Abstract
The extracytoplasmic function sigma factor AlgU of Pseudomonas aeruginosa is responsible for alginate overproduction, leading to mucoidy and chronic infections of cystic fibrosis patients. We investigated here the role of AlgU in the formation of nonmucoid biofilms. The algU mutant of P. aeruginosa PAO1 (PAOU) showed a dramatic impairment in biofilm formation under dynamic conditions. PAOU was defective both in cell attachment to glass and in development of robust, shear-resistant biofilms. This was explained by an impaired production of extracellular matrix, specifically of the exopolysaccharide Psl, as revealed by microscopy and enzyme-linked immunosorbent assay. Complementing the algU mutation with a plasmid-borne algU gene restored wild-type phenotypes. Compared with that in PAO1, expression of the psl operon was reduced in the PAOU strain, and the biofilm formation ability of this strain was partially restored by inducing the transcription of the psl operon. Furthermore, expression of the lectin-encoding lecA and lecB genes was reduced in the PAOU strain. In agreement with the requirement of LecB for type IV pilus biogenesis, PAOU displayed impaired twitching motility. Collectively, these genetic downregulation events explain the biofilm formation defect of the PAOU mutant. Promoter mapping indicated that AlgU is probably not directly responsible for transcription of the psl operon and the lec genes, but AlgU is involved in the expression of the ppyR gene, whose product was reported to positively control psl expression. Expressing the ppyR gene in PAOU partially restored the formation of robust biofilms.
Pseudomonas aeruginosa is a Gram-negative bacterium found in various environments, especially in soil, and is an opportunistic pathogen, responsible in particular for chronic lung infections of cystic fibrosis (CF) patients (21). P. aeruginosa frequently adopts a biofilm lifestyle, both in the environment and in the course of pathogenesis (15, 23). Biofilms are microbial communities of cells that are attached to a substratum, an interface, or each other and are embedded in a matrix of extracellular polymeric substances that they have produced (15). Maintenance of long-term infections in the CF lung is associated with the conversion of P. aeruginosa to a mucoid phenotype, which results from overproduction of the exopolysaccharide alginate (21, 48). Alginate overproduction leads to highly structured biofilms and is responsible for some biofilm properties, such as increased resistance to the antibiotic tobramycin (24). In contrast, alginate is not a major component of the extracellular matrix when biofilms are formed by nonmucoid P. aeruginosa strains, such as PAO1 and PA14 (69). Consistently, alginate production is not critical for biofilm formation by these strains (17, 57). Biofilm formation by nonmucoid P. aeruginosa strains is of primary importance for pathogenesis, since (i) this leads to seeding dispersal of large numbers of cells that might subsequently initiate an infection, (ii) virulent phenotypes might survive and expand within biofilms, and (iii) high cell densities reached in biofilms might activate the quorum-sensing (QS) network which controls the production of virulence factors (23). Furthermore, CF patients are initially colonized by motile, nonmucoid P. aeruginosa strains, which attach to mucin-covered epithelial cells and likely develop as a biofilm before switching to a mucoid phenotype (48).
The extracytoplasmic function (ECF) sigma factor AlgU (also known as AlgT, RpoE, σE, and σ22) is responsible for transcription of the alginate biosynthetic operon (45). Conversion to mucoidy is due in most cases to spontaneous mutations in mucA, leading to hyperactivity of AlgU, which in turn results in alginate operon overexpression (21, 48). AlgU therefore plays a key role in the formation of mucoid P. aeruginosa biofilms. AlgU is also involved in other traits, such as resistance to various stresses (oxidative, heat, hyperosmotic, and cell wall stresses) and control of some virulence factor synthesis (2, 16, 45, 67, 68). Surprisingly, few studies have addressed the role of AlgU in nonmucoid P. aeruginosa, even though the sigma factor is expressed in these cells (50, 67). Here we examined the potential role of AlgU in the formation of nonmucoid P. aeruginosa biofilms. We observed that AlgU is critical for the formation of robust biofilms, and we identified genetic and biochemical defects related to biofilm formation and virulence in the algU mutant.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The strains and plasmids used in this study are listed in Table 1. Escherichia coli cells were grown in Luria-Bertani (LB) broth at 37°C. P. aeruginosa cells were grown in liquid at 37°C with shaking, using LB or PPGAS medium (20 mM NH4Cl, 20 mM KCl, 120 mM Tris-HCl, 1.6 mM MgSO4, 0.5% glucose, 1% peptone [65]). The solid media were LB agar (15 g liter−1) and Pseudomonas isolation agar (PIA), in which the antimicrobial agent Irgasan inhibits bacteria other than Pseudomonas spp. (Gibco-BRL, Grand Island, NY). When necessary, the following antibiotics were used at the indicated concentrations (μg ml−1): carbenicillin (Cb), 600; gentamicin (Gm), 50 and 400, in liquid and solid media, respectively; tetracycline (Tc), 150; kanamycin (Kan), 500; and chloramphenicol (Cm), 200 for P. aeruginosa. Ampicillin (Ap), Gm, and Cm were used at 100, 15, and 30 μg ml−1 for E. coli.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| Strains | ||
| P. aeruginosa strains | ||
| PAO1 | Wild type | 28 |
| PAOU | PAO1 ΔalgU::lox | This study |
| PAOU PBAD::psl | PAOU araC PBAD::psl | This study |
| E. coli strains | ||
| Top10 | Electrocompetent cells used as cloning host | Invitrogen, Paisley, United Kingdom |
| S17-1 | recA pro (RP4-2Tet::Mu Kan::Tn7), donor and helper strain for conjugation | 53 |
| Plasmids | ||
| pCR2.1-TOPO | E. coli cloning vector for PCR products; Apr | Invitrogen, Paisley, UK |
| pSMC21 | gfp expression plasmid; Kanr | 6, 11 |
| pEX100Tlink | Suicide vector; Apr | 47 |
| pUCGmlox | Source of lox-aacC1-lox cassette; Apr Gmr | 47 |
| pCM157 | cre expression vector; Tcr | 37 |
| pEXalgU | pEX100Tlink with disrupted algU; Apr | This study |
| pEXUGL | pEXalgU with lox-aacC1-lox cassette; Apr Gmr | This study |
| pBBR1MCS-5 | E. coli-P. aeruginosa shuttle vector; Gmr | 33 |
| pBAUC | pBBR1MCS-5 with a 911-bp fragment including algU | This study |
| pMA9 | pEX18Gm containing the 5′ end of pslA and its upstream region (positions −912 to 591 relative to pslA), with the psl promoter (positions −267 to −54) replaced by araC-PBAD; Gmr | 36 |
| pMMB207-PA2663 | Ptac::PA2663 in pMMB207; Cmr | 3 |
Biofilm formation.
