Abstract
The intracellular signaling molecule cyclic-di-GMP (c-di-GMP) has been shown to influence surface-associated behaviors of Pseudomonas aeruginosa, including biofilm formation and swarming motility. Previously, we reported a role for the bifA gene in the inverse regulation of biofilm formation and swarming motility. The bifA gene encodes a c-di-GMP-degrading phosphodiesterase (PDE), and the ΔbifA mutant exhibits increased cellular pools of c-di-GMP, forms hyperbiofilms, and is unable to swarm. In this study, we isolated suppressors of the ΔbifA swarming defect. Strains with mutations in the pilY1 gene, but not in the pilin subunit pilA gene, show robust suppression of the swarming defect of the ΔbifA mutant, as well as its hyperbiofilm phenotype. Despite the ability of the pilY1 mutation to suppress all the c-di-GMP-related phenotypes, the global pools of c-di-GMP are not detectably altered in the ΔbifA ΔpilY1 mutant relative to the ΔbifA single mutant. We also show that enhanced expression of the pilY1 gene inhibits swarming motility, and we identify residues in the putative VWA domain of PilY1 that are important for this phenotype. Furthermore, swarming repression by PilY1 specifically requires the diguanylate cyclase (DGC) SadC, and epistasis analysis indicates that PilY1 functions upstream of SadC. Our data indicate that PilY1 participates in multiple surface behaviors of P. aeruginosa, and we propose that PilY1 may act via regulation of SadC DGC activity but independently of altering global c-di-GMP levels.
Pseudomonas aeruginosa forms surface-attached communities known as biofilms, and this microbe is also capable of surface-associated motility, including twitching and swarming. The mechanism by which cells regulate and coordinate these various surface-associated behaviors, or how these microbes transition from one surface behavior to another, has yet to be elucidated. Given that P. aeruginosa is capable of such diverse surface-associated lifestyles, this Gram-negative organism serves as a useful model to address questions regarding the regulation of surface-associated behaviors.
Recent studies indicate that biofilm formation and swarming motility by P. aeruginosa are inversely regulated via a common pathway (12, 27, 37). Important factors that influence early biofilm formation by P. aeruginosa strain PA14 include control of flagellar motility and the robust production of the Pel exopolysaccharide (EPS). Swarming occurs when cells move across a hydrated, viscous semisolid surface, and like biofilm formation, flagellar function is important for this surface-associated motility. Additionally, swarming requires production of rhamnolipid surfactant acting as a surface-wetting agent (25, 58). In contrast to biofilm formation, swarming motility is enhanced in strains which are defective for the production of Pel EPS (12).
The inverse regulation of biofilm formation and swarming motility is reminiscent of the regulation of sessile and motile behaviors that occurs in a wide range of bacterial species via the intracellular signaling molecule cyclic-di-GMP (c-di-GMP) (17, 24, 50, 51, 56). High levels of this signaling molecule promote sessile behaviors and inhibit motility, whereas low levels of c-di-GMP favor motile behaviors (8, 9, 22, 56). Recently, we reported that the BifA phosphodiesterase, which catalyzes the breakdown of c-di-GMP, inversely regulates biofilm formation and swarming motility (27). In addition, Merritt et al. reported that SadC, a diguanylate cyclase (DGC) which synthesizes c-di-GMP, participates with BifA to modulate cellular c-di-GMP levels and thus regulate biofilm formation and swarming motility (37).
Consistent with a role for BifA as a c-di-GMP phosphodiesterase, ΔbifA mutants exhibit increased cellular pools of c-di-GMP relative to the wild type (WT) (27). Phenotypically, ΔbifA mutants form hyperbiofilms and are unable to swarm. The hyperbiofilm phenotype of the ΔbifA mutant results largely from increased synthesis of the pel-derived polysaccharide; that is, the ΔbifAΔpel double mutant shows a marked decrease in biofilm formation compared to the ΔbifA mutant (27). Interestingly, elevated Pel polysaccharide production alone is not sufficient to explain the swarming defect of the ΔbifA mutant, as the ΔbifAΔpel double mutant recovers only minimal swarming ability (27). These data indicate that high levels of c-di-GMP inhibit swarming motility in a largely Pel-independent manner.
To better understand how elevated c-di-GMP levels in the cell inhibit swarming motility, we exploited the swarming defect of the ΔbifA mutant, and using a genetic screen, we identified suppressors in the ΔbifA background that restored the ability to swarm. Here we report a role for the PilY1 protein in repression of swarming motility in the ΔbifA mutant background. Our data are consistent with a model in which PilY1 functions upstream of the c-di-GMP diguanylate cyclase SadC to regulate swarming motility by P. aeruginosa.
MATERIALS AND METHODS
Strains and media.
The strains, plasmids, and primers used in this study are listed in Table 1. The P. aeruginosa PA14 and Escherichia coli DH5α and S17-1 λpir strains were routinely cultured on lysogeny broth (LB) medium, which was solidified with 1.5% agar when necessary. Gentamicin (Gm) was used at from 25 to 50 μg/ml for P. aeruginosa and at 10 μg/ml for E. coli. Ampicillin (Ap) was used at 150 μg/ml and nalidixic acid (NA) at 20 μg/ml for E. coli. For all phenotypic assays with P. aeruginosa, M63 minimal salts medium supplemented with MgSO4 (1 mM), glucose (0.2%), and Casamino Acids (CAA) (0.5%) was used, except where noted. For expression plasmids harboring the pBAD promoter, arabinose was added to cultures at a 0.2% final concentration, unless noted otherwise.
TABLE 1.
Strains, plasmids, and primers
| Strain, plasmid, or primer | Relevant genotype, description, or sequencea | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | supE44 ΔlacU169(φ80lacZΔM15) hsdR17 thi-1 relA1 recA1 | Life Technologies |
| S17-1 (λpir) | thi pro hsdR−M+ ΔrecA RP4-2::TcMu-Km::Tn7 | 57 |
| P. aeruginosa strains | ||
| PA14 | Wild type | 47 |
| PA14 ΔbifA | PA14 with unmarked deletion of bifA | 27 |
| PA14 ΔpelA | PA14 with unmarked deletion of pelA | 19 |
| PA14 ΔbifA ΔpelA | PA14 ΔpelA with unmarked deletion of bifA | This study |
| PA14 ΔbifA pilY1::Mar 57E12 | PA14 ΔbifA with mariner insertion in pilY1; Gmr | This study |
| PA14 ΔbifA pilY1::Mar 68C1 | PA14 ΔbifA with mariner insertion in pilY1; Gmr | This study |
| PA14 ΔbifA pilY1::Mar 52H5 | PA14 ΔbifA with mariner insertion in pilY1; Gmr | This study |
| PA14 ΔbifA ΔsadA::Mar 51G2 | PA14 ΔbifA with mariner insertion in sadA; Gmr | This study |
| PA14 ΔbifA ΔpvrS::Mar 57B1 | PA14 ΔbifA with mariner insertion in pvrS; Gmr | This study |
| PA14 ΔbifA ΔpvrS::Mar 39D11 | PA14 ΔbifA with mariner insertion in pvrS; Gmr | This study |
| PA14 ΔbifA ΔpvrS::Mar PA14 23H6 | PA14 ΔbifA with mariner insertion in pvrS; Gmr | This study |
| ΔbifA ΔpvrS::Mar PA14 58A9 | PA14 ΔbifA with mariner insertion in pvrS; Gmr | This study |
| ΔbifA ΔpvrS::Mar PA14 39H1 | PA14 ΔbifA with mariner insertion in pvrS; Gmr | This study |
| PA14 ΔpilY1 | PA14 with unmarked deletion of pilY1 | This study |
| PA14 ΔbifA ΔpilY1 | PA14 ΔbifA with unmarked deletion of pilY1 | This study |
| PA14 ΔpilA | PA14 with unmarked deletion of pilA | This study |
| PA14 ΔbifA ΔpilA | PA14 ΔbifA with unmarked deletion of pilA | This study |
| PA14 ΔbifA ΔpilY1 ΔpilA | PA14 ΔbifA ΔpilY1 with unmarked deletion of pilA | This study |
| PA14 ΔsadC | PA14 with unmarked deletion of sadC | 37 |
| PA14 ΔwspR | PA14 with unmarked deletion of wspR | This study |
| PA14 ΔPA1107 | PA14 with unmarked deletion of PA1107 | This study |
| Plasmids | ||
| pEX18-bifA | Vector for knockout of bifA gene; Gmr | 27 |
| pMQ72 | Shuttle vector for cloning in yeast and for arabinose-inducible gene expression; Gmr | 55 |
| pRapA | P. fluorescens rapA gene cloned in pMQ72; Gmr | 38 |
| pBT20 | Vector carrying mariner transposon; Apr Gmr (marker on transposon) | 30 |
| pMQ30 | Shuttle vector for yeast cloning and Gram-negative allelic replacement; Gmr | 55 |
| pMQ30-pilY1 | Vector for knockout of pilY1 gene; Gmr | This study |
| pMQ30-pilA | Vector for knockout of pilA gene; Gmr | This study |
| pEX19Gm-wspR | Vector for knockout of wspR gene; Gmr | 22 |
| pMQ80 | Shuttle vector for cloning in yeast and for arabinose-inducible gene expression; Gmr | 55 |
| pPilY1His | His-tagged pilY1 gene cloned in pMQ80; Gmr | This study |
| pSadC | HA-tagged sadC gene cloned in pMQ72; Gmr | 37 |
| pPilY1HisΔPilC | His-tagged pilY1 gene lacking the PilC domain in pMQ80; Gmr | This study |
| pPilY1His-MTAA | His-tagged pilY1 gene with D309A/G310A mutations in pMQ80; Gmr | This study |
| Primers | ||
| pilY1 KO-1 | tgtaaaacgacggccagtgccaagcttgcatgcctgACGGTGTCTGCTTGCGCTATC | |
| pilY1 KO-2 | GACGTAAGGGGTTCATGTTCAAGAGGCGAACAGGATGCTCAG | |
| pilY1 KO-3 | CTGAGCATCCTGTTCGCCTCTTGAACATGAACCCCTTACGTC | |
| pilY1 KO-4 | ccatgattacgaattcgagctcggtacccggggatccCAGCCCCAACAGTTCCAACTC | |
| pilY1 KO conf 5′ | AGACTATCGCTGCGGCAACTG | |
| pilY1 KO conf 3′ | CAGGATGCCGTGAAGGAACTG | |
| pilY1 comp 5′ | tctccatacccgtttttttgggctagcgaattcgaaggagatatacatATGATCCACCAGATTACC | |
| pilY1 comp 3′ | tcctttactcatatgtatatctccttccccgggtaccCTGCCCATCGACTACTACCAG | |
| pilY1-C6His A | AGGGGTTCATGTTCAGTGGTGATGGTGGTGGTGTTTCTCCTCGACGACCCG | |
| pilY1-C6His B | GTCGAGGAGAAACACCACCACCATCACCACTGAACATGAACCCCTTACGTC | |
| pilA KO-1 | tgtaaaacgacggccagtgccaagcttgcatgcctgTCGATGGACTTGGTCTTCGAC | |
| pilA KO-2 | GGCAGGCATATTGCGGTCCTGAGCTTTCATGTATCTCTC | |
| pilA KO-3 | GAGAGATACATGAAAGCTCAGGACCGCAATATGCCTGCC | |
| pilA KO-4 | ccatgattacgaattcgagctcggtacccggggatccCCTGACCAAGGATGTCAG | |
| pilA KO conf 5′ | GATCCGCATCACGATCTTCTC | |
| pilA KO conf 3′ | GAACTCAGCCTGGACGACATG | |
| pvrR RT 5′ | GTCGAAGATCAGCCGTTTCAG | |
| pvrR RT 3′ | GTCGAACCTGTCCTGCTTCAG | |
| sadR RT 5′ | TCATTCGGTGATCCTCAGCAG | |
| sadR RT 3′ | GAGGTCGCCGAGAAAATTGAG | |
| pelA RT 5′ | GCTACGTGCCGTTCGTCAGCACC | |
| pelA RT 3′ | CAGGCCGCCGAGGTAGACGTG | |
| pilY1 RT 5′ | TGCACCCTGAGCATCCTGTTC | |
| pilY1 RT 3′ | GGCCATACTGCCTGAGTCATC | |
| Del-PilC 5′ | CGGGTTTTGGCTAATAAGCCGCCAAGTGGCTAACTGACTTCC | |
| Del-PilC 3′ | GGAAGTCAGTTAGCCACTTGGCGGCTTATTAGCCAAAACCCG | |
| MTDGmutF | AGCTATCACATCATGATGACCGCCGCCATCTGGAACGGTCGGAACGTC | |
| MTDGmutR | GACGTTCCGACCGTTCCAGATGGCGGCGGTCATCATGATGTGATAGCT | |
| TPLmutF | AACAGCCCGGCCAGCGGCGGTGCTCCTCTGCATGCGGCTCTTG | |
| TPLmutR | CAAGAGCCGCATGCAGAGGAGCACCGCCGCTGGCCGGGCTGTT | |
| PCRmut1 Rev | GAGCGAATAGGAAAGACCTAG | |
| P730 | GCAACTCTCTACTGTTTCTCC | |
| PCRmut2 For | ACTTGGCAACACATGGTCAAC | |
| P883 | AGAATTGGGACAACTCCAGTG | |
| PCRmut seq1 | CCTGGCTTTCAAATACTGGAC | |
| PCRmut seq2 | AGTCTCCACGAGAGTTGATAG |
In primer sequences, lowercase letters indicate sequence homology to the cloning vector, uppercase letters indicate a Pseudomonas gene-specific sequence, boldface indicates a His tag sequence, and italics indicate a 5′ sequence flanking a deletion mutation.
