Abstract
The Stagonospora nodorum StuA transcription factor gene SnStuA was identified by homology searching in the genome of the wheat pathogen Stagonospora nodorum. Gene expression analysis revealed that SnStuA transcript abundance increased throughout infection and in vitro growth to peak during sporulation. To investigate its role, the gene was deleted by homologous recombination. The growth of the resulting mutants was retarded on glucose compared to the wild-type growth, and the mutants also failed to sporulate. Glutamate as a sole carbon source restored the growth rate defect observed on glucose, although sporulation remained impaired. The SnstuA strains were essentially nonpathogenic, with only minor growth observed around the point of inoculation. The role of SnstuA was investigated using metabolomics, which revealed that this gene's product played a key role in regulating central carbon metabolism, with glycolysis, the TCA cycle, and amino acid synthesis all affected in the mutants. SnStuA was also found to positively regulate the synthesis of the mycotoxin alternariol. Gene expression studies on the recently identified effectors in Stagonospora nodorum found that SnStuA was a positive regulator of SnTox3 but was not required for the expression of ToxA. This study has uncovered a multitude of novel regulatory targets of SnStuA and has highlighted the critical role of this gene product in the pathogenicity of Stagonospora nodorum.
Proteins with the conserved APSES (ASM-1, Phd1, StuA, EFG1, and Sok2) domain (1) belong to a class of transcription factors unique to the ascomycetes that regulate developmental differentiation (3, 5, 7, 15, 18, 20, 21, 33, 35). The APSES domain was modeled as a basic helix-loop-helix (bHLH)-like structure and binds to a specific stress response element (STRE) with the consensus sequence (A/T)CGCG(T/A)N(A/C) (7). The stunted protein StuA, which has a conserved APSES domain, has been well characterized in Aspergillus nidulans. A stuA knockout mutant of A. nidulans demonstrated that it regulates cellular differentiation during the late stages of the life cycle of the fungus. The stuAp deletion mutant exhibited various abnormal structural developments and reduced conidiospores compared to those of the wild type (15). The sexual reproduction cycle was also affected in stuA deletion mutants, as they failed to produce any ascospores and did not develop any of the cells and tissues distinctive of the sexual cycle of A. nidulans (15). Additionally, overexpression of StuAp was shown to promote extensive vegetative proliferation at the expense of conidiospore formation (35), and restoring the level of StuAp expression promoted increasing conidiospore complexity (3). Gel retardation analysis demonstrated that StuA binds to STREs found upstream of critical developmental regulators, such as AbaA and BrlA (7).
Similarly, in Aspergillus fumigatus, Fusarium oxysporum, and Glomerella cingulata, deletion of the StuA ortholog resulted in mutants with reduced and abnormal conidiospore formation (18, 21, 33). In studies of G. cingulata, the stuA deletion mutants failed to infect intact apple fruit due to failure to generate normal turgor pressure within the wild-type-appearing appressorium (33). However, in studies of A. fumigatus and F. oxysporum, the deletion mutant still caused disease (18, 21).
The dothideomycete Stagonospora nodorum is the causal agent of glume blotch disease of wheat and is responsible for significant yield losses throughout the world. A recently commissioned pathometry report in Australia determined that glume blotch accounted for in excess of AUD 100 million in yield losses per annum (16). The disease is typically initiated through the germination of sexual ascospores on wheat seedlings during the start of the growing season. The ensuing lesions produce asexual pycnidiospores, housed within pycnidia. The infection cycle continues via the splash dispersal of the pycnidiospores from one leaf to another, keeping pace with the growth of the host, until the glumes develop and become infected (23). Consequently, asexual sporulation of this polycyclic disease is critical for the pathogen to inflict yield loss.
Previous studies in S. nodorum have identified multiple genes and metabolic pathways implicated in asexual sporulation. Mutants of S. nodorum lacking mannitol 1-phosphate dehydrogenase were unable to sporulate in the absence of exogenous mannitol (25, 28, 29). A similar study also found that S. nodorum mutants impaired in the ability to anabolize trehalose also suffered sporulation defects (14). Various signaling genes have also been shown to be required for sporulation (24, 26, 30). The short-chain dehydrogenase Sch1 was recently demonstrated to have a role in the structural formation of the pycnidia (31). Strains of S. nodorum lacking sch1 surprisingly produced vast quantities of the mycotoxin alternariol, although this appeared to be not related to the mutant's inability to sporulate (32). Consequently, a solid platform of data is being developed to understand the intricacies of asexual sporulation in S. nodorum.
The previously demonstrated roles of StuA orthologs for sporulation in other fungi led to the question of its requirement for asexual development in S. nodorum. Consequently, the role of the S. nodorum StuA transcription factor gene SnStuA in the S. nodorum pathosystem was investigated using a reverse genetics approach. Functional analyses of S. nodorum strains lacking SnStuA revealed the gene to have a required role in asexual sporulation, as well as a multitude of other processes.
MATERIALS AND METHODS
Strains and culture conditions.
S. nodorum SN15 was obtained and cultured on V8PDA medium or minimal medium as described previously (31).
Infection assays.
Detached-leaf assays were performed as described in reference 26.
Construct development and transformation.
The SnstuA deletion construct was made using overlap PCR as previously described (24). SnStuAKO5′F (5′-GCCTTTCTTCAGCAGCTTC-3′) and SnStuAKO5′R (5′-TGTGACTTTTGGTTACGCCGTCTATAGTGCGAGATTCCCGAG-3′) were used to amplify a 720-bp region upstream of SnstuA, while SnStuAKO3′F (5′-CTCCTATGAGTCGTTTACCCAGAAATGATGCATATCCCGGC-3′) and SnStuAKO3′R (5′-GGAAAGCACGAGCTGAAA-3′) amplified a 743-bp region downstream. These flanks were fused with a phleomycin resistance cassette by using overlap PCR, resulting in a deletion construct of 3.8 kb. The deletion cassette was transformed into S. nodorum protoplasts as previously described (27) and screened for the use of primers designed outside the flanking DNA (SnStuAKOscr-F [5′-CGTGAACCGCGTAGTCTATT-3′] and SnStuAKOscr-R [5′-ACTTCACTGACACAGCATGG-3′]). The copy number of the transformed construct was determined as previously described (22).
Microscopy.
Tissues for histological examination were fixed overnight in formal acetic alcohol and dehydrated in an ascending series of ethanol concentrations (50% to 100%), followed by a clearing in chloroform. The tissue was then infiltrated with molten paraffin wax (Paraplast). The paraffin blocks were then sectioned at 5 to 7 μm, and the sections were allowed to dry onto the slides at 60°C overnight. The wax was removed from the sections by treating the slides with two changes of xylene. The sections were treated with 1% toluidine blue stain where appropriate, and a coverslip was applied to the sections. The sections were examined using an Olympus BX51 compound microscope.
Infected leaf materials were stained with trypan blue as described by Solomon et al. (24). Briefly, leaf lesions were boiled with trypan blue stain for 5 min, followed by destaining and washing in 100% ethanol. The stained tissues were examined using an Olympus BX51 compound microscope.