Under static conditions, P. aeruginosa strains containing pSMC21 (Table 1) were inoculated at a turbidity at 600 nm of 0.1 into fresh medium from an overnight culture. After 8 h at 37°C with shaking (mid-exponential phase), 8 ml of the culture was transferred to a 15-ml Corning tube containing a glass slide and incubated for 16 h without agitation at 37°C. pSMC21 is maintained in the absence of selection pressure (6, 11), and no antibiotic was used during biofilm growth. The biofilms were then either observed by confocal laser scanning microscopy (CLSM) or fixed as described below for subsequent observation by scanning electron microscopy (SEM).
Under dynamic conditions, P. aeruginosa biofilms were grown at 37°C in a three-channel flow cell with individual channel dimensions of 1 by 4 by 40 mm. The system was assembled and prepared as previously described (59). The substratum was an st1 microscope glass coverslip (VWR International, Fontenay sous Bois, France). P. aeruginosa cells containing pSMC21 were grown for 24 h in PPGAS with kanamycin (and gentamicin for the pBAUC-harboring strain), washed, and adjusted to a turbidity at 600 nm of 0.1 in 0.9% NaCl. The bacterial suspension was injected into a channel, and bacteria were allowed to adhere to the coverslip for 2 h. A flow (2.5 ml h−1) of PPGAS medium without antibiotic was then applied. Bacterial attachment and biofilm formation were observed by CLSM after 15 min and 24 or 48 h of flow, respectively. Polysaccharides produced in biofilm were stained with 0.3 mg ml−1 calcofluor white (Sigma-Aldrich, St.-Quentin Fallavier, France) in 0.9% NaCl just before CLSM observation. When the strains did not contain pSMC21 [PAOU PBAD::psl and PAOU(pMMB207-PA2663)], precultures were grown without antibiotic (PAOU PBAD::psl) or with chloramphenicol [PAOU(pMMB207-PA2663)], bacteria were washed, and biofilms were grown for 24 h under dynamic conditions without antibiotic, as described above. Biofilms were then stained for 10 min with 5 μM Syto 61 Red (Invitrogen, Paisley, United Kingdom) before CLSM observation. Arabinose (0.2%) was included in the PPGAS medium during growth of PAOU PBAD::psl biofilms.
Assessment of biofilm stability.
We used the method described by Friedman and Kolter (17) to assess biofilm stability. Briefly, biofilms were grown for 48 h at 37°C in 96-well polyvinyl chloride (PVC) microtiter plates without shaking. Biofilms were then either gently washed by submerging the microtiter plate in water or harshly washed with running water and were stained with crystal violet, which was quantified by turbidity measurement at 600 nm. Alternatively, bacteria in fresh PPGAS were inoculated into flow cell channels and biofilms were grown for 16 h at 37°C without medium flow to allow biofilm formation under static conditions. PPGAS flows at increasing rates (0.6, 1.2, 2.5, 3.6, 7.3, 12.0, and 20.0 ml h−1) were then successively applied as follows: the 0.6-ml h−1 flow was applied for 5 min, the flow was turned off to observe biofilms by CLSM, the 1.2-ml h−1 flow was applied for 5 min, and so on.
Image acquisition and analyses.
CLSM observations were performed with a TCS-SP2 microscope (Leica Microsystems, Heidelberg, Germany), using a 63× immersion objective. Excitation wavelengths for green fluorescent protein (GFP), calcofluor white, and Syto 61 Red were 488, 400, and 633 nm, respectively. Emissions wavelengths were 500 to 550, 410 to 450, and 645 nm, respectively. Calcofluor white produced a blue light, which we artificially converted to red during image acquisition for better contrast. Images were taken every micrometer at all biofilm depths, at 10 random positions. Three-dimensional (3D) images were obtained using Leica LAS AF software, and biofilm parameters (biovolume and thickness) were quantified with COMSTAT (25).