Saccharomyces cerevisiae strain InvSc1 (Invitrogen), used for plasmid construction via in vivo homologous recombination, was grown with yeast extract-peptone-dextrose (1% Bacto yeast extract, 2% Bacto peptone, and 2% dextrose) (55). Selections with InvSc1 were performed using synthetic defined agar-uracil (catalog no. 4813-065; Qbiogene).
mariner mutagenesis of the ΔbifA mutant.
The donor strain E. coli S17-1 λ pir carrying the pBT20 plasmid and the P. aeruginosa ΔbifA mutant recipient strain were conjugated as follows. From overnight LB-grown cultures (with the appropriate antibiotic), 1 ml of each strain was centrifuged to pellet the cells. Cell pellets were washed twice with LB and resuspended in 100 μl of fresh LB. The cell suspensions of each strain were then mixed, and 60 μl of the mixture was plated to two LB plates and the conjugation was allowed to proceed for 1 h at 30°C. The conjugation products were harvested, resuspended in 100 μl of LB, and then plated to LB agar plates containing Gm, as well as NA to select against E. coli growth. Libraries of mutants were generated by inoculating colonies into 96-well plates containing LB medium supplemented with Gm, and plates were incubated overnight at 37°C.
Mutant strains in library plates were screened for recovery of swarming motility by inoculating swarm motility plates (see below) using a multichannel pipette to deliver cells from the wells of the plate to the swarm plate. WT and ΔbifA mutant controls were included on each plate. Plates were incubated at 37°C for 12 to 16 h, and mutant strains were assessed for swarming motility.
Approximately 5,500 mariner transposon mutants in the ΔbifA mutant background were screened. DNA sequences flanking the transposon insertions in mutants of interest were determined using an arbitrary PCR method, as described previously (11, 43). PCR products were sequenced at the Molecular Biology and Proteomics Core at Dartmouth College. The resulting DNA sequences were aligned to the PA14 genomic sequence using the NCBI BLAST program.
Construction of in-frame deletion mutants and plasmids.
The ΔbifA ΔpelA double mutant was constructed by deletion of bifA in the ΔpelA mutant (provided by R. Kolter) using the previously described pEX18Gm-bifA construct (27). The pilY1 deletion construct was generated using primers pilY1 KO-1 with pilY1 KO-2 and pilY1 KO-3 with pilY1 KO-4, and these 1-kb fragments were cloned into the pMQ30 vector via homologous recombination in the yeast S. cerevisiae InvSc1 as described previously (55). The resulting pMQ30-pilY1 vector was used to transform E. coli S17-1 and introduced into the P. aeruginosa WT and ΔbifA strain backgrounds via conjugation. Integrants were isolated on Gm and NA and subjected to sucrose selection. The resolved integrants were confirmed by PCR using primers pilY1 KO conf 5′ and 3′. The resulting internal deletion in pilY1 removes approximately 3.4 kb, resulting in a deletion from amino acid 26 to 1158 (up to the stop codon) of the 1158-amino-acid full-length PilY1 protein.
The pMQ30-pilA knockout construct was generated using primers pilA KO-1 with pilA KO-2 and pilA KO-3 with pilA KO-4, and the PCR fragments were cloned into pMQ30 by homologous recombination in yeast. Construction of the pilA deletion mutation using pMQ30-pilA in P. aeruginosa proceeded as described above and was confirmed by PCR using the pilA KO conf 5′ and pilA KO conf 3′ primers. The resulting internal deletion in pilA removes approximately 450 base pairs, resulting in a deletion of amino acids 5 to 157 of the 179-amino-acid full-length PilA protein. The ΔbifA ΔpilY1 ΔpilA triple mutant was constructed by deletion of the pilA gene in the ΔbifA ΔpilY1 double mutant.
The wspR deletion was constructed using the pEX19Gm-wspR plasmid (provided by C. Harwood). The construct was introduced into WT P. aeruginosa, and deletion mutants were isolated as described above.
The pPilY1 complementation construct was generated by PCR amplification using the high-fidelity Phusion polymerase (Finnzyme, Espoo, Finland). Primer pilY1 comp 5′ was used with pilY1-C6His B and pilY1 comp 3′ was used with pilY1-C6His A, and these fragments were cloned into pMQ80 by homologous recombination in yeast to generate an arabinose-inducible C-terminally His-tagged version of the pilY1 gene.
Deletion of the PilC domain from the pilY1 gene was performed by PCR amplification using Del-PilC 5′ with the pilY1 comp 5′ primer and Del-PilC 3′ with the pilY1 comp 3′ primer and using the pPilY1 construct as the template for these reactions to incorporate the C-terminal His tag. These PCR fragments were cloned into pMQ80 in yeast.
Mutagenesis of pilY1. (i) Random PCR mutagenesis of pilY1ΔPilC.
Using the pPilY1ΔPilC construct as a template for PCR, linear DNA fragments comprised of overlapping halves of the pilY1ΔPilC mutant coding region (∼1,200 bp each) were amplified by PCR using Phusion polymerase (Finnzyme). The primers p730 and PCRmut1Rev were used to amplify the N-terminal half of the pilY1ΔPilC gene, and primers PCRmut2For and p883 were used to amplify the C-terminal half of the gene. These linear DNA fragments served as templates for the subsequent mutagenic PCR, which was performed as reported previously (10), with the following modification: 0.125 mM MnCl was added (instead of 0.5 mM) to lower the frequency of mutations to ∼2 mutations per 1,200 bp (as determined by sequencing a number of PCR products at both MnCl concentrations).
Mutagenized PCR products were gel purified using the Qiagen gel extraction kit. To reconstruct the intact pilY1ΔPilC gene, a mutated N-terminal fragment was paired with a nonmutated C-terminal region of the gene (and vice versa) for cloning together into the pMQ80 vector via homologous recombination in yeast. This also served to focus sequencing of the mutations generated by mutagenesis to only one half of the pilY1ΔPilC gene. The resulting plasmids were purified and electroporated first into E. coli DH5α and then into PA14 for greater efficiency. The resulting colonies were picked into a 96-well plate to grow overnight at 37°C in LB medium supplemented with Gm. The resulting cultures, as well as the control strains (WT carrying either vector alone, pPilY1ΔPilC or pPilY1), were then inoculated onto swarm motility plates containing 0.2% arabinose using a multichannel pipette. Strains carrying mutated versions of pPilY1ΔPilC that lost swarming inhibition were subjected to dot blot analysis (described below) to check for protein stability. Strains expressing stable PilY1ΔPilC proteins were retested by repeating the plasmid isolation, transforming PA14, and again testing for swarming. Mutants were sequenced with the primers p730, p883, pPCRmut seq1, and pPCRmut seq2.
(ii) Site-directed mutagenesis of pilY1.
To identify regions for mutagenesis, PilY1 homologs were identified by BLAST and aligned with the PA14 PilY1 protein using ClustalW2. Regions of amino acid conservation were identified, and two such regions were targeted for mutagenesis. The threonine at position 270 was changed to alanine by site-directed mutagenesis using primer pTPLmutF with pilY1-comp 5′ and primer pTPLmutR with pilY1-comp 3′. Amino acids D310 and G311 were both replaced with alanines via the same method, using primer MTDGmutF with pilY1-comp 5′ and primer MTDGmutR with pilY1-comp 3′. Products were cloned into the pMQ80 vector via yeast-based homologous recombination and transformed into PA14. The mutations were verified by sequencing using primers p730 and pPCRmut1Rev. Strains expressing these mutant versions of PilY1 were tested in swarming assays as described below.
Motility assays.
Swim (0.3% agar) and twitch (1.5% agar) motility plates consisted of M63 medium supplemented with glucose, MgSO4 and CAA. Swim and twitch assays were performed as previously reported (43, 61). Swarm motility plates were comprised of M8 medium supplemented with MgSO4, glucose, and CAA and solidified with 0.5% agar. For testing individual strains, 2 μl of LB-grown overnight cultures was inoculated onto the surface of the swarm plates and incubated for 12 to 16 h at 37°C. For screening large numbers of strains, cells from overnight cultures grown in 96-well plates were transferred with a multichannel pipette to the surface of swarm plates.
Biofilm formation assays.
Biofilm formation in 96-well microtiter plates was assayed and quantified as previously described (13, 42). All biofilm assays were performed using M63 minimal medium supplemented with glucose, MgSO4, and CAA.
CR binding assays.
Congo red (CR) binding assays were performed as reported previously (12, 19, 20).
qRT-PCR.
Strains were diluted from LB-grown overnight cultures 1:100 into M63 minimal medium supplemented with glucose, MgSO4, and CAA and grown to an optical density at 600 nm (OD600) of 0.6. RNA was isolated, and cDNA was prepared as previously described (28). Quantitative reverse transcription-PCR (qRT-PCR) was performed using an ABI 7500 Fast System and analyzed using ABI Fast System software version 1.4. Expression levels were quantified in picograms of input cDNA using a standard curve method for absolute quantification, and these values were normalized to rplU expression. Results shown are based on the average from two independent experiments with three replicates per sample. The primers used are listed in Table 1.
In vivo quantification of c-di-GMP. (i) Whole-cell labeling.
Whole-cell [32P]orthophosphate labeling, acid extraction, and two-dimensional (2D) thin-layer chromatography (TLC) analysis of c-di-GMP for quantification were performed as reported previously (22, 38).
(ii) Mass spectrometry.