Metabolomics.
The fungus was grown on minimal medium with 30 mM glucose as the carbon source. Agar cultures were grown for 1 to 2 weeks with a 12-h white-light regimen at 22°C, and 50-ml liquid cultures were grown in the dark at 22°C with shaking at 150 rpm. Mycelia were harvested as quickly as possible and rinsed in 50% methanol to wash away possible medium carryover. The fungal material was frozen in liquid nitrogen and freeze-dried overnight in a Maxi-Dry Lyo freeze drier (Heto, Allerod, Denmark) (13). Six to eight biological replicates were used for each strain per sampling time point.
Metabolites were extracted and derivatized as previously described (32). The freeze-dried sample (1 to 5 mg) was mixed with 700 μl −40°C methanol in a tube and snap-frozen using liquid nitrogen. The tube was thawed on ice and centrifuged at 20,000 × g for 30 s. The supernatant was transferred to a fresh tube, and the extraction procedure was repeated again with a fresh aliquot of 700-μl −40°C methanol, snap-frozen, thawed on ice, and centrifuged at 20,000 × g. Ribitol (50 μl) at a concentration of 0.2 mg/ml was added to the pooled supernatant as an internal standard prior to lyophilization using a speed vac concentrator (13). The dried metabolites were mixed with 25 μl methoxyamine hydrochloride (20 mg/ml in pyridine) and incubated for 90 min at 30°C with shaking at 1,200 rpm. N-Trimethylsilyl-N-methyl trifluoroacetamide (MSTFA) (40 μl) was added to the tube, and the contents incubated at 37°C for 30 min.
Derivatized metabolites (1 μl) were injected in a 20:1 split ratio for gas chromatography-mass spectrometry (GC-MS) analysis. The GC-MS equipment consisted of an Agilent 7680 autosampler, an Agilent 6890 gas chromatograph, and an Agilent 5973N quadrupole mass spectrometer (Agilent, Palo Alto, CA). The GC-MS system was autotuned using perfluorotributylamine (PFTBA). For polarity separation by gas chromatography, a 30-m Varian VF-5ms column with a 10-m integrated Varian EZ-Guard column was used (Varian, Palo Alto, CA). The injection temperature was 230°C, the interface temperature was 300°C, and the ion source temperature was 230°C. Helium was used as the carrier gas, and the flow rate was retention time locked to elute mannitol-trimethylsilyl (TMS) at 30.6 min. The temperature gradient consisted of an initial temperature of 70°C, increasing at 1°C per minute for 5 min before increasing to a final temperature of 300°C at a temperature ramp rate of 5.6°C per minute. AnalyzerPro (SpectralWorks Ltd., Runcorn, United Kingdom) was used to analyze the mass spectra and chromatograms. Metabolite peak areas representing the abundance of the metabolites were normalized to the ribitol internal standard and the weight of the sample, and the data were cross-referenced against the target component library using the MatrixAnalyser function (13).
Peaks that could not be matched to the target component library were named unknown metabolites and were labeled according to the following format: “Unknown_retention time_retention index_base peak.” The processed metabolomics data were subjected to a principal component analysis (The Unscrambler; CAMO) subsequent to the data transformation [x = log(x + 1)].
RNA extraction and gene expression analysis.
For determining the expression of SnstuA in vitro, mycelia were scraped from plates at 4 days and 16 days postinoculation (dpi). In planta samples were collected at 3 dpi, 5 dpi, 7 dpi, and 10 dpi in biological triplicate. For measuring the expression of the SnTox3 and ToxA host-selective toxins, the fungal strains were grown on Fries medium as previously described (12). Fungal tissue for RNA extraction was collected after 3 days of growth in triplicate. RNA from all samples was extracted using Trizol reagent (Invitrogen Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. Contaminating genomic DNA in the RNA samples was removed using DNA-Free reagent (Ambion, Austin, TX). The reverse transcription of mRNA to cDNA was performed using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) according to the manufacturer's instructions.
Quantitative PCR (qPCR) was performed using 0.1-ml strip tubes (Corbett Research, San Francisco, CA) and 20-μl reaction mixtures, each of which consisted of 10 μl IQ SYBR supermix (Bio-Rad, Hercules, CA), 2.5 μl (2.5 μM) each of the forward and reverse gene-specific primers (Geneworks, Adelaide, SA, Australia), and 5 μl of the cDNA sample. The primers used were as follows: Tox3qPCRf (AATGTCGACCGTTTTGACC), Tox3qPCRr (GGTTGCCGCAGTTGATATAA), ToxAqPCRf (CGATCCCGGTTACGAAAT), ToxAqPCRr (TTGACATGCAGCTTCCCT), ActinqPCRf (AGTCGAAGCGTGGTATCCT), and ActinqPCRr (ACTTGGGGTTGATGGGAG). Thermal cycling was performed in a RotorGene RG-3000 (Corbett Research) using the following program: 95°C, 15 min, and 40 cycles of 95°C, 15 s; 57°C, 15 s; and 72°C, 20 s. Readings from the fluorescent dye were acquired using a 470-nm excitation filter and detected at 510 nm during the annealing stage. The amplification plot and cycle threshold (CT) values were examined using RotorGene software version 6 (24).
RESULTS
SNOG_14941, a StuA ortholog.
The gene SNOG_14941, encoding a protein with a conserved APSES domain, was found within the genome of S. nodorum and named SnStuA. Multiple sequence alignment of SnStuA against StuAp from A. nidulans and FoStuA from F. oxysporum showed a highly conserved APSES domain (see Fig. S1 in the supplemental material).
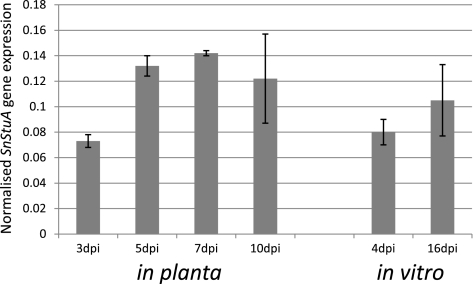
The transcript level of SnStuA was analyzed using qPCR to ensure that SnStuA was not a pseudogene. The expression data from the in vitro and in planta samples showed that the expression of SnStuA was upregulated toward the late stages of the life cycle of the pathogen, correlating with the development of pycnidia (Fig. 1).
Fig. 1.
Expression of SnStuA as determined by qPCR. Values shown represent the expression of SnStuA normalized against the expression of γ-actin. Biological triplicates were assayed using technical replicates. Standard error bars are shown.
Deletion of SnStuA.
To investigate the properties of SnStuA in S. nodorum, the gene was deleted using homologous recombination. Flanking regions of approximately 900 bp were amplified on both sides of the SnStuA gene and fused to a phleomycin cassette to create a deletion construct. The construct, conferring resistance to phleomycin, was used to replace the SnStuA gene via homologous recombination (see Fig. S2A in the supplemental material).
