransformants expressing the introduced homeodomain protein gene. Therefore, we considered the possibility that both homeodomain protein gene (A3-hox1 and A3-hox2) are needed for hook cell fusion and separately introduced both hox genes into the A4 strain. The cotransformants expressing both introduced hox genes demonstrated a significantly increased ratio of clamps among total clamplike cells (ca. 50%). Similar results were also detected in cotransformants introduced with A3-hox1 and A3-hox2 gene fragments that are linked together like the native genes. This also excluded the possibility that the promoter region of the homeodomain protein gene not only exists in the homologous spacer region between A3-hox1 and A3-hox2 but also in the opposite homeodomain protein gene region, since A3-hox1 or A3-hox2 containing part of the promoter region can be expressed at a low level. That similar results were obtained when the two combined hox genes were used for transformation excluded a possible problem caused by the promoter.
When we connected the sip promoter to the coding region of the A3-hox1 and A3-hox2 genes and introduced the fused fragment into the A4 strain NGW19-6, more than 85% of the clamplike cells in transformants were true clamps, and each cell contained two nuclei. The real-time RT-PCR results indicated that the promoter activity of sip is higher than that of the homeodomain protein gene in P. microspora. Based on these results, we concluded that complete clamp formation is controlled by the expression level of homeodomain protein genes and that altered expression of A-mating-type genes is sufficient to drive true clamp formation.
In Shox1-1, Shox1-2, Shox2-1, and Shox2-2 strains, only the A3-hox1 or the A3-hox2 gene was under the control of the sip promoter. However, the amount of A4-hox1 and A4-hox2 gene expression increased, surpassing the corresponding levels in the host strain NGW19-6 (A4) and the wild-type strain (NGW19-6 × NGW12-163). There are two possible reasons why the A4-hox1 and A4-hox2 gene expression were increased: (i) it could be caused by two nuclei in the same cell, or (ii) it could be caused by the self-regulation of homeodomain protein genes. These reasons may also explain the increased gene expression of the A4-hox1 gene in Hox2-hox1-1, Hox2-hox1-2, Hox1-hox1-3, and Hox1,2-3 strains.
Although a pair of homeodomain protein genes is needed for clamp-cell formation in P. microspora, only ca. 50% clamps were detected in the cotransformants. In the wild-type dikaryon, most clamplike cells were clamps (Table 3). These findings raise the question of how clamp formation is completed in the wild-type dikaryon. Perhaps in the wild-type dikaryon, clamp formation can also be achieved by changing expression levels, but the wild-type situation is still not completely determined and requires further research.
When protein-protein interactions of homeodomain proteins in Schizophyllum commune were tested by using filter and liquid assays for β-galactosidase, moderate β-galactosidase activities (∼0.44 U/h per cell in yeast strain HF7c and >1.3 U/h per cell in yeast strain FY526) between two homeodomain proteins from different Aα backgrounds were detected, confirming the ability of two different mating proteins to form a heterodimer (23). In our present research with yeast two-hybrid systems, we used an α-galactosidase assay, which is more sensitive than a β-galactosidase assay, to detect the Hox protein interaction. However, a low-intensity interaction between A4-Hox2 and A3-Hox1 (∼0.033 mU/ml per cell in yeast strain Y187), A3-Hox2 and A4-Hox1 (∼0.023 mU/ml per cell in yeast strain Y187) was detected by using the α-galactosidase assay. Thus, we hypothesized that it is possible that the low interaction intensity between homeodomain proteins from two compatible monokaryon strains requires more heterodimer production. To achieve this requirement, the gene dosage and the overexpression of homeodomain protein genes may induce the nearly 100% clamp formation in P. microspora. Also, heterodimers between homeodomain proteins may regulate the expression of homeodomain protein genes and promote clamp formation.
In tetrapolar mushrooms, fusion is clearly a function of the pheromone receptor signaling pathway. However, using transformation studies, we confirmed that the bipolar mushroom P. microspora does not use pheromone receptors to specify the mating type and that fusion of the hook cell is somehow accomplished via HD protein expression changes. The mating system of P. microspora is similar to semicompatible crosses with different A loci and common B loci (A≠ B=) in a tetrapolar mushroom (18), because during mating crosses the nucleus migration that is controlled by B loci in tetrapolar mushroom is very slow in this species (data not shown) and monokaryotized mycelia can easily be isolated from the peripheral growing zone in a dikaryotic colony (24). If this species evolves from a tetrapolar mushroom with semicompatible crosses, it is possible that in the tetrapolar mushroom B loci control the expression of A loci, which affect the fusion of the hook cell, whereas this species has common B loci and has to increase the expression amount of A loci by other means.
In tetrapolar mushrooms, a heterodimer of compatible HD1 and HD2 proteins is assumed to be a transcription factor that binds unique target sites within the promoters of genes that commit cells to a new developmental pathway. Although we know that the overexpression of homeodomain protein genes may induce nearly 100% clamp formation in P. microspora, we do not know whether the genes regulated by the heterodimer of homeodomain proteins have corresponding changes in expression. Our future research will address this question so that we may understand the gene regulation of clamp cell formation with homeodomain protein genes.
ACKNOWLEDGMENT
This research was supported in part by a grant from the Global COE Program.
Footnotes
Published ahead of print on 7 May 2010.
REFERENCES
- 1.Aimi T., Yoshida R., Ishikawa M., Bao D., Kitamoto Y. 2005. Identification and linkage mapping of the genes for the putative homeodomain protein (hox1) and the putative pheromone receptor protein homologue (rcb1) in a bipolar basidiomycete, Pholiota nameko. Curr. Genet. 48:184–194 [DOI] [PubMed] [Google Scholar]
- 2.Bakkeren G., Kronstad J. W. 1994. Linkage of mating-type loci distinguishes bipolar from tetrapolar mating in basidiomycetous smut fungi. Proc. Natl. Acad. Sci. U. S. A. 92:7085–7089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banham A. H., Asante-Owusu R. N., Gottgens B., Thompson S. A. J., Kingsnorth C. S., Mellor E. J. C., Casselton L. A. 1995. An N-terminal dimerization domain permits homeodomain proteins to choose compatible partners and initiate sexual development in the mushroom Coprinus cinereus. Plant Cell 7:773–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnboim H. C. 1983. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 100:243–255 [DOI] [PubMed] [Google Scholar]
- 5.Casselton L. A., Challen M. P. 2006. The mating type genes of the basidiomycetes, p. 357–374 InKües U., Fischer R. (ed.) The mycota: growth, differentiation, and sexuality, 2nd ed., vol. 1, pt. 17 Springer, New York, NY [Google Scholar]
- 6.Casselton L. A., Kües U. 2007. The origin of multiple mating types in the model mushrooms Coprinopsis cinerea and Schizophyllum commune, p. 283–300 InHeitman J., Kronstand J. W., Taylor J. W., Casselton L. A. (ed.), Sex in fungi ASM Press, Washington, DC [Google Scholar]
- 7.Dellaporta S. L., Wood J., Hicks J. B. 1983. A plant DNA minipreparation, version II. Plant Mol. Biol. Reprod. 1:19–21 [Google Scholar]
- 8.Doudrick R. L., Anderson N. A. 1989. Incompatibility factors and mating competence of two Laccaria spp. (Agaricales) associated with black spruce in northern Minnesota. Phytopathology 79:694–700 [Google Scholar]
- 9.Fowler T. J., Mitton M. F., Rees E. I., Raper C. 2004. Crossing the boundary between the B alpha and B beta mating-type loci in Schizophyllum commune. Fungal Genet. Biol. 41:89–101 [DOI] [PubMed] [Google Scholar]
- 10.Frankel C., Ellingboe A. H. 1977. Sexual incompatibility factors and somatic recombination in Schizophyllum commune. Genetics 85:427–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fries N., Mueller G. M. 1984. Incompatibility systems, cultural features and species circumscriptions in the ectomycorrhizal genus Laccaria (Agaricales). Mycologia 76:633–642 [Google Scholar]
- 12.Harper J. W., Adami G. R., Wei N., Keyomarsi K., Elledge S. J. 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805–816 [DOI] [PubMed] [Google Scholar]
- 13.Hiscock S. J., Ursula Kües U., Dickinson H. G. 1996. Molecular mechanisms of self-incompatibility in flowering plants and fungi—different means to the same end. Trends Cell Biol. 6:411–450 [DOI] [PubMed] [Google Scholar]
- 14.Ito H., Fukada Y., Murata K., Kimura A. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwasa M., Tanabe S., Kamada T. 1998. The two nuclei in the dikaryon of the homobasidiomycete Coprinus cinereus change position after each conjugate division. Fungal Genet. Biol. 23:110–116 [DOI] [PubMed] [Google Scholar]
- 16.James T. Y., Srivilai P., Kües U., Vilgalys R. 2006. Evolution of the bipolar mating system of the mushroom Coprinellus disseminatus from its tetrapolar ancestors involves loss of mating-type-specific pheromone receptor function. Genetics 172:1877–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamper J., Reichamann M., Romeis T., Bolker M., Kahmann R. 1995. Multiallelic recognition: nonself-dependent dimerization of the bE and bW homeodomain proteins in Ustilago maydis. Cell 81:73–83 [DOI] [PubMed] [Google Scholar]