Cultures were grown in LB for 13 h before being back diluted into 50 ml of M63-glucose-CAA medium to an OD600 of 0.05. Subcultured strains were grown with shaking at 37°C for ∼4 h until the OD600 was 0.4, at which point 40 ml of cultured cells was pelleted by centrifugation for 3 min at 10,000 × g and 25°C. The culture broth supernatant was discarded, and 250 μl of cold (−20°C) extraction solvent (methanol-acetonitrile-water at a volumetric ratio of 40:40:20, acidified with formic acid to final concentration of 0.1 N) was immediately added to the cell pellet to quench metabolism and initiate c-di-GMP extraction. Extraction was allowed to proceed for 30 min at −20°C. The mixture was then transferred to a 1.5-ml Eppendorf tube and microcentrifuged for 5 min at 4°C and maximum speed (13,000 rpm). The supernatant was set aside in a fresh tube on ice. The cell pellet was then resuspended in an additional 125 μl of extraction solvent, and the mixture was incubated at −20°C for 15 min and then microcentrifuged as above. The resulting supernatant was combined with the prior one, and the combined supernatants were microcentrifuged again to ensure complete removal of debris. The final solution was then neutralized by addition of 4 μl of 15% (NH4)2HCO3. Note that the extracts were collected from pelleted cells; thus, although our results were reproducible across multiple independent experiments, they may be affected by metabolic events occurring during centrifugation.
The cellular extracts were analyzed using liquid chromatography-tandem mass spectrometry (LC-MS/MS) on a Finnigan TSQ Quantum DiscoveryMax triple-quadrupole mass spectrometer (Thermo Fisher Scientific, San Jose, CA) coupled with an LC-20AD high-pressure liquid chromatography (HPLC) system (Shimadzu, Columbia, MD). The mass spectrometry parameters were as follows: spray voltage, 3,000 V; nitrogen as the sheath gas at 30 lb/in2 and auxiliary gas at 10 lb/in2; argon as the collision gas at 1.5 millitorr; and capillary temperature, 325°C. Reversed-phase liquid chromatography separation was achieved on a Synergi Hydro-RP column (4-μm particle size, 150 by 2 mm; Phenomenex, Torrance, CA). Solvent A was 10 mM tributylamine plus 15 mM acetic acid in 97:3 water-methanol. Solvent B was methanol. The gradient was as follows: t = 0, 0% solvent B; t = 5 min, 0% solvent B; t = 10 min, 20% solvent B; t = 20 min, 20% solvent B; t = 35 min, 65% solvent B; t = 38 min, 95% solvent B; t = 42 min, 95% solvent B; t = 43 min, 0% solvent B; and t = 50 min, 0% solvent B. Each sample had a running time of 50 min. Other liquid chromatography parameters were as follows: autosampler temperature, 4°C; column temperature, 25°C; injection volume, 20 μl; and solvent flow rate, 200 μl/min.
The nucleotide c-di-GMP was detected by using selected reaction monitoring (SRM) in negative ionization mode. The scan time for each SRM was 0.1 s, with a scan width of 1 m/z. The LC-MS/MS parameters for c-diGMP were as follows: parent ion m/z 689 forms product ion m/z 344 at a collision energy of 32 eV, eluting at a retention time of 32.1 min.
Assessment of PilY1 protein expression and stability. (i) SDS-PAGE and Western blotting.
Strains were cultured overnight at 37°C in LB and then diluted 1:100 into 5 ml of M63 minimal medium supplemented with glucose, MgSO4, and Casamino Acids (and arabinose at the indicated concentrations where necessary) and grown to an OD600 of 0.6. Cells were centrifuged at 16,000 × g to pellet them, the supernatants were removed, and the pellets were resuspended in 1× sodium dodecyl sulfate (SDS) gel loading buffer with 100 mM dithiothreitol (DTT). Samples were boiled for 10 min and resolved by SDS-polyacrylamide gel electrophoresis (PAGE) in a 7.5% polyacrylamide gel (Bio-Rad, Hercules, CA). Proteins were transferred to a nitrocellulose membrane and probed with either PilY1 antiserum to detect the PilY1 protein or an anti-penta-His antibody (Qiagen, Valencia, CA) to detect the His-tagged version of PilY1 where appropriate.
Western blots were developed with the Western Lightning ECL detection kit (Perkin-Elmer, Boston, MA) according to the manufacturer's instructions.
(ii) Dot blot analysis.
Strains carrying mutated versions of pPilY1ΔPilC that lost swarming inhibition when expressed in the WT were cultured overnight at 37°C in 96-well plates containing LB supplemented with Gm and then subcultured 1:100 in M63 minimal medium supplemented with glucose, MgSO4 and CAA, and 0.2% arabinose and grown for another 6 h. The cultures were combined with 6x SDS protein loading buffer and boiled for 10 min, and 5 μl of each sample was spotted onto a nitrocellulose membrane and allowed to dry. Membranes were probed with the anti-penta-His antibody (Qiagen), and antibodies were detected in the same manner described above for Western blots.
Mutants of interest were further examined by SDS-PAGE to resolve and specifically detect the PilY1 variant, as described above.
PilY1 cellular localization experiments.
Cell fractionations were performed as previously reported (27, 40) with modifications. LB-grown overnight cultures were diluted (1:100) into M63 supplemented with glucose, MgSO4, and CAA and grown for ∼5 h (OD600 of ∼0.6) at 37°C with shaking. Bacterial cells were harvested by centrifugation at 5,520 × g for 15 min at 4°C. Thirty milliliters of culture supernatant for each strain was collected and filtered through a 0.22-μm filter and concentrated approximately 300-fold using Amicon Ultra-15 filter devices (10,000-molecular-weight cutoff [MWCO]) (Millipore Corporation, Billerica, MA) according to the manufacturer's instructions. The resulting fractions were designated the supernatant (Sup).
The harvested cell pellets were resuspended in Tris-buffered saline (5 ml). Cell suspensions were vortexed vigorously for 2 min, followed by centrifugation at 4,400 × g. The supernatants were collected, filtered through a 0.22-μm filter, and concentrated approximately 100-fold using the Amicon Ultra-15 (10,000-MWCO) filter devices according to the manufacturer's instructions. The resulting supernatant was designated the cell-associated (CA) fraction, as defined previously (38).
The cell pellets were then resuspended in buffer A (200 mM Tris-HCl [pH 7.5], 20 mM EDTA [pH 8.0]) with 1× Complete protease inhibitors [Roche Diagnostics Corp., Indianapolis, IN]) and lysed in a French pressure cell. Samples were centrifuged at 9,300 × g for 10 min at 4°C to remove unbroken cells, and supernatants were collected as whole-cell lysates. Samples were further fractionated to isolate the inner and outer membrane fractions, as described previously (27).
For Western blots of cellular fractions, samples from each fraction were normalized to the same concentration, and equivalent amounts of total protein were mixed with a 2× SDS loading buffer containing dithiothreitol (200 mM). Concentrations of total protein in each fraction were determined using a Bio-Rad DC protein assay kit (Bio-Rad, Hercules, CA) according to the manufacturer's instructions. Samples were resolved by SDS-PAGE using 7.5% polyacrylamide gels (Bio-Rad). Proteins were transferred to a nitrocellulose membrane and probed with one of four antibodies: (i) PilY1 antiserum to detect the PilY1 protein (41), (ii) anti-penta-His antibody (Qiagen, Valencia, CA) to detect His-tagged PilY1 where appropriate, (iii) SecY antiserum (23) to detect the inner membrane protein SecY (1), and (iv) antiserum to detect the outer membrane protein OprF (7, 21, 34). Western blots were developed as described above.
RESULTS
A ΔbifA mutant does not swarm, likely due to high cellular c-di-GMP pools.
In previous studies, we reported that the bifA gene (PA4367) encodes a phosphodiesterase (PDE) that is able to degrade c-di-GMP and influence surface-associated behaviors of Pseudomonas aeruginosa (27). Our studies showed that deletion of the bifA gene leads to an increase in cellular c-di-GMP pools and that the ΔbifA mutant exhibits a hyperbiofilm phenotype as well as a complete loss of swarming motility (27).
We predicted that the nonswarming phenotype of the ΔbifA mutant results from the enhanced production of the pel-derived polysaccharide. However, a pelA mutation in the ΔbifA mutant background resulted in only a minor improvement in the swarming defect, indicating that excess Pel polysaccharide production is not the primary contributor to the ΔbifA swarming impairment (Fig. 1 A). This finding is consistent with a previous report from our group (27). Thus, we postulated that high levels of c-di-GMP inhibit swarming motility of the ΔbifA mutant via one or more Pel-independent factors.
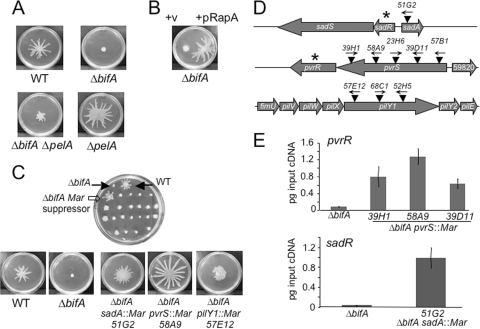
FIG. 1.
Genetic analyses of the ΔbifA swarming defect. (A) Impact of Pel polysaccharide on the ΔbifA swarming impairment. Representative images of swarms formed by the WT, the ΔbifA mutant, the ΔbifA ΔpelA double mutant, and the ΔpelA mutant swarms after 16 h at 37°C on 0.5% swarming agar are shown. (B) Impact of expression of the RapA PDE on swarming motility of the ΔbifA mutant. Representative images of swarms of the ΔbifA mutant carrying the pMQ72 vector control (left) and pMQ72 expressing the RapA c-di-GMP phosphodiesterase (right) are shown. The plates contained 0.2% (wt/vol) arabinose and were incubated for 16 h at 37°C. (C) Genetic screen for suppressors of the ΔbifA swarming impairment. The top image shows a swarm plate from the screen of ΔbifA mariner transposon mutants, with filled arrows indicating the WT control (top right) and the ΔbifA mutant (top left). The open arrow indicates a candidate ΔbifA mariner suppressor mutant. The lower panel shows typical images of swarms of the WT, the ΔbifA mutant, and the ΔbifA sadA::Mar, ΔbifA pvrS::Mar, and ΔbifA pilY1::Mar suppressor mutants. The plates were incubated for 16 h at 37°C. (D) Genetic loci of ΔbifA swarm suppressors. The schematic diagram shows the approximate locations of mariner transposon mutations (▾) in the sadA, pvrS, and pilY1 genes. Arrows above transposons indicate the direction of transcription from the mariner Ptac promoter. Asterisks denote genes encoding predicted c-di-GMP phosphodiesterases. (E) qRT-PCR analysis of pvrR expression in ΔbifA pvrS::Mar suppressor mutants (top panel) and sadR expression in the ΔbifA sadA::Mar suppressor mutant (bottom panel) relative to the ΔbifA single mutant. Expression is plotted as picograms input cDNA for each strain. Error bars indicate standard deviations.
To first confirm that the accumulation of excess c-di-GMP does, in fact, inhibit swarming motility in the ΔbifA mutant, we tested whether expression of a nonendogenous PDE in the ΔbifA mutant could rescue the swarming defect. We expressed the Pseudomonas fluorescens rapA gene, encoding a PDE previously shown to possess c-di-GMP-degrading activity (38), under the control of the arabinose-inducible pBAD promoter. As shown in Fig. 1B, expression of pRapA in the ΔbifA mutant background is able to rescue the swarming defect of the ΔbifA mutant. These results indicate that degradation of c-di-GMP in the ΔbifA mutant is able to restore swarming motility and supports the hypothesis that excess c-di-GMP inhibits swarming motility in the ΔbifA mutant.
Identification of suppressors of the ΔbifA swarming motility defect.
To better understand how c-di-GMP negatively affects swarming motility, we sought to identify those factors required for swarming inhibition in the ΔbifA mutant. To this end, we performed mariner transposon mutagenesis of the ΔbifA mutant and screened for restoration of swarming motility. We screened approximately 5,500 mariner transposon mutants in the ΔbifA mutant background on swarm agar plates (Fig. 1C, top panel) and isolated suppressor strains with mutations in four different genes.