The transformants were screened using primers spanning the flanking region of the deletion construct (see Fig. S2B in the supplemental material). The PCR product from an SnStuA deletion mutant produced a 4.7-kb product, while an ectopic strain and the wild-type strain produced 4-kb products. A transformation efficiency of 4.3% was observed with three knockout mutants identified out of the 69 transformants screened. qPCR was used to determine the copy number of the knockout construct in selected transformants as previously described (22). Two of the three deletion mutants were shown to have single insertions and were subsequently named SnstuA-48 and SnstuA-54. An ectopic strain, SnStuA-5, was also chosen as a control for subsequent testing. Despite multiple attempts, complementation of the SnStuA mutants was unsuccessful. Protoplasts are typically generated from 1-day old mycelia grown in flask inoculated with spores. As described below, the mutants did not sporulate, and thus, flasks were inoculated with homogenized mycelial fragments. Previous such attempts in this laboratory to generate protoplasts from flasks inoculated with mycelia have proven unsuccessful, and it appears that such an approach is not technically possible (31).
Phenotypic characterization of the deletion mutants.
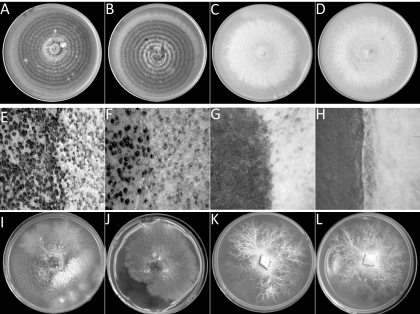
The growth of the mutant strains was examined on the complete medium V8PDA. The mutants (SnstuA-48 and SnstuA-54) appeared to grow more slowly than the wild type and the ectopic mutant on complex medium. After 14 days of growth, pycnidia were readily apparent on the wild-type and ectopic strains, while the SnstuA strains produced thick white aerial hyphae but no evidence of sporulation (Fig. 2 A to D). A closer examination under the dissecting microscope showed that the wild-type and ectopic strains produced thin aerial mycelia and dark brown melanized pycnidia with pink cirrus oozing out of the bursting pycnidia. The deletion mutants did not differentiate any observable reproductive structures and produced a thick layer of aerial mycelia (Fig. 2E to H).
Fig. 2.
Photographs showing the morphology of the S. nodorum strains SN15 (A, E, and I), SnStuA-5 (B, F, and J), SnstuA-48 (C, G, and K), and SnstuA-54 (D, H, and L). The top four panels represent growth on V8PDA, the middle four panels show sporulation on V8PDA, and the bottom four panels show growth of the strains on minimal medium.
The growth of the strains was also examined on defined minimal medium (Fig. 2I to L). The wild-type and ectopic strains grew more slowly and produced less biomass than was observed on the complex medium. Sectoring was also evident, as is usually observed when growing S. nodorum on minimal medium. The deletion mutants produced white mycelia and exhibited a vastly different morphology than the wild-type strain or their own growth on the complex medium. Instead of the usual circular ring growth, major hyphae grew from the inoculation spot and smaller lateral hyphae developed from these major hyphae, forming branchlike structures.
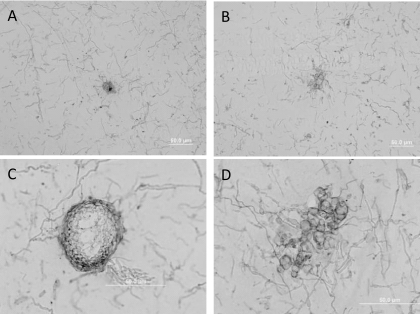
The sporulation defect of the mutants was further assessed using microscopy. Figure 3 shows light-microscope images of asexual development of the wild-type and the SnStuA-48 mutant in vitro. Preliminary observations at 40× magnification suggested that both strains produce mycelial knots, the precursor developmental stage to mature pycnidia. Clear structural differences were evident, though, at increased magnification. The wild-type pycnidia appeared as previously described, with a uniform pycnidial wall and subparietal layer (31). The structure observed in the SnstuA-48 mutant lacked the uniformity of that in the wild type and appeared to be more a conglomeration of swollen hyphae than a mature pycnidium. No pycnidial wall or subparietal layer appears to have differentiated, and there was no evidence of pycnidiospores.
Fig. 3.
Light-microscope images of agar cross sections showing fungal culture morphology when grown on minimal medium. (A and C) Mature pycnidium from S. nodorum SN15 at 40× (A) and 100× (C) magnification. (B and D) Abnormal structures produced during pycnidium development from S. nodorum SnstuA-48 mutant at 40× (B) and 100× (D) magnification.
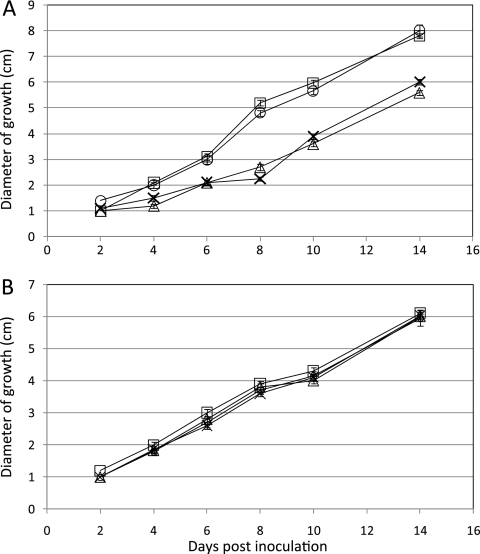
The ability of the SnStuA mutants to utilize a range of carbon sources was also assessed. A routine assay typically undertaken on mutants is to examine their ability to utilize different carbon sources. The phenotypic characterization of the strains described above showed that the deletion mutants grew more slowly than the wild-type and ectopic strains on sucrose and glucose as carbon sources. This trend appeared to be consistent with the results for nearly all other carbon sources tested, including fructose, mannitol, and glycerol (data not shown). Growth on glutamate, though, appeared to be different, with comparable growth rates measured for all strains, suggesting that SnStuA has a regulatory role in carbon metabolism (Fig. 4).
Fig. 4.
Line graphs representing the growth of the S. nodorum strains SN15 (○), SnStuA-5 (□), SnsntuA-48 (▵), and SnstuA-54 (×) on 30 mM glucose (A) or glutamate (B). Standard error bars are shown (n = 4).
SnStuA is required for pathogenicity.
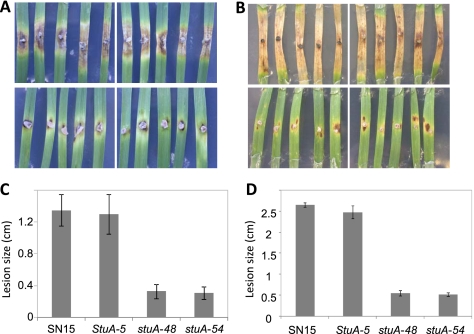
Pathogenicity assays with S. nodorum are typically undertaken using a whole-plant spray or detached-leaf assay. Due to the lack of spores produced by the SnStuA mutants, agar blocks of growing fungi were cut from plates and used to inoculate detached leaves to determine the requirement of SnStuA for pathogenicity (Fig. 5). At 7 dpi, the lesions caused by the wild-type and ectopic strains were spreading rapidly and ranged between 0.7 and 1 cm in length. The lesions induced by the mutant strains were much smaller and averaged only 0.16 cm.