To observe biofilms by SEM, samples were fixed in 3% glutaraldehyde overnight at 4°C, washed three times for 10 min in 0.1 M phosphate buffer, and then dehydrated by soaking for 10 min in increasing concentrations of alcohol, i.e., 70%, 95%, and absolute ethanol. The samples were dried in ethanol in a CPD 030 critical point dryer (Bal-Tec, France), using CO2 as a transitional fluid until the critical point was reached. The samples were mounted on aluminum stubs and coated for 120 s at 20 mA with gold-palladium alloy, using a model 501 sputter coater (Edwards Pirani, United Kingdom), and were observed with a JEOL 6460LV microscope (JEOL Ltd., Tokyo, Japan) at magnifications of ×8,500 to ×10,000. The voltage was kept at 10 kV or 15 kV and at an average distance from the electron gun of about 10 mm.
Nucleic acid procedures.
Restriction enzymes, T4 DNA ligase, and alkaline phosphatase were purchased from Invitrogen (Paisley, United Kingdom). PCRs were performed using Failsafe PCR reagent with 2× premix D (Tebu-Bio, Epicentre Biotechnologies, Madison, WI). Plasmids and RNAs were purified using a QIAprep Spin miniprep kit and an RNeasy Midi kit (Qiagen, Hilden, Germany). E. coli (commercial electrocompetent Top10 cells [Invitrogen, Paisley, United Kingdom] or S17-1 cells) and P. aeruginosa cells were transformed by electroporation as previously described (55) or by conjugation as follows. Exponential-phase cultures of E. coli S17-1 carrying the plasmid to transfer and the recipient strain P. aeruginosa PAO1 were mixed on LB agar medium without NaCl and incubated overnight at 37°C. Bacteria were then resuspended in liquid LB and plated onto solid PIA medium (which inhibits the growth of E. coli but not of P. aeruginosa) containing the appropriate antibiotic.
algU inactivation and complementation of the mutation.
The algU mutant of P. aeruginosa PAO1 was obtained by allelic exchange with a truncated version of algU carrying the Gm resistance gene aacC1 framed by lox sequences, using plasmid pEXUGL. The full cre-lox gene deletion procedure used here was described by Quenée et al. (47). To construct pEXUGL, the algU-flanking regions were PCR amplified with primer pairs AlgU1-AlgU2 and AlgU3-AlgU4 (Table 2). After PstI digestion, the two fragments were ligated with each other, generating a 546-bp deletion within the algU coding sequence. This fragment was cloned into the EcoRI-HindIII sites of the suicide vector pEX100Tlink (47), resulting in plasmid pEXalgU (Table 1). The lox-aacC1-lox cassette of pUCGmlox (47) was then subcloned as a PstI fragment between the 5′ and 3′ parts of algU in pEXalgU. The resulting plasmid, pEXUGL (Table 1), was introduced into the E. coli donor/helper strain S17-1, from which it was transferred by conjugation into P. aeruginosa PAO1. After mating, Gm-resistant colonies grown on PIA plates were counterselected on 5% sucrose-LB agar plates to force the excision of the vector, followed by a search for Gm-resistant (presence of aacC1) and Cb-sensitive (loss of vector) colonies. The excision of the lox-flanked aacC1 gene was then catalyzed by the Cre recombinase encoded by pCM157 (37) to yield an antibiotic resistance-free mutant and to avoid the putative polar effects resulting from aacC1 insertion (47). Primers AlgU1 and AlgU4 were used to verify the construction by PCR. The mutant strain selected for further studies was named PAOU.
TABLE 2.
Primers used in this study
| Primer use and name | Sequence (5′-3′) | Reference |
|---|---|---|
| Construction of algU mutant and complementing plasmid | ||
| AlgU1 | ATATGAATTCTGGAGCCCTAGTATATAGAAGGG | This study |
| AlgU2 | ATTACTGCAGGTTAGCATGAAAGCTCCTCT | This study |
| AlgU3 | AAGCAATCGACAAAGCTCTG | This study |
| AlgU4 | ATATAAGCTTAAGCAAAAGCAACAGGGA | This study |
| AUCP1 | ATTAGGATCCTGTCATCGGCAGGCGTCATCA | This study |
| AUCP2 | ATTAGAATTCTGGCATTTGCCGCTGTGTCAG | This study |
| Quantification of mRNA levels by qRT-PCR | ||
| 16SF | CAGGATTAGATACCCTGGTAGTCCAC | 10 |
| 16SR | GACTTAACCCAACATCTCACGACAC | 10 |
| PelA1 | TGTCGGGTATCTGAAAGAGCAG | This study |
| PelA2 | GACCGACAGATAGGCGAAGG | This study |
| PslA1 | GTTCTGCCTGCTGTTGTTCATG | This study |
| PslA2 | AGGTAGGGAAACAGGCCCAG | This study |
| LecA1 | TGGAAAGGTGAGGTTCTGGC | This study |
| LecA2 | AATCGACGTTACCTGCCCTG | This study |
| LecB3 | ACCAATAACGCCGTCATCG | This study |
| LecB4 | CTGTACCTTGCCACTGCTGC | This study |
| Fhp1 | AATTACCTGCATGACCGGGTC | This study |
| Fhp2 | CGGAAACAGATCCAGCTCGT | This study |
| PpyR1 | TTTCCTCGACCGCTACCTGA | This study |
| PpyR2 | AGCGCATGGGAAAACACC | This study |
| Promoter mapping by 5′-RACE PCR | ||
| gsp1lecB | ATTTGCTGTGCCGAGACCAGA | This study |
| gsp2lecB | TTATTGGTGCTTTGCCCGC | This study |
| neslecB | AGGCGGTGACGCCGAACCGG | This study |
To complement the mutation, a 911-bp DNA fragment including the full-length algU gene and its P1, P2, and P3 promoters (52) was amplified by PCR with primers AUCP1 and AUCP2 (Table 2). The amplicon was inserted into pBBR1MCS-5 (33) after EcoRI/BamHI digestion, generating pBAUC (Table 1), which was introduced into P. aeruginosa PAOU.