We mapped these transposon mutations in the suppressors to sadA, pvrS, pilY1, and sadB (Fig. 1D). The swarm phenotypes of three representative ΔbifA suppressor mutants are shown in Fig. 1C, bottom panel. The ΔbifA sadA::Mar mutant exhibits a pattern of swarming motility distinct from that of the WT, with shorter and more numerous tendrils. We isolated five independent pvrS mutants as suppressors of the ΔbifA swarming defect (Fig. 1D). For the representative ΔbifA pvrS::Mar mutant (58A9) shown in Fig. 1C, the extent of swarming of the mutant is greater than that of the WT. The remaining four pvrS::Mar mutants suppress the ΔbifA mutant phenotype to various degrees, which likely reflects the different positions of the transposon insertion in each mutant (see below). The three independent ΔbifA pilY1::Mar suppressor mutants isolated exhibit an unusual swarm morphology relative to the WT in that the mutants lack distinct tendril formation and instead show a more uniform swarming pattern radiating from the origin of inoculation (Fig. 1C). The fourth gene, sadB, was previously shown to be required for inhibition of swarming by the ΔbifA mutant (27). Given that mutations in the sadB gene completely suppress the swarming defects of the ΔbifA mutant, the isolation of sadB mutants served to validate the screen.
Enhanced expression of genes encoding putative PDEs suppresses the ΔbifA swarming defect.
Of the genes identified as suppressors in this screen, two genes, sadA and pvrS, are located directly adjacent to genes that code for proteins containing EAL motifs and thus are predicted to encode c-di-GMP phosphodiesterases (sadR and pvrR, respectively) (Fig. 1D). The sadA gene (also known as rocA) encodes a response regulator that has been previously shown, together with the adjacent genes, sadR and sadS, to be involved in biofilm formation as well as regulation of Cup fimbrial gene expression (28, 30). The pvrS gene encodes a sensor histidine kinase and is adjacent to the response regulator gene pvrR, which has been shown to influence the frequency of formation of antibiotic-resistant small-colony variants of P. aeruginosa (18).
Given the proximity of the transposon insertions in sadA and pvrS to a PDE-encoding gene and the fact that overexpression of the RapA PDE in the ΔbifA mutant is able to rescue the swarming defect, we hypothesized that the phenotypic suppression of the ΔbifA swarming defect observed for the ΔbifA sadA::Mar and ΔbifA pvrS::Mar mutants was due to enhanced expression of sadR and pvrR rather than to inactivation of either sadA or pvrS. This also seemed a reasonable prediction based on the fact that the mariner transposon used in these studies harbors an outward-directed Ptac promoter and has been shown in previous studies to enhance expression of genes adjacent to transposon insertion sites (30). Furthermore, in a recent study, overexpression of pvrR led to a reduction in intracellular c-di-GMP levels, with a corresponding impact on c-di-GMP-sensitive phenotypes (36).
As shown in Fig. 1D, the Ptac promoter for several of the transposon insertion alleles is oriented toward the adjacent PDE-encoding gene. In cases where the opposite orientation is found, it is possible that the promoter driving expression of the aaC1 gene, encoding resistance to gentamicin, is able to suitably activate transcription of adjacent genes. To address the possibility that sadR and pvrR are overexpressed in these mutants, we performed qRT-PCR to examine expression of the pvrR gene in several of the ΔbifA pvrS::Mar mutants, as well as expression of the sadR gene in the ΔbifA sadA::Mar 51G2 mutant (Fig. 1E). The results show that expression of the pvrR gene is elevated in each of the ΔbifA pvrS::Mar mutants, ranging from 8-fold to 17-fold higher than that in the ΔbifA mutant (Fig. 1E, top panel). Likewise, expression of the sadR gene in the ΔbifA sadA::Mar 51G2 mutant is elevated approximately 30-fold relative to that in the ΔbifA single mutant (Fig. 1E, bottom panel).
To further confirm that inactivation of either the sadA or pvrS gene is not responsible for suppression of the ΔbifA swarming impairment, we tested the ΔbifA ΔsadA double mutant and a ΔbifA mutant containing a single crossover insertion in the pvrS gene and found that they are indistinguishable from the ΔbifA single mutant in swarming motility, CR binding, and biofilm formation assays (data not shown). Taking these results together with the data from the RT-PCR experiments, we infer that suppression of the swarming motility defect exhibited by the ΔbifA sadA::Mar and ΔbifA pvrS::Mar mutants occurs via overexpression of the adjacent PDE-encoding genes, sadR and pvrR, respectively. These data are consistent with our observation that overexpression of a heterologous PDE (i.e., RapA) also restores swarming to a ΔbifA mutant (Fig. 1B).
Mutations in the pilY1 gene suppress multiple ΔbifA mutant defects.
We isolated three independent pilY1 mutants as suppressors of the ΔbifA swarming defect (Fig. 1D). The pilY1 gene encodes a protein involved in type IV pilus biogenesis and twitching motility, a form of surface-associated motility of P. aeruginosa that is distinct from swarming motility. While studies from our laboratory and others have previously shown that pili are not absolutely required for swarming motility by this bacterium (49, 58), it has been reported that pili participate in the patterning of swarming motility (31).
To better understand the potential role of the pilY1 gene in c-di-GMP-mediated swarming suppression, we further characterized the ability of ΔbifA pilY1::Mar suppressors to affect each of the ΔbifA mutant phenotypes resulting from the accumulation of c-di-GMP. We observed that ΔbifA pilY1::Mar mutants show markedly decreased biofilm formation (Fig. 2A, bottom row, and B) and CR binding (a measure of Pel polysaccharide production) relative to those of the ΔbifA single mutant (Fig. 2A, middle row). Furthermore, we generated an in-frame deletion of the pilY1 gene and observed the same suppression of these ΔbifA mutant phenotypes as we observed for the mariner suppressor mutants (Fig. 2A, fourth column).
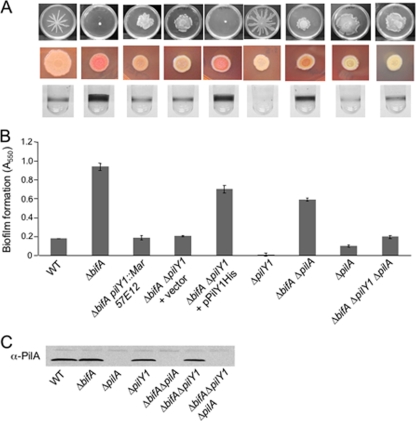
FIG. 2.
Phenotypes of the bifA suppressor mutants. (A) pilY1 mutations suppress multiple defects of the ΔbifA mutant. Top panel, typical swarm images for (left to right) the WT, the ΔbifA mutant, a representative ΔbifA pilY1::Mar mutant (57E12), the ΔbifA ΔpilY1 double mutant carrying either empty vector or the pPilY1His complementing plasmid, and the ΔpilY1 mutant. Also shown are swarm images of the ΔbifA ΔpilA double mutant, the ΔpilA single mutant, and the ΔbifA ΔpilA ΔpilY1 triple mutant. The plates were incubated for 16 h at 37°C. The middle panel shows CR binding for each strain. CR assay plates were incubated for 24 h at 37°C followed by 48 h at room temperature. The bottom panel shows representative wells of a 96-well dish from a biofilm assay for each strain. Strains were grown in M63 with glucose, MgSO4, and CAA for 24 h at 37°C prior to crystal violet staining. (B) Quantification of crystal violet-stained biofilms. Crystal violet was solubilized in 30% glacial acetic acid, and the absorbance was measured at 550 nm. Error bars indicate standard deviations. (C) Western blot analysis showing PilA protein in crude lysates prepared from the indicated strain.
Given that the pilY1 gene maps to a region containing other pilus-related genes and has been shown to be cotranscribed as a unit with members of this locus (fimU-pilVWXY1Y2E) (6) (Fig. 1D), we assessed whether suppression of the ΔbifA mutant defects was due to inactivation of pilY1 alone, or possibly due to polar effects of the ΔpilY1 mutation, by generating a His-tagged version of the pilY1 gene on a multicopy plasmid under the control of the pBAD promoter. Introduction of this construct (pPilY1His) into the ΔbifA ΔpilY1 mutant background restored the hyperbiofilm and hyper-CR binding phenotypes as well as swarming inhibition to the same extent observed for the ΔbifA single mutant (Fig. 2A and B). This and all complementation studies with pPilY1His were performed in the absence of added arabinose and thus rely upon the basal expression of the pBAD promoter to drive PilY1 production. These results indicate that mutation of pilY1 alone is responsible for the observed suppression of the ΔbifA mutant phenotypes.
Finally, we also constructed a ΔpilY1 single mutant strain. This strain is defective for twitching motility (not shown), has reduced CR binding, and shows both increased swarming motility (a 2-fold increase in percent surface coverage of the swarm plate) compared to the WT and an altered swarming pattern. Thus, the ΔpilY1 single mutant does have a small but reproducible positive impact on swarming motility, consistent with a role for PilY1 in repressing swarming motility.
Loss of type IV pili is not sufficient for full suppression of ΔbifA defects.
Both the ΔpilY1 and ΔbifA ΔpilY1 mutants fail to twitch in twitching motility assays, and we can complement these twitching defects using the pilY1 complementation construct described above (data not shown). Given that loss of pilY1 function leads to disruption of type IV pilus biogenesis, it is formally possible that suppression of the ΔbifA mutant defects is due to loss of surface piliation and/or twitching motility. To test this possibility, we introduced a deletion mutation of the pilA gene, which encodes the major pilin subunit of P. aeruginosa, into the ΔbifA mutant background and assessed the suppression of each of the ΔbifA mutant phenotypes in this ΔbifA ΔpilA double mutant compared to the ΔbifA ΔpilY1 double mutant (Fig. 2A and B).
The data show that elimination of pilA function in the ΔbifA mutant background does lead to partial suppression of the swarming, CR binding, and biofilm formation phenotypes. However, the suppression observed for the ΔbifA ΔpilA mutant is not nearly as robust as that observed for the ΔbifA ΔpilY1 double mutant in each of the assays tested. These results indicate that it is not loss of surface pili per se that leads to suppression of the ΔbifA mutant defects but that loss of PilY1 function in particular is responsible for full suppression of the ΔbifA mutant defects.
Although the ΔbifA ΔpilY1 and the ΔbifA ΔpilA double mutants both lack functional surface pili (as determined by the twitching motility assay), they differ in that the ΔbifA ΔpilY1 mutant produces pilin whereas the ΔbifA ΔpilA mutant does not (Fig. 2C). To control for the possibility that a potential buildup of pilin in cells of the ΔbifA ΔpilY1 mutant contributes to suppression of the swarming defect, we constructed a ΔbifA ΔpilY1 ΔpilA triple mutant and found no difference in the swarming, biofilm, and CR binding phenotypes compared with those of the ΔbifA ΔpilY1 mutant (Fig. 2A to C). Together, these data indicate that it is loss of PilY1 function and not loss of pili that suppresses the ΔbifA swarming defect.
Global c-di-GMP pools are not detectably altered by deletion of the pilY1 gene.
Deletion of the pilY1 gene in the ΔbifA mutant background suppresses all of the c-di-GMP-related phenotypes of the ΔbifA mutant, suggesting that such a mutation might lead to a decrease in the cellular pools of c-di-GMP in the ΔbifA mutant. To test this notion, we examined levels of c-di-GMP in ΔbifA and ΔbifA ΔpilY1 mutant cells grown in the presence of radiolabeled inorganic phosphate, followed by nucleotide extraction, analysis by 2D TLC, and autoradiography (22, 38).
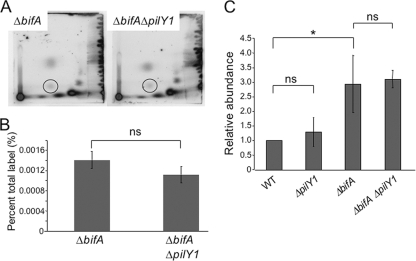
Inspection of the two-dimensional TLC autoradiographs revealed no detectable difference in the levels of c-di-GMP between the two strains (Fig. 3A). Quantification of c-di-GMP and normalization to total 32Pi incorporation (for which there is no significant difference between the two strains [P = 0.55, n = 3]) showed no significant difference between the ΔbifA single mutant and ΔbifA ΔpilY1 double mutant (P > 0.05) (Fig. 3B). These data suggest that the pilY1 mutation has little impact on the global levels of c-di-GMP in cells of the ΔbifA mutant.
FIG. 3.
Quantification of global c-di-GMP pools. (A) Shown are autoradiographs of representative two-dimensional TLC plates used to separate [32P]orthophosphate-labeled, acid-extracted whole-cell extracts prepared from the ΔbifA mutant and the ΔbifA ΔpilY1 double mutant. The circles indicate the position of c-di-GMP. (B) Quantification of c-di-GMP levels determined by analyzing autoradiographs using the Storm 860 and ImageQuant software (v5.1). The graph shows a representative experiment with results for three replicates of each strain plotted as a percentage of the total 32P label incorporated into c-di-GMP. ns, no statistically significant difference between the ΔbifA mutant and the ΔbifA ΔpilY1 double mutant using the criterion of a P value of <0.05. (C) Quantification of c-di-GMP levels as measured using liquid chromatography-tandem mass spectrometry. The graph depicts the relative abundance of c-di-GMP for three replicates. *, statistically significant difference; ns, no significant difference (using the criterion of a P value of <0.05 by the t test). Error bars indicate standard deviations.
To further investigate c-di-GMP cellular pools, we used liquid chromatography-mass spectrometry (LC-MS) to quantify the levels of c-di-GMP in these strains. Strains were grown to OD600 of ≈0.6 prior to acid extraction of nucleotides, followed by LC-MS analysis. Consistent with our previous studies, the ΔbifA mutant accumulates approximately 3-fold more c-di-GMP than the WT (Fig. 3C) (27). Consistent with our observations using the whole-cell labeling method described above, mutation of pilY1 has no detectable impact on c-di-GMP levels in either the WT or the ΔbifA mutant background.
Expression of the pel polysaccharide genes is not altered by deletion of the pilY1 gene.
Mutation of the pilY1 gene in the ΔbifA mutant background suppresses the hyper-CR binding phenotype of the ΔbifA mutant and thus abrogates the increased production of Pel polysaccharide. It is plausible that pilY1 affects Pel polysaccharide production by influencing pel gene expression, and if so, we would predict that pel gene expression is reduced in the ΔbifA ΔpilY1 mutant relative to the ΔbifA single mutant. Thus, we assessed pel gene expression in the WT, ΔbifA mutant, and ΔbifA ΔpilY1 mutant backgrounds. Consistent with our previous studies (27), we observe no statistically significant difference (using the criterion of a P value of <0.05 [n = 3]) in pelA expression between the WT (0.012 ± 0.001 pg input cDNA) and the ΔbifA mutant (0.013 ± 0.004 pg input cDNA). Furthermore, our data show that the pilY1 mutation confers no significant change in pelA gene expression in the ΔbifA ΔpilY1 (0.01 ± 0.002 pg input cDNA) mutant relative to the ΔbifA single mutant. These data indicate that excess Pel polysaccharide production in the ΔbifA mutant is likely not due to a pilY1-mediated increase in pel transcription.
Enhanced expression of pilY1 represses swarming in a pilus-independent manner.
The identification of the pilY1 gene as a suppressor of the ΔbifA swarming defect indicates that PilY1 is important for inhibition of swarming motility in the ΔbifA mutant. Additionally, a pilY1 single mutant is a hyperswarmer relative to the WT (Fig. 2A), suggesting that the pilY1 gene can repress swarming in the WT. Based on these observations, we assessed whether the pilY1 gene could act as a repressor of swarming when expressed from a multicopy plasmid in the WT background.
As shown in Fig. 4A (left panel), arabinose-induced expression of the pPilY1His plasmid was indeed able to repress swarming motility by the WT. Based on the swarm suppression results with the ΔbifA ΔpilA mutant above indicating that the pilus might affect pilY1 function, we next asked whether pilY1-mediated repression of swarming required functional pili. As shown in Fig. 4A (right panel), overexpression of pilY1 in the ΔpilA mutant was still able to repress swarming motility. These results further confirm that the pilY1 gene is able to function as a repressor of swarming motility and indicate that pilin production is not required for this effect.
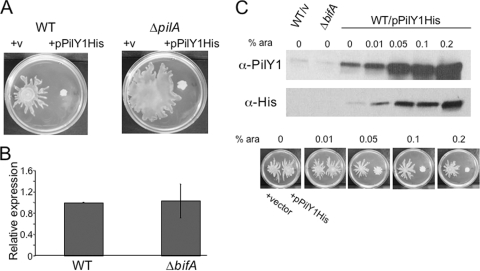
FIG. 4.
Increased expression of PilY1 represses swarming motility. (A) Enhanced expression of pilY1 represses swarming in a pilus-independent manner. Shown are representative images of the WT (left panel) and the ΔpilA mutant (right panel) carrying either empty vector or the pPilY1His arabinose-inducible expression plasmid. Swarm plates contained 0.2% arabinose and were incubated for 16 h at 37°C. (B) qRT-PCR analysis of pilY1 expression in the ΔbifA mutant relative to the WT. Expression data were normalized to the WT, and the results shown are the average from two independent experiments. Error bars indicate standard deviations. (C) Assessment of PilY1 levels and impact on swarming phenotype. Shown in the upper panels are Western blots showing PilY1 protein levels in the WT carrying empty vector, the ΔbifA mutant, and the WT carrying the pPilY1His plasmid. Concentrations of arabinose (wt/vol) added during culturing of each sample are indicated. PilY1 protein was detected using anti-PilY1 antibodies (top panel) or anti-His antibody to detect His-tagged PilY1 (middle panel) expressed from the pPilY1His plasmid. Shown in the lower panel are swarm plates containing arabinose at the indicated concentrations. On the left side of each plate is the WT carrying the empty vector control, and on the right is the WT carrying pPilY1His.
Expression of pilY1 in a ΔbifA mutant.
Our data thus far suggest that PilY1 functions to repress swarming motility. Thus, one possibility is that levels of c-di-GMP modulate pilY1 gene expression and that the high levels of c-di-GMP in a ΔbifA mutant could enhance pilY1 expression, thus providing an explanation for the loss of swarming motility upon loss of BifA function. To address this hypothesis, we examined the levels of pilY1 gene expression in the WT and ΔbifA mutant backgrounds by qRT-PCR. Expression of the pilY1 gene is not different in the ΔbifA mutant and the WT, suggesting that elevated levels of c-di-GMP do not affect pilY1 transcription (Fig. 4B).
To address whether c-di-GMP affects pilY1 expression in a posttranscriptional manner, we examined whether levels of PilY1 protein are altered in the ΔbifA mutant relative to the WT using a polyclonal PilY1 antibody. As shown on a representative Western blot in Fig. 4C (top panel), the levels of PilY1 protein in the WT (lane 1) and the ΔbifA mutant (lane 2) are comparable. Quantification of PilY1 protein levels on a Western blot reveals no significant difference between the WT and ΔbifA mutant backgrounds (P = 0.78, n = 3 replicates each), indicating that c-di-GMP exerts no detectable posttranscriptional effects on PilY1 levels.
To further assess whether the levels of PilY1 in the ΔbifA background would be sufficient to repress swarming, we utilized the pPilY1 arabinose-inducible pMQ80 construct to titrate the levels of PilY1 in the WT and determine the impact on swarming motility. When the pilY1 gene is expressed from the pPilY1His plasmid in the WT strain (WT/pPilY1His) in the absence of arabinose (Fig. 4C, lane 3), we observe a 13-fold increase in PilY1 protein levels relative to PilY1 levels in the ΔbifA mutant (lane 2), as detected using the PilY1 antibody (top panel). Importantly, we do not observe an impact on swarming motility (bottom left panel) under these conditions, indicating that this increase in PilY1 above the levels in the ΔbifA mutant is not sufficient to repress swarming motility of the WT.
For robust repression of WT swarming motility, arabinose was added to a final concentration of 0.2%. Under these conditions, a substantial induction of PilY1 protein expression occurs (Fig. 4C, lane 7) and swarming motility of the WT strain is repressed (bottom right panel). Using intermediate arabinose concentrations of between 0 and 0.2%, we can estimate a minimum level of PilY1 protein that is sufficient to repress swarming in the WT. At 0.01% arabinose, we still do not reliably observe swarming repression at levels of PilY1 that are ∼15-fold above that in the ΔbifA mutant. However, we begin to observe swarming repression with the addition of 0.05% arabinose, which results in PilY1 levels that are ∼30-fold higher than those in the ΔbifA mutant. Together, these data argue that swarming repression in the ΔbifA mutant is not due primarily to increased expression of the PilY1 protein.
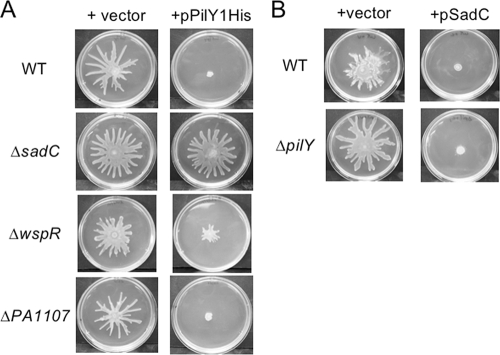
PilY1 functions genetically upstream of the DGC SadC and specifically requires SadC, but not other DGCs, for repression of swarming motility.
Previous work in our laboratory has shown that the SadC protein, a diguanylate cyclase (DGC), functions in a genetic pathway with the BifA protein to modulate levels of c-di-GMP in the cell and thereby influence both biofilm formation and swarming motility (37). As shown in those studies, mutating the sadC gene in the ΔbifA mutant background led to suppression of the ΔbifA mutant swarming motility defect, as well as partial suppression of the hyper-biofilm formation and CR binding phenotypes (37), results that are similar to those observed for the ΔbifA ΔpilY1 double mutant. Furthermore, overexpression of the sadC gene in the WT leads to inhibition of swarming motility as well as stimulation of biofilm formation and CR binding (37). Given the similarities between the effects of the sadC and pilY1 genes on surface-associated behaviors, we wondered whether these two genes might function together in a pathway to regulate biofilm formation and swarming motility. To determine the potential epistatic relationship between the pilY1 and sadC genes, we first asked whether the sadC gene is required for PilY1-mediated repression of swarming. When overexpressed in the ΔsadC mutant, pPilY1His fails to repress swarming, indicating that sadC functions downstream of pilY1 in the repression of swarming (Fig. 5A, second row).
FIG. 5.
PilY1 functions genetically upstream of the SadC diguanylate cyclase. (A) Shown are representative swarms for the WT and the ΔsadC, ΔwspR, and ΔPA1107 DGC mutants carrying either the empty vector (left column) or the pPilY1His plasmid (right column). Plates contained arabinose at a concentration of 0.2%. (B) Swarms of the WT and ΔpilY1 mutant when carrying either the vector (left column) or pSadC expression plasmid (right column). Plates contained arabinose at a concentration of 0.2%.
Given that there are 16 predicted DGCs encoded by the P. aeruginosa PA14 genome, we assessed whether this requirement by pilY1 for a downstream DGC was specific for SadC or whether other DGCs might also fulfill this function. In particular, we tested two additional DGC mutants for their impacts on pilY1-mediated swarming repression. The wspR gene encodes a CheY-like response regulatory component of a chemosensory signal transduction pathway (16). The WspR protein contains a GGDEF domain and was shown to possess DGC activity and to be required for the hyperbiofilm phenotype of a wspF mutant (16, 22). The PA1107 gene also encodes a GGDEF domain-containing protein and was shown previously to possess DGC activity and to stimulate hyperbiofilm formation upon overexpression (29). As shown in Fig. 5A, enhanced expression of pPilY1His is still able to repress swarming in both the ΔwspR and ΔPA1107 mutant backgrounds, indicating a specific interaction between the pilY1 gene and sadC-encoded DGC in the repression of swarming.
To further confirm that the pilY1 gene functions upstream of the sadC gene in a genetic pathway, we performed the reciprocal experiments to those described above. That is, we overexpressed the sadC gene in the ΔpilY1 mutant to determine whether pilY1 function is required for repression of swarming by sadC. Using an arabinose-inducible hemagglutinin (HA)-tagged version of the sadC gene (pSadC), we found that sadC overexpression was still able to repress swarming to the same extent in the ΔpilY1 mutant background as in the WT (Fig. 5B), indicating that pilY1 likely functions upstream of sadC in swarm repression.
Cellular localization of PilY1.
According to the signal peptide prediction program SignalP, the PilY1 protein harbors an N-terminal signal peptide resembling a Sec secretion system signal peptide for transport across the inner membrane. The bacterial localization prediction tool PSORTB predicts that the PilY1 protein localizes to the outer membrane and also is secreted to the extracellular environment. Consistent with these predictions, previous studies using the P. aeruginosa PAK strain showed that PilY1 was found in the insoluble membrane fraction as well as the pilus-associated extracellular shear fraction (3). Results from a phoA fusion screen to identify exported proteins in P. aeruginosa PAO1 indicated that PilY1 is transported across the cytoplasmic membrane (32).
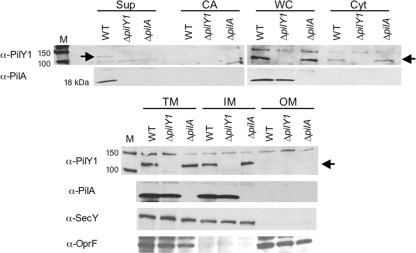
To determine the cellular localization of PilY1 in P. aeruginosa strain PA14, we performed cellular fractionations of WT cells and detected PilY1 using a polyclonal PilY1 antibody. In WT cells, we observe PilY1 in the whole-cell (WC) lysate and in the cytosolic (Cyt) fraction as well as in the total membrane (TM) and inner membrane (IM) fractions (Fig. 6). We do not detect PilY1 in the outer membrane (OM) fraction. Given predictions that PilY1 is a secreted protein, we tested whether PilY1 was present in the supernatant (Sup) fraction and the so-called cell-associated (CA) fraction, which refers to proteins weakly bound to the cell surface (see Materials and Methods for details). We can detect PilY1 in the supernatant after concentrating these samples approximately 300-fold. However, we are unable to detect PilY1 in the CA fractions.
FIG. 6.
PilY1 cellular localization. Cellular fractions of the WT, ΔpilY1 mutant, and ΔpilA mutant were separated by SDS-PAGE. Fractions are indicated as supernatant (Sup), cell-associated (CA), whole-cell (WC), soluble cytoplasmic (Cyt), total membrane (TM), inner membrane (IM), and outer membrane (OM) fractions. The cell-associated fraction refers to proteins weakly associated with the cell surface that can be released by brief vortexing. Western analysis was performed using the following antibodies as indicated: anti-PilY1, anti-SecY, anti-OprF or anti-PilA antibody. SecY (∼50 kDa) served as a control for inner membrane localization, and OprF (∼37 kDa) served as an outer membrane marker. The protein size marker lanes are indicated (M), with sizes in kDa. The arrow indicates the position of the PilY1 protein at ∼110 kDa. The molecular mass of WT PilY1 remains constant across all cellular fractions, as confirmed by SDS-PAGE and Western blotting (not shown).
The integrity of the inner and outer membrane fractions was confirmed by Western blotting using antibodies to SecY, an inner membrane protein (1), and OprF, an outer membrane protein (7, 21, 34).
Based on our findings indicating that loss of pilin production by mutation of pilA partially suppresses the ΔbifA swarming defect, we hypothesized that pilin production and/or pilus formation might affect PilY1 cellular localization. Thus, we tested whether PilY1 localization is altered in a ΔpilA mutant relative to the WT. As shown in Fig. 6 (compare lane 3 to lane 1), we observe that the supernatant fraction of the ΔpilA mutant lacks detectable PilY1 relative to the WT. The remaining cellular fractions contain levels of PilY1 that are comparable in the ΔpilA mutant and the WT. These data suggest that loss of pilin (and/or pilus formation) affects localization of PilY1 to the supernatant fraction. Furthermore, it is plausible that the absence of PilY1 from the supernatant fraction of the ΔpilA mutant contributes to the partial suppression of swarming observed in the ΔbifA ΔpilA double mutant. However, we cannot exclude the possibility that loss of surface pili alone contributes to the partial swarming restoration observed for the ΔbifA ΔpilA double mutant.
Structure/function analysis of PilY1.
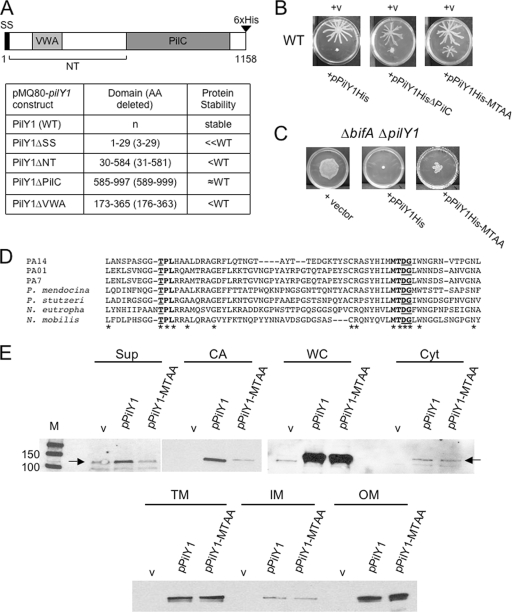
To better understand the potential mechanism by which PilY1 represses swarming motility, we undertook a structure/function analysis of the protein. A schematic diagram of the predicted PilY1 structure is shown in Fig. 7A. At the N terminus, the PilY1 protein has a Sec secretion signal for transport across the inner membrane. Besides other PilY1 homologs from various species, the remainder of the N-terminal region bears little resemblance to other proteins and lacks any obvious protein motifs. However, a 200-amino-acid region within the N terminus shows weak similarity to the von Willebrand factor A (VWA) domain. Most VWA domains that have been well characterized to date belong to extracellular eukaryotic proteins, including cell adhesion and extracellular matrix (ECM) proteins, and are involved in protein-protein interactions (14, 15, 59). However, many intracellular eukaryotic proteins with a diverse array of functions, including transcription, DNA repair, and proteolysis, also contain VWA domains (62). Recently, VWA domains in prokaryotic proteins have also been identified (45), although the precise role of this domain in these proteins has not yet been established, with the exception of a recent study showing a role for the VWA domain of the Streptococcus agalactiae PilA protein, a pilus-associated adhesin, in adherence of this organism to epithelial cells (26).
FIG. 7.
Structure/function analysis of PilY1. (A) A schematic diagram of the PilY1 protein. Numbers indicate amino acids, and predicted regions of PilY1 are denoted as follows: signal sequence (SS), N terminus (NT), von Willebrand factor A (VWA), and the domain with similarity to PilC of Neisseria species (PilC). The table lists the pilY1 deletion constructs made in the pMQ80 vector (left column), the amino acids deleted from PilY1 for each construct (middle column), and the protein stability (right column) for each of the expressed proteins relative to the WT protein as assessed by Western blot analysis using the anti-His antibody. WT PilY1 and all PilY1 variants are C-terminally His tagged as indicated in the diagram. (B) Representative images of swarms for the WT carrying either the vector (v), pPilY1His (left), pPilY1HisΔPilC (middle), or the pPilY1His-MTAA mutant construct (right). Plates contained 0.2% arabinose and were incubated for 16 h at 37°C. (C) Partial complementation of the ΔbifA ΔpilY1 mutant by pPilY1His-MTAA. Shown are images of swarms for the ΔbifA ΔpilY1 mutant carrying the vector (left), pPilY1His (middle), or pPilY1His-MTAA (right). Plates contained 0.2% arabinose and were incubated 16 h at 37°C. (D) CLUSTALW-generated amino acid sequence alignment of the PilY1 protein from representative strains listed on the left: P. aeruginosa strains PA14, PAO1, and PA7; Pseudomonas mendocina, Pseudomonas stutzeri, Nitrosomonas eutropha, and Nitrococcus mobilis. Asterisks indicate identical amino acids conserved across the PilY1 proteins shown in the alignment. Bold indicates conserved regions (“TPL” and “MTDG”), with underlined amino acids mutated to alanine residues. (E) Western blot analysis showing cellular localization of PilY1 in the WT carrying either the vector, pPilY1, or pPilY1-MTAA. Cellular fractions are as follows: supernatant (Sup), cell-associated (CA), whole-cell (WC), soluble cytoplasmic (Cyt), total membrane (TM), inner membrane (IM), and outer membrane (OM) fractions. Fractions were separated by SDS-PAGE, and Western blots were probed with the anti-PilY1 polyclonal antibody. The protein size marker (M) is indicated on the top left panel with sizes in kDa. Note that the PilY1 protein expressed from its native promoter can be detected in the TM and IM fractions (Fig. 6) but is not observed in those fractions in this figure due to the shorter exposure times used to detect the PilY1His protein expressed from the arabinose-inducible plasmid. Furthermore, endogenously expressed PilY1 was not detected in the OM fraction (Fig. 6) but can be detected when PilY1 is overexpressed from this plasmid.
The most obvious domain of PilY1 lies within the C-terminal half of this protein, which shares similarity (24% identity and 46% similarity) with the C terminus of the PilC proteins of pathogenic Neisseria species. PilC of Neisseria has been shown to associate with the bacterial cell surface, including localization to the tip of the pilus, and is required for bacterial adherence to epithelial cells (39, 46, 52, 53). A recent crystal structure-based analysis of the PilC-like domain of PilY1 has shown this domain to be essential for regulating pilus extension and retraction mechanisms involved in twitching motility in a calcium-dependent manner (41).
To test the functions of these various PilY1 domains in swarming inhibition as well as twitching motility, we generated deletion constructs of the pilY1 gene (Fig. 7A) and expressed the His-tagged PilY1 variants from each of these constructs in the WT background. However, all of the deletion constructs except PilY1ΔPilC produced proteins that appeared to be unstable relative to full-length PilY1His protein as assessed by Western blotting (not shown), making it difficult to draw any firm conclusions regarding the functions of these domains.
Thus, we proceeded to characterize the PilY1ΔPilC construct. The results show that the PilY1ΔPilC protein is able to inhibit swarming motility as well as the full-length PilY1 protein, indicating that the PilC-like region of the PilY1 protein is dispensable for swarming inhibition (Fig. 7B). Expression of the PilY1ΔPilC protein in the WT background has no impact on twitching motility. In complementation assays, expression of the PilY1ΔPilC protein is unable to complement the twitching defect of the ΔpilY1 mutant (not shown), a result consistent with recent findings regarding the essential role of this domain in twitching motility (41).