Fig. 5.
Pathogenicity assays at 7 (A and C) and 14 (B and D) dpi. The images of diseased leaves represent infections of the S. nodorum strains SN15 and SnStuA-5 in the top two panels and SnstuA-48 and SnstuA-54 in the bottom panels. The histograms represent the sizes of the lesions corresponding with the images above. Standard error bars are shown (n = 6).
At 14 dpi, the wild-type and ectopic strains caused extended light-brown necrosis across the whole leaf, and pycnidia were observed. Average lesion sizes of around 2 cm in length were typically recorded for these inoculations. The lesions on the leaves inoculated with the SnStuA mutants had not progressed much further than at 7 dpi, with the average length being only around 0.22 cm. Furthermore, the deletion mutant infections showed no sign of sporulation. The possibility that the reduced pathogenicity of the SnStuA mutants was due to their inability to penetrate the leaf surface was examined by inoculating wounded leaves (see Fig. S3 in the supplemental material). The state of the infections on the wounded leaves correlated with that observed on intact leaves, suggesting that SnStuA is not required for the penetration stage of infection.
StuA has a central regulatory role in carbon metabolism.
The characterization of the SnStuA mutants described above highlighted a number of phenotypic impairments that may contribute to a reduced level of pathogenic fitness. To attempt to identify the role of SnStuA in each of these phenotypes, a comparative metabolomics approach was undertaken. The use of metabolite profiling to dissect phenotypes is rapidly gaining momentum and has been successfully applied to S. nodorum (13, 14, 32). Each of the four strains described above was grown for 14 days on minimal medium plates with glucose as the carbon source, and the metabolites were extracted as described in Materials and Methods. Samples from eight biological replicates for each strain were processed and analyzed using GC-MS for metabolite detection. The resulting data were processed using AnalyzerPro (SpectralWorks, United Kingdom) prior to being subjected to multivariate analysis.
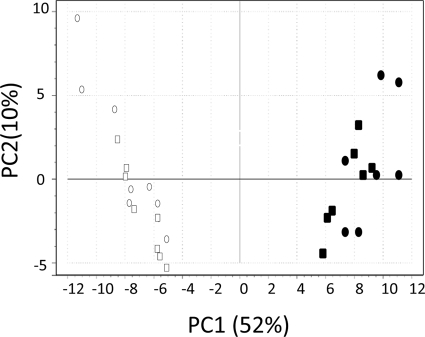
Principal component analysis (PCA) was applied to gain insight into the nature of the multivariate metabolite data set (4, 11). PCA identifies and ranks major sources of variance within datasets and allows clustering of biological samples into both expected and unexpected groups based on similarities and differences in the parameters measured (4). The score plot generated from the PCA is shown in Fig. 6. Principal component 1 (PC1) accounts for 52% of the variation, with the strains lacking StuA clearly clustering independently from the wild-type and ectopic strains. PC2 represents 10% of the variation and appears not significant for this data set.
Fig. 6.
Score plot generated from the PCA. S. nodorum SN15 (○), SnStuA-5 (□), SnstuA-48 (•), and SnstuA-54 (▪).
An analysis of the factor loadings from the PCA score plot and subsequent analysis of variance identified 62 peaks from the GC-MS traces that were significantly differentially abundant when comparing normalized peak areas of the wild-type and ectopic strains to those of the SnStuA mutants (Table 1). These 62 peaks represent 56 compounds of various classes. Analysis of the data revealed that deletion of SnStuA has a significant effect on primary metabolism during sporulation. All amino acids detected were significantly less abundant in the SnStuA mutants, while all sugars and sugar alcohols were present at higher levels. Similarly, organic acids linked with the tricarboxylic acid (TCA) cycle (pyruvic acid and lactic acid) were also more abundant in the SnStuA deletion strains. γ-Aminobutyric acid and γ-hydroxybutyric acid, though, were present at lower levels in the mutants. Nearly all compounds not assigned to specific classes were more abundant in the wild-type strain, with the exception of phosphoric acid. These include allantoin, myoinositol, and the recently identified mycotoxin in S. nodorum, alternariol. The final class of compounds listed on Table 1 are those that remain unidentified after the GC-MS analysis. The most striking of these are unknowns 39.33_2484_319 and 35.27_2194_319. These compounds are highly abundant in the mutants and are present at levels comparable to that of glucose, while being barely detectable in the wild-type strain. There were also several other unknown compounds that shared a 319 m/z base peak that were significantly more abundant in the mutants. While the identity of these compounds at present remains elusive, some information can be attained from their fragmentation pattern. This is discussed further below.
Table 1.