psl operon and ppyR gene expression in PAOU.
To construct a PAOU strain containing an inducible psl operon, the pMA9 plasmid (36) was introduced by conjugation into PAOU and a double-crossover recombinant was selected as previously described (27). This led to the replacement of the psl promoter region on the PAOU chromosome by a DNA fragment including the araC gene and the PBAD promoter, allowing psl operon induction by the addition of arabinose (0.2%) to the culture medium (36). The recombinant strain was named PAOU PBAD::psl (Table 1). To express ppyR, the pMMB207-PA2663 plasmid containing ppyR under the control of the Ptac promoter (3) was introduced into PAOU by conjugation.
mRNA assay by quantitative reverse transcription-PCR (qRT-PCR).
RNA quantification was performed as previously described (22), using bacteria grown for 8 h in liquid PPGAS medium. The primers used are given in Table 2. 16S rRNA was used as an endogenous control (10). PCRs were performed in triplicate, and the standard deviations were <0.15 threshold cycle (CT). The relative quantifications were obtained as previously described (5), using the comparative CT (2−ΔΔCT) method (34).
Promoter mapping by 5′-RACE PCR.
Total RNAs were isolated from P. aeruginosa PAO1 grown in PPGAS medium by use of a MasterPure RNA purification kit (Epicentre Biotechnologies, Madison, WI). The 5′ end of lecB mRNA was amplified using a system for 5′ rapid amplification of cDNA ends (5′-RACE) (version 2.0; Invitrogen, Paisley, United Kingdom) according to the manufacturer's instructions. The primers used for cDNA synthesis and for the first and second PCRs are listed in Table 2. The final PCR product of 5′-RACE amplifications was cloned into the pCR2.1-TOPO vector (Invitrogen) and sequenced (Cogenics, Takeley, United Kingdom).
Quantification of the exopolysaccharide Psl.
Psl was extracted from the cell surface, and its levels were evaluated by an enzyme-linked immunosorbent assay (ELISA) as described in previous work (7).
Twitching motility tests.
Twitching assays were performed by inoculating bacteria by stabbing to the bottom of a petri dish through a layer of PPGAS agar (10 g liter−1) (49). After 24 h of incubation at 37°C, the agar was removed, the petri dish was rinsed with water, and the cells were stained with 1% crystal violet. Each assay was performed at least in triplicate.
RESULTS
The algU mutant is impaired in attachment to glass and formation of flow-resistant biofilms.
In preliminary experiments, two rich media were compared for biofilm formation, namely, the commonly used LB broth and PPGAS, which is limited in phosphate and thereby promotes rhamnolipid production (65). Biofilms of wild-type P. aeruginosa PAO1 grown on glass slides for 16 h at 37°C under static conditions were about 1.5-fold thicker in PPGAS than in LB broth (about 9 versus 6 μm), with similar cell densities (data not shown). Since growth rates did not differ significantly between LB and PPGAS liquid cultures, PPGAS seemed more favorable for biofilm formation and was therefore selected for the subsequent experiments. To test the role of AlgU in biofilm formation by nonmucoid P. aeruginosa, we constructed a P. aeruginosa PAOU mutant in which 546 bp of the 582-bp sequence of algU were deleted (Table 1). This mutant is devoid of antibiotic resistance, allowing its transformation by the GFP-encoding plasmid pSMC21 (11) (Table 1) in order to visualize biofilm-grown cells by CLSM. The algU mutation did not affect the growth rate in liquid PPGAS at 37°C (data not shown). To grow biofilms on a microscope glass coverslip in flow cells, bacteria in 0.9% NaCl were allowed to attach to the coverslip for 2 h, and a flow (2.5 ml h−1) of PPGAS was applied. Attached bacteria were observed by CLSM after 15 min of medium flow, revealing that there was 34-fold reduced coverage of the surface with PAOU compared to that with PAO1 (Fig. 1A; Table 3). The algU mutant thus displayed dramatically reduced attachment properties. After 24 h of PPGAS flow, a PAOU biofilm defect was particularly obvious: PAOU failed to produce a biofilm covering the glass surface, whereas the PAO1 biofilm covered the surface (Fig. 1A). To complement the PAOU mutant, we constructed the low-copy-number plasmid pBAUC (Table 1), which carries the PAO1 algU gene under the control of three (P1, P2, and P3) of its five promoters (52). The cloning of a larger fragment, including algU and all five promoters, was unsuccessful. The presence of pBAUC in strain PAOU increased its attachment ability 11-fold and allowed PAOU to form biofilms similar to those of PAO1 (Fig. 1A; Table 3). This verified that the absence of AlgU was responsible for the PAOU biofilm phenotypes.