Given that the N-terminal half of the PilY1 protein is sufficient to inhibit swarming motility, we focused on this region for further mutagenesis experiments. We first undertook a random PCR mutagenesis approach by mutagenizing the PilY1ΔPilC construct and screening for loss of swarming inhibition when expressed in the WT background. We screened approximately 1,000 strains expressing mutant variants of PilY1, and about 30% of those mutants recovered swarming motility. However, upon closer investigation using Western dot blot analysis, we found that only 3% of these swarmer strains produced a detectable PilY1ΔPilC protein, indicating that the vast majority of the mutations were likely causing PilY1 protein instability. Furthermore, when those constructs making apparently stable PilY1ΔPilC protein were retransformed into WT P. aeruginosa, the swarming phenotypes were not reproducible, indicating that the mutation in the original mutant strains was not linked to the plasmid. Thus, we chose to take a more directed mutagenesis approach.
Alignment of the PilY1 protein across pseudomonads as well as other bacterial species using the ClustalW2 sequence alignment tool indicated there were a number of highly conserved amino acid residues throughout the N terminus of the protein, and we used this alignment as a guide to generate site-directed mutations. A subset of the highly conserved amino acids found in the alignment fall within the putative VWA-like region of PilY1. The alignment of PilY1 proteins spanning two of the conserved sites within the VWA-like region is shown in Fig. 7D.
To determine whether either of these conserved regions is important for PilY1 function in swarming repression, we changed the threonine in the conserved “TPL site” to an alanine in one construct and changed the aspartic acid and glycine in the conserved “MTDG site” to alanine residues in a second construct. We then overexpressed these His-tagged mutant PilY1 proteins (PilY1-APL and PilY1-MTAA) in the WT background and assessed swarming motility. As shown in Fig. 7B (right panel), expression of the PilY1-MTAA mutant protein led to only partial swarming inhibition relative to the complete inhibition observed for the WT protein (left panel), whereas the PilY1-APL mutant protein behaved like the WT in regard to swarming inhibition (data not shown). The stability of the PilY1-MTAA protein was comparable to that of the WT PilY1 protein as assessed by Western blotting (Fig. 7E). These data indicate a potential role for the conserved aspartic acid and glycine amino acids in swarming inhibition by PilY1. Furthermore, these data are consistent with our observations showing that the PilY1-MTAA construct only partially complements the swarming phenotype of the ΔbifA ΔpilY1 double mutant (Fig. 7C). Similarly, the PilY1-MTAA construct only partially complements the twitching phenotype of the ΔpilY1 mutant (not shown).
Our observations above indicate that the PilY1-MTAA mutant protein shows only partial function in swarming inhibition. To further investigate this, we determined whether the cellular localization of PilY1-MTAA differs from that of WT PilY1 when overexpressed in the WT background. When expression is induced with arabinose, we observe elevated levels of WT PilY1 in all of the cellular fractions relative to the WT carrying vector alone (Fig. 7E). In contrast, the levels of PilY1-MTAA in the Sup and CA fractions are greatly reduced relative to those fractions containing WT PilY1. In all other cellular fractions, the levels of PilY1-MTAA are comparable to those of the WT PilY1. These data indicate that localization of the PilY1-MTAA mutant protein is altered in both the Sup and CA fractions and suggest that the mutated amino acids may affect proper extracellular localization of the PilY1-MTAA protein. Taken together with the results showing an absence of PilY1 in the ΔpilA mutant Sup fraction relative to the WT, these data support the notion that extracellular localization of PilY1 may contribute to its function in swarming inhibition.
DISCUSSION
The mechanism(s) by which the intracellular signaling molecule c-di-GMP negatively affects swarming motility of P. aeruginosa is not well understood. In this study, we chose to address this question using a genetic screen to identify suppressors of a mutant that is defective for swarming motility, a defect that we show is most likely due to excess accumulation of c-di-GMP (Fig. 1). In previous work, we reported that the bifA gene encodes a c-di-GMP-degrading phosphodiesterase and that the ΔbifA mutant accumulates elevated levels of c-di-GMP relative to the WT, forms hyperbiofilms, and is completely defective for swarming motility (27). Here we demonstrate that the swarming defect of the ΔbifA mutant can be rescued by heterologous expression of the RapA c-di-GMP phosphodiesterase from P. fluorescens, thereby supporting the hypothesis that elevated c-di-GMP levels repress swarming by P. aeruginosa and demonstrating a proof of concept for the ΔbifA suppressor screen to identify factors that participate in c-di-GMP-mediated swarming repression.
As further validation for c-di-GMP-mediated repression of swarming and the utility of our screen, we isolated several ΔbifA suppressor strains with transposon mutations that lead to the overexpression of two different genes predicted to encode c-di-GMP PDEs, namely, SadR and PvrR, which have both been shown to be associated with c-di-GMP-degrading activity (29, 36, 48). From these results, we surmise that increased expression of these genes leads to an increase in PDE activity in the ΔbifA mutant, and presumably decreased c-di-GMP levels, with subsequent restoration of swarming motility.
Using this suppressor screen, we also found that pilY1 mutants suppressed the loss of swarming motility of the ΔbifA mutant, and we present data showing that the PilY1 protein, a component of the pilus biogenesis machinery, is able to repress swarming motility. That is, in addition to suppressing the swarming impairment of the ΔbifA mutant, a ΔpilY1 single mutant is a hyperswarmer relative to the WT (Fig. 2). Furthermore, overexpression of PilY1 in the WT background represses swarming motility (Fig. 4).
When interpreting data involving overexpression of a gene, caution is warranted, given that the elevated levels of PilY1 observed in our studies might not be physiologically relevant and may give rise to misleading secondary effects. However, our observations that PilY1 overexpression requires the DGC SadC to exert its effects on swarming, but not other DGCs with known roles in regulating surface behaviors, argue for a level of specificity that one would not expect if PilY1 overexpression led to nonspecific effects on the cell. PilY1 overexpression has no impact on twitching motility, which also argues against a broad cellular impact caused by PilY1 overexpression. Given these data, we postulate that overexpression of PilY1 allows for activation of this specific SadC-dependent signaling pathway in the absence of a putative input signal, and as such, it is a useful tool to determine what additional factors might be required for PilY1-mediated swarming repression. One set of candidate factors includes the minor pilins encoded at the pilY1 locus itself, namely, the fimU, pilV, pilW, pilX, pilY2, and pilE products. The minor pilins are required for pilus biogenesis, but little is known about their precise roles in pilus assembly and function (3, 4, 5, 54, 63). Given that pilY1 and the minor pilin genes are cotranscribed as an operon (6), it is tempting to speculate that the minor pilins not only are involved in pilus assembly/function but also play a role, together with PilY1, in swarming repression.
Recent studies provide evidence that pilY1 gene expression may be differentially regulated under different environmental conditions. The pilY1 operon has been shown to be under the control of the AlgR transcriptional regulator (6, 33), and that AlgR-dependent regulation of this operon is growth phase specific (33). Interestingly, expression of a pilY1 luciferase gene fusion was found to be induced in the presence of subinhibitory concentrations of aminoglycoside antibiotics (35). However, our observations indicate that pilY1 expression is not altered at least under conditions of elevated c-di-GMP in the ΔbifA mutant. Furthermore, our data examining the cellular localization of PilY1 suggest that it is not necessarily the total abundance but rather the localization of PilY1 to a particular subcellular fraction that may be important for its role in swarming repression. More specifically, mutations that decrease the extracellular localization of PilY1 correlate with a decrease in repression of swarming motility. For example, mutation of the pilA gene, encoding the structural subunit of the pilus, leads to a reduction in the supernatant levels of PilY1 relative to those in the WT, which correlates with a partial suppression of the ΔbifA swarming impairment. However, functional pili are not absolutely required for PilY1-mediated repression of swarming. Furthermore, a stable PilY1 mutant protein containing alanine substitutions of two amino acids in a conserved region of the putative VWA domain (PilY1-MTAA) shows decreased levels in both the cell-associated and supernatant fractions relative to WT PilY1, and this mutant protein is unable to fully complement the ΔbifA ΔpilY1 double mutant for its swarming phenotype, nor is it able to repress swarming as well as WT PilY1 when overexpressed in the WT background. Together, these data suggest that extracellular secretion and proper localization of PilY1 may be important for swarming repression.
Mutation of the pilY1 gene in the ΔbifA background not only suppresses the swarming defect but also eliminates both hyper-biofilm formation and hyper-CR binding, thus affecting all of the c-di-GMP-sensitive phenotypes of the ΔbifA mutant. Based on these data, we predicted that cellular c-di-GMP levels in the ΔbifA ΔpilY1 double mutant would be reduced relative to those in the ΔbifA single mutant. However, using two different methods to measure in vivo c-di-GMP levels, we found no difference in the pools of c-di-GMP between these two strains. It is formally possible that PilY1 exerts its effects via a c-di-GMP-independent mechanism, although the experiments with SadC argue against such a model. An alternative possibility to explain our results is the notion that PilY1 does not affect the overall global levels of c-di-GMP but rather exerts a more localized or specific effect on this signaling molecule and that changes in such a local pool of c-di-GMP would be difficult to detect when measuring global pools of this signal using current methods. Indeed, our data do suggest that PilY1 might affect the activity of SadC but not other DGCs, and as such they are consistent with the possibility of SadC-mediated localized alterations in the c-di-GMP level.
This notion of “local action” has been postulated to explain how c-di-GMP can specifically modulate a particular cellular process, given that the genomes of many bacterial species encode a large number of both DGCs and PDEs (24). That is, rather than a common global pool of signaling molecule regulating cellular behaviors, a local pool would be generated, controlled, and/or responded to by a subset of spatially localized enzymes and effectors to influence a cellular process. This model seems particularly compelling for Caulobacter crescentus, for example, where the DGC activity of PleD is spatially restricted to the cell pole where it controls the swarmer-to-stalked cell transition during cell differentiation (2, 44, 60), but it may represent a more general theme of c-di-GMP signaling in many bacterial species (24). In the case of PilY1, it may be that the specificity we observe between PilY1 and the SadC DGC enables local control of c-di-GMP levels to specifically influence swarming motility of P. aeruginosa.
Acknowledgments
We thank R. Kolter for providing the ΔpelA mutant strain and C. Harwood for the pEX19Gm-wspR deletion plasmid. We also thank M. Wolfgang for the generous gift of PilY1 polyclonal antibody.
This work was supported by NIH training grant T32 GM08704 (predoctoral fellowship) to J.H.M., NIH training grant T32 AI07363 (predoctoral fellowship) to A.E.B., NIH grant R01AI083256 to G.A.O, and National Science Foundation CAREER Award MCB-0643859 and National Institute of General Medical Sciences Center for Quantitative Biology/National Institutes of Health grant P50 GM-071508 to J.D.R.
Footnotes
Published ahead of print on 16 March 2010.
REFERENCES
- 1.Akiyama, Y., and K. Ito. 1985. The SecY membrane component of the bacterial protein export machinery: analysis by new electrophoretic methods for integral membrane proteins. EMBO J. 4:3351-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldridge, P., R. Paul, P. Goymer, P. Rainey, and U. Jenal. 2003. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol. Microbiol. 47:1695-1708. [DOI] [PubMed] [Google Scholar]
- 3.Alm, R. A., J. P. Hallinan, A. A. Watson, and J. S. Mattick. 1996. Fimbrial biogenesis genes of Pseudomonas aeruginosa: pilW and pilX increase the similarity of type 4 fimbriae to the GSP protein-secretion systems and pilY1 encodes a gonococcal PilC homologue. Mol. Microbiol. 22:161-173. [DOI] [PubMed] [Google Scholar]
- 4.Alm, R. A., and J. S. Mattick. 1995. Identification of a gene, pilV, required for type 4 fimbrial biogenesis in Pseudomonas aeruginosa, whose product possesses a pre-pilin-like leader sequence. Mol. Microbiol. 16:485-496. [DOI] [PubMed] [Google Scholar]
- 5.Alm, R. A., and J. S. Mattick. 1996. Identification of two genes with prepilin-like leader sequences involved in type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J. Bacteriol. 178:3809-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belete, B., H. Lu, and D. J. Wozniak. 2008. Pseudomonas aeruginosa AlgR regulates type IV pilus biosynthesis by activating transcription of the fimU-pilVWXY1Y2E operon. J. Bacteriol. 190:2023-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benz, R., and R. E. Hancock. 1981. Properties of the large ion-permeable pores formed from protein F of Pseudomonas aeruginosa in lipid bilayer membranes. Biochim. Biophys. Acta 646:298-308. [DOI] [PubMed] [Google Scholar]
- 8.Bobrov, A. G., O. Kirillina, and R. D. Perry. 2005. The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol. Lett. 247:123-130. [DOI] [PubMed] [Google Scholar]
- 9.Boles, B. R., and L. L. McCarter. 2002. Vibrio parahaemolyticus scrABC, a novel operon affecting swarming and capsular polysaccharide regulation. J. Bacteriol. 184:5946-5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cadwell, R. C., and G. F. Joyce. 1994. Mutagenic PCR. PCR Methods Appl. 3:S136-S140. [DOI] [PubMed] [Google Scholar]
- 11.Caetano-Anolles, G. 1993. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 3:85-92. [DOI] [PubMed] [Google Scholar]
- 12.Caiazza, N. C., J. H. Merritt, K. M. Brothers, and G. A. O'Toole. 2007. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189:3603-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caiazza, N. C., and G. A. O'Toole. 2004. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J. Bacteriol. 186:4476-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colombatti, A., and P. Bonaldo. 1991. The superfamily of proteins with von Willebrand factor type A-like domains: one theme common to components of extracellular matrix, hemostasis, cellular adhesion, and defense mechanisms. Blood 77:2305-2315. [PubMed] [Google Scholar]
- 15.Colombatti, A., P. Bonaldo, and R. Doliana. 1993. Type A modules: interacting domains found in several non-fibrillar collagens and in other extracellular matrix proteins. Matrix 13:297-306. [DOI] [PubMed] [Google Scholar]
- 16.D'Argenio, D. A., M. W. Calfee, P. B. Rainey, and E. C. Pesci. 2002. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol. 184:6481-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Argenio, D. A., and S. I. Miller. 2004. Cyclic di-GMP as a bacterial second messenger. Microbiology 150:2497-2502. [DOI] [PubMed] [Google Scholar]
- 18.Drenkard, E., and F. M. Ausubel. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740-743. [DOI] [PubMed] [Google Scholar]
- 19.Friedman, L., and R. Kolter. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675-690. [DOI] [PubMed] [Google Scholar]
- 20.Friedman, L., and R. Kolter. 2004. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hancock, R. E., G. M. Decad, and H. Nikaido. 1979. Identification of the protein producing transmembrane diffusion pores in the outer membrane of Pseudomonas aeruginosa PA01. Biochim. Biophys. Acta 554:323-331. [DOI] [PubMed] [Google Scholar]
- 22.Hickman, J. W., D. F. Tifrea, and C. S. Harwood. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. U. S. A. 102:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinsa, S. M., and G. A. O'Toole. 2006. Biofilm formation by Pseudomonas fluorescens WCS365: a role for LapD. Microbiology 152:1375-1383. [DOI] [PubMed] [Google Scholar]
- 24.Jenal, U. 2004. Cyclic di-guanosine-monophosphate comes of age: a novel secondary messenger involved in modulating cell surface structures in bacteria? Curr. Opin. Microbiol. 7:185-191. [DOI] [PubMed] [Google Scholar]
- 25.Kohler, T., L. K. Curty, F. Barja, C. van Delden, and J. C. Pechere. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 182:5990-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konto-Ghiorghi, Y., E. Mairey, A. Mallet, G. Dumenil, E. Caliot, P. Trieu-Cuot, and S. Dramsi. 2009. Dual role for pilus in adherence to epithelial cells and biofilm formation in Streptococcus agalactiae. PLoS Pathog. 5:e1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuchma, S. L., K. M. Brothers, J. H. Merritt, N. T. Liberati, F. M. Ausubel, and G. A. O'Toole. 2007. BifA, a cyclic-di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189:8165-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuchma, S. L., J. P. Connolly, and G. A. O'Toole. 2005. A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 187:1441-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulasekara, H., V. Lee, A. Brencic, N. Liberati, J. Urbach, S. Miyata, D. G. Lee, A. N. Neely, M. Hyodo, Y. Hayakawa, F. M. Ausubel, and S. Lory. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. U. S. A. 103:2839-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulasekara, H. D., I. Ventre, B. R. Kulasekara, A. Lazdunski, A. Filloux, and S. Lory. 2005. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol. 55:368-380. [DOI] [PubMed] [Google Scholar]
- 31.Leech, A. J., and J. S. Mattick. 2006. Effect of site-specific mutations in different phosphotransfer domains of the chemosensory protein ChpA on Pseudomonas aeruginosa motility. J. Bacteriol. 188:8479-8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewenza, S., J. L. Gardy, F. S. Brinkman, and R. E. Hancock. 2005. Genome-wide identification of Pseudomonas aeruginosa exported proteins using a consensus computational strategy combined with a laboratory-based PhoA fusion screen. Genome Res. 15:321-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lizewski, S. E., J. R. Schurr, D. W. Jackson, A. Frisk, A. J. Carterson, and M. J. Schurr. 2004. Identification of AlgR-regulated genes in Pseudomonas aeruginosa by use of microarray analysis. J. Bacteriol. 186:5672-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacEachran, D. P., S. Ye, J. Bomberger, D. A. Hogan, B. A. Stanton, and G. A. O'Toole. 2007. The Pseudomonas aeruginosa secreted protein, PA2934, decreases apical membrane expression of the cystic fibrosis transmembrane conductance regulator. Infect. Immun. 75:3902-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marr, A. K., J. Overhage, M. Bains, and R. E. Hancock. 2007. The Lon protease of Pseudomonas aeruginosa is induced by aminoglycosides and is involved in biofilm formation and motility. Microbiology 153:474-482. [DOI] [PubMed] [Google Scholar]
- 36.Meissner, A., V. Wild, R. Simm, M. Rohde, C. Erck, F. Bredenbruch, M. Morr, U. Romling, and S. Haussler. 2007. Pseudomonas aeruginosa cupA-encoded fimbriae expression is regulated by a GGDEF and EAL domain-dependent modulation of the intracellular level of cyclic diguanylate. Environ. Microbiol. 9:2475-2485. [DOI] [PubMed] [Google Scholar]
- 37.Merritt, J. H., K. M. Brothers, S. L. Kuchma, and G. A. O'Toole. 2007. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J. Bacteriol. 189:8154-8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monds, R. D., P. D. Newell, R. H. Gross, and G. A. O'Toole. 2007. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol. Microbiol. 63:656-679. [DOI] [PubMed] [Google Scholar]
- 39.Nassif, X., J.-L. Beretti, J. Lowy, P. Stenberg, P. O'Gaora, P. Pfeifer, S. Normark, and M. So. 1994. Roles of pilin and PilC adhesion of Neisseria meningitidis to human epithelial cells. Proc. Natl. Acad. Sci. U. S. A. 91:3769-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nunn, D., and S. Lory. 1993. Cleavage, methylation, and localization of the Pseudomonas aeruginosa export proteins XcpT, -U, -V, and -W. J. Bacteriol. 175:4375-4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orans, J., M. D. Johnson, K. A. Coggan, J. R. Sperlazza, R. W. Heiniger, M. C. Wolfgang, and M. R. Redinbo. 2010. Crystal structure analysis reveals Pseudomonas PilY1 as an essential calcium-dependent regulator of bacterial surface motility. Proc. Natl. Acad. Sci. U. S. A. 107:1065-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 43.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 44.Paul, R., S. Weiser, N. C. Amiot, C. Chan, T. Schirmer, B. Giese, and U. Jenal. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18:715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ponting, C. P., L. Aravind, J. Schultz, P. Bork, and E. V. Koonin. 1999. Eukaryotic signalling domain homologues in archaea and bacteria. Ancient ancestry and horizontal gene transfer. J. Mol. Biol. 289:729-745. [DOI] [PubMed] [Google Scholar]
- 46.Rahman, M., H. Kallstrom, S. Normark, and A.-B. Jonsson. 1997. PilC of pathogenic Neisseria is associated with the bacterial cell surface. Mol. Microbiol. 25:11-25. [DOI] [PubMed] [Google Scholar]
- 47.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 48.Rao, F., Y. Yang, Y. Qi, and Z. X. Liang. 2008. Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J. Bacteriol. 190:3622-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rashid, M. H., and A. Kornberg. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 97:4885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Römling, U., M. Gomelsky, and M. Y. Galperin. 2005. c-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol. 57:629-639. [DOI] [PubMed] [Google Scholar]
- 51.Ross, P., H. Weinhouse, Y. Aloni, D. Michaeli, P. Weinberger-Ohana, R. Mayer, S. Braun, E. de Vroom, G. A. van der Marel, J. H. van boom, and M. Benziman. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279-281. [DOI] [PubMed] [Google Scholar]
- 52.Rudel, T., I. Schleuerflug, and T. Meyer. 1995. Neisseria PilC protein identified as type-4 pilus tip-located adhesin. Nature 373:357-359. [DOI] [PubMed] [Google Scholar]
- 53.Rudel, T., J. P. M. van Putten, C. P. Gibbs, R. Haas, and T. F. Meyer. 1992. Interaction of two variable proteins (PilE and PilC) required for pilus-mediated adherence of Neisseria gonorrhoeae to human epithelial cells. Mol. Microbiol. 6:3439-3450. [DOI] [PubMed] [Google Scholar]
- 54.Russell, M. A., and A. Darzins. 1994. The pilE gene product of Pseudomonas aeruginosa, required for pilus biogenesis, shares amino acid sequence identity with the N-termini of type 4 prepilin proteins. Mol. Microbiol. 13:973-985. [DOI] [PubMed] [Google Scholar]
- 55.Shanks, R. M., N. C. Caiazza, S. M. Hinsa, C. M. Toutain, and G. A. O'Toole. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl. Environ. Microbiol. 72:5027-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simm, R., M. Morr, A. Kader, M. Nimtz, and U. Romling. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53:1123-1134. [DOI] [PubMed] [Google Scholar]
- 57.Simon, R., U. Priefer, and Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology 1:784-789. [Google Scholar]
- 58.Toutain, C. M., M. E. Zegans, and G. A. O'Toole. 2005. Evidence for two flagellar stators and their role in the motility of Pseudomonas aeruginosa. J. Bacteriol. 187:771-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tuckwell, D. 1999. Evolution of von Willebrand factor A (VWA) domains. Biochem. Soc. Trans. 27:835-840. [DOI] [PubMed] [Google Scholar]
- 60.Wheeler, R. T., and L. Shapiro. 1999. Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol. Cell 4:683-694. [DOI] [PubMed] [Google Scholar]
- 61.Whitchurch, C. B., M. Hobbs, S. P. Livingston, V. Krishnapillai, and J. S. Mattick. 1991. Characterization of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialized protein export system widespread in eubacteria. Gene 101:33-44. [DOI] [PubMed] [Google Scholar]
- 62.Whittaker, C. A., and R. O. Hynes. 2002. Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol. Biol. Cell 13:3369-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winther-Larsen, H. C., M. Wolfgang, S. Dunham, J. P. van Putten, D. Dorward, C. Lovold, F. E. Aas, and M. Koomey. 2005. A conserved set of pilin-like molecules controls type IV pilus dynamics and organelle-associated functions in Neisseria gonorrhoeae. Mol. Microbiol. 56:903-917. [DOI] [PubMed] [Google Scholar]