Normalized metabolite abundances
| Metabolite and IDa | Normalized abundanceb (mean ± SD) in strain: |
|||
|---|---|---|---|---|
| SN15 | SnStuA-5 | SnstuA-48 | SnstuA-54 | |
| Amino acids | ||||
| l-Glutamine 3TMS_27.96_1779 | 4.17 ± 0.89 | 3.29 ± 0.67 | 1.19 ± 0.87 | 1.02 ± 0.82 |
| l-Glutamic acid 3TMS_24.85_1623_246 | 2.40 ± 0.63 | 2.03 ± 0.40 | 0.93 ± 0.36 | 0.79 ± 0.38 |
| l-Tyrosine 3TMS_30.91_1933_218 | 0.60 ± 0.13 | 0.53 ± 0.12 | 0.18 ± 0.05 | 0.12 ± 0.06 |
| l-Serine 3TMS_18.99_1363_204 | 0.46 ± 0.13 | 0.38 ± 0.16 | 0.19 ± 0.06 | 0.16 ± 0.08 |
| l-Threonine 3TMS_19.60_1387_218 | 0.43 ± 0.12 | 0.40 ± 0.12 | 0.08 ± 0.03 | 0.07 ± 0.02 |
| l-Lysine 4TMS_30.58_1913_174 | 0.37 ± 0.17 | 0.21 ± 0.08 | 0.06 ± 0.02 | 0.04 ± 0.02 |
| l-Histidine 3TMS_30.58_1913_154 | 0.35 ± 0.07 | 0.22 ± 0.07 | ND | ND |
| l-Glycine 3TMS_17.61_1308_174 | 0.32 ± 0.07 | 0.27 ± 0.06 | 0.18 ± 0.05 | 0.16 ± 0.02 |
| l-Isoleucine 2TMS_17.29_1295_158 | 0.31 ± 0.06 | 0.26 ± 0.10 | 0.09 ± 0.02 | 0.07 ± 0.01 |
| l-Proline 2TMS_17.41_1300_142 | 0.23 ± 0.09 | 0.18 ± 0.07 | 0.03 ± 0.02 | ND |
| l-Asparagine 3TMS_25.87_1674_116 | 0.21 ± 0.08 | 0.22 ± 0.11 | 0.02 ± 0.02 | 0.02 ± 0.01 |
| l-Threonine 2TMS_17.44_1301_117 | 0.20 ± 0.04 | 0.19 ± 0.04 | 0.04 ± 0.02 | 0.04 ± 0.02 |
| l-Aspartic acid 3TMS_22.72_1516_232 | 0.18 ± 0.08 | 0.12 ± 0.02 | 0.05 ± 0.02 | 0.05 ± 0.02 |
| l-Serine 2TMS_16.38_1258_132 | 0.16 ± 0.03 | 0.15 ± 0.03 | 0.07 ± 0.02 | 0.07 ± 0.03 |
| Sugars/sugar alcohols | ||||
| Glucose 5TMS_30.21_1884_319 | 0.19 ± 0.07 | 0.17 ± 0.10 | 9.36 ± 1.38 | 6.53 ± 1.85 |
| Arabitol 5TMS_26.75_1719_217 | 0.51 ± 0.21 | 0.77 ± 0.28 | 5.90 ± 0.86 | 4.26 ± 1.10 |
| Glucose 5TMS_30.39_1902_319 | 0.02 ± 0.02 | 0.02 ± 0.02 | 2.02 ± 0.27 | 1.20 ± 0.34 |
| Thrietol 4TMS_22.21_1496_217 | ND | 0.01 ± 0.01 | 1.29 ± 0.20 | 1.02 ± 0.21 |
| Erythritol 4TMS_21.21_1492_217 | ND | ND | 1.26 ± 0.21 | 0.95 ± 0.23 |
| Glucopyranose 5TMS_31.49_1967_204 | 0.10 ± 0.09 | 0.08 ± 0.05 | 1.08 ± 0.39 | 1.10 ± 0.32 |
| Fructose 5TMS_29.59_1861_103 | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.28 ± 0.06 | 0.19 ± 0.05 |
| Fructose 5TMS_29.77_1870_103 | ND | ND | 0.15 ± 0.03 | 0.11 ± 0.03 |
| d-(−)-Ribose 4TMS_25.97_1679_103 | ND | ND | 0.05 ± 0.01 | 0.03 ± 0.01 |
| Organic acids | ||||
| Fumaric acid 2TMS_18.89_1359_245 | 0.09 ± 0.03 | 0.09 ± 0.03 | 0.25 ± 0.06 | 0.24 ± 0.07 |
| Pyruvic acid 1TMS_10.23_1049_174 | ND | ND | 0.25 ± 0.02 | 0.14 ± 0.04 |
| Malic acid 3TMS_22.01_1484_147 | 0.08 ± 0.02 | 0.08 ± 0.02 | 0.21 ± 0.05 | 0.22 ± 0.06 |
| Lactic acid 2TMS_10.93_1060_147 | 0.06 ± 0.01 | 0.05 ± 0.01 | 0.18 ± 0.04 | 0.13 ± 0.03 |
| GABA 3TMS_22.93_1526_174 | 0.16 ± 0.06 | 0.08 ± 0.03 | 0.01 ± 0.01 | ND |
| γ-Hydroxybutyric acid 2TMS_15.97_1242_1 | 0.11 ± 0.08 | 0.15 ± 0.06 | ND | ND |
| 2-Ketoglutaric acid 2TMS_24.04_1582 | ND | ND | 0.08 ± 0.02 | 0.06 ± 0.02 |
| Other classes of compounds | ||||
| Phosphoric acid 3TMS_16.72_1272_299 | 4.07 ± 0.93 | 3.75 ± 0.75 | 6.54 ± 1.50 | 5.98 ± 1.05 |
| Alternariol 3TMS_45.58_2959_459 | 2.06 ± 0.82 | 1.42 ± 0.67 | ND | ND |
| Allantoin 4TMS_29.92_1878_331 | 1.04 ± 0.39 | 0.91 ± 0.35 | 0.44 ± 0.17 | 0.37 ± 0.19 |
| Allantoin 5TMS_30.25_1894_518 | 0.44 ± 0.18 | 0.37 ± 0.16 | 0.09 ± 0.06 | 0.06 ± 0.04 |
| Tryptamine_24.44_1602_188 | 0.34 ± 0.10 | 0.32 ± 0.11 | 0.08 ± 0.04 | 0.08 ± 0.04 |
| N-Acetylglutamic acid 2TMS_23.05_1532_8 | 0.25 ± 0.07 | 0.20 ± 0.06 | 0.09 ± 0.02 | 0.08 ± 0.02 |
| Myoinositol 6TMS_32.32_2017_318 | 0.18 ± 0.04 | 0.16 ± 0.05 | 0.02 ± 0.01 | 0.03 ± 0.01 |
| Myoinositol 6TMS_33.36_2080_217 | 0.18 ± 0.04 | 0.16 ± 0.05 | 0.01 ± 0.01 | 0.02 ± 0.01 |
| β-Alanine 3TMS_20.61_1428_174 | 0.09 ± 0.02 | 0.07 ± 0.02 | 0.02 ± 0.01 | 0.02 ± 0.01 |
| Ornithine 4TMS_28.66_1815_142 | 0.03 ± 0.01 | 0.04 ± 0.02 | ND | ND |
| Inosine 4TMS_40.65_2579_217 | ND | ND | 0.02 ± 0.01 | 0.02 ± 0.01 |
| Unknowns | ||||
| Unknown_39.33_2484_319 | 0.03 ± 0.01 | 0.04 ± 0.01 | 8.00 ± 1.39 | 7.45 ± 1.26 |
| Unknown_35.27_2194_319 | ND | ND | 7.68 ± 3.61 | 8.27 ± 2.10 |
| Unknown_27.63_1763_357 | 2.23 ± 0.59 | 2.05 ± 0.60 | 0.36 ± 0.09 | 0.33 ± 0.06 |
| Unknown_34.09_2123_319 | ND | ND | 1.00 ± 0.27 | 0.86 ± 0.18 |
| Unknown_34.00_2117_319 | ND | ND | 0.72 ± 0.62 | 0.62 ± 0.12 |
| Unknown_30.02_1883_204 | 0.07 ± 0.04 | 0.04 ± 0.02 | 0.56 ± 0.24 | 0.54 ± 0.27 |
| Unknown_37.61_2361_217 | ND | ND | 0.54 ± 0.18 | 0.37 ± 0.06 |
| Unknown_40.14_2542_287 | ND | ND | 0.43 ± 0.11 | 0.39 ± 0.13 |
| Unknown_32.39_2021_204 | ND | ND | 0.39 ± 0.06 | 0.23 ± 0.07 |
| Unknown_52.11_3560_307 | 0.34 ± 0.17 | 0.39 ± 0.13 | ND | ND |
| Unknown_37.74_2370_217 | ND | ND | 0.35 ± 0.11 | 0.25 ± 0.04 |
| Unknown_24.44_1603_211 | 0.34 ± 0.10 | 0.32 ± 0.11 | 0.08 ± 0.04 | 0.08 ± 0.04 |
| Unknown_28.85_1824_231 | 0.29 ± 0.07 | 0.22 ± 0.06 | 0.12 ± 0.04 | 0.12 ± 0.04 |
| Unknown_34.66_2157_159 | 0.26 ± 0.16 | 0.24 ± 0.13 | ND | ND |
| Unknown_44.27_2846_402 | 0.15 ± 0.08 | 0.25 ± 0.17 | ND | ND |
| Unknown_51.36_3487_469 | 0.13 ± 0.05 | 0.19 ± 0.09 | ND | ND |
| Unknown_33.85_2109_357 | ND | ND | 0.18 ± 0.05 | 0.14 ± 0.07 |
| Unknown_35.85_2234_299 | 0.07 ± 0.02 | 0.05 ± 0.02 | 0.14 ± 0.03 | 0.12 ± 0.02 |
| Unknown_29.78_1871_319 | ND | ND | 0.14 ± 0.03 | 0.11 ± 0.02 |
| Unknown_48.39_3199_363 | 0.12 ± 0.03 | 0.08 ± 0.03 | 0.03 ± 0.01 | 0.02 ± 0.01 |
| Unknown_28.99_1831_428 | 0.05 ± 0.02 | 0.05 ± 0.02 | ND | ND |
All metabolites listed are differentially abundant in the wild-type strain and the SnstuA mutants (P < 0.05). Metabolites in boldface are significantly more abundant in the mutant strains than in the wild type, while those not in boldface are less abundant in the mutants than in the wild type. GC-MS data are included in the identification (ID) code in the format retention time_retention index_base peak.