FIG. 1.
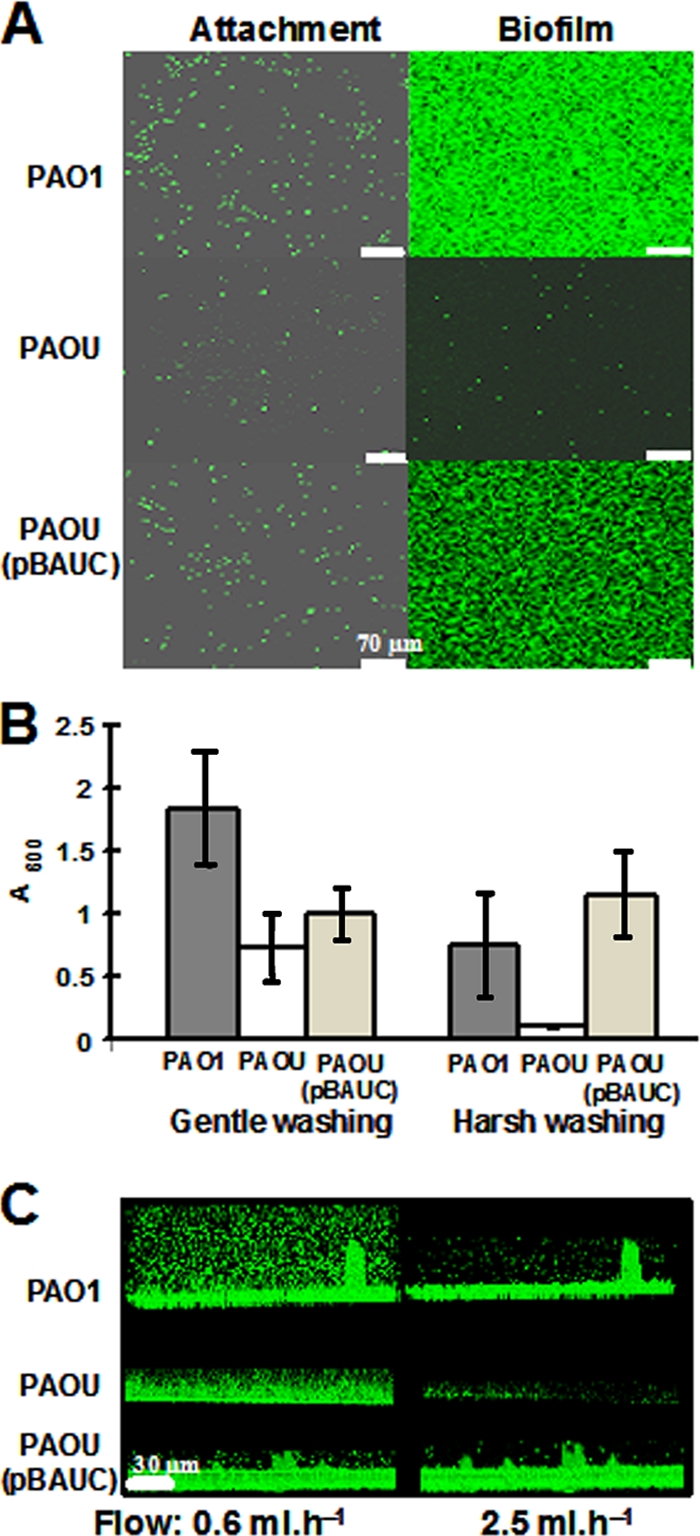
Attachment of P. aeruginosa and biofilm growth and robustness. (A) The indicated strains containing pSMC21 were allowed to attach to the glass surface before application of a medium flow for 15 min (left; attachment) and 24 h (right; biofilm). Each maximal projection is representative of five CLSM observations. (B) Stability of biofilms grown under static conditions in 96-well PVC microtiter plates. Biofilms were quantified by crystal violet staining after gentle washing by immersion into water or harsh washing with running water. Each value is the average for three independent experiments. (C) Side views of biofilms of the indicated strains containing pSMC21. Biofilms were grown under static conditions, and medium flows of 0.6 ml h−1 (left) and 2.5 ml h−1 (right) were applied for 5 min before CLSM observations. These data are representative of two distinct experiments.
TABLE 3.
Quantification of bacterial attachment and biofilm parametersc
| Strain | Attachmenta (%) | 24-h biofilmb |
48-h biofilmb |
||
|---|---|---|---|---|---|
| Biovolume (μm3/μm2) | Avg thickness (μm) | Biovolume (μm3/μm2) | Avg thickness (μm) | ||
| PAO1 | 2.41 | 6.35 | 7.70 | 4.52 | 8.06 |
| PAOU | 0.07 | 0.12 | 0.16 | 0.53 | 8.09 |
| PAOU(pBAUC) | 0.77 | 4.92 | 6.53 | 2.87 | 8.02 |
| PAOU PBAD::psl + arabinose | ND | 2.31 | 3.31 | ND | ND |
| PAOU(pMMB207-PA2663) | ND | 0.52 | 1.44 | ND | ND |
Percentage of glass surface covered by bacteria after static adhesion step and 15 min of medium flow.
