Abstract
Development in ciliated protozoa involves extensive genome reorganization within differentiating macronuclei, which shapes the somatic genome of the next vegetative generation. Major events of macronuclear differentiation include excision of internal eliminated sequences (IESs), chromosome fragmentation, and genome amplification. Proteins required for these events include those with homology throughout eukaryotes as well as proteins apparently unique to ciliates. In this study, we identified the ciliate-specific Defective in IES Excision 5 (DIE5) genes of Paramecium tetraurelia (PtDIE5) and Tetrahymena thermophila (TtDIE5) as orthologs that encode nuclear proteins expressed exclusively during development. Abrogation of PtDie5 protein (PtDie5p) function by RNA interference (RNAi)-mediated silencing or TtDie5p by gene disruption resulted in the failure of developing macronuclei to differentiate into new somatic nuclei. Tetrahymena ΔDIE5 cells arrested late in development and failed to complete genome amplification, whereas RNAi-treated Paramecium cells highly amplified new macronuclear DNA before the failure in differentiation, findings that highlight clear differences in the biology of these distantly related species. Nevertheless, IES excision and chromosome fragmentation failed to occur in either ciliate, which strongly supports that Die5p is a critical player in these processes. In Tetrahymena, loss of zygotic expression during development was sufficient to block nuclear differentiation. This observation, together with the finding that knockdown of Die5p in Paramecium still allows genome amplification, indicates that this protein acts late in macronuclear development. Even though DNA rearrangements in these two ciliates look to be quite distinct, analysis of DIE5 establishes the action of a conserved mechanism within the genome reorganization pathway.
The biology of ciliates offers an extreme case of differential regulation of separate copies of the genome (see reference 34). These protists possess two morphologically and functionally distinct types of nuclei coexisting within a common cell (reviewed in references 20 and 43). The somatic macronucleus is polygenomic and transcriptionally active, whereas the germinal micronucleus is diploid and transcriptionally inert throughout the vegetative growth cycle.
When ciliates undergo development during the sexual phase of the life cycle, the existing somatic macronuclei disappear and new micro- and macronuclei arise from germ line-derived precursors. These genetically identical precursor nuclei are formed upon the conjugation of two mating-compatible partners, which induces meiosis of germ line micronuclei. The products of meiosis and a postmeiotic division are two haploid gametic nuclei in each conjugate, one of which is exchanged between partners. The migratory pronucleus fuses with the stationary copy to form a diploid zygotic nucleus that subsequently divides and differentiates. The new micronuclei are maintained in a silent state, while the differentiating macronuclei acquire extensive chromatin modifications that mediate regulated gene expression. In some ciliate species, nuclear differentiation can alternatively occur through a self-fertilization process called autogamy, which occurs without cell pairing and exchange of genetic material, but nonetheless starts with meiosis and leads to the production of new micro- and macronuclei.
Macronuclear differentiation includes genome-wide DNA rearrangements that extensively remodel the developing somatic chromosomes. The processes of chromosome fragmentation and DNA elimination dramatically alter the genome found in the mature somatic macronucleus after conjugation. Different ciliate species eliminate anywhere from 15% to 90% of the germ line (micronucleus-limited) DNA during this process (reviewed in references 43 and 57). In addition to these physical alterations, the genome is endoreplicated to tens or even thousands of copies per macronucleus, a ploidy level that varies between species.
The DNA segments eliminated from the developing macronucleus are called internal eliminated sequences (IESs), which can vary in structure both within a species and between different species. In Paramecium tetraurelia, IESs are generally short (26 to 883 bp) and have a modestly conserved 8-bp inverted repeat that has some similarity to the termini of the mariner/Tc1 superfamily of transposable elements (22). Excision of these IESs is precise, leaving a single copy of the 5′-TA-3′ dinucleotide in the macronucleus-retained sequences. The mechanism generates double-stranded breaks at both ends, followed by joining of the macronucleus-destined DNA (see references 5 and 19). On the other hand, IESs in another ciliate, Tetrahymena thermophila, are relatively large (0.6 to >20 kb) and show substantial heterogeneity in their excision boundaries. The elimination of the Tetrahymena IESs shows some similarity to the imprecise elimination of transposon-like sequences from the developing genome of Paramecium (24). Unlike the Paramecium IESs, those in Tetrahymena show no obviously conserved sequence characteristics at their excision boundaries. The model proposed for their excision also differs from that of Paramecium. Evidence from the characterization of rearrangement intermediates suggests that a double-stranded break at one end of the IES is followed by transesterification of the freed end to the other boundary, generating the macronuclear junction (45, 46).
Despite substantial variability in the size, structure, and excision mechanisms of IESs in different ciliate species, there is a growing body of molecular evidence that IES excision in both Tetrahymena and Paramecium involves an RNA interference (RNAi)-like pathway (17, 34, 36, 38, 56). This pathway is more extensively characterized for Tetrahymena. A large pool of germ line-derived small (27- to 30-nucleotide [nt]) RNAs is generated by cleavage of RNA produced by extensive, bidirectional transcription within meiotic micronuclei (11, 36). These “scan RNAs” (scnRNAs), generated by the Dicer-like protein Dcl1p (30, 37), direct methylation of histone H3 on lysine 9 (K9) and/or lysine 27 (K27), marking the IESs at the beginning of nuclear differentiation for later removal from the genome (26, 50). A role for small RNAs has also been identified in Paramecium and possibly in Stylonychia, but it is less clear if these act by directing specific chromatin modifications on or near IESs (17, 21, 25). Further studies and comparative analyses should reveal additional biochemical pathways that mediate these remarkable genome reorganization processes. For example, a recent study discovered a domesticated piggyBac transposase in Paramecium that is required to remove the IESs from the developing macronuclei and reported that a similar, developmentally expressed transposase gene is contained within the Tetrahymena genome (4).
In this report, we show that a gene named Defective IES Excision 5 (DIE5) is expressed during sexual reproduction and is required for removal of germ line-specific sequences from the Paramecium genome. Although significant sequence identity with DIE5 could not be found in most other eukaryotes, we identified a candidate homologue in Tetrahymena thermophila (TtDIE5) that is also developmentally regulated. Disruption of TtDIE5 revealed that it is also required for IES excision and chromosome breakage. Further investigations demonstrated that loss of TtDie5p does not appear to disturb well-characterized steps in macronuclear development, such as small-RNA accumulation and formation of DNA rearrangement foci. Comparison of our results for both species revealed a conserved role for Die5p in the formation of the ciliate macronucleus yet also uncovered significant differences in the action of Die5p between these organisms. We believe this type of comparative study has the power to discern key conserved features of ciliate genome reorganization.
MATERIALS AND METHODS
Cell lines and culture.
Paramecium tetraurelia stock d4-110 (hr-b/hr-b) was used to generate cultures undergoing synchronized conjugation for RNA isolation and whole-cell PCR analysis. Strains d4-502 (pwA-502/pwA-502; nd6-1/nd6-1) and a3093 (pwB-96/pwB-96; nd9-c/nd9-c1) (from Mihoko Takahashi, University of Tsukuba) were used for genetic analysis. Elsewhere, nd6 (nd6-1/nd6-1) was used as a wild-type control. Paramecia were cultured at 27°C as described by Sonneborn (49) in 1.25 to 2.5 g Austrian winter pea (Outsidepride) in 800 ml double-distilled water (ddH2O) buffered with K-DS (4 mM sodium citrate, 2.8 mM sodium phosphate dibasic, 1.2 mM potassium phosphate monobasic, 1.5 mM calcium chloride) supplemented with 1.25 mg/liter stigmasterol and inoculated with Klebsiella pneumoniae 1 to 2 days prior to use (53). Tetrahymena thermophila stocks B2086, CU427, and CU428.1 were cultured axenically in Neff's (0.5% dextrose, 0.25% yeast extract, 0.25% proteose peptone, 3.3 mM FeCl3) or SPP (0.2% dextrose, 0.1% yeast extract, 1% proteose peptone, 0.003% Sequestrene) medium at 30°C as described previously (18, 42).
For conjugation, mating-reactive Paramecium cells in starvation medium were mixed at a density of >2,000 cells/ml and incubated at 27°C. Conjugating cells were enriched for by the methods described by Yang and Takahashi (55) and Vosskühler and Tiedtke (54), which resulted in cultures 80 to 99% pure for conjugating Paramecium. Conjugating Tetrahymena cells were prepared for mating by washing out growth medium and culturing them in 10 mM Tris-HCl (pH 7.4) for >6 h. Mixing cultures of complementary mating types at equal cell density produced cultures with mating efficiencies of 80 to 95% (29).
Total RNA isolation.
Total RNA was isolated from 50 to 100 ml of Paramecium cell culture (100 to 1,000 cells/ml) with an RNeasy minikit (Qiagen) supplemented by a QIAshredder for cell homogenization and an RNase-free DNase set (Qiagen) for elimination of genomic DNA. All products were used according to the manufacturer's directions. RNA was extracted from 10 to 20 ml Tetrahymena cell culture (2 × 105 cells/ml), using RNAzol extraction (14). Total RNA was used in reverse transcription (RT)-PCR to monitor expression in wild-type and ΔDIE5 lines, using TtDIE5 oligos 5′-GTTTATGTTTTCTAATTGAGCTTT-3′ and 5′CTGGTATAATCATTAATGCTCG-3′ or HHP1 (Tetrahymena HP1 gene) oligonucleotides 5′-GGAGCTTCAACTCATTAAACACG-3′ and 5′-TCGGGAGAAGCATACTTAGCA-3′, which amplify 252-bp or 371-bp cDNA products, respectively.
Microinjection and observation of GFP fluorescence.
Paramecium plasmids containing the green fluorescent protein (GFP)-DIE5 fusions were derived from pZC′ΔRI (kindly provided by Eric Meyer, CNRS, Paris, and Jean Cohen, CNRS, Gif-sur-Yvette). Plasmid p5AGN5At contains a 652-bp upstream region, the GFP gene, and the full-length open reading frame (ORF) of DIE5a (we designated the gene identified by differential display as DIE5a and its paralog as DIE5b) followed by the 330-bp DIE5a downstream region. Plasmid sequences are available on request. Approximately 2 pl of plasmid solution (∼5 μg/μl) in distilled water was injected into the macronuclei of Paramecium cells as previously described (32). GFP-expressing cells were fixed and stained with propidium iodide (Vector Laboratories, Burlingame, CA) as described previously (32). Confocal microscopy was performed with a Bio-Rad MRC 1024 UV/Vis system.
To examine Die5p localization in Tetrahymena, the TtDIE5 coding region (genome coordinates CH445530:323,516 to 324,279) was amplified and cloned into the pENTR-D plasmid to create pENTR-DIE5, which is compatible with Gateway recombination cloning (Invitrogen). Subsequently, LR Clonase II was used to recombine the DIE5 coding sequence into a destination vector containing an MTT1-inducible GFP expression cassette cloned upstream of a cycloheximide-resistant rpl29 allele. This construct was linearized with HindIII in the flanking rpl29 sequences and introduced into starved Tetrahymena cells by biolistic transformation. Transformants were selected in SPP medium containing 12.5 μg/ml cycloheximide. To induce GFP-DIE5 expression, 0.08 μg/ml CdCl2 was added to mating cells 3.5 h postmixing. Cells at 6 h to 14 h postmixing were fixed in 2% paraformaldehyde, counter stained with DAPI (4′,6-diamidino-2-phenylindole), and visualized, using a Nikon model Eclipse E600 microscope outfitted with a QImaging Retiga EX CCD camera driven by Openlab image acquisition software (Improvision).
RNAi and phenotypic observation.
A cDNA fragment corresponding to the region of DIE5a between a HincII site and the polyA addition site was cloned into pL4440 (52). (Note that a 23-bp segment in this region perfectly matches DIE5b and therefore likely silences both paralogs). RNAi experiments were performed by feeding Paramecium Escherichia coli producing double-stranded RNA as previously described (http://Paramecium.cgm.cnrs-gif.fr/RNAi/). Conjugation or autogamy of RNAi-treated cells was induced within 48 h of this feeding. Conjugating pairs were isolated in fresh culture medium for phenotypic analysis. For genetic studies, exconjugants were isolated and grown separately for ∼10 cell divisions prior to observation of phenotypes. To observe the phenotypes of the F2 generation, ∼10 starved F1 cells were transferred to fresh culture fluid and allowed to grow for additional cell divisions, and their phenotype was scored after autogamy (self-fertilization). Nuclear DNA was stained with propidium iodide in Vectashield after cells were fixed with 4% paraformaldehyde as described previously (32).
Micronuclear and macronuclear ΔDIE5 strains.
A Tetrahymena DIE5 knockout construct was generated, using a MultiSite Gateway cloning kit (Invitrogen). DIE5 upstream (814 bps; nts 322753 to 323567 of contig CH445530) and downstream (761 bps; nts 324295 to 325056 of contig CH445530) flanking sequences were amplified by PCR and cloned into the Gateway donor vectors, pDONR-P4-P1R and pDONR-P2R-P3, respectively, using BP recombinase. The MTT1-NEO (2,079 bps) selection cassette derived from pMNBL (47) was amplified and cloned into donor plasmid pENTR-D by a topoisomerase-mediated reaction. These three donor plasmids were mixed in equal molar ratios with the destination vector pDEST-R4-R3 and LR Clonase Plus, and the resulting recombination created the gene disruption vector pKO-TtDIE5.
Plasmid pKO-TtDIE5 was digested with StuI and introduced by biolistic transformation into either starved CU427 and CU428 populations (macronuclear transformation) or mating B2086 and CU428 populations (germ line transformation) 2.5 to 3.5 h postmixing as described previously (7, 8). Putative transformants with DIE5 disrupted (ΔDIE5) within either their macronuclei or both their macro- and micronuclei were selected by growth in the presence of paromomycin. Micronuclear knockouts were verified by crossing original transformants to CU427 to test for segregation of the MTT1-NEO cassette among the cycloheximide-resistant progeny. The heterozygous micronuclear ΔDIE5 transformants were crossed to the star strains B*VI and B*VII to generate homozygous micronuclear ΔDIE5 and ΔDIE5 micronuclei/wild-type macronuclei heterokaryons. For both somatic and germ line transformants, which initially contained a mixture of wild-type and ΔDIE5 alleles in their macronuclei, cells were subcloned and cultured in growth medium containing increasing concentrations of paromomycin until only mutant alleles remained, thus producing complete macronuclear-knockout strains. The elimination of the DIE5 gene in the macronucleus and micronucleus was confirmed by PCR and Southern blot hybridization analysis as described previously (30).
Whole-cell PCR amplification.
PCR amplifications were performed on whole-cell Paramecium as described previously (32) (see Fig. 3).
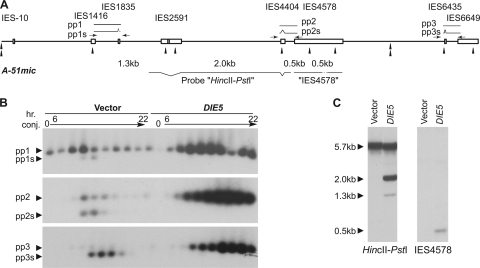
Fig. 3.
Silencing of DIE5 inhibits excision of IESs. (A) A partial map of the micronuclear version of the A-51 allele showing locations of IESs (boxes), SspI recognition sites (arrowheads), and sizes of fragments generated by SspI digestion. Positions of expected PCR products (pp1 to pp3) and probes for Southern hybridization are also indicated. Arrows show positions of the primers relative to IESs not drawn to scale. (B) Whole-cell semiquantitative PCRs of RNAi-treated exconjugants, using one primer in the macronuclear sequence and the other primer inside the IESs. Each lane represents whole-cell PCR products taken at 2-h intervals from 6 to 22 h after induction of conjugation. The predicted PCR products correspond to pp1 to pp3 in panel A. Due to excision of smaller IESs during rearrangement, two bands are expected for each primer set. (C) SspI-digested total genomic Southern blots of RNAi-treated exautogamous cells probed with either a HincII-PstI fragment or IES4578, as indicated in panel A. Total DNA (∼10 μg) was isolated from an exautogamous cell culture when about 50% of control cells (RNAi using empty vector) had undergone the first cell division. Most IESs in the micronuclear version of the A-51 allele contain SspI sites (single arrow heads in panel A), while only one site is present in the macronuclear-destined sequence of the A-51 allele (the double arrow head in panel A). Thus, for probe HincII-PstI, 1.3- and 2.0-kb fragments are expected for unprocessed DNA, while a 5.7-kb fragment is expected for the processed DNA, including abundant old macronuclear DNA in exconjugants. Probe IES4578 contains only the IES sequence and detects 0.5-kb fragments if the IES is present at high levels in exautogamous cells.
Northern and Southern blot analyses.
DNA from Paramecium cultures was isolated as previously described (23). Tetrahymena genomic DNA was isolated from 1 × 106 to 2 × 106 cells, using a Wizard genomic DNA isolation kit (Promega), followed by resuspension in 10 mM Tris-HCl by incubation at 65°C for 1 h or at 4°C overnight. Northern and Southern blots were performed as described previously (30). Probes for Northern hybridization were derived from cloned cDNA fragments containing whole Paramecium or Tetrahymena DIE5 ORFs. Plasmids used to generate Southern probes were pSA2.1HP for the 2.1-kb HincII and PstI or 1.5-kb BglII-PstI fragments of the macronuclear A-51 allele or p4578c containing a 787-bp fragment of the A-51 allele generated by PCR, using a forward primer (5′-GGATCTGTTGATCAACTAG-3′) and a reverse primer (5′-CTGATAGCGTATTTGGATTAG-3′) with total genomic DNA from exconjugant cells (see probes in Fig. 3A). This reaction amplifies the circularized IES4578 of the A-51 allele that is present transiently in the genomic DNA of cells during sexual reproduction. To examine the Tetrahymena DIE5 locus in the knockout lines, isolated genomic DNA was digested with BstBI and separated on 0.9% agarose gel at 40 V overnight. The probe used for analysis of the knockout lines was isolated from pDONR-Die5, created for generating the pKO-TtDie5 plasmid. To assess failure of chromosome breakage, total genomic DNA isolated from wild-type or ΔDIE5 cells after 16 h of mating was digested with EcoRI, fractionated, and probed with a 0.8-kbp fragment that spans the EcoRI site at position 335013 of chromosomal scaffold CH445662.
Fluorescence microscopy.
For examining the nuclear morphology of knockout strains and for cellular localization of GFP-Die5p, cells were fixed in 2% paraformaldehyde and stained with DAPI (1 μg/ml) for 10 to 30 min. Cells were then immobilized under 22- by 22-mm coverslips in 5 μl of 2% methylcellulose. For the visualization of DNA elimination structures, an integrative PDD1-YFP fusion construct was introduced into Tetrahymena cells by biolistic transformation. Conjugating transformants induced with 0.05 μg/ml CdCl2 were fixed with 2% paraformaldehyde at 14 h postmixing and were counter-stained with DAPI. For histone modification analysis, 9-h conjugating cells were fixed with Schaudinn's fixative (2 parts saturated mercuric chloride to 1 part 95% ethanol) and dehydrated with methanol. The cells were then rehydrated with Tris-buffered saline (TBS) and blocked in 1% bovine serum albumin (BSA) plus 0.01% Tween 20. Anti-H3K9me2 rabbit polyclonal (Upstate Biotechnology) and anti-H3K27me3 mouse monoclonal (Abcam) antibodies were used at 1:500 dilution for immunostaining. Secondary antibodies used were Alexa Fluor 488-conjugated anti-rabbit and anti-mouse antibodies (1:1,000; Invitrogen).
Nucleotide sequence accession numbers.
The nucleotide sequences of the Paramecium DIE5a and DIE5b genes are present in the GenBank database under accession numbers 124427424 and 124429605, respectively. The Tetrahymena DIE5 gene is found on genomic scaffold scf_8254365 under GenBank accession number CH445530. The Tetrahymena DIE5 gene is designated TTHERM_00686240, and the protein ID in GenBank is EAS04981.1. Preliminary Paramecium and Tetrahymena genome sequence data were obtained from Genoscope (http://www.genoscope.cns.fr/) and the J. Craig Venter Institute (formerly the Institute for Genomic Research; http://www.jcvi.org/), respectively (2, 13).
RESULTS
Paramecium DIE5 encodes a novel, developmentally expressed nuclear protein.
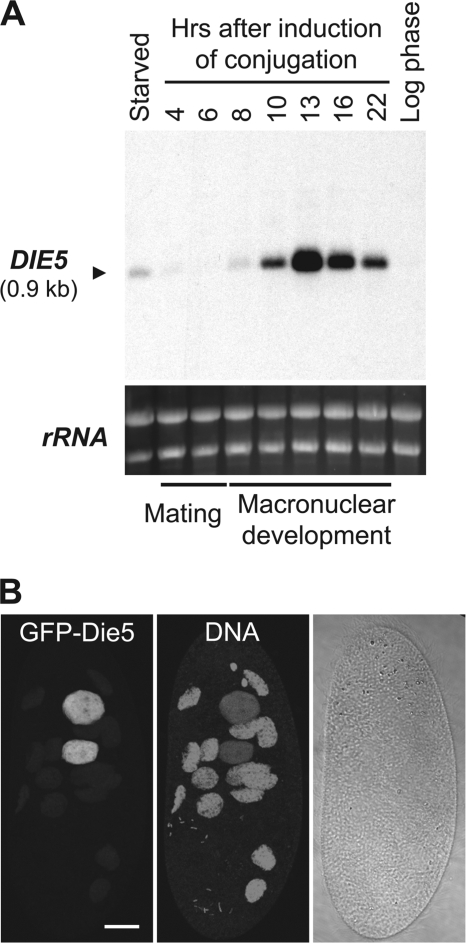
As the genome remodeling that creates the somatic macronucleus is a major event in Paramecium development, we used differential display to identify proteins expressed exclusively during conjugation, as these are candidates that promise to be important for this nuclear differentiation (32). Upon further examination of the expression of individual candidate genes by Northern blot analysis, one in particular exhibited a dramatic increase in its mRNA abundance at 13 h into conjugation (Fig. 1A), and overall its expression closely corresponded with the known timing of IES excision (10 to 22 h, with a peak at 14 h) (6, 23). We also detected a low level of expression in starved cell populations, but this is likely derived from developing cells spontaneously undergoing autogamy (self-fertilization).
Fig. 1.
DIE5 is a developmentally regulated gene encoding a nuclear protein in Paramecium. (A) Northern blot of total RNA (20 μg per lane) from Paramecium probed with Paramecium DIE5a. Ethidium bromide staining of rRNA was used as a loading control. (B) Cellular localization of GFP-Die5a expressed from an extrachromosomal DNA driven by the DIE5a promoter. Fluorescent images are projections of optical sections obtained by confocal microscopy. The GFP fluorescence exclusively localized to the new macronuclei, which show weaker propidium iodide staining (DNA) than the old macronuclear fragments. The image corresponds to a cell approximately 14 h after the start of mating. The bar corresponds to 20 μm.
We named this promising candidate Defective IES Excision 5 (DIE5), due to its gene knockdown phenotype described below. DIE5 is predicted to encode a 199-amino-acid (aa) protein with a molecular mass of 24 kDa. A nearly identical protein is encoded by a second locus in the Paramecium genome (http://www.genoscope.cns.fr/externe/GenomeBrowser/Paramecium/). This copy is clearly a DIE5 paralog derived from the recent whole-genome duplication in the Paramecium lineage (2), as the two genes are remarkably similar (86% nucleotide identity; 98% amino acid identity) and both loci contain homologous copies of the neighboring gene, NMD3, present in the same orientation. Therefore, following the nomenclature convention for Paramecium genes (1), we designated the gene identified by differential display as DIE5a and its paralog as DIE5b. (We refer to both genes below simply as DIE5, given that 196 of 199 aa are conserved between them, and we used the DIE5a sequence in the design of all functional experiments described.) Analysis of the Paramecium tetraurelia Die5p amino acid sequence revealed no conserved protein domains or motifs except for two classical nuclear localization signals (NLSs); however, an identifiable homologue was found in the genome of the ciliate, Tetrahymena thermophila (described below). (When necessary to distinguish the Paramecium and Tetrahymena genes, we add the prefix Pt or Tt before DIE5). Thus, DIE5 appears to be a novel gene that is conserved within the oligohymenophora lineage.
The two putative NLSs, along with the timing of expression, suggested to us that DIE5 may encode a nuclear protein that participates in macronuclear differentiation. To further investigate this possibility, we examined Die5p localization by fusing GFP to its N terminus in a transgene expressed from the DIE5 promoter. Paramecium cells containing this transgene showed no detectable GFP fluorescence during logarithmic growth, starvation, or the early stages of conjugation. However, consistent with DIE5 mRNA expression (Fig. 1A), GFP-Die5p was observed in cells by 7 h after the initiation of mating. The fusion protein was found to localize exclusively within the developing macronuclei but not in the fragments of old macronuclei of conjugating or autogamous cells (Fig. 1B and data not shown). Thus, PtDIE5 encodes a developmentally expressed protein that appears to act in differentiating macronuclei.
Silencing of PtDIE5 inhibits formation of a functional macronucleus.
To investigate whether the expression timing and localization of Die5p is indicative of a function in macronuclear development, we knocked down DIE5 expression, using RNAi-mediated gene silencing, by feeding Paramecium cells E. coli expressing double-stranded RNA corresponding to a fragment of DIE5 (16). To assess the level of knockdown achieved by this approach, we monitored protein expression of GFP-Die5p in transformed cells, using GFP-specific antibodies, which provided a proxy for endogenous expression. Western blot analysis showed as substantial reduction in GFP-Die5p levels, to ∼35% of that observed upon feeding cells E. coli transformed with the empty RNAi vector (data not shown).
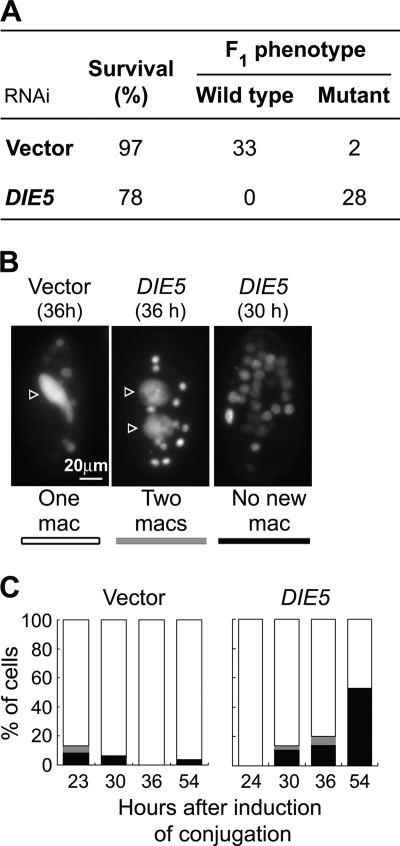
To determine whether DIE5 is essential to complete development, we knocked down expression in mating cells that were genetically marked to allow true progeny to be distinguished from the parental lines. The two parental cell lines used in this experiment were each homozygous for a recessive allele at different loci (pwA or pwB; see Materials and Methods). Since successful conjugation of these cells generates F1 progeny that are heterozygous at all loci, these F1 exhibit a wild-type phenotype (genotype PWA/pwA, PWB/pwB). Both DIE5 RNAi-treated and control cultures (i.e., cells fed bacteria containing the empty RNAi vector) produced high percentages of viable cells (78 and 97%, respectively), but only the control cells gave rise to true progeny that were phenotypically wild-type, whereas all viable cells of DIE5 RNAi-treated cells exhibited the parental mutant phenotypes (Fig. 2A). Thus, loss of Die5p resulted in failure to form new macronuclei.
Fig. 2.
Silencing of DIE5 disrupts nuclear events during conjugation. (A) Progeny from conjugation treated with RNAi. Survival (%) and phenotype of the marker genes are shown. Both parental strains for conjugation were homozygous for Mendelian recessive mutant alleles of different marker genes (pwA and pwB; required for ciliary reversal); thus, successful conjugation should produce the wild-type phenotype of backward swimming, while failure in either nuclear exchange, fertilization, or formation of the new macronucleus should result in the mutant phenotype, i.e., no backward swimming upon stimulation. (B) Representative DAPI-stained Paramecium exconjugants with normal and defective cytological phenotypes observed after RNAi treatments. Each picture contains a single cell with or without the new macronucleus (arrowheads) and old macronuclear fragments. Phenotypic classes were assigned to white, gray, or black, as indicated below the pictures and plotted in the graphs in panel C. mac, macronucleus.
We visually followed the DIE5 RNAi-treated conjugants throughout development to ascertain whether the lack of sexual progeny was caused by a failure in prezygotic events (e.g., meiosis, nuclear exchange, or karyogamy) or in events associated with postzygotic differentiation of the new macronucleus. Control matings that produced wild-type cells generated exconjugants with two macronuclear anlagen that eventually segregated to the daughter cells at the first postmating cell division (∼18 h after induction of conjugation). In contrast, cytological observations of DIE5 RNAi-treated cells (examined 30 and 36 h after induction of conjugation) showed a dramatic reduction in cells with differentiating macronuclei, as only 2 to 8% of exconjugants had two new macronuclei, while 12 to 23% had no macronuclear anlagen at all (Fig. 2B and C). The no-macronucleus cells are expected to include those that did not survive conjugation (lethal). The increase in no-macronucleus cells at 54 h may also include cells that entered autogamy shortly after conjugation. This is not possible in a normal mating but could occur in cells that have undergone macronuclear regeneration. Despite these nuclear abnormalities, most DIE5 RNAi-treated conjugants proceeded through the first postconjugative cell division. These results are substantially different from those for silencing UBA2 (which encodes a SUMO-activating enzyme), where most cells were arrested with two micronuclei and two macronuclei (32). Together with the genetic analysis of exconjugants, these results reveal that knockdown of DIE5 expression blocks the completion of macronuclear development.
Despite the defects observed upon DIE5 knockdown, these cells still exhibited a high level of viability upon exit from conjugation. This observation suggests that DIE5 RNAi treatment likely induced the alternative developmental pathway of parental macronuclear regeneration (MR), which can occur in Paramecium when new macronuclei fail to form. This pathway has been observed upon silencing of other genes during conjugation (32, 41). Normally in wild-type cells, old macronuclear fragments remain in the exconjugants and are transcriptionally active for several postconjugative cell divisions. DNA replication no longer occurs, though, and these fragments are typically lost within 8 to 10 cell divisions (either actively or by dilution). When the new macronucleus is incapable of division after sexual reproduction, one or more of the parental macronuclear fragments regenerate into a single macronucleus that is again capable of DNA replication, amitotic division, and transcription. After MR, the macronuclear genotype is the same as that of the parental lines (e.g., a mutant for pwA or pwB), while the micronuclear genotype is heterozygous, as nuclear exchange occurs between conjugates. To determine whether MR had occurred, several cells that survived DIE5 knockdown during conjugation were followed into autogamy to reveal their micronuclear genotypes. The resulting F2 lines of the DIE5 RNAi F1 survivors produced wild-type (as well as mutant) progeny, which demonstrates that the F1 cell lines contained heterozygous micronuclei. Thus, the prezygotic events of conjugation (i.e., meiosis and nuclear exchange) must have occurred normally in the DIE5 RNAi-treated cells, and the surviving F1 were the result of MR.
DIE5 is required for Paramecium IES excision.
The failure to form mature macronuclei after DIE5 RNAi treatment despite successful formation of the zygotic nucleus raised the possibility that this protein is important for the removal of IESs. To examine the effect of DIE5 knockdown on IES excision, we first employed a PCR-based approach to specifically amplify micronucleus-derived sequences. By locating one PCR primer for each amplicon within an IES and its partner in the flanking macronucleus-destined DNA, we could distinguish DNA in the macronuclear anlagen (and new micronuclei) from the abundant, rearranged DNA of the old macronuclear fragments. The abundance of the resulting PCR products (named “pp1,” “pp2,” and “pp3” in Fig. 3A) should increase as anlagen DNA is amplified and decrease as the IESs containing the primers are excised (Fig. 3B, vector lanes). In addition, as small IESs are contained within the PCR amplicons, we could follow their fates, as their excision prior to removal of the IESs containing the primer sites generated smaller products observed as faster-migrating bands in the gels (pp1s, pp2s, and pp3s in Fig. 3A and B). RNAi of DIE5 showed a gradual increase in the amount of full-length (IES-containing) PCR product over the developmental time course, indicating that loss of DIE5 does not inhibit developmental DNA amplification (Fig. 3B). Nevertheless, we saw no evidence of shorter products, which are readily detectable in control cells, that would indicate IES excision had occurred.
To further demonstrate that loss of DIE5 blocked IES excision, we used Southern blot hybridization to analyze the rearrangement status of the new macronuclei. For this study, total DNA was isolated from large cultures of postautogamous cells at a point when about 50% of well-fed control cells (treated with the empty RNAi vector) had undergone the first cell division. The DNA was digested with SspI, which has frequent recognition sites in IESs but only one in the coding region of the A-51 allele. The probe containing macronuclear DNA from the A gene (HincII-PstI in Fig. 3A) detected only the macronuclear form of this region (indicated by the 5.7-kbp band in Fig. 3C) in DNA from control cells (Fig. 3C, labeled vector). In contrast, two additional bands of 1.3 and 2.0 kb were observed in DNA from DIE5 RNAi-treated cells. These are the sizes expected for amplification of the unprocessed A-51 gene (Fig. 3C). Furthermore, using a probe for IES4578 (Fig. 3A) that contained only micronucleus-limited DNA, we detected a single 500-bp DNA fragment, which corresponds to the IES-containing locus in DNA from DIE5 RNAi-treated cells but not from control cells (Fig. 3C). The unprocessed DNA can be observed in these experiments because amplification of DNA continues in the developing macronuclei despite the absence of IES excision. Together the results show that knockdown of DIE5 expression inhibits IES excision in the developing macronuclei of Paramecium, yet the DNA is amplified to levels comparable to that for normal developing macronuclei.
Tetrahymena DIE5 is required for macronuclear differentiation.
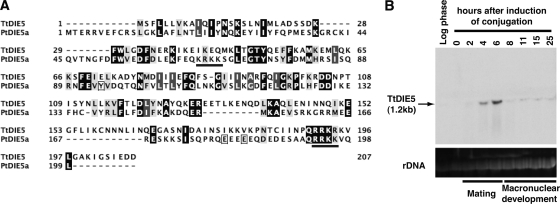
The PtDie5p sequence contained no conserved domains that offered clues to its biochemical function; however, we did find a putative DIE5 homologue (TtDIE5) encoded within the Tetrahymena thermophila genome. These two ciliate proteins are similar in size (199 and 207 aa, respectively) and share 21% amino acid identity throughout their coding regions (Fig. 4A). In addition, when the sequences were analyzed for predicted secondary structures using PSIPRED (33), a common domain structure that predicted beta sheets and alpha helices in the form β1α1α2β3β4 was revealed within the first 110 aa of both proteins (data not shown). Intriguingly, our Northern blot analysis revealed that TtDIE5 is expressed exclusively during conjugation, providing data to support that this Tetrahymena gene may have a function similar to that of its Paramecium counterpart. TtDIE5 expression was first detected 4 h into conjugation, which corresponds to the end of meiosis, and peaked at 6 h, when new macronuclei first emerge. Expression levels declined slightly at 8 h but continued at a low level until the end of macronuclear development (about 15 h after cells first paired) (Fig. 4B). The expression pattern we observed is nearly identical to recently published microarray data (35).
Fig. 4.
Tetrahymena homologue (TtDIE5) is developmentally expressed. (A) Pairwise alignment of Paramecium Die5 protein from paralog a (PtDIE5a) and Tetrahymena Die5p sequences. The boxes around three amino acids indicate differences between the two Paramecium sequences (the differences are C, D, D, respectively). The underlined regions indicate potential nuclear localization signals in the Paramecium sequence. (B) Northern blot analysis of total RNA (20 μg per lane) extracted from growing (log), starved, or conjugating cells (between 2 and 25 h after mixing populations of compatible mating types). Ethidium bromide staining of rRNA was used as a loading control.
Our RNAi knockdown experiments showed that Die5p is essential for macronuclear differentiation in Paramecium. If TtDIE5 is a homologue of the Paramecium protein, it is likely to have an essential role in Tetrahymena development as well. To investigate this possibility and learn more about Die5p function, we used homologous gene replacement to disrupt TtDIE5. A knockout construct (Fig. 5A) consisting of the neo3 selectable cassette (45) flanked on each side by DNA sequences from immediately upstream and downstream of the TtDIE5 locus was introduced by biolistic transformation into starved or conjugating Tetrahymena cells to disrupt the macronuclear or micronuclear copies, respectively. Paromomycin-resistant transformants were obtained by both strategies and were subsequently cultured to generate full macronuclear knockouts or homozygous micronuclear-knockout lines (see below and Materials and Methods). Genetic crosses of strains lacking all macronuclear DIE5 copies produced viable progeny (data not shown), indicating that DIE5 expression prior to transcriptional activation of developing macronuclei is not required to complete conjugation. In contrast, attempts to generate complete knockout strains (lacking DIE5 in both micro- and macronuclei) by crossing heterozygous germ line (micronuclear) knockout strains were unsuccessful. One-quarter of the progeny resulting from these crosses should have been homozygous knockouts (in both nuclei), yet only homozygous wild type or heterozygous knockout strains were found among the >30 viable progeny screened (data not shown). This finding provided the first indication that TtDie5p is critical for development, as is the Paramecium protein.
Fig. 5.
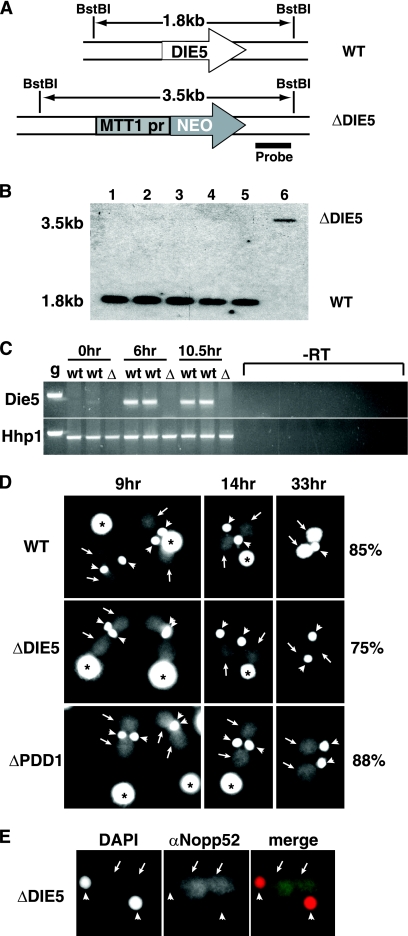
TtDIE5 zygotic expression is essential to complete conjugation. (A) Diagram of the gene disruption construct showing replacement of the coding sequence with the neo3 selectable cassette (47). Restriction enzyme sites (BstBI) and the region corresponding to the radiolabeled probe fragment used for Southern blot analysis are indicated. WT, wild type; MTT1pr, metallothionein gene 1 promoter. (B) Southern blot hybridization of DNA isolated from the wild type (lane 1), DIE5 micronuclear-knockout lines (lanes 2 to 5), and the DIE5 complete knockout line (lane 6) using the probe shown in panel A. Longer exposure of the blot reveals the 3.5-kb band in DIE5 micronuclear-knockout samples corresponding to the two copies of DIE5 disrupted by the neo3 cassette in their micronuclei. (C) RT-PCR for expression of DIE5 transcripts in conjugating wild-type (wt) and DIE5 complete knockout (Δ) cells at indicated time points. The bottom panel shows control RT-PCR with HHP1 primers. Both DIE5 and HHP1 primers span an intron of their respective genes. g, Tetrahymena genomic DNA used as a control for amplification. (D) Fluorescent images of representative DAPI-stained wild-type (WT), micronuclear-knockout (ΔDIE5), and ΔPDD1 strains at 9 h, 14 h, and 33 h postmixing. For each mating strain, the percentage of cells exhibiting their respective arrest phenotype at 33 h is indicated on the right. Asterisks, arrows, and arrowheads indicate old/parental macronuclei, new macronuclei, and the micronuclei, respectively. (E) The nuclear envelope remains intact in arrested ΔDIE5 micronuclear-knockout cells (ΔDIE5). Postconjugative ΔDIE5mic cells were fixed with 2% paraformaldehyde and stained with αNopp52 antibody and DAPI. New macronuclei and micronuclei are indicated as described for panel D.
As strains lacking all macronuclear DIE5 copies grew normally and produced viable progeny when mated, the inability of heterozygous DIE5 micronuclear knockouts to produce homozygous knockout progeny must result from the loss of zygotic DIE5 expression during macronuclear differentiation. To allow us to further investigate Die5p's role during Tetrahymena development, we generated homozygous micronuclear-knockout heterokaryon strains of different mating types by performing genomic exclusion crosses between heterozygous micronuclear knockouts and two different star strains, B*VI and B*VII, which have defective micronuclei. These abortive matings resulted in the transfer of a haploid micronucleus from the DIE5 knockout to its star strain partner without inducing new macronuclear development, such that after pair separation and micronuclear endoreplication, both exconjugants were homozygous in their germ line. The resulting paromomycin-sensitive exconjugants (the star strain partner) with homozygous knockout micronuclei and wild-type macronuclei were identified by genomic locus PCR (data not shown) and Southern blot analysis (Fig. 5B) and then verified by genetic crosses (data not shown). Herein we refer to these cell lines as DIE5 micronuclear knockouts or ΔDIE5mic (these cells were used for most experiments; therefore, all figures labeled ΔDIE5 refer to micronuclear knockouts). Phenotypic assortment of the paromomycin-resistant, homozygous, micronuclear-knockout exconjugants allowed us to generate complete (micro-/macronuclear) knockout strains (referred to herein as DIE5 complete knockouts). We confirmed the loss of all DIE5 copies and expression during conjugation by Southern blot analyses and RT-PCR (Fig. 5B and C), respectively. The ability to generate DIE5 complete knockouts confirms that Die5p is dispensable for vegetative growth.
ΔDIE5mic strains are paromomycin sensitive but are homozygous for the DIE5::NEO3 allele in their silent micronuclei, so that when mated, their progeny, if viable, would be paromomycin-resistant. To measure the ability of these germ line knockout strains to produce ΔDIE5 progeny, >106 postconjugative cells were cultured in growth medium containing paromomycin (plus CdCl2). In most trials, all cells died upon the addition of the drug, providing further support that zygotic expression of DIE5 is essential for the completion of conjugation. We did obtain paromomycin-resistant cells for some matings (up to two survivors per 106 mating pairs). We interpret this to mean that some cells express sufficient Die5p from their wild-type parental macronuclei to provide for the essential functions of this protein late in macronuclear differentiation. To further test this possibility, we created an N-terminal GFP-TtDie5p transgene expressed from the strong, cadmium-inducible MTT1 promoter, integrating the construct upstream of the macronuclear rpL29 genomic locus in the ΔDIE5mic strains. Expression of this transgene by the addition of cadmium during conjugation increased progeny survival by 3 to 4 orders of magnitude. Thus, inducing additional Die5p from the parental macronucleus partially rescued the loss of DIE5 zygotic expression from macronuclear anlagen, which provided clear evidence that loss of DIE5 expression is the cause of lethality upon mating ΔDIE5mic cells and that the GFP-TtDie5p fusion is functional. We were also able to increase survival of ΔDIE5mic conjugates by introducing an rDNA-based expression vector by electroporation that carries the GFP-TtDIE5 transgene at ∼9 h into conjugation, an observation which further supports that this protein is required relatively late in macronuclear development.
Tetrahymena conjugation can be readily staged by the configuration of nuclei within mating pairs or exconjugants (31). To begin to ascertain why mating ΔDIE5mic cells fail to produce viable progeny, we fixed and DAPI-stained cells to monitor their development. For most of conjugation, the progression of ΔDIE5mic conjugates was indistinguishable from that of wild-type mating cells as they completed meiosis, karyogamy of gametic nuclei, and the subsequent nuclear divisions to generate macronuclear anlagen (Fig. 5D). Mating pairs separated, and their nuclear morphology appeared normal until ∼14 h of conjugation, after which it became evident that most knockout cells arrested their development. The last visible event to be triggered during conjugation is the elimination of one of the two micronuclei in each conjugant. Wild-type cells remain with the characteristic nuclear configuration of one micronucleus and two new macronuclei (Fig. 5D, 33 h) until they are fed, at which point they divide their one remaining micronucleus and undergo a specialized postconjugative cytokinesis to partition one micro- and one macronucleus to each daughter. In contrast, the ΔDIE5mic conjugates failed to trigger micronuclear resorption, arresting with two micro- and macronuclei, and were unable to divide when returned to growth medium.
The two micro-/two macronuclei arrest phenotype has been described for knockouts of several genes required for programmed DNA rearrangement, including ΔPDD1 strains (Fig. 5D, bottom panels). Such mutant strains not only fail to eliminate one micronucleus, but unlike wild-type cells, stop amplification of the DNA in the new macronuclei (Fig. 5D, compare the DAPI-staining intensity of ΔPDD1 cells to that of the wild type at 33 h). Intriguingly, not only did ΔDIE5mic exconjugates stop anlagen DNA amplification, but the majority of cells (75%) actually lost most of the DNA content of the anlagen, as indicated by the loss of DAPI staining between 14 and 33 h postmixing (Fig. 5D). The macronuclear structure appeared to remain intact, as immunofluorescence to detect a nucleolar protein, NOPP52, revealed the integrity of the nuclear compartment (Fig. 5E). We are unsure of the mechanism of DNA loss, as attempts to detect chromosome degradation by the presence of unprotected ends revealed that unlike the degrading old macronucleus, these new macronuclei do not label in terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays (data not shown). Nevertheless, the loss of macronuclei in the ΔDIE5mic exconjugates is consistent with the Paramecium DIE5 RNAi phenotype in which anlagen form but fail to become the macronuclei of the surviving exconjugants.
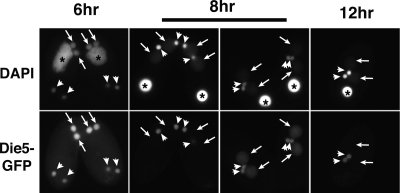
As the DIE5 expression profile and knockdown/knockout phenotypes were very similar in both Paramecium and Tetrahymena, we expected that TtDIE5 would also encode a protein localized primarily to macronuclear anlagen. However, we were surprised when we examined the localization of the GFP-TtDie5p fusion used in the rescue experiments described above. Although GFP-TtDie5p was detected in the developing macronuclei during early stages of macronuclear development (Fig. 6, 6-h to 8-h arrows), this localization was rapidly lost as conjugation progressed, before the period when the essential zygotic expression would occur. In contrast, we observed the GFP fluorescence within micronuclei through all stages of conjugation (Fig. 6, arrowheads). Therefore, while Die5p is a developmentally expressed nuclear protein in both ciliates, their localization patterns suggest some differentiation in their action.
Fig. 6.
Cellular localization of GFP-Die5p in conjugating Tetrahymena cells at 6 h, 8 h, and 12 h. The cells were fixed with 2% paraformaldehyde and counterstained with DAPI. Asterisks, arrows, and arrowheads indicate old/parental macronuclei, new macronuclei, and the micronuclei, respectively.
Zygotic expression of DIE5 is required for programmed DNA rearrangement in Tetrahymena.
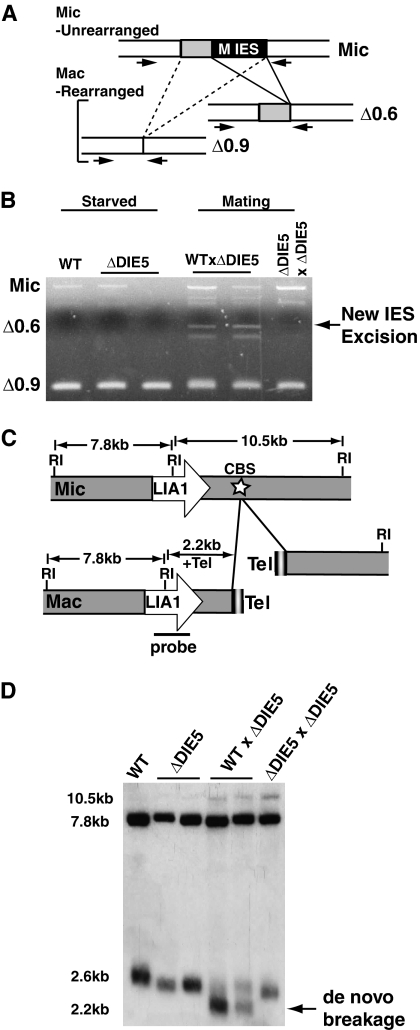
Transcription of TtDIE5 was induced by 4 h into conjugation (Fig. 4A) and peaked at 6 h; thus, it was somewhat unexpected that we found that the macronuclear copies were dispensable. Additionally, complete-knockout cells lacking DIE5 from both macro- and micronuclei exhibit the same phenotype as the germ line knockouts, which further confirms that the critical TtDIE5 expression occurs from macronuclear anlagen. This suggests that DIE5 functions at late stages of Tetrahymena conjugation when DNA rearrangement occurs. To determine whether zygotic Die5p is required for Tetrahymena DNA rearrangements, we monitored IES excision and chromosome breakage in ΔDIE5mic exconjugants. To assess the process of IES excision, we examined the elimination of the M IES, which is a >1-kbp sequence located on micronuclear chromosome 4 (3, 9). This IES has two equally used left deletion boundaries that are 300-bp apart, such that successful IES excision generates two alternative products through the elimination (Δ) of either 0.6 kbp or 0.9 kbp from the locus (Fig. 7A). These two forms can be easily distinguished by a PCR-based IES excision assay, using primers designed to amplify across the IES. By crossing ΔDIE5mic lines together or with wild-type strain B2086, each of which contains only the MΔ0.9-kbp rearranged form in their macronuclei, we could test for appearance of the MΔ0.6-kbp deletion as an indicator of new rearrangement in the anlagen. Whereas the slower-migrating PCR product indicative of the MΔ0.6-kbp deletion was detected when each ΔDIE5mic line was crossed to B2086, we saw no evidence of M-element rearrangement in crosses of the two ΔDIE5mic strains (Fig. 7B).
Fig. 7.
Germ line TtDIE5 is required for Tetrahymena-programmed DNA rearrangement. Total genomic DNA was isolated from starved wild-type (WT) and DIE5 micronuclear-knockout strains (ΔDIE5), as well as from cells 16 h after crosses of DIE5 micronuclear knockouts to the wild type (WT × ΔDIE5) and two DIE5 micronuclear knockouts (ΔDIE5 × ΔDIE5). The DNA was used for PCR-based IES excision assays (A and B) or Southern blot analyses for chromosome breakage (C and D). (A) Schematic of PCR-based IES excision assay strategy. Arrows denote forward and reverse primers used to amplify across the M element. Alternative rearrangement products resulting from deletion of 0.6-kbp (Δ0.6) or 0.9-kbp (Δ0.9) are shown. (B) M-element excision PCR. Arrow indicates new IES excision. (C) Diagram shows the macronuclear chromosomal scaffold surrounding the LIA1 gene, which lies within 2.2 kbp of a chromosomal-breakage sequence (CBS) (white star). Relevant EcoRI (RI) restriction sites used for the Southern blot analysis are shown. The probe spans the central EcoRI site and detects a 7.8-kbp fragment common to both nuclei as well as to either the 10.5-kbp micronucleus-specific fragment or a 2.5- to 2.6-kbp macronucleus-specific fragment (2.2 kbp of unique sequence plus 300 to 400 bp of telomeric DNA). Tel, telomere. (D) Southern blot analysis to assess chromosome breakage. Arrow indicates the product of de novo breakage.
Since RNAi knockdown of Paramecium DIE5 blocked chromosome fragmentation (data not shown) as well as IES excision (Fig. 3), we tested whether ΔTtDIE5mic knockouts also fail to fragment chromosomes. We isolated DNA from postconjugative ΔDIE5mic cells and used Southern blot analysis to examine chromosome fragmentation at a site which lies 2.2 kbp downstream of the LIA1 gene (Fig. 7C) (30). During conjugation, breakage occurs at this sequence, followed by the addition of 300 bp to 400 bp of telomeric DNA during growth. Restriction digestion of genomic DNA with EcoRI allows simultaneous detection of the unprocessed micronuclear form of this locus at 10.5 kbp, the fragmented chromosome of parental macronuclei with fully elongated telomeres that migrates at 2.5 to 2.6 kbp, and the 2.2-kbp form that results from de novo breakage and minimal telomere addition (Fig. 7C). Whereas this 2.2-kbp fragment was easily observed in genomic DNA samples from wild-type matings, the DNA from ΔDIE5mic conjugants showed no evidence of chromosome fragmentation, as the predominant band observed was ∼2.5 kbp, which represents DNA from parental macronuclei of unmated cells in the population (Fig. 7D). These results together with experiments with Paramecium (data not shown) indicate that DIE5 is required for IES excision and chromosome fragmentation in both ciliates.
Critical events leading to DNA rearrangement are unaffected in ΔDIE5.
IES excision is guided by small RNAs. In Tetrahymena, it is known that these small RNAs target H3K9 and K27 methylation to IES chromatin, which is bound by the Pdd1 and Pdd3 chromodomain-containing proteins that are reorganized into distinct foci that are hypothesized to be the site of IES excision (reviewed in reference 10). The timing of these events has been well described, and proteins required for many of these steps have been identified (11, 26, 36, 50). To investigate the role of DIE5 in Tetrahymena DNA rearrangement, we examined these events to determine whether they are perturbed by the loss of DIE5.
Production of developmental-specific small RNAs occurs during meiosis, before the emergence of zygotic expression. As zygotic, not somatic, expression of DIE5 is essential for conjugation in Tetrahymena, we expected that DIE5 would not be required for their biogenesis or accumulation. We isolated small RNAs from conjugating complete-DIE5-knockout cells, and as predicted, we found that the levels of total small RNAs and those homologous to the M IES observed were comparable in wild-type and ΔDIE5mic conjugating cells (data not shown).
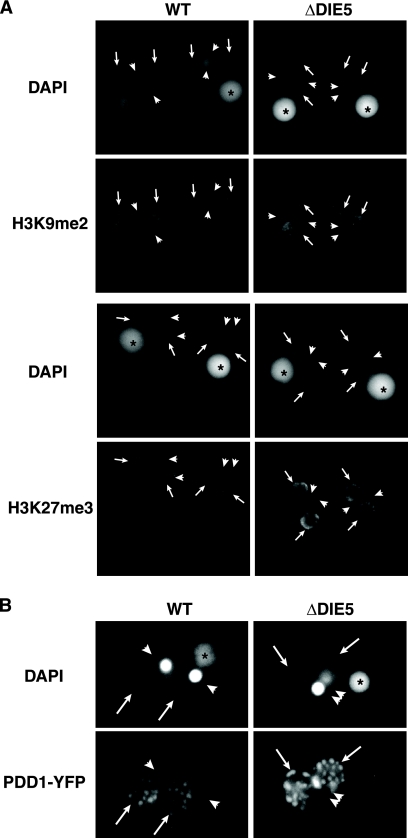
We also examined whether H3K9 and H3K27 methylation was perturbed by the loss of DIE5. Although establishment of these marks occurs during the time that we observed GFP-TtDie5p to localize to developing macronuclei, deposition of these modifications in conjugating DIE5 knockout cells appeared to be unaffected (Fig. 8A). Therefore, Die5p is not required for the establishment of these heterochromatic modifications in Tetrahymena, further suggesting that the essential function of DIE5 occurs at later stages of conjugation.
Fig. 8.
Germ line knockout of TtDIE5 does not inhibit critical events leading to IES excision. (A) H3K27me3 and H3K9me2 histone mark deposition is unaffected in TtDIE5 germ line knockouts. Nine-hour-conjugating wild type (WT) and micronuclear-knockout (ΔDIE5) cells were fixed with Schaudinn's fixative and stained with either H3K27me3 or H3K9me2 antibodies at 9 h into conjugation. The cells were counter stained with DAPI. (B) Formation of PDD1 foci is not disrupted in ΔDIE5mic cells. Conjugating wild-type and DIE5 micronuclear-knockout cells (ΔDIE5) expressing an inducible PDD1-YFP transgene were fixed at 14 h with 2% paraformaldehyde and counter stained with DAPI. Asterisks, arrows, and arrowheads indicate old/parental macronuclei, new macronuclei, and the micronuclei, respectively.
The chromodomain-containing protein Pdd1p is an essential component of the DNA rearrangement machinery and a major constituent of DNA rearrangement foci (12, 27). Disruption of genes required for IES excision can block the formation of these foci (26, 44). To investigate chromatin reorganization upon loss of Die5p, we examined the localization of a Pdd1p-YFP fusion expressed in ΔDIE5mic cells. Even though these mutant lines fail to excise IESs, Pdd1p foci in the developing macronuclei appear to form, as we observed in wild-type conjugants, albeit with a slight delay in their maturation (Fig. 8B). Given that all events prior to IES excision described to date appear normal in ΔDIE5 knockouts, we suspect that Die5p acts after these known events.
DISCUSSION
Orthologous DIE5 genes in Paramecium and Tetrahymena are essential for macronuclear development.
Research over the past 10 years has revealed that some key components of the genome reorganization pathway in ciliates are well-known proteins in other eukaryotic organisms. Examples include proteins of the RNA interference pathway, histone-modifying enzymes, and transposases (4, 26, 30, 36). Other studies have identified proteins with recognizable domains, and yet clear orthologs cannot be identified, even in other ciliate genomes (58). DIE5 is a member of a third group, proteins that are both conserved among ciliates (Paramecium and Tetrahymena) but novel to this class of organisms. This group may contain core components that account for the unusual precision and efficiency of ciliate genome reorganization. Our results provide strong evidence that Paramecium and Tetrahymena encode orthologous Die5 proteins. Although the amino acid identity between the proteins is modest (21%), each is the top reciprocal BLAST hit in comparisons of their respective genomes. Additionally, the amino acid identity is not centered in a single region, as expected for a shared domain; rather, it is spread across the entire coding region. Both are small, nuclear proteins (199 aa and 207 aa in Paramecium and Tetrahymena, respectively) expressed exclusively during conjugation. Finally, RNAi knockdown (Paramecium) and gene disruption (Tetrahymena) demonstrate that these Die5 proteins are essential for genome rearrangements in their respective species. Together the data demonstrate that DIE5 encodes a conserved component of the macronuclear development pathway in ciliates.
Die5 is one of a few conserved ciliate proteins known to be required for DNA rearrangements.
Previous studies have identified several developmentally regulated proteins that are required for genome rearrangements in Paramecium or Tetrahymena. These include chromatin-associated proteins (27, 28, 39, 40, 48, 58), a transposase (4), components of the RNAi pathway (30, 36, 37) and SUMO pathways (32), and a putative RNA binding protein (41). Of these identified components, only the RNAi-associated proteins, piggyMac transposase, and SUMO components show clear homologues between these different ciliates. In contrast, the Pdd and Lia (localized in macronuclear anlagen) DNA rearrangement proteins of Tetrahymena do not have obvious homologues in Paramecium (58). This search for homologues is complicated by the significant sequence diversity between Paramecium and Tetrahymena, a point that was evident even before complete-genome sequences were available (reviewed in reference 15). Thus, failure to detect homologues based on primary sequence data is not definitive, and one must consider the possibility that protein three-dimensional structure is maintained with minimal sequence identity. Functional homology is the most relevant criteria, but few of the proteins involved in these rearrangements have known biochemical roles. For those that do, such as the Tetrahymena chromodomain-containing Pdd1 and Pdd3 proteins that have been shown to associate with IES chromatin-containing histone H3 methylated on lysine 27 and/or lysine 9, respectively, evidence for an analogous role in Paramecium DNA rearrangements is lacking (26, 50). The Nowa proteins in Paramecium are required for elimination of germ line transposons and a subset of IES that are controlled by maternal effects (41). The N-terminal domains contain repeated elements with similarity to RNA binding motifs in other species, and evidence of nucleic acid binding activity was obtained. While there are Tetrahymena proteins, including CnjBp (41, 51), that share similar glycine-rich repeats, none are clearly identifiable as homologues. Whereas the Pdd, Lia, and Nowa proteins reinforce the divergence between the DNA rearrangement machinery in the two species, Die5p reveals a novel connection between the two systems.
DIE5 is required late in macronuclear development.
Although the novel sequence of Die5p limits speculation on its biochemical function, the molecular events that are disrupted (or not disrupted) by inhibiting DIE5 expression in two species argue for late action in macronuclear development. First of all, prezygotic events of meiosis and pronuclear exchange occur normally upon DIE5 RNAi silencing in Paramecium. The survivors of conjugation upon DIE5 knockdown are heterokaryons with heterozygous micronuclei (with alleles from both parents), but they have macronuclei regenerated from fragments of their parental macronuclei. This phenotype has been observed upon knockdown of other genes involved in macronuclear development (32, 41). Likewise, early-stage events of scnRNA biogenesis (data not shown) and accumulation of specific chromatin modifications (Fig. 8) associated with DNA elimination appear unaltered upon disruption of DIE5 in Tetrahymena. This is in contrast to disruption of RNAi components (e.g., DCL1 or TWI1) that are defective in these events.
Even relatively late-stage events prior to IES excision were not disrupted upon loss of DIE5 expression. Macronuclear anlagen form and substantial DNA amplification occurs in RNAi-treated Paramecium even though IES excision is inhibited. In Tetrahymena ΔDIE5mic conjugants, Pdd1p foci formed as expected for wild-type cells (Fig. 8B). These observations, together with the fact that we saw no phenotype when DIE5 was disrupted only from the parental macronucleus, suggest that its essential role occurs after activation of zygotic expression of the macronuclear anlagen. Most striking is our observation that Tetrahymena heterokaryons with wild-type macronuclei but homozygous for the ΔDIE5 allele in their micronuclei fail to excise IESs and arrest prior to the elimination of one of the two micronuclei. These results clearly demonstrate a late role for Die5p in macronuclear differentiation.
While loss of DIE5 expression blocked IES excision in both ciliates studied here, we did observe some differences in terminal phenotypes, in particular the degree of anlagen DNA amplification. DIE5 RNAi-treated Paramecium appeared to extensively amplify their anlagen DNA, whereas ΔDIE5mic Tetrahymena cells arrested their anlagen differentiation at a low amplification level. We favor the explanation that this most likely represents distinct differences in nuclear differentiation events (e.g., macronuclear regeneration occurs in Paramecium but not in Tetrahymena) in these evolutionarily diverse organisms. Nevertheless, we cannot rule out that such phenotypic differences may result from low levels of PtDie5p remaining after RNAi silencing. What is clear is that RNAi silencing that was robust enough to prevent IES excision in Paramecium to the same extent as was observed in Tetrahymena ΔDIE5mic conjugants elicited differential effects on anlagen DNA amplification.
The surprising observation that anlagen DNA is degraded in Tetrahymena ΔDIE5 conjugants has not been observed in other DNA rearrangement mutants (12, 30, 36, 40). The mechanism of degradation is not understood, and we were unable to demonstrate labeling in TUNEL assays. It is interesting to note that anlagen DNA degradation appears to be the outcome in Paramecium DIE5 RNAi silencing, but as mentioned previously, these cells survive by regenerating a functional macronucleus from an old macronuclear fragment. This is possible in Paramecium because macronuclear fragments persist throughout conjugation and continue transcription for multiple cell divisions after conjugation. In contrast, degradation of the old macronucleus in Tetrahymena is complete prior to formation of the new genome and leaves no recovery pathway if development is defective. Further analysis will be required to establish whether the degradation of Tetrahymena anlagen DNA has a similar molecular basis as the fate of Paramecium anlagen in DIE5-silenced cells.
The significance of TtDie5p localization to the micronuclei but not anlagen at late time points is unclear. This may reflect the limitation of using a macronuclear GFP-DIE5 transgene that cannot be expressed after degradation of the old macronucleus. Alternatively, the localization could be evidence of an unexpected role of Die5p from the newly formed micronucleus. Unusual observations such as these underscore the importance of investigating the role of conserved proteins such as Die5p in the DNA rearrangement process to elucidate the evolution and regulation of this massive genome-remodeling process and its connections to large-scale genome reorganizations in other species.
ACKNOWLEDGMENTS
We thank Charles H. Rexer for initial work toward characterizing TtDIE5. We also thank Genoscope and the J. Craig Venter Institute for preliminary Paramecium and Tetrahymena sequence data, respectively.
A.M. and J.D.F. were supported by a National Science Foundation grant (MCB-9506009). D.L.C. and A.W.-Y.S. were supported by National Institutes of Health research grant GM06593.
Footnotes
Published ahead of print on 21 May 2010.
REFERENCES
- 1.Allen S. L., Altschuler M. I., Bruns P. J., Cohen J., Doerder F. P., Gaertig J., Gorovsky M., Orias E., Turkewitz A. 1998. Proposed genetic nomenclature rules for Tetrahymena thermophila, Paramecium primaurelia and Paramecium tetraurelia on behalf of the Seventh International Meeting on Ciliate Molecular Biology Genetics Nomenclature. Genetics 149:459–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aury J. M., Jaillon O., Duret L., Noel B., Jubin C., Porcel B. M., Segurens B., Daubin V., Anthouard V., Aiach N., Arnaiz O., Billaut A., Beisson J., Blanc I., Bouhouche K., Camara F., Duharcourt S., Guigo R., Gogendeau D., Katinka M., Keller A. M., Kissmehl R., Klotz C., Koll F., Le Mouel A., Lepere G., Malinsky S., Nowacki M., Nowak J. K., Plattner H., Poulain J., Ruiz F., Serrano V., Zagulski M., Dessen P., Betermier M., Weissenbach J., Scarpelli C., Schachter V., Sperling L., Meyer E., Cohen J., Wincker P. 2006. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444:171–178 [DOI] [PubMed] [Google Scholar]
- 3.Austerberry C. F., Allis C. D., Yao M. C. 1984. Specific DNA rearrangements in synchronously developing nuclei of Tetrahymena. Proc. Natl. Acad. Sci. U. S. A. 81:7383–7387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baudry C., Malinsky S., Restituito M., Kapusta A., Rosa S., Meyer E., Betermier M. 2009. PiggyMac, a domesticated piggyBac transposase involved in programmed genome rearrangements in the ciliate Paramecium tetraurelia. Genes Dev. 23:2478–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bétermier M. 2004. Large-scale genome remodelling by the developmentally programmed elimination of germ line sequences in the ciliate Paramecium. Res. Microbiol. 155:399–408 [DOI] [PubMed] [Google Scholar]
- 6.Bétermier M., Duharcourt S., Seitz H., Meyer E. 2000. Timing of developmentally programmed excision and circularization of Paramecium internal eliminated sequences. Mol. Cell. Biol. 20:1553–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruns P. J., Cassidy-Hanley D. 2000. Biolistic transformation of macro- and micronuclei. Methods Cell Biol. 62:501–512 [DOI] [PubMed] [Google Scholar]
- 8.Cassidy-Hanley D., Bowen J., Lee J. H., Cole E., VerPlank L. A., Gaertig J., Gorovsky M. A., Bruns P. J. 1997. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics 146:135–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cassidy-Hanley D., Yao M. C., Bruns P. J. 1994. A method for mapping germ line sequences in Tetrahymena thermophila using the polymerase chain reaction. Genetics 137:95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chalker D. L. 2008. Dynamic nuclear reorganization during genome remodeling of Tetrahymena. Biochim. Biophys. Acta 1783:2130–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chalker D. L., Yao M. C. 2001. Nongenic, bidirectional transcription precedes and may promote developmental DNA deletion in Tetrahymena thermophila. Genes Dev. 15:1287–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coyne R., Nikiforov M., Smothers J., Allis C., Yao M. 1999. Parental expression of the chromodomain protein Pdd1p is required for completion of programmed DNA elimination and nuclear differentiation. Mol. Cell 4:865–872 [DOI] [PubMed] [Google Scholar]
- 13.Eisen J. A., Coyne R. S., Wu M., Wu D., Thiagarajan M., Wortman J. R., Badger J. H., Ren Q., Amedeo P., Jones K. M., Tallon L. J., Delcher A. L., Salzberg S. L., Silva J. C., Haas B. J., Majoros W. H., Farzad M., Carlton J. M., Smith R. K., Jr., Garg J., Pearlman R. E., Karrer K. M., Sun L., Manning G., Elde N. C., Turkewitz A. P., Asai D. J., Wilkes D. E., Wang Y., Cai H., Collins K., Stewart B. A., Lee S. R., Wilamowska K., Weinberg Z., Ruzzo W. L., Wloga D., Gaertig J., Frankel J., Tsao C. C., Gorovsky M. A., Keeling P. J., Waller R. F., Patron N. J., Cherry J. M., Stover N. A., Krieger C. J., del Toro C., Ryder H. F., Williamson S. C., Barbeau R. A., Hamilton E. P., Orias E. 2006. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 4:e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan Q., Sweeney R., Yao M. C. 2000. Creation and use of antisense ribosomes in Tetrahymena thermophila. Methods Cell Biol. 62:533–547 [DOI] [PubMed] [Google Scholar]
- 15.Frankel J. 2000. Cell biology of Tetrahymena thermophila. Methods Cell Biol. 62:27–125 [DOI] [PubMed] [Google Scholar]
- 16.Galvani A., Sperling L. 2002. RNA interference by feeding in Paramecium. Trends Genet. 18:11–12 [DOI] [PubMed] [Google Scholar]
- 17.Garnier O., Serrano V., Duharcourt S., Meyer E. 2004. RNA-mediated programming of developmental genome rearrangements in Paramecium tetraurelia. Mol. Cell. Biol. 24:7370–7379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorovsky M. A., Yao M. C., Keevert J. B., Pleger G. L. 1975. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 9:311–327 [DOI] [PubMed] [Google Scholar]
- 19.Gratias A., Betermier M. 2003. Processing of double-strand breaks is involved in the precise excision of paramecium internal eliminated sequences. Mol. Cell. Biol. 23:7152–7162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jahn C. L., Klobutcher L. A. 2002. Genome remodeling in ciliated protozoa. Annu. Rev. Microbiol. 56:489–520 [DOI] [PubMed] [Google Scholar]
- 21.Juranek S. A., Rupprecht S., Postberg J., Lipps H. J. 2005. snRNA and heterochromatin formation are involved in DNA excision during macronuclear development in stichotrichous ciliates. Eukaryot. Cell 4:1934–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klobutcher L. A., Herrick G. 1995. Consensus inverted terminal repeat sequence of Paramecium IESs: resemblance to termini of Tc1-related and Euplotes Tec transposons. Nucleic Acids Res. 23:2006–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ku M., Mayer K., Forney J. D. 2000. Developmentally regulated excision of a 28-base-pair sequence from the Paramecium genome requires flanking DNA. Mol. Cell. Biol. 20:8390–8396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Mouël A., Butler A., Caron F., Meyer E. 2003. Developmentally regulated chromosome fragmentation linked to imprecise elimination of repeated sequences in paramecia. Eukaryot. Cell 2:1076–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lepère G., Nowacki M., Serrano V., Gout J. F., Guglielmi G., Duharcourt S., Meyer E. 2009. Silencing-associated and meiosis-specific small RNA pathways in Paramecium tetraurelia. Nucleic Acids Res. 37:903–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y., Taverna S. D., Muratore T. L., Shabanowitz J., Hunt D. F., Allis C. D. 2007. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev. 21:1530–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madireddi M. T., Coyne R. S., Smothers J. F., Mickey K. M., Yao M.-C., Allis C. D. 1996. Pdd1p, a novel chromodomain-containing protein, links heterochromatin assembly and DNA elimination in Tetrahymena. Cell 87:75–84 [DOI] [PubMed] [Google Scholar]
- 28.Madireddi M. T., Davis M., Allis D. 1994. Identification of a novel polypeptide involved in the formation of DNA-containing vesicles during macronuclear development in Tetrahymena. Dev. Biol. 165:418–431 [DOI] [PubMed] [Google Scholar]
- 29.Malavé T. M., Forney J. D. 2004. Identification of a developmentally regulated translation elongation factor 2 in Tetrahymena thermophila. Gene 326:97–105 [DOI] [PubMed] [Google Scholar]
- 30.Malone C. D., Anderson A. M., Motl J. A., Rexer C. H., Chalker D. L. 2005. Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Mol. Cell. Biol. 25:9151–9164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martindale D. W., Allis C. D., Bruns P. 1982. Conjugation in Tetrahymena thermophila: a temporal analysis of cytological stages. Exp. Cell Res. 140:227–236 [DOI] [PubMed] [Google Scholar]
- 32.Matsuda A., Forney J. D. 2006. The SUMO pathway is developmentally regulated and required for programmed DNA elimination in Paramecium tetraurelia. Eukaryot. Cell 5:806–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGuffin L. J., Bryson K., Jones D. T. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404–405 [DOI] [PubMed] [Google Scholar]
- 34.Meyer E., Chalker D. L. 2007. Epigenetics of ciliates, p. 127–150 InAllis C. D., Jenuwein T., Reinberg D., Caparros M.-L. A. E. (ed.), Epigenetics Cold Spring Harbor Press, Cold Spring Harbor, NY: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miao W., Xiong J., Bowen J., Wang W., Liu Y., Braguinets O., Grigull J., Pearlman R. E., Orias E., Gorovsky M. A. 2009. Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS One 4:e4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mochizuki K., Fine N. A., Fujisawa T., Gorovsky M. A. 2002. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell 110:689–699 [DOI] [PubMed] [Google Scholar]
- 37.Mochizuki K., Gorovsky M. A. 2005. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev. 19:77–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mochizuki K., Gorovsky M. A. 2004. Small RNAs in genome rearrangement in Tetrahymena. Curr. Opin. Genet. Dev. 14:181–187 [DOI] [PubMed] [Google Scholar]
- 39.Nikiforov M., Gorovsky M., Allis C. 2000. A novel chromodomain protein, Pdd3p, associates with internal eliminated sequences during macronuclear development in Tetrahymena thermophila. Mol. Cell. Biol. 20:4128–4134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nikiforov M., Smothers J., Gorovsky M., Allis C. 1999. Excision of micronuclear-specific DNA requires parental expression of Pdd2p and occurs independently from DNA replication in Tetrahymena thermophila. Genes Dev. 13:2852–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nowacki M., Zagorski-Ostoja W., Meyer E. 2005. Nowa1p and Nowa2p: novel putative RNA binding proteins involved in trans-nuclear crosstalk in Paramecium tetraurelia. Curr. Biol. 15:1616–1628 [DOI] [PubMed] [Google Scholar]
- 42.Orias E., Hamilton E. P., Orias J. D. 2000. Tetrahymena as a laboratory organism: useful strains, cell culture, and cell line maintenance. Methods Cell Biol. 62:189–211 [DOI] [PubMed] [Google Scholar]
- 43.Prescott D. M. 1994. The DNA of ciliated protozoa. Microbiol. Rev. 58:233–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rexer C. H., Chalker D. L. 2007. Lia1p, a novel protein required during nuclear differentiation for genome-wide DNA rearrangements in Tetrahymena thermophila. Eukaryot. Cell 6:1320–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saveliev S. V., Cox M. M. 1996. Developmentally programmed DNA deletion in Tetrahymena thermophila by a transposition-like reaction pathway. EMBO J. 15:2858–2869 [PMC free article] [PubMed] [Google Scholar]
- 46.Saveliev S. V., Cox M. M. 2001. Product analysis illuminates the final steps of IES deletion in Tetrahymena thermophila. EMBO J. 20:3251–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shang Y., Song X., Bowen J., Corstanje R., Gao Y., Gaertig J., Gorovsky M. A. 2002. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc. Natl. Acad. Sci. U. S. A. 99:3734–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smothers J. F., Madireddi M. T., Warner F. D., Allis C. D. 1997. Programmed DNA degradation and nucleolar biogenesis occur in distinct organelles during macronuclear development in Tetrahymena. J. Eukaryot. Microbiol. 44:79–88 [DOI] [PubMed] [Google Scholar]
- 49.Sonneborn T. M. 1970. Gene action in development. Proc. R. Soc. Lond. B Biol. Sci. 176:347–366 [DOI] [PubMed] [Google Scholar]
- 50.Taverna S. D., Coyne R. S., Allis C. D. 2002. Methylation of histone h3 at lysine 9 targets programmed DNA elimination in tetrahymena. Cell 110:701–711 [DOI] [PubMed] [Google Scholar]
- 51.Taylor F. M., Martindale D. W. 1993. Retroviral-type zinc fingers and glycine-rich repeats in a protein encoded by cnjB, a Tetrahymena gene active during meiosis. Nucleic Acids Res. 21:4610–4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Timmons L., Fire A. 1998. Specific interference by ingested dsRNA. Nature 395:854. [DOI] [PubMed] [Google Scholar]
- 53.Tsukii Y. 1994. Evolution of mitochondrial DNA in Paramecium. Jpn. J. Genet. 69:685–696 [Google Scholar]
- 54.Voskühler C., Tiedtke A. 1993. Magnetic separation of phagosomes of defined age from Tetrahymena thermophila. J. Eukaryot. Microbiol. 40:556–562 [Google Scholar]
- 55.Yang X., Takahashi M. 1999. Disturbance of the determination of germinal and somatic nuclei by heat shock in Paramecium caudatum. J. Eukaryot. Microbiol. 46:49–55 [DOI] [PubMed] [Google Scholar]
- 56.Yao M. C., Chao J. L. 2005. RNA-guided DNA deletion in Tetrahymena: an RNAi-based mechanism for programmed genome rearrangements. Annu. Rev. Genet. 39:537–559 [DOI] [PubMed] [Google Scholar]
- 57.Yao M. C., Duharcourt S., Chalker D. L. 2002. Genome-wide rearrangements of DNA in ciliates, p. 730–758 InCraig N., Craigie R., Gellert M., Lambowitz A. (ed.), Mobile DNA II Academic Press, New York, NY [Google Scholar]
- 58.Yao M. C., Yao C. H., Halasz L. M., Fuller P., Rexer C. H., Wang S. H., Jain R., Coyne R. S., Chalker D. L. 2007. Identification of novel chromatin-associated proteins involved in programmed genome rearrangements in Tetrahymena. J. Cell Sci. 120:1978–1989 [DOI] [PubMed] [Google Scholar]