- 18.Kothe E. 1999. Mating types and pheromone recognition in homobasidiomycete Schizophyllum commune. Fungal Genet. Biol. 27:146–152 [DOI] [PubMed] [Google Scholar]
- 19.Kropp B. R., Fortin J. A. 1988. The incompatibility system and relative ectomycorrhizal performance of monokaryons and reconstituted dikaryons of Laccaria bicolor. Can. J. Bot. 68:289–294 [Google Scholar]
- 20.Kües U., Asante-Owusu R. N., Mutasa E. S., Tymon A. M., Pardo E. H., O'Shea S. F., Gottgens B., Casselton L. A. 1994. Two classes of homeodomain proteins specify the multiple A mating types of the mushroom Coprinus cinereus. Plant Cell 6:1467–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kües U., Casselton L. A. 1992. Homeodomains and regulation of sexual development in basidiomycetes. Trends Genet. 8:154–155 [DOI] [PubMed] [Google Scholar]
- 22.Lazo P. S., Ochoa A. G., Gascon S. 1978. α-galactosidase (melibiase) from Saccharomyces Carlsbergenesis: structural and kinetic properties. Arch. Biochem. Biophys. 191:316–324 [DOI] [PubMed] [Google Scholar]
- 23.Magae Y., Novotny C., Ullrich R. 1995. Interaction of the Aα Y mating-type and Z mating-type homeodomain proteins of Schizophyllum commune detected by the two-hybrid system. Biochem. Biophys. Res. Commun. 211:1071–1076 [DOI] [PubMed] [Google Scholar]
- 24.Masuda P., Yamanaka K., Sato Y., Kitamoto Y. 1995. Nuclear selection in monokaryotization of dikaryotic mycelia of Pholiota nameko as described by leading and following nuclei. Mycoscience 36:413–420 [Google Scholar]
- 25.Niculita-Hirzel H., Labbé J., Kohler A., le Tacon F., Martin F., Sanders I. R., Kües U. 2008. Gene organization of the mating type regions in the ectomycorrhizal fungus Laccaria bicolor reveals distinct evolution between the two mating type loci. New Phytol. 180:329–342 [DOI] [PubMed] [Google Scholar]
- 26.O'Shea S. F., Chaure P. T., Halsall J. H., Olesnicky N. S., Leibbrandt A., Connerton I. F., Casselton L. A. 1998. A large pheromone and receptor gene complex determines multiple B mating type specificities in Coprinus cinereus. Genetics 148:1081–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raper J. R. 1966. Genetics of sexuality in higher fungi, p. 1–283 The Ronald Press, New York, NY [Google Scholar]
- 28.Raper C. A. 1983. Controls for development and differentiation of the dikaryon in basidiomycetes, p. 195–236 InBennett J. W., Ciegler A. (ed.), Secondary metabolism and differentiation in fungi, vol. 15 Marcel Dekker, Inc., New York, NY [Google Scholar]
- 29.Ratanatragooldacha S., Hui C., Kumata A., Kitamoto Y. 2002. The constitution of incompatibility factor and mating characteristics of spore isolates in a bipolar mushroom, Pholiota nameko. Mycoscience 43:113–117 [Google Scholar]
- 30.Riquelme M., Challen M. P., Casselton L. A., Brown A. J. 2005. The origin of multiple B mating specificities in Coprinus cinereus. Genetics 170:1105–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen G. P., Park D. C., Ullrich R. C., Novotny C. P. 1996. Cloning and characterization of a Schizophyllum gene with A beta 6 mating type activity. Curr. Genet. 29:136–142 [DOI] [PubMed] [Google Scholar]
- 32.Stankis M. M., Specht C. A., Yang H., Giasson L., Ullrich R. C., Novotny C. P. 1992. The A alpha mating locus of Schizophyllum commune encodes two dissimilar multiallelic homeodomain proteins. Proc. Natl. Acad. Sci. U. S. A. 89:7169–7173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yi R., Tachikawa T., Ishikawa M., Mukaiyama H., Bao D., Aimi T. 2009. Genomic structure of the A mating-type locus in a bipolar basidiomycete, Pholiota nameko. Mycol. Res. 113:240–248 [DOI] [PubMed] [Google Scholar]
- 34.Yi R., Tachikawa T., Mukaiyama H., Mochida Y., Ishikawa M., Aimi T. 2009. DNA-mediated transformation system in a bipolar basidiomycete, Pholiota microspora (P. nameko). Mycoscience 50:123–129 [Google Scholar]
- 35.Vaillancourt L. J., Raudaskoski M., Specht C. A., Raper C. A. 1997. Multiple genes encoding pheromones and a pheromone receptor define the B1 mating-type specificity in Schizophyllum commune. Genetics 146:541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wendland J., Vaillancourt L. J., Hegner J., Lengeler K. B., Laddison K. J., Specht C. A., Raper C. A., Kothe E. 1995. The mating-type locus B alpha 1 of Schizophyllum commune contains a pheromone receptor gene and putative pheromone genes. EMBO J. 14:5271–5278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitehouse H. L. K. 1949. Multiple allelomorph heterothallism in the fungi. New Phytol. 48:212–244 [Google Scholar]