Biofilms were grown under a flow of PPGAS medium.
Standard deviations were <10% for all values, except for the average thickness of the PAOU(pMMB207-PA2663) biofilm, which was 1.44 ± 0.47 μm. ND, not determined.
Because the biofilm formation defect of PAOU after 24 h of medium flow was severe, we hypothesized that this was due not only to its reduced attachment ability but also to impaired stability of the PAOU biofilm community. To examine biofilm formation under static conditions and to assess the stability of these biofilms, cells were grown in a microtiter plate for 48 h without shaking. Biofilms were then either gently washed by submerging the plate in water or harshly washed with running water, as described by Friedman and Kolter (17). A gentle washing revealed that PAOU was able to form biofilms whose cell population was reduced only about 2-fold compared with that of PAO1 (Fig. 1B). Harsh washings led to the removal of most (about 85%) of the cells from PAOU biofilms, whereas it removed about 60% of cells from PAO1 biofilms (Fig. 1B). Biofilms formed by PAOU(pBAUC) cells were particularly resistant (Fig. 1B), confirming a relationship between AlgU and biofilm stability.
To further investigate their stability, biofilms were allowed to form under static conditions in flow cell channels containing PPGAS medium for 16 h before application of a PPGAS flow of increasing strength. Low shear-flow rates (0.6 ml h−1) showed that all three strains had developed biofilms of similar thicknesses (Fig. 1C). However, PAO1 and PAOU(pBAUC) biofilms resisted flows of 2.5 ml h−1 (Fig. 1C) and even 20 ml h−1 (not shown). Importantly, a 2.5-ml h−1 flow rate was sufficient to dissociate cells from PAOU biofilms (Fig. 1C). This showed that AlgU is required for the formation of robust, shear-resistant biofilms.
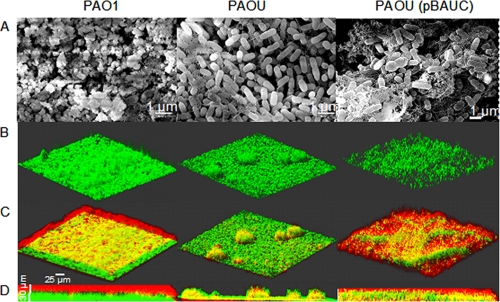
We used SEM to observe the surfaces of biofilms grown on glass slides. An abundant extracellular matrix masked PAO1 cells, whereas cells were visible within the rare PAOU-attached aggregates (Fig. 2 A). Matrix production by PAOU increased upon complementation with pBAUC (Fig. 2A). We used the polysaccharide-binding dye calcofluor white, which stains P. aeruginosa biofilm matrix (40), to compare biofilms grown for 48 h under dynamic conditions. This allowed PAOU to form a thin biofilm layer with dispersed microcolonies, whereas PAO1 and PAOU(pBAUC) biofilms were more homogeneous and had higher biovolumes than PAOU biofilms (Fig. 2B and D; Table 3). PAOU microcolonies were thicker than PAO1 and PAOU(pBAUC) biofilms (Fig. 2D), leading to similar average thicknesses for 48-h biofilms of all three strains (Table 3). Consistent with the SEM images, calcofluor white revealed that PAO1 and PAOU(pBAUC) biofilms were covered by polysaccharidic matrixes, whereas PAOU biofilms were almost devoid of exopolysaccharides (Fig. 2C and D). AlgU therefore appears to be required for synthesis of major polysaccharidic components of PAO1 biofilm matrix.
FIG. 2.
Polysaccharide matrix in P. aeruginosa biofilms. (A) SEM observations of static biofilms of the indicated strains. Magnification, ×8,500 to ×10,000. (B) 3D images resulting from CLSM observations of 48-h biofilms grown as described in the legend to Fig. 1A. The three strains contained pSMC21, and bacteria are shown in green. (C) Polysaccharides of the same biofilms as in panel B, stained with calcofluor white. This dye produced a blue light, which we artificially converted to red in our images for better contrast. Yellow results from the green (bacteria) and red (polysaccharides) overlay. (D) Side views of biofilms shown in panel C. All images are representative of five observations.
Finally, it should be noted that we also observed a biofilm formation defect with strain PAOU for static biofilms grown in LB and minimal M9 glucose media: no biofilm, but only dispersed PAOU cells, remained attached to glass slides after washing (data not shown). This indicated that AlgU modulation of biofilm formation occurs under several environmental conditions.
The algU mutant is defective in production of the Psl exopolysaccharide, a key matrix component.
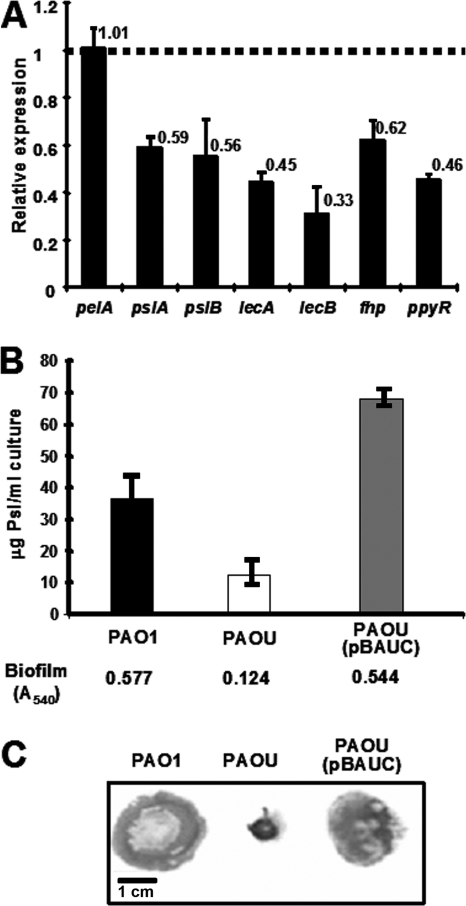
Prior studies showed that alginate is not a major component of nonmucoid biofilms (69). Since the PAOU biofilms appeared to have reduced exopolysaccharides, we hypothesized that AlgU controls production of Psl and/or Pel, two other PAO1 polysaccharides linked to biofilm formation (17, 18, 51). We examined pslA, pslB (the first two genes of the psl operon), and pelA (the first gene of the pel operon) mRNA levels by qRT-PCR. The latter was performed on RNAs extracted from bacteria harvested after 8 h of growth in liquid PPGAS cultures, since we used bacteria at this growth stage for static biofilm formation. Whereas the pelA mRNA level was unaltered, pslA and pslB mRNA levels were 1.7- and 1.8-fold lower, respectively, in PAOU than in PAO1 (Fig. 3 A). This indicated that the algU mutant had reduced psl transcription compared with that of wild-type cells. Unfortunately, we were not able to analyze gene expression in biofilm-grown cells because PAOU biofilms did not yield a sufficient biomass to extract adequate amounts of RNA.
FIG. 3.
Alterations in gene expression, Psl exopolysaccharide production, and twitching motility in the absence of AlgU. (A) Expression levels of pelA, pslA, pslB, lecA, lecB, fhp, and ppyR genes in the algU mutant PAOU relative to those in PAO1. pslA and pslB are the first two genes of the psl operon, and fhp and ppyR are the first two genes of the putative fhp-ppyR-PA2662 operon. RNAs were extracted after 8 h of growth in liquid PPGAS medium and were assayed by qRT-PCR. A relative value of <1 indicates a reduction of gene expression in PAOU. 16S rRNA was used as an endogenous control to normalize the RNA input and reverse transcription efficiency. PCRs were performed in triplicate, and the standard deviations were <0.15 CT. The values are the means for at least two independent experiments. (B) Quantification of Psl production by ELISA. PAO1, PAOU, and PAOU(pBAUC) cells were grown, and surface exopolysaccharide was extracted and quantified by ELISA using a Psl-specific antibody, as outlined previously (7). Shown below the graph are rapid-attachment biofilm values (7) obtained for strains used for Psl enumeration (means for three replicates). (C) Twitching motilities of the indicated strains were examined at the petri dish-PPGAS agar interface. The pictures are representative of at least three tests.
To directly test Psl levels in the P. aeruginosa strains, PAO1, PAOU, and PAOU(pBAUC) cells were cultured, cell surface polysaccharides were extracted, and amounts of cell-associated Psl were evaluated by ELISA as previously described (7). Consistent with the above qRT-PCR data, Psl synthesis was reduced 2.8-fold in PAOU compared with that in PAO1, and complementation resulted in enhanced Psl levels (Fig. 3B). In prior studies, 2- to 3-fold changes in psl transcription or Psl synthesis resulted in profound changes in biofilm integrity and architecture (36, 58). For strain PAO1, psl mutants were severely compromised in biofilm formation (8, 29, 38, 42). Psl was recently shown to be anchored on the cell surface in a helical pattern, promoting cell-cell interaction and assembly of a matrix which holds bacteria in the biofilm and on the surface (35). The instability displayed by PAOU biofilms exposed to a medium flow is therefore consistent with the observed Psl production defect. Finally, the psl operon was placed under the control of the arabinose-inducible promoter PBAD in the PAOU background. Inducing psl expression restored the ability of PAOU to form flow-resistant biofilms in 24 h, as illustrated by the biovolume and average thickness values, which were both about 20-fold higher with the PAOU PBAD::psl strain than with PAOU (Table 3). This result strengthens the notion that psl downregulation contributes to the algU mutant phenotype. However, the biofilm formation was restored less efficiently by inducing psl expression than by algU complementation, since the biovolume and average thickness of PAOU PBAD::psl biofilms were both about 2-fold lower than those of PAOU(pBAUC) biofilms (Table 3). This suggested that psl downregulation might not be the only cause of the PAOU phenotype.
Lectin-encoding genes are underexpressed in the absence of AlgU.
We examined if the expression of other genes known to be involved in biofilm formation is compromised in the absence of AlgU. We observed that lecA and lecB mRNA levels were reduced (2.2- and 3.0-fold, respectively) in the algU mutant compared to those in PAO1 (Fig. 3A), which was not reported in published microarray studies (16, 60, 67, 68). The lecA and lecB genes each encode a lectin, namely, galactophilic LecA (PA-IL) and l-fucose-specific LecB (PA-IIL), respectively. Since single lecA and lecB mutants of PAO1 showed impaired abilities to form biofilms on abiotic surfaces (14, 62), the downregulation of lecA and lecB genes in the algU mutant likely contributes to its biofilm formation defect. Both lectins were proposed to mediate the adhesion of P. aeruginosa to other bacterial cells of the same or different species (14, 62), although this role remains debated (56). LecB was shown to be required for posttranscriptional type IV pilus biogenesis, without impairing transcription of pil genes encoding pilus components (56). LecB was therefore necessary for twitching motility, which occurs on solid surfaces or interfaces by extension and retraction of type IV pili (54). A lecB mutant displayed the same biofilm formation defect as a nonpiliated mutant, leading Sonawane et al. (56) to conclude that pilus biogenesis explains the role of LecB in biofilm formation. Consistently, the twitching ability of the PAOU strain was strongly reduced compared to that of the parental PAO1 strain and the complemented mutant (Fig. 3C). This effect did not appear to be due to transcriptional effects on pilA, which encodes the major pilus subunit, since the mRNA level of pilA was higher (1.7-fold) in PAOU than in PAO1 after 8 h of growth (not shown). These observations suggested that lecB expression was reduced sufficiently in the algU mutant to impair type IV pilus assembly. This, in turn, could contribute to the attachment and biofilm stability defects of this strain, since type IV pili are involved both in the initial attachment step, by directly binding surfaces, and in formation of biofilm structures through twitching motility (19, 30, 31, 41). In contrast, LecA is not involved in type IV pilus biogenesis, and its role in biofilm formation remains unclear (56).
Promoter mapping and regulatory pathway controlling psl expression.
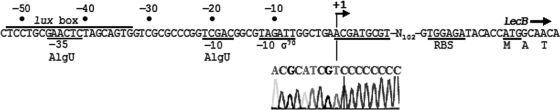
AlgU-dependent promoters possess the consensus sequence GAACTT-N16/17-TCCAA-N5/6-+1 (16). Overhage et al. (42) and Winzer et al. (66) previously mapped the transcription start sites of pslA and lecA, respectively. For each promoter, a single +1 base was identified, 41 bp and 70 bp upstream of the translation start sites of pslA and lecA, respectively (42, 66). However, there are no obvious matches to the AlgU consensus promoter element at the appropriate position relative to these +1 bases. To our knowledge, there has been no mapping study reported for the lecB promoter, prompting us to identify its transcriptional start site by 5′-RACE PCR. The +1 base is located 124 bp upstream of the translation initiation codon (Fig. 4). The GAACTc-N19-TCgAc sequence (lowercase bases differ from the consensus) is similar to the consensus for AlgU-dependent promoters, but the position of the transcriptional start site (17 nucleotides downstream) leaves uncertain the functionality of this putative promoter. The TAGATT sequence lying around position −10 rather suggests that the lecB promoter depends on σ70 (consensus −10 sequence, TATAAT [45]) or on RpoS, which recognizes a similar −10 element and is required for LecB synthesis (66). Interestingly, we observed a lux box-like sequence, which could allow binding of LasR and RhlR QS regulators (64), between positions −52 and −33 (Fig. 4). This is consistent with the report that LecB production requires RhlR and its cognate signal molecule, N-butanoyl-l-homoserine lactone (C4-HSL) (66). The presence of a lux box was highlighted at a similar position (−53 to −34 relative to the +1 base) in the promoter region of lecA, whose expression also depends on RhlR/C4-HSL (66).
FIG. 4.
Mapping of lecB promoter. The transcriptional start site (+1) was identified by 5′-RACE PCR, as shown by the relevant part of the chromatogram displaying the PCR product sequence. RBS is the ribosome binding site, and the first three codons of lecB are translated. Sequences similar to −35 and −10 consensus sequences of AlgU- and σ70-dependent promoters are indicated, and the lux box is a potential binding sequence for quorum-sensing regulatory proteins LasR and/or RhlR.
These data suggest that AlgU is not directly responsible for transcription of the psl operon or of the lecA and lecB genes. A dramatic reduction in psl gene expression was reported for the ppyR mutant of P. aeruginosa PAO1 compared to the parental strain (3). The fhp and PA2662 genes flank ppyR, and the short sequences separating these genes (54 bp between fhp and ppyR and 12 bp between ppyR and PA2662) suggest that they are cotranscribed (www.pseudomonas.com). Since fhp was the most strongly overexpressed gene (60-fold) in mucoid strains (16), we hypothesized that expression of the fhp-ppyR-PA2662 operon would be reduced in the algU mutant. This was confirmed by qRT-PCR: the mRNA levels of fhp and ppyR genes were reduced 1.6- and 2.2-fold, respectively, in the PAOU strain compared to PAO1 (Fig. 3A). Consequently, our data suggest that AlgU stimulates the expression of the fhp-ppyