Abstract
The localization and local translation of mRNAs constitute an important mechanism to promote the correct subcellular targeting of proteins. mRNA localization is mediated by the active transport of mRNPs, large assemblies consisting of mRNAs and associated factors such as RNA-binding proteins. Molecular motors move mRNPs along the actin or microtubule cytoskeleton for short-distance or long-distance trafficking, respectively. In filamentous fungi, microtubule-based long-distance transport of vesicles, which are involved in membrane and cell wall expansion, supports efficient hyphal growth. Recently, we discovered that the microtubule-mediated transport of mRNAs is essential for the fast polar growth of infectious filaments in the corn pathogen Ustilago maydis. Combining in vivo UV cross-linking and RNA live imaging revealed that the RNA-binding protein Rrm4, which constitutes an integral part of the mRNP transport machinery, mediates the transport of distinct mRNAs encoding polarity factors, protein synthesis factors, and mitochondrial proteins. Moreover, our results indicate that microtubule-dependent mRNA transport is evolutionarily conserved from fungi to higher eukaryotes. This raises the exciting possibility of U. maydis as a model system to uncover basic concepts of long-distance mRNA transport.
In order to compartmentalize functions, eukaryotic cells need to sort their proteins to distinct subcellular sites. A widespread mechanism for the spatiotemporal regulation of protein expression is localized translation, i.e., the concerted action of mRNA localization and confined translation. Thereby, the correct subcellular localization of translation products is promoted, and the deleterious mislocalization of proteins is prevented (5, 37).
Most commonly, mRNA localization is mediated by active transport along the actin or microtubule cytoskeleton for short-distance or long-distance mRNA transport, respectively. Transported mRNAs contain specific cis-acting sequences that function as zipcodes to determine the correct subcellular destination. These RNA elements are recognized by RNA-binding proteins that combine with accessory factors to form higher-order ribonucleoprotein complexes, designated mRNPs (40, 82). Adaptor proteins are thought to connect mRNPs to molecular motors that actively transport them along the cytoskeleton to their final destination (88, 92). Commonly, premature translation is inhibited during mRNP transport by specific inhibitors. Upon arrival mRNAs are offloaded and kept in place by anchoring factors. The local phosphorylation of RNA-binding proteins then triggers unloading and the release of translational inhibitor (39, 68). When the formation of transport-competent mRNPs fails, mRNAs are translated at wrong locations, leading to the mislocalization of the encoded proteins. An example of the importance of mRNA localization is the local synthesis of morphogens during oogenesis and embryogenesis in Drosophila melanogaster, which determines the two main body axes of developing embryos (55, 59).
In fungi, actin-dependent transport was quite extensively studied for Saccharomyces cerevisiae and was recently discovered in filaments of Candida albicans (25, 65, 68). Examples of long-distance mRNA transport along microtubules have so far been reported only for the corn pathogen Ustilago maydis. Study of the role of RNA-binding proteins during filamentous growth and pathogenic development revealed that microtubule-dependent mRNP transport is essential for the fast polar growth of infectious hyphae (6, 7, 26, 45). In this review we will introduce the basic aspects of short- and long-distance mRNA transports in fungal and animal models. In addition, we will shortly address polar growth and microtubule-dependent transport in filamentous fungi. This will be the foundation to present recent advances in the microtubule-dependent transport of mRNAs in U. maydis.
ACTIN-DEPENDENT mRNA TRANSPORT IN HIGHER EUKARYOTES
Many studies of actin-dependent mRNA transport in higher eukaryotes were done with migrating cells, which are an excellent model system to study the role of mRNA transport (62, 72). In chicken fibroblasts, β-actin mRNA localizes to the lamellipodia, where its translation allows actin polymerization at the leading edge (18). The transport of β-actin mRNA is at least in part actin dependent and requires the molecular motor myosin II-B (50). A key factor for localization is the RNA-binding protein ZBP1 (zipcode-binding protein 1) that associates with cargo mRNA during transcription (66). ZBP1 recognizes a specific zipcode in the 3′ untranslated region (UTR) of β-actin mRNA that is necessary and sufficient for peripheral localization (44). ZBP1 is also crucial for the translational repression of its target mRNA, which is released at its destination by the local phosphorylation of the RNA-binding protein via Src kinase (39). mRNA anchoring most likely involves the concerted action of cortical actin and the translation elongation factor EF1α (56). In essence, this actin-dependent mRNA transport system results in the local synthesis of β-actin at the site of demand, enabling the efficient polymerization of cytoskeletal filaments during cell migration (18).
ACTIN-DEPENDENT mRNA TRANSPORT IN FUNGI
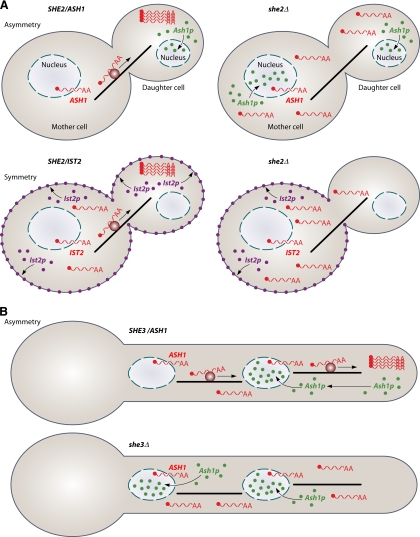
In fungi, the best-studied example of mRNA transport is the localization of ASH1 mRNA during the mating-type switching of S. cerevisiae (24, 65, 68, 97). Ash1p is a transcriptional repressor that prevents mating-type switching in daughter cells upon division. Its asymmetric distribution is efficiently achieved by the transport of ASH1 mRNA to the distal pole of the emerging bud. Here it is locally translated, ensuring that Ash1p first encounters the nucleus of the daughter cell. In the absence of mRNA transport, the protein accumulates in both nuclei, and mating-type switching is no longer asymmetric (Fig. 1A) (40, 65).
Fig. 1.
Actin-dependent mRNA transport in fungi. (A) Schematic representation of mRNA transport in S. cerevisiae during budding. (Top) mRNA transport results in the asymmetric distribution of the encoded protein. ASH1 mRNA (red wavy line) is transported along the actin cytoskeleton (black bar) from the mother to the daughter cell. Active transport is mediated by the SHE machinery (dark red circle), consisting of the key RNA-binding protein She2p, the potential adapter She3p, and the myosin motor Myo4p. Upon translation at the distal pole of the daughter cell, the transcription factor Ash1p predominantly enters the nucleus of the daughter cell. Genetic ablation (she2Δ) causes a mislocalization of mRNAs and translated proteins (right). (Bottom) mRNA transport is important for the symmetric distribution of the encoded protein. IST2 mRNA is transported by the same machinery to the daughter cell. The translation product localizes to the cell membrane. In the absence of mRNA transport (she2Δ), Ist2p localizes predominantly in the membrane of the mother cell (right). (B) mRNA transport in filaments of C. albicans. (Top) ASH1 mRNA is most likely transported by a related SHE machinery to the hyphal tip. Thereby, Ash1p is targeted to the first nucleus within the filament. (Bottom) In the absence of mRNA transport (she3Δ), Ash1p enters both nuclei.
Intensive studies over the last two decades have uncovered the basic mechanisms of this mRNA transport process. mRNA is bound directly by She2p, which recognizes four defined zipcodes in the ASH1 mRNA (9, 57). She2p is connected to the molecular motor Myo4p via the putative adaptor protein She3p, which results in the transport of mature mRNPs along the actin cytoskeleton. During transport translation is inhibited by the combined action of She2p, Khd1p, and Puf6p. At the distal pole, the membrane-associated kinase Yck1p phosphorylates Khd1p to reduce its RNA binding capacity (69). Thus, translational repression is released, leading to local protein synthesis (65, 68).
In recent years, however, this transport process turned out to be more complex than initially anticipated. Besides ASH1 mRNA, the responsible machinery transports more than 20 additional mRNAs encoding predominantly membrane-associated proteins such as Ist2p (77, 87). In the absence of mRNP transport, Ist2p is restricted to the plasma membrane of the mother cell (42), suggesting that mRNP transport provides the daughter cell with IST2 mRNA to guarantee the symmetric distribution of the encoded protein (Fig. 1A). Thus, mRNP transport is used to generate symmetry as well as asymmetry in S. cerevisiae (42, 77).
In addition, polarity factors such as the small G protein Rho3p are important for mRNA transport (3). Indeed, mRNAs encoding polarity factors localize to the daughter cell pole prior to protein localization and polarized growth. This indicates an intimate relationship between the localization of polarity factors and mRNA transport, and vice versa (2).
Furthermore, the cotransport of ASH1 mRNPs with tubules of the endoplasmic reticulum (ER) was described to be an alternative transport route. In this scenario, She2p might serve as the membrane anchor (74). Such membrane hitchhiking might also occur in other systems (17, 31).
In the human pathogen C. albicans, it was recently discovered that actin-dependent mRNA transport regulates hyphal morphology and invasive growth. Using She3 as a molecular handle for affinity purification, a set of about 40 transported mRNAs was identified. A comparison with S. cerevisiae revealed that cargo mRNAs diverged considerably, reflecting the individual growth needs of these two species. Investigations of ASH1 mRNA localization in this species revealed that mRNAs accumulate at hyphal tips and that the encoded transcription factor localizes predominantly to the apical nucleus. The genetic ablation of She3 caused mislocalization toward basal nuclei (Fig. 1B) (25). Thus, actin-dependent mRNA transport is important to support local expression in fungal filaments.
MICROTUBULE-DEPENDENT mRNA TRANSPORT IN HIGHER EUKARYOTES
Until recently, microtubule-dependent mRNA transport was known only for higher eukaryotes, where mRNA localization and confined translation are important during developmental and neuronal processes. This has been intensively studied during oogenesis and embryogenesis in D. melanogaster (37). Indeed, ca. 70% of expressed mRNAs exhibit distinct localization patterns in fly embryos (51). The most prevalent mechanism of mRNA localization is the long-distance trafficking of mRNPs along microtubules. Active transport is mediated by microtubule-dependent molecular motors such as plus-end-directed kinesins and minus-end-directed dyneins (35, 88).
At early stages of oogenesis, maternal mRNAs that are transcribed in ovarian nurse cells are delivered to the developing oocyte, most likely along microtubules (16, 64). Within oocytes a distinct localization of bicoid and oskar mRNAs, which encode key determinants of embryonic development, is essential to organize both main body axes (5, 82). oskar mRNA localizes to the posterior pole of the oocyte. Active transport along microtubules of oskar mRNPs containing the RNA-binding protein Staufen was shown previously to depend on the integrity of the kinesin heavy chain (67, 98). An indirect effect could not be completely ruled out, however, since kinesin is also crucial for the transport of dynein to the plus ends (14, 41). Surprisingly, the oskar mRNPs travel in all possible directions within the oocyte. However, detailed quantification experiments of the net movement of these particles led to the hypothesis that a biased random walk along a weakly polarized cytoskeleton results in the posterior localization of oskar mRNAs over time (98).
bicoid mRNA localizes to the anterior pole of the oocyte. The encoded protein is important for the anteroposterior axis but exerts its function only after fertilization. Localization is achieved by continuous active transport along microtubules in a dynein-dependent fashion (95). Apparently, bicoid mRNPs travel on a specific subpopulation of microtubules toward the anterior pole (5, 95), a concept that is consistent with recent findings from other animal systems (61, 63).
During early embryogenesis, pair-rule transcripts such as hairy apically localize in the syncytial blastoderm embryo by microtubule-dependent transport. hairy mRNPs shuttle bidirectionally along microtubules whose minus ends face toward the apical cytoplasm. RNA zipcodes are recognized by Egl, which interacts with the dynein cofactor BicD (23). The number of zipcodes within a message might determine the amount of recruited Egl that in turn activates dynein via BicD. Thus, zipcode-containing mRNPs exhibit an increase in frequency, velocity, and duration of minus-end-directed transport, leading ultimately to the apical delivery of the mRNP (15, 23).
In essence, these examples of microtubule-dependent mRNA transport demonstrate common molecular mechanisms of short-distance and long-distance transport, in which RNA-binding proteins constitute key players. Importantly, the same mechanisms underlie microtubule-dependent mRNA transport in neurons. Here localized translation has been implicated in the growth of axons in developing neurons (54) and in synaptogenesis and synaptic plasticity in dendrites (19, 72), and associated defects are implicated in human pathologies (19, 20).
MICROTUBULE-DEPENDENT TRANSPORT IN FILAMENTOUS FUNGI
The establishment and maintenance of a defined axis of polarity determines the growth and shape of filamentous fungi. Hyphae expand at the apical pole and are partitioned by septa that function as molecular barriers (27, 33, 79, 84). Hyphal tip growth is supported by local exocytosis of vesicles involving the Spitzenkörper, a potential vesicle supply center positioned in the apical dome that contains exocytotic as well as endocytotic vesicles (32, 34). The long-distance transport of vesicles is thought to supply the Spitzenkörper during hyphal growth. Specialized vesicles such as chitosomes might contain enzymes and building blocks for hyphal tip expansion (4, 71).
Active transport by molecular motors constitutes an essential feature of polar growth. This is mediated by a highly ordered cytoskeletal network consisting of actin microfilaments and microtubules for short-distance and long-distance transport, respectively. Potential cargos that might be delivered to the region of active growth are enzymes for cell wall biosynthesis as well as landmark and signaling proteins. Whereas the actin cytoskeleton is absolutely essential for polar growth, the microtubule cytoskeleton confers speed and directionality (27, 33, 71, 79). For instance, in Aspergillus nidulans and Neurospora crassa the loss of microtubules causes a destabilization of the Spitzenkörper position and aberrant hyphal growth. Consistently, the genetic ablation of conventional kinesin causes a significant reduction of filamentous growth rates (70, 76). Potential cargos for this mode of transport are microtubule plus-end-binding proteins, vesicles (27, 46, 86), and even mRNAs (see below).
The corn pathogen U. maydis has become an important model fungus. Besides the study of the resulting plant pathology (13), U. maydis has the potential to make important contributions to the fields of DNA repair, signaling, molecular transport, and RNA biology (10, 26, 60, 80). An important breakthrough was the sequencing of the genome including manual refinement (43). This laid the foundation to establish techniques such as DNA microarray profiling, proteomics, and efficient gene-targeting strategies. Cells and filaments of this fungus are also particularly suitable for microscopic live imaging of vesicles, proteins, and RNA (12, 13, 45, 73, 80).
A switch from yeast-like to polar growth is essential for the pathogenicity of U. maydis, since the latter represents the infectious form of the fungus. Initially, two mating-compatible sporidia recognize each other using a pheromone/pheromone receptor system (11, 85). The conjugation tubes then grow toward each other and fuse at their tips (78), resulting in the formation of a filamentously growing dikaryon. These filaments grow with a defined axis of polarity, expanding at the apical pole and inserting retraction septa at the basal pole (Fig. 2A) (52, 81).
Fig. 2.
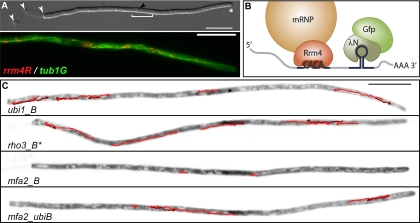
Rrm4-dependent mRNA transport in hyphae of U. maydis. (A, top) Monokaryotic filament of U. maydis growing with a defined axis of polarity. The filament expands at the hyphal tip (asterisk) and inserts retraction septa at the basal pole (white arrowheads). The elongated nucleus (white bracket; the black arrowhead indicates the nucleolus) is positioned in the center of the filament. (Bottom) Rrm4-Rfp-containing mRNP particles (red) colocalize with microtubules (green, Tub1-Gfp decorated). Since colocalization is nearly complete, almost all Rrm4 particles appear orange. (B) Schematic representation of the λN-Gfp RNA live-imaging system. Rrm4 (red) target mRNAs containing CA-rich zipcodes are labeled by the recruitment of λN-Gfp (green) to its specific binding sites (box B, hairpin) in the 3′ UTR of the message. (C) Filaments of strains expressing different candidate mRNAs. ubi1_B and rho3_B are direct targets of Rrm4, whereas mfa2_B serves as a control. mfa2_ubiB is a hybrid RNA consisting of mfa2 and the CA-rich 3′ UTR of ubi1 as well as box B binding sites. Inverted frames were taken from fluorescence time-lapse movies. The movement of directed mRNA particles is tracked by red arrows (dark and light colors are used to mark overlapping particle tracks). (Reprinted from reference 45 with permission of the publisher.)
Investigation of a temperature-sensitive mutant defective in cell wall organization revealed that the potential t-SNARE Yup1 is important for hyphal growth and morphology (93). Yup1 is transported as part of putative endosomes along microtubules, suggesting that the long-distance transport of vesicles supports membrane recycling during polar growth (79, 93). Vesicle trafficking is mediated by the concerted action of plus-end-directed Kin3, an UNC-104/KIF1-like Kinesin-3, and minus-end-directed split dynein Dyn1/2 (83). This results in the bidirectional shuttling of vesicles along polarized microtubules (94).
The conventional kinesin Kin1 (52) transports dynein to a loading zone at the plus ends of microtubules for retrograde transport (53). Upon the disruption of the microtubules using inhibitors or upon the deletion of kin1, infectious filaments are shorter, and a substantial number of filaments exhibit bipolar growth, stressing the importance of active microtubule-dependent transport by molecular motors (28, 75). At present, most research is focused on uncovering the molecular cargo(s) in order to close the link between long-distance vesicle trafficking and polar growth.
MICROTUBULE-DEPENDENT TRANSPORT OF DISTINCT mRNAs IN U. MAYDIS
Research over the past 7 years revealed that as well as vesicle trafficking, mRNA transport is important for the polar growth of U. maydis. We discovered that the deletion of rrm4, encoding an RNA-binding protein, resulted in defects in filamentous growth that correlated with reduced virulence. No aberrant phenotype was observed during sporidial growth, indicating that posttranscriptional regulation via Rrm4 is of particular importance during filament formation (7, 26). The protein contains three N-terminal RNA recognition motifs (RRMs) and a C-terminal PABC [poly(A)-binding C terminus] domain. The latter has been described to function as a protein-protein interaction domain in the human poly(A)-binding protein (47, 48). Analysis of the subcellular localization of Rrm4 revealed that the protein shuttles in particles along microtubules (Fig. 2A). These Rrm4-containing particles move with high processivity toward both poles, where they change directionality without apparent delay (6).
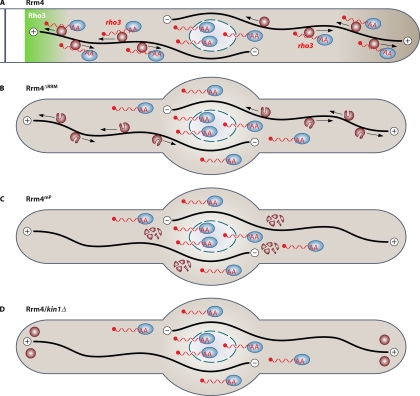
Mutations in the PABC domain lead to a loss of function and the accumulation of large immobile particles (6), indicating that this domain is essential for the formation of mobile transport units (Fig. 3). RNA binding appears to be crucial, since mutations in the first RRM also abolish function (6). Motor-mediated active shuttling of Rrm4-containing particles appears to be important for function, since a loss of conventional kinesin results in the accumulation of Rrm4 at poles of bipolar growing filaments (6). These results are consistent with the hypothesis that Rrm4 mediates microtubule-dependent mRNA transport (97).
Fig. 3.
Model depicting microtubule-dependent mRNA transport in U. maydis. (A) Rrm4-containing particles (dark red circles) transport mRNAs (red wavy lines) carrying the poly(A)-binding protein (blue ovals) bidirectionally along microtubules (black lines). The transport of rho3 mRNA might support a predominant accumulation of Rho3 at hyphal septa (45). (B) Deletion of RRM1 to RRM3 in Rrm4 causes a loss-of-function phenotype: filaments grow mostly bipolar, and target mRNAs are no longer transported along microtubules. However, the mutated Rrm4 version is still part of shuttling particles, indicating that Rrm4 is an integral part of the transport machinery and does not only hitchhike like the poly(A)-binding protein. (C) Mutations in the PABC domain of Rrm4 inhibit particle formation, thus abolishing microtubule-dependent mRNA transport. (D) Loss of the conventional kinesin Kin1 results in the accumulation of Rrm4-containing particles at both poles. Also in this case, the transport of mRNAs does not occur.
This idea was also supported by analyses of the subcellular localization of the poly(A)-binding protein, which is generally associated with the vast majority of mRNAs in eukaryotes (36). In U. maydis, the poly(A)-binding protein Pab1 colocalizes with Rrm4 in vivo in almost all shuttling particles, which is indicative of the presence of transported mRNPs (Fig. 3A). Interestingly, in the absence of Rrm4, Pab1 is no longer present in shuttling particles. Hence, Rrm4-containing mRNPs appear to be the main units of microtubule-dependent mRNA transport (45).
For the identification of target RNAs of Rrm4 we performed in vivo UV cross-linking and immunoprecipitation (CLIP) experiments (89, 91). By UV irradiation, RNAs can be cross-linked in vivo to proteins in close vicinity, generating a snapshot of RNA-protein interactions (90). CLIP experiments identified about 50 potential mRNAs encoding proteins from distinct functional categories such as translation proteins, mitochondrial proteins, and polarity factors (45). Pattern search algorithms revealed the presence of a CA-rich motif as a potential binding site. In this context, it is interesting that the human RRM-containing protein hnRNP L was previously described to bind CA-rich sequences that function as splice enhancers (38). Moreover, CA-rich sequences have been implicated in mRNA transport in Xenopus laevis (1).
In order to verify that these potential target mRNAs are also present in shuttling mRNPs, we established RNA live imaging for U. maydis based on a modified version of the λN-Gfp RNA reporter system (21). This technology was first described for S. cerevisiae (8) and comprises the expression of a heterologous RNA-binding protein fused to a fluorescent protein in combination with the insertion of its cognate binding sites into the mRNA of interest. Thereby, the fluorescence-tagged RNA-binding protein is recruited to specific mRNAs, and its subcellular localization can be monitored in real time (Fig. 2B) (21, 49).
Using the most abundant target mRNA, ubi1, which encodes a natural fusion protein of ubiquitin and the ribosomal protein Rpl40, we demonstrated that this mRNA accumulates in shuttling particles (Fig. 2C) (45). The same holds true for rho3 mRNA. In contrast, shuttling particles are observed to a significantly lesser extent when visualizing the control mRNA mfa2, which encodes a mating pheromone. Importantly, the 3′ UTR of ubi1, which contains a CA-rich sequence, is sufficient to increase the amount and processivity of shuttling mfa2 particles when inserted into the 3′ UTR (Fig. 2C). Thus, this regulatory sequence fulfills the criteria to serve as a zipcode of mRNA transport. These results indicate that although, in principle, all mRNAs can be transported along microtubules, target mRNAs of Rrm4 are transported more frequently and over longer distances. Consistently, RNA live imaging in mammalian cells revealed that regardless of any specific cytoplasmic localization, mRNAs moved along microtubules. However, the presence of the β-actin mRNA zipcode augmented both the frequency and length of mRNP movement (29). In accordance, both localizing and nonlocalizing transcripts were transported on microtubules in embryos of D. melanogaster. Again, mRNA zipcodes increased the frequency and duration of directed movement, potentially via the recruitment of additional microtubule-dependent motors (15). Thus, mRNA transport is not exclusive but exhibits a preference for distinct mRNAs. In U. maydis, this might be important to distribute most mRNAs within filaments and at the same time to enhance the subcellular localization of specific target mRNAs.
Combining protein and RNA live imaging revealed that shuttling ubi1 mRNA particles colocalize with those containing Rrm4. Consistently, the deletion of rrm4 results in the loss of shuttling mRNPs (45). Finally, it was demonstrated that the removal of the RRMs alone is sufficient to cause a loss of shuttling mRNPs. However, the Rrm4 variant without RNA-binding domains itself is still able to shuttle along microtubules. Thus, Rrm4 is an integral part of the transport machinery and does not hitchhike on the transported mRNAs, as seen for the poly(A)-binding protein (Fig. 3B) (45). In essence, our data from two independent in vivo approaches converge to demonstrate that Rrm4 mediates microtubule-dependent mRNA transport of distinct mRNAs in infectious filaments (Fig. 3).
Why are distinct mRNAs transported over long distances in U. maydis? The most parsimonious explanation is that mRNA transport is needed to support the symmetric and/or asymmetric localization of the encoded proteins. This is in close analogy to short-distance transport in S. cerevisiae and C. albicans (Fig. 1) (25, 40). For instance, the transport of rho3 mRNA might support the predominant localization of Rho3 at retraction septa (Fig. 3A) (45). In the case of mRNAs encoding mitochondrial proteins, transport might be important to distribute these mRNAs throughout the infectious filament to particularly support protein import into mitochondria that are far from the nucleus. This is consistent with observations of S. cerevisiae, where local translation in the vicinity of mitochondria depends on regulatory RNA sequences within the 3′ UTR (30, 58, 97). In the future, it will be important to address these different possibilities and to more specifically uncover the mechanisms of transport. This will include the identification of potential adapter and scaffold proteins as well as the responsible molecular motors.
CONCLUSIONS
Study of the formation of infectious filaments in U. maydis revealed that microtubule-dependent mRNA transport is important for polar growth. Although the key RNA-binding protein is specific for basidiomycetes, the underlying molecular concepts of long-distance transport are remarkably similar to molecular events occurring in higher eukaryotes. (i) mRNPs shuttle bidirectionally along microtubules, and this movement is mediated by active transport through microtubule-dependent motors such as kinesins (6). (ii) The key RNA-binding protein recognizes zipcodes within target mRNAs (45). (iii) mRNA transport activity can be modulated. In particular, RNA binding of Rrm4 is increased during the switch toward filamentous growth in U. maydis, analogous to the observation that the synaptic stimulation of neurons increases dendritic mRNA transport (6, 22). (iv) Target mRNAs encode similar proteins, such as the polarity factors Rho3 in U. maydis and RhoA in human neurons, where local translation is involved in growth cone collapse (96).
These examples demonstrate that U. maydis is emerging as a promising model system for long-distance mRNA transport. Thus, once again, fungi serve to uncover basic concepts of biological processes.
ACKNOWLEDGMENTS
We acknowledge N. McGlincy and laboratory members for critical reading of the manuscript. We are grateful to J. Koepke and S. Baumann for unpublished images shown in Fig. 2A.
Our research was supported by grants from the MPG and DFG to M.F.
Footnotes
Published ahead of print on 14 May 2010.
REFERENCES
- 1.Andken B. B., Lim I., Benson G., Vincent J. J., Ferenc M. T., Heinrich B., Jarzylo L. A., Man H. Y., Deshler J. O. 2007. 3′-UTR SIRF: a database for identifying clusters of short interspersed repeats in 3′ untranslated regions. BMC Bioinformatics 8:274–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aronov S., Gelin-Licht R., Zipor G., Haim L., Safran E., Gerst J. E. 2007. mRNAs encoding polarity and exocytosis factors are co-transported with cortical ER to the incipient bud in yeast. Mol. Cell. Biol. 27:3441–3455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aronov S., Gerst J. E. 2004. Involvement of the late secretory pathway in actin regulation and mRNA transport in yeast. J. Biol. Chem. 279:36962–36971 [DOI] [PubMed] [Google Scholar]
- 4.Bartnicki-Garcia S. 2006. Chitosomes: past, present and future. FEMS Yeast Res. 6:957–965 [DOI] [PubMed] [Google Scholar]
- 5.Becalska A. N., Gavis E. R. 2009. Lighting up mRNA localization in Drosophila oogenesis. Development 136:2493–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becht P., König J., Feldbrügge M. 2006. The RNA-binding protein Rrm4 is essential for polarity in Ustilago maydis and shuttles along microtubules. J. Cell Sci. 119:4964–4973 [DOI] [PubMed] [Google Scholar]
- 7.Becht P., Vollmeister E., Feldbrügge M. 2005. Role for RNA-binding proteins implicated in pathogenic development of Ustilago maydis. Eukaryot. Cell 4:121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertrand E., Chartrand P., Schaefer M., Shenoy S. M., Singer R. H., Long R. M. 1998. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2:437–445 [DOI] [PubMed] [Google Scholar]
- 9.Böhl F., Kruse C., Frank A., Ferring D., Jansen R. P. 2000. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J. 19:5514–5524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bölker M. 2001. Ustilago maydis—a valuable model system for the study of fungal dimorphism and virulence. Microbiology 147:1395–1401 [DOI] [PubMed] [Google Scholar]
- 11.Bölker M., Urban M., Kahmann R. 1992. The a mating type locus of U. maydis specifies cell signaling components. Cell 68:441–450 [DOI] [PubMed] [Google Scholar]
- 12.Brachmann A., König J., Julius C., Feldbrügge M. 2004. A reverse genetic approach for generating gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 272:216–226 [DOI] [PubMed] [Google Scholar]
- 13.Brefort T., Doehlemann G., Mendoza-Mendoza A., Reissmann S., Djamei A., Kahmann R. 2009. Ustilago maydis as a pathogen. Annu. Rev. Phytopathol. 47:423–445 [DOI] [PubMed] [Google Scholar]
- 14.Brendza R. P., Serbus L. R., Saxton W. M., Duffy J. B. 2002. Posterior localization of dynein and dorsal-ventral axis formation depend on kinesin in Drosophila oocytes. Curr. Biol. 12:1541–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bullock S. L., Nicol A., Gross S. P., Zicha D. 2006. Guidance of bidirectional motor complexes by mRNA cargoes through control of dynein number and activity. Curr. Biol. 16:1447–1452 [DOI] [PubMed] [Google Scholar]
- 16.Clark A., Meignin C., Davis I. 2007. A dynein-dependent shortcut rapidly delivers axis determination transcripts into the Drosophila oocyte. Development 134:1955–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen R. S. 2005. The role of membranes and membrane trafficking in RNA localization. Biol. Cell 97:5–18 [DOI] [PubMed] [Google Scholar]
- 18.Condeelis J., Singer R. H. 2005. How and why does beta-actin mRNA target? Biol. Cell 97:97–110 [DOI] [PubMed] [Google Scholar]
- 19.Dahm R., Kiebler M., Macchi P. 2007. RNA localisation in the nervous system. Semin. Cell Dev. Biol. 18:216–223 [DOI] [PubMed] [Google Scholar]
- 20.Dahm R., Macchi P. 2007. Human pathologies associated with defective RNA transport and localization in the nervous system. Biol. Cell 99:649–661 [DOI] [PubMed] [Google Scholar]
- 21.Daigle N., Ellenberg J. 2007. LambdaN-GFP: an RNA reporter system for live-cell imaging. Nat. Methods 4:633–636 [DOI] [PubMed] [Google Scholar]
- 22.Dictenberg J. B., Swanger S. A., Antar L. N., Singer R. H., Bassell G. J. 2008. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev. Cell 14:926–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dienstbier M., Boehl F., Li X., Bullock S. L. 2009. Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev. 23:1546–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du T. G., Schmid M., Jansen R. P. 2007. Why cells move messages: the biological functions of mRNA localization. Semin. Cell Dev. Biol. 18:171–177 [DOI] [PubMed] [Google Scholar]
- 25.Elson S. L., Noble S. M., Solis N. V., Filler S. G., Johnson A. D. 2009. An RNA transport system in Candida albicans regulates hyphal morphology and invasive growth. PLoS Genet. 5:e1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feldbrügge M., Zarnack K., Vollmeister E., Baumann S., Koepke J., König J., Münsterkötter M., Mannhaupt G. 2008. The posttranscriptional machinery of Ustilago maydis. Fungal Genet. Biol. 45:S40–S46 [DOI] [PubMed] [Google Scholar]
- 27.Fischer R., Zekert N., Takeshita N. 2008. Polarized growth in fungi—interplay between the cytoskeleton, positional markers and membrane domains. Mol. Microbiol. 68:813–826 [DOI] [PubMed] [Google Scholar]
- 28.Fuchs U., Manns I., Steinberg G. 2005. Microtubules are dispensable for the initial pathogenic development but required for long-distance hyphal growth in the corn smut fungus Ustilago maydis. Mol. Biol. Cell 16:2746–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fusco D., Accornero N., Lavoie B., Shenoy S. M., Blanchard J. M., Singer R. H., Bertrand E. 2003. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr. Biol. 13:161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia M., Darzacq X., Delaveau T., Jourdren L., Singer R. H., Jacq C. 2007. Mitochondria-associated yeast mRNAs and the biogenesis of molecular complexes. Mol. Biol. Cell 18:362–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerst J. E. 2008. Message on the Web: mRNA and ER co-trafficking. Trends Cell Biol. 18:68–76 [DOI] [PubMed] [Google Scholar]
- 32.Gierz G., Bartnicki-Garcia S. 2001. A three-dimensional model of fungal morphogenesis based on the vesicle supply center concept. J. Theor. Biol. 208:151–164 [DOI] [PubMed] [Google Scholar]
- 33.Harris S. D. 2006. Cell polarity in filamentous fungi: shaping the mold. Int. Rev. Cytol. 251:41–77 [DOI] [PubMed] [Google Scholar]
- 34.Harris S. D., Read N. D., Roberson R. W., Shaw B., Seiler S., Plamann M., Momany M. 2005. Polarisome meets Spitzenkörper: microscopy, genetics, and genomics converge. Eukaryot. Cell 4:225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirokawa N., Noda Y. 2008. Intracellular transport and kinesin superfamily proteins, KIFs: structure, function, and dynamics. Physiol. Rev. 88:1089–1118 [DOI] [PubMed] [Google Scholar]
- 36.Hogan D. J., Riordan D. P., Gerber A. P., Herschlag D., Brown P. O. 2008. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 6:e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holt C. E., Bullock S. L. 2009. Subcellular mRNA localization in animal cells and why it matters. Science 326:1212–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hui J., Hung L. H., Heiner M., Schreiner S., Neumüller N., Reither G., Haas S. A., Bindereif A. 2005. Intronic CA-repeat and CA-rich elements: a new class of regulators of mammalian alternative splicing. EMBO J. 24:1988–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hüttelmaier S., Zenklusen D., Lederer M., Dictenberg J., Lorenz M., Meng X., Bassell G. J., Condeelis J., Singer R. H. 2005. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature 438:512–515 [DOI] [PubMed] [Google Scholar]
- 40.Jansen R. P. 2001. mRNA localization: message on the move. Nat. Rev. Mol. Cell Biol. 2:247–256 [DOI] [PubMed] [Google Scholar]
- 41.Januschke J., Gervais L., Dass S., Kaltschmidt J. A., Lopez-Schier H., St. Johnston D., Brand A. H., Roth S., Guichet A. 2002. Polar transport in the Drosophila oocyte requires dynein and kinesin I cooperation. Curr. Biol. 12:1971–1981 [DOI] [PubMed] [Google Scholar]
- 42.Jüschke C., Ferring D., Jansen R. P., Seedorf M. 2004. A novel transport pathway for a yeast plasma membrane protein encoded by a localized mRNA. Curr. Biol. 14:406–411 [DOI] [PubMed] [Google Scholar]
- 43.Kämper J., Kahmann R., Bölker M., Ma L. J., Brefort T., Saville B. J., Banuett F., Kronstad J. W., Gold S. E., Müller O., Perlin M. H., Wösten H. A., de Vries R., Ruiz-Herrera J., Reynaga-Pena C. G., Snetselaar K., McCann M., Pérez-Martín J., Feldbrügge M., Basse C. W., Steinberg G., Ibeas J. I., Holloman W., Guzman P., Farman M., Stajich J. E., Sentandreu R., González-Prieto J. M., Kennell J. C., Molina L., Schirawski J., Mendoza-Mendoza A., Greilinger D., Münch K., Rössel N., Scherer M., Vranes M., Ladendorf O., Vincon V., Fuchs U., Sandrock B., Meng S., Ho E. C., Cahill M. J., Boyce K. J., Klose J., Klosterman S. J., Deelstra H. J., Ortiz-Castellanos L., Li W., Sanchez-Alonso P., Schreier P. H., Häuser-Hahn I., Vaupel M., Koopmann E., Friedrich G., Voss H., Schlüter T., Margolis J., Platt D., Swimmer C., Gnirke A., Chen F., Vysotskaia V., Mannhaupt G., Güldener U., Münsterkötter M., Haase D., Oesterheld M., Mewes H. W., Mauceli E. W., DeCaprio D., Wade C. M., Butler J., Young S., Jaffe D. B., Calvo S., Nusbaum C., Galagan J., Birren B. W. 2006. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444:97–101 [DOI] [PubMed] [Google Scholar]
- 44.Kislauskis E. H., Li Z., Singer R. H., Taneja K. L. 1993. Isoform-specific 3′-untranslated sequences sort alpha-cardiac and beta-cytoplasmic actin messenger RNAs to different cytoplasmic compartments. J. Cell Biol. 123:165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.König J., Baumann S., Koepke J., Pohlmann T., Zarnack K., Feldbrügge M. 2009. The fungal RNA-binding protein Rrm4 mediates long-distance transport of ubi1 and rho3 mRNAs. EMBO J. 28:1855–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Konzack S., Rischitor P. E., Enke C., Fischer R. 2005. The role of the kinesin motor KipA in microtubule organization and polarized growth of Aspergillus nidulans. Mol. Biol. Cell 16:497–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kozlov G., De Crescenzo G., Lim N. S., Siddiqui N., Fantus D., Kahvejian A., Trempe J. F., Elias D., Ekiel I., Sonenberg N., O'Connor-McCourt M., Gehring K. 2004. Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase. EMBO J. 23:272–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kozlov G., Trempe J. F., Khaleghpour K., Kahvejian A., Ekiel I., Gehring K. 2001. Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc. Natl. Acad. Sci. U. S. A. 98:4409–4413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Larson D. R., Singer R. H., Zenklusen D. 2009. A single molecule view of gene expression. Trends Cell Biol. 19:630–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Latham V. M., Yu E. H., Tullio A. N., Adelstein R. S., Singer R. H. 2001. A Rho-dependent signaling pathway operating through myosin localizes beta-actin mRNA in fibroblasts. Curr. Biol. 11:1010–1016 [DOI] [PubMed] [Google Scholar]
- 51.Lécuyer E., Yoshida H., Parthasarathy N., Alm C., Babak T., Cerovina T., Hughes T. R., Tomancak P., Krause H. M. 2007. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131:174–187 [DOI] [PubMed] [Google Scholar]
- 52.Lehmler C., Steinberg G., Snetselaar K. M., Schliwa M., Kahmann R., Bölker M. 1997. Identification of a motor protein required for filamentous growth in Ustilago maydis. EMBO J. 16:3464–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lenz J. H., Schuchardt I., Straube A., Steinberg G. 2006. A dynein loading zone for retrograde endosome motility at microtubule plus-ends. EMBO J. 25:2275–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin A. C., Holt C. E. 2007. Local translation and directional steering in axons. EMBO J. 26:3729–3736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lipshitz H. D. 2009. Follow the mRNA: a new model for Bicoid gradient formation. Nat. Rev. Mol. Cell Biol. 10:509–512 [DOI] [PubMed] [Google Scholar]
- 56.Liu G., Grant W. M., Persky D., Latham V. M., Jr., Singer R. H., Condeelis J. 2002. Interactions of elongation factor 1alpha with F-actin and beta-actin mRNA: implications for anchoring mRNA in cell protrusions. Mol. Biol. Cell 13:579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Long R. M., Gu W., Lorimer E., Singer R. H., Chartrand P. 2000. She2p is a novel RNA-binding protein that recruits the Myo4p-She3p complex to ASH1 mRNA. EMBO J. 19:6592–6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Margeot A., Garcia M., Wang W., Tetaud E., di Rago J. P., Jacq C. 2005. Why are many mRNAs translated to the vicinity of mitochondria: a role in protein complex assembly? Gene 354:64–71 [DOI] [PubMed] [Google Scholar]
- 59.Martin K. C., Ephrussi A. 2009. mRNA localization: gene expression in the spatial dimension. Cell 136:719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mazloum N., Holloman W. K. 2009. Brh2 promotes a template-switching reaction enabling recombinational bypass of lesions during DNA synthesis. Mol. Cell 36:620–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Messitt T. J., Gagnon J. A., Kreiling J. A., Pratt C. A., Yoon Y. J., Mowry K. L. 2008. Multiple kinesin motors coordinate cytoplasmic RNA transport on a subpopulation of microtubules in Xenopus oocytes. Dev. Cell 15:426–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mili S., Macara I. G. 2009. RNA localization and polarity: from A(PC) to Z(BP). Trends Cell Biol. 19:156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mili S., Moissoglu K., Macara I. G. 2008. Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature 453:115–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mische S., Li M., Serr M., Hays T. S. 2007. Direct observation of regulated ribonucleoprotein transport across the nurse cell/oocyte boundary. Mol. Biol. Cell 18:2254–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Müller M., Heuck A., Niessing D. 2007. Directional mRNA transport in eukaryotes: lessons from yeast. Cell. Mol. Life Sci. 64:171–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oleynikov Y., Singer R. H. 2003. Real-time visualization of ZBP1 association with beta-actin mRNA during transcription and localization. Curr. Biol. 13:199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Palacios I. M., St. Johnston D. 2002. Kinesin light chain-independent function of the kinesin heavy chain in cytoplasmic streaming and posterior localisation in the Drosophila oocyte. Development 129:5473–5485 [DOI] [PubMed] [Google Scholar]
- 68.Paquin N., Chartrand P. 2008. Local regulation of mRNA translation: new insights from the bud. Trends Cell Biol. 18:105–111 [DOI] [PubMed] [Google Scholar]
- 69.Paquin N., Ménade M., Poirier G., Donato D., Drouet E., Chartrand P. 2007. Local activation of yeast ASH1 mRNA translation through phosphorylation of Khd1p by the casein kinase Yck1p. Mol. Cell 26:795–809 [DOI] [PubMed] [Google Scholar]
- 70.Requena N., Alberti-Segui C., Winzenburg E., Horn C., Schliwa M., Philippsen P., Liese R., Fischer R. 2001. Genetic evidence for a microtubule-destabilizing effect of conventional kinesin and analysis of its consequences for the control of nuclear distribution in Aspergillus nidulans. Mol. Microbiol. 42:121–132 [DOI] [PubMed] [Google Scholar]
- 71.Riquelme M., Bartnicki-García S., González-Prieto J. M., Sánchez-León E., Verdín-Ramos J. A., Beltrán-Aguilar A., Freitag M. 2007. Spitzenkörper localization and intracellular traffic of green fluorescent protein-labeled CHS-3 and CHS-6 chitin synthases in living hyphae of Neurospora crassa. Eukaryot. Cell 6:1853–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rodriguez A. J., Czaplinski K., Condeelis J. S., Singer R. H. 2008. Mechanisms and cellular roles of local protein synthesis in mammalian cells. Curr. Opin. Cell Biol. 20:144–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schink K. O., Bölker M. 2009. Coordination of cytokinesis and cell separation by endosomal targeting of a Cdc42-specific guanine nucleotide exchange factor in Ustilago maydis. Mol. Biol. Cell 20:1081–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schmid M., Jaedicke A., Du T. G., Jansen R. P. 2006. Coordination of endoplasmic reticulum and mRNA localization to the yeast bud. Curr. Biol. 16:1538–1543 [DOI] [PubMed] [Google Scholar]
- 75.Schuchardt I., Assmann D., Thines E., Schuberth C., Steinberg G. 2005. Myosin-V, kinesin-1, and kinesin-3 cooperate in hyphal growth of the fungus Ustilago maydis. Mol. Biol. Cell 16:5191–5201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seiler S., Nargang F. E., Steinberg G., Schliwa M. 1997. Kinesin is essential for cell morphogenesis and polarized secretion in Neurospora crassa. EMBO J. 16:3025–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shepard K. A., Gerber A. P., Jambhekar A., Takizawa P. A., Brown P. O., Herschlag D., DeRisi J. L., Vale R. D. 2003. Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc. Natl. Acad. Sci. U. S. A. 100:11429–11434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Snetselaar K. M. 1993. Microscopic observation of Ustilago maydis mating interactions. Exp. Mycol. 17:345–355 [Google Scholar]
- 79.Steinberg G. 2007. Hyphal growth: a tale of motors, lipids, and the Spitzenkörper. Eukaryot. Cell 6:351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Steinberg G. 2007. Tracks for traffic: microtubules in the plant pathogen Ustilago maydis. New Phytol. 174:721–733 [DOI] [PubMed] [Google Scholar]
- 81.Steinberg G., Schliwa M., Lehmler C., Bölker M., Kahmann R., McIntosh J. R. 1998. Kinesin from the plant pathogenic fungus Ustilago maydis is involved in vacuole formation and cytoplasmic migration. J. Cell Sci. 111:2235–2246 [DOI] [PubMed] [Google Scholar]
- 82.St. Johnston D. 2005. Moving messages: the intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 6:363–375 [DOI] [PubMed] [Google Scholar]
- 83.Straube A., Hause G., Fink G., Steinberg G. 2006. Conventional kinesin mediates microtubule-microtubule interactions in vivo. Mol. Biol. Cell 17:907–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sudbery P., Court H. 2007. Polarised growth in fungi, p. 137–166 InHoward R. J., Gow N. A. R. (ed.), The Mycota VIII Springer-Verlag, Berlin, Germany [Google Scholar]
- 85.Szabó Z., Tönnis M., Kessler H., Feldbrügge M. 2002. Structure-function analysis of lipopeptide pheromones from the plant pathogen Ustilago maydis. Mol. Genet. Genomics 268:362–370 [DOI] [PubMed] [Google Scholar]
- 86.Takeshita N., Vienken K., Rolbetzki A., Fischer R. 2007. The Aspergillus nidulans putative kinase, KfsA (kinase for septation), plays a role in septation and is required for efficient asexual spore formation. Fungal Genet. Biol. 44:1205–1214 [DOI] [PubMed] [Google Scholar]
- 87.Takizawa P. A., DeRisi J. L., Wilhelm J. E., Vale R. D. 2000. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science 290:341–344 [DOI] [PubMed] [Google Scholar]
- 88.Tekotte H., Davis I. 2002. Intracellular mRNA localization: motors move messages. Trends Genet. 18:636–642 [DOI] [PubMed] [Google Scholar]
- 89.Ule J. 2009. High-throughput sequencing methods to study neuronal RNA-protein interactions. Biochem. Soc. Trans. 37:1278–1280 [DOI] [PubMed] [Google Scholar]
- 90.Ule J., Jensen K., Mele A., Darnell R. B. 2005. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods 37:376–386 [DOI] [PubMed] [Google Scholar]
- 91.Ule J., Jensen K. B., Ruggiu M., Mele A., Ule A., Darnell R. B. 2003. CLIP identifies Nova-regulated RNA networks in the brain. Science 302:1212–1215 [DOI] [PubMed] [Google Scholar]
- 92.Vale R. D. 2003. The molecular motor toolbox for intracellular transport. Cell 112:467–480 [DOI] [PubMed] [Google Scholar]
- 93.Wedlich-Söldner R., Bölker M., Kahmann R., Steinberg G. 2000. A putative endosomal t-SNARE links exo- and endocytosis in the phytopathogenic fungus Ustilago maydis. EMBO J. 19:1974–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wedlich-Söldner R., Straube A., Friedrich M. W., Steinberg G. 2002. A balance of KIF1A-like kinesin and dynein organizes early endosomes in the fungus Ustilago maydis. EMBO J. 21:2946–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Weil T. T., Forrest K. M., Gavis E. R. 2006. Localization of bicoid mRNA in late oocytes is maintained by continual active transport. Dev. Cell 11:251–262 [DOI] [PubMed] [Google Scholar]
- 96.Wu K. Y., Hengst U., Cox L. J., Macosko E. Z., Jeromin A., Urquhart E. R., Jaffrey S. R. 2005. Local translation of RhoA regulates growth cone collapse. Nature 436:1020–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zarnack K., Feldbrügge M. 2007. mRNA trafficking in fungi. Mol. Genet. Genomics 278:347–359 [DOI] [PubMed] [Google Scholar]
- 98.Zimyanin V. L., Belaya K., Pecreaux J., Gilchrist M. J., Clark A., Davis I., St. Johnston D. 2008. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell 134:843–853 [DOI] [PMC free article] [PubMed] [Google Scholar]