Values are in arbitrary units. ND, not determined.
There were also many other compounds that were not significantly different in their abundance in the wild-type strain and the SnStuA mutants. Notable compounds in this category include mannitol and trehalose, both previously having been shown to be required for asexual sporulation in S. nodorum (14, 28).
SnStuA regulates SnTox3 expression.
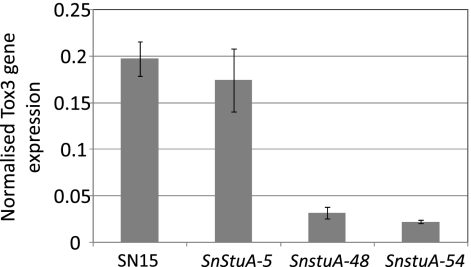
Each of the phenotypic assays described above provides further evidence toward understanding the role of SnStuA. However, the reason(s) for the significantly reduced pathogenicity remain unclear. Recent studies in S. nodorum have highlighted the critical role of necrotrophic effectors in pathogenicity (8, 9, 12). The expression of the genes encoding each of the known effectors was examined in the SnStuA deletion strains and compared to the level in the wild type to determine if SnStuA has a regulatory role over these important pathogenicity proteins. After three days of growth on the effector induction growth medium Fries, the expression of SnTox3 was approximately 6-fold less in the SnStuA mutants than in the wild type (Fig. 7). ToxA transcript levels were considerably lower than those of SnTox3 but were not significantly different in any of the strains (data not shown).
Fig. 7.
Histogram representing the expression of Tox3 normalized against the γ-actin level. Standard error bars are shown (n = 3).
DISCUSSION
Transcription factors with the conserved APSES domain are unique to fungi and have been shown to regulate cellular differentiation (5). An ortholog of the stunted proteins (StuA) was identified in S. nodorum, and the gene expression pattern suggested that this transcription factor might be involved in sporulation. To investigate the role of this transcription factor in S. nodorum, a gene deletion approach was undertaken and the resulting strains characterized.
In vitro growth observations showed that the SnStuA deletion mutants remained in a perpetual vegetative state and did not sporulate when grown in vitro. Detached-leaf assays showed that the deletion mutants developed lesions at vastly lower rates than the wild-type and also failed to sporulate. However, the connection between the low growth rate in vitro and the reduced pathogenicity is unclear. Similar studies in other fungal pathogens have shown that deletion of the genes encoding StuA homologs resulted in defects in asexual or/and sexual reproduction (3, 5, 7, 15, 18, 20, 21, 33, 35).
The StuA protein has been extensively studied in A. nidulans (3, 7, 15, 20, 21, 35). A reduction in conidiation was observed in the StuAp deletion mutant, with the StuA protein being shown to work together with the bristle (brlA) and abacus (abaA) genes for asexual development (15). There is evidence to suggest that the mechanisms of asexual sporulation in A. nidulans do differ from those in S. nodorum. For example, extensive homology searching of the S. nodorum genome has failed to find any orthologs of BrlA or AbaA. Sequence homology between transcription factors, even among closely related species, is often very low. However, another gene that is involved in asexual reproduction in A. nidulans, fluG, has been identified in S. nodorum and was found not to be required for asexual reproduction (P. S. Solomon, unpublished data). Further evidence of different asexual sporulation mechanisms in S. nodorum and A. nidulans is the requirement of mannitol for sporulation of S. nodorum. Mutants of A. nidulans lacking mannitol anabolic activity maintained the ability to sporulate, although the resulting conidia suffered increased susceptibility to stresses (19).
In F. oxysporum, StuA is encoded by FoSTUA, and deletion of the gene led to reduced conidiation both in planta and in vitro (18). In G. cingulata, deletion of GcSTUA significantly reduced the formation of conidia and abolished the production of perithecia (33). In A. fumigatus, deletion of StuA resulted in mutants that showed reduction in conidiation and abnormal asexual structures. Additionally, the abnormal conidia exhibited higher germination rates than the wild-type (21). Thus, the deletion of StuA in most pathogens has not abolished sporulation but led to a reduction whereby viable spores were still produced. Conversely, the deletion of SnStuA in S. nodorum produced a nonsporulating mutant. Microscopic examination of the SnStuA mutants during attempted sporulation did reveal that the strains were capable of producing mycelial knots, but these were unable to develop further into mature pycnidia (6). A previous study in S. nodorum identified a very similar sporulation phenotype for strains lacking a Ca2+/calmodulin-dependent protein kinase (CpkA), although a link (if any) between SnStuA and CpkA has yet to be established (24).
Pathogenicity assays revealed that the SnStuA mutants were significantly less pathogenic than the wild-type strain. The possibility that this was due to impaired penetration was investigated by infecting wounded leaves; this was found to have no effect on pathogenicity, implying that SnStuA is not required for the fungus to breach the leaf surface. In the fungal pathogens Magnaporthe grisea and Glomerella cingulata, infection has also been reduced with the deletion of StuA (17, 33). Characterization of the mutants revealed that the reduced pathogenicity was due to insufficient development of turgor pressure in the appressorium, although mobilization of glycogen and triacylglycerol appeared normal. It should be noted, though, that unlike the pathogens listed above, S. nodorum does not rely on turgor pressure to breach the leaf surface.
A key finding from this study was the regulation of tox3 gene expression by SnStuA. Tox3 is a recently described effector protein that interacts with Snn3 in wheat to cause disease (12). The expression of a second effector-encoding gene, ToxA, was unaffected in the SnStuA mutants, implying selected regulation of the effector genes by SnStuA. However, it is unlikely that the reduced expression of Tox3 was the sole cause of the pathogenicity defect, as this strain of S. nodorum also produces ToxA and the wheat line infected harbors the ToxA susceptibility gene, Tsn1; this combination alone is sufficient to cause disease (8).
Metabolomics is a powerful technique now gaining acceptance as a tool to dissect phenotypes. Previous studies in S. nodorum have used metabolomics to great effect for dissecting osmotic stress and trehalose synthesis, as well as mycotoxin synthesis (13, 14, 32). Metabolomics was chosen to characterize the SnStuA mutation in this study for deliberate reasons. First, it is now well established that S. nodorum is highly reliant on particular primary metabolic pathways for asexual sporulation. Metabolomics offers the chance to precisely dissect these known pathways at the metabolite level during sporulation. Second, glutamate complemented the growth rate defect in the SnStuA mutants, implying it has a role in central primary metabolism.
The primary finding from the metabolomics studies was the increased abundance of sugars present in the mutants, while all detectable amino acids were less abundant. Intermediates of the TCA cycle, along with pyruvate and lactate, were also increased in the mutant compared to their levels in the wild type. These data suggest that the metabolism of fermentable carbon sources is negatively affected in the SnstuA background. This statement is corroborated by the fact that the growth rate of the mutants was restored to wild-type levels when glutamate was supplied as a sole carbon source. Glutamate enters carbon metabolism through α-ketoglutarate in the TCA cycle and, thus, appears to bypass the problems associated with typical sugar metabolism through glycolysis and into the TCA cycle in the SnStuA mutants. One possible explanation for the inhibition of glycolysis and the accumulation of sugars could be the inhibition of both pyruvate kinase and phosphofructokinase, the key regulatory control points of glycolysis. Pyruvate kinase and phosphofructokinase are inhibited by high concentrations of ATP, leading to feedback inhibition of the glycolytic pathway (2). The metabolomics data strongly suggest that ATP levels in the SnStuA mutants are high compared to the level in the wild-type strain, as evidenced by the detection of significantly higher levels of phosphoric acid, a known marker for ATP levels. Growth on glutamate would of course bypass the glycolysis inhibition and allow wild-type growth, which has been demonstrated. It is interesting to note that lower growth rates have also been observed from StuA deletion mutants of M. grisea and F. oxysporum (17, 18), suggesting that StuA may regulate glycolysis and/or the TCA cycle in these pathogens as well.
A further significant result to have emerged from the metabolomics was that mannitol and trehalose levels were unchanged in the SnStuA mutants. Both compounds are known to be required for sporulation, but they appear to be independent of the sporulation defect in these mutants. Another interesting result was the decrease in the abundance of alternariol in the mutants. Alternariol was recently identified in S. nodorum during a metabolomics screening of a short-chain dehydrogenase mutant (sch1) (32). Mutants of S. nodorum lacking Sch1 were also unable to sporulate but produced significantly increased amounts of the mycotoxin, although no conclusive links between the presence or absence of alternariol and sporulation have been demonstrated (31). Studies in Aspergillus fumigatus have demonstrated StuA-dependent regulation of genes involved in secondary metabolism and mycotoxin synthesis, namely, gliP and clusters (10, 34). It is also intriguing to note that yet another nonsporulating mutant has altered alternariol synthesis; however, its role in asexual development (if any) remains unclear.
The other notable result from the metabolite profiling was the substantial increase in abundance of the two unknown peaks 39.33_2484_319 and 35.27_2194_319. These peaks are massively abundant and appear to be among the most concentrated soluble polar metabolites in the mutants. While the identity of these peaks remains unknown, a base peak of 319 m/z is a strong indication that these are a sugar or sugar alcohol. Such an identity would fit elegantly with the metabolite patterns observed for the other sugars detected. Various standards of different sugars and polyols have been scrutinized, but the identity of these two peaks remains elusive. It also cannot be excluded that these two peaks represent one compound, as can often happen as a result of the derivatization process during sample preparation.
In conclusion, this paper has presented several novel findings regarding the role and regulation of the SnStuA transcription factor in the wheat pathogen Stagonospora nodorum. Studies are now ongoing using additional functional genomics techniques to further elucidate the regulatory role of SnStuA.
Supplementary Material
ACKNOWLEDGMENT
This research was supported by the Grains Research and Development Corporation, Australia (grant number UMU00022).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 21 May 2010.
REFERENCES
- 1.Aramayo R., Peleg Y., Addison R., Metzenberg R. 1996. Asm-1+, a Neurospora crassa gene related to transcriptional regulators of fungal development. Genetics 144:991–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg J. M., Tymoczko J. L., Stryer L. 2007. Biochemistry W. H. Freeman and Company, New York, NY [Google Scholar]
- 3.Busby T. M., Miller K. Y., Miller B. L. 1996. Suppression and enhancement of the Aspergillus nidulans medusa mutation by altered dosage of the bristle and stunted genes. Genetics 143:155–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desbrosses G. G., Kopka J., Udvardi M. K. 2005. Lotus japonicus metabolic profiling. Development of gas chromatography-mass spectrometry resources for the study of plant-microbe interactions. Plant Physiol. 137:1302–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doedt T., Krishnamurthy S., Bockmuhl D. P., Tebarth B., Stempel C., Russell C. L., Brown A. J., Ernst J. F. 2004. APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol. Biol. Cell 15:3167–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Douaiher M. N., Halama P., Janex-Favre M. C. 2004. The ontogeny of Stagonospora nodorum pycnidia in culture. Sydowia 56:39–50 [Google Scholar]
- 7.Dutton J. R., Johns S., Miller B. L. 1997. StuAp is a sequence-specific transcription factor that regulates developmental complexity in Aspergillus nidulans. EMBO J. 16:5710–5721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friesen T. L., Stukenbrock E. H., Liu Z., Meinhardt S., Ling H., Faris J. D., Rasmussen J. B., Solomon P. S., McDonald B. A., Oliver R. P. 2006. Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 38:953–956 [DOI] [PubMed] [Google Scholar]
- 9.Friesen T. L., Zhang Z., Solomon P. S., Oliver R. P., Faris J. D. 2008. Characterization of the interaction of a novel Stagonospora nodorum host-selective toxin with a wheat susceptibility gene. Plant Physiol. 146:682–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gravelat F. N., Doedt T., Chiang L. Y., Liu H., Filler S. G., Patterson T. F., Sheppard D. C. 2008. In vivo analysis of Aspergillus fumigatus developmental gene expression determined by real-time reverse transcription-PCR. Infect. Immun. 76:3632–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joliffe I. T. 2005. Principal component analysis Springer-Verlag, New York, NY [Google Scholar]
- 12.Liu Z., Faris J. D., Oliver R. P., Tan K. C., Solomon P. S., McDonald M. C., McDonald B. A., Nunez A., Lu S., Rasmussen J. B., Friesen T. L. 2009. SnTox3 acts in effector triggered susceptibility to induce disease on wheat carrying the Snn3 gene. PLoS Pathog. 5:e1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lowe R. G., Lord M., Rybak K., Trengove R. D., Oliver R. P., Solomon P. S. 2008. A metabolomic approach to dissecting osmotic stress in the wheat pathogen Stagonospora nodorum. Fungal Genet. Biol. 45:1479–1486 [DOI] [PubMed] [Google Scholar]
- 14.Lowe R. G. T., Lord M., Rybak K., Trengove R. D., Oliver R. P., Solomon P. S. 2009. Trehalose biosynthesis is involved in sporulation of Stagonospora nodorum. Fungal Genet. Biol. 46:381–389 [DOI] [PubMed] [Google Scholar]
- 15.Miller K. Y., Wu J., Miller B. L. 1992. StuA is required for cell pattern formation in Aspergillus. Genes Dev. 6:1770–1782 [DOI] [PubMed] [Google Scholar]
- 16.Murray G. M., Brennan J. P. 2009. Estimating disease losses to the Australian wheat industry. Australas. Plant Pathol. 38:558–570 [Google Scholar]
- 17.Nishimura M., Fukada J., Moriwaki A., Fujikawa T., Ohashi M., Hibi T., Hayashi N. 2009. Mstu1, an APSES transcription factor, is required for appressorium-mediated infection in Magnaporthe grisea. Biosci. Biotechnol. Biochem. 73:1779–1786 [DOI] [PubMed] [Google Scholar]
- 18.Ohara T., Tsuge T. 2004. FoSTUA, encoding a basic helix-loop-helix protein, differentially regulates development of three kinds of asexual spores, macroconidia, microconidia, and chlamydospores, in the fungal plant pathogen Fusarium oxysporum. Eukaryot. Cell 3:1412–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruijter G. J. G., Bax M., Patel H., Flitter S. J., van de Vondervoort P. J. I., de Vries R. P., vanKuyk P. A., Visser J. 2003. Mannitol is required for stress tolerance in Aspergillus niger conidiospores. Eukaryot. Cell 2:690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scherer M., Wei H., Liese R., Fischer R. 2002. Aspergillus nidulans catalase-peroxidase gene (cpeA) is transcriptionally induced during sexual development through the transcription factor StuA. Eukaryot. Cell 1:725–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheppard D. C., Doedt T., Chiang L. Y., Kim H. S., Chen D., Nierman W. C., Filler S. G. 2005. The Aspergillus fumigatus StuA protein governs the up-regulation of a discrete transcriptional program during the acquisition of developmental competence. Mol. Biol. Cell 16:5866–5879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Solomon P. S., Ipcho S. V. S., Hane J. K., Tan K.-C., Oliver R. P. 2008. A quantitative PCR approach to determine gene copy number. Fungal Genet. Rep. 55:5–8 [Google Scholar]
- 23.Solomon P. S., Lowe R. G. T., Tan K. C., Waters O. D. C., Oliver R. P. 2006. Stagonospora nodorum: cause of stagonospora nodorum blotch of wheat. Mol. Plant Pathol. 7:147–156 [DOI] [PubMed] [Google Scholar]
- 24.Solomon P. S., Rybak K., Trengove R. D., Oliver R. P. 2006. Investigating the role of calcium/calmodulin-dependent protein kinases in Stagonospora nodorum. Mol. Microbiol. 62:367–381 [DOI] [PubMed] [Google Scholar]
- 25.Solomon P. S., Tan K. C., Oliver R. P. 2005. Mannitol 1-phosphate metabolism is required for sporulation in planta of the wheat pathogen Stagonospora nodorum. Mol. Plant Microbe Interact. 18:110–115 [DOI] [PubMed] [Google Scholar]
- 26.Solomon P. S., Tan K. C., Sanchez P., Cooper R. M., Oliver R. P. 2004. The disruption of a Gα subunit sheds new light on the pathogenicity of Stagonospora nodorum on wheat. Mol. Plant Microbe Interact. 17:456–466 [DOI] [PubMed] [Google Scholar]
- 27.Solomon P. S., Thomas S. W., Spanu P., Oliver R. P. 2003. The utilisation of di/tripeptides by Stagonospora nodorum is dispensable for wheat infection. Physiol. Mol. Plant Pathol. 63:191–199 [Google Scholar]
- 28.Solomon P. S., Waters O. D. C., Jörgens C. I., Lowe R. G. T., Rechberger J., Trengove R. D., Oliver R. P. 2006. Mannitol is required for asexual sporulation in the wheat pathogen Stagonospora nodorum (glume blotch). Biochem. J. 399:231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Solomon P. S., Waters O. D. C., Oliver R. P. 2007. Decoding the mannitol enigma in filamentous fungi. Trends Microbiol. 15:257–262 [DOI] [PubMed] [Google Scholar]
- 30.Solomon P. S., Waters O. D. C., Simmonds J., Cooper R. M., Oliver R. P. 2005. The Mak2 MAP kinase signal transduction pathway is required for pathogenicity in Stagonospora nodorum. Curr. Genet. 48:60–68 [DOI] [PubMed] [Google Scholar]
- 31.Tan K. C., Heazlewood J. L., Millar A. H., Thomson G., Oliver R. P., Solomon P. S. 2008. A signaling-regulated, short-chain dehydrogenase of Stagonospora nodorum regulates asexual development. Eukaryot. Cell 7:1916–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan K. C., Trengove R. D., Maker G. L., Oliver R. P., Solomon P. S. 2009. Metabolite profiling identifies the mycotoxin alternariol in the pathogen Stagonospora nodorum. Metabolomics 5:330–335 [Google Scholar]
- 33.Tong X., Zhang X., Plummer K. M., Stowell K. M., Sullivan P. A., Farley P. C. 2007. GcSTUA, an APSES transcription factor, is required for generation of appressorial turgor pressure and full pathogenicity of Glomerella cingulata. Mol. Plant Microbe Interact. 20:1102–1111 [DOI] [PubMed] [Google Scholar]
- 34.Twumasi-Boateng K., Yu Y., Chen D., Gravelat F. N., Nierman W. C., Sheppard D. C. 2009. Transcriptional profiling identifies a role for BrlA in the response to nitrogen depletion and for StuA in the regulation of secondary metabolite clusters in Aspergillus fumigatus. Eukaryot. Cell 8:104–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu J., Miller B. L. 1997. Aspergillus asexual reproduction and sexual reproduction are differentially affected by transcriptional and translational mechanisms regulating stunted gene expression. Mol. Cell. Biol. 17:6191–6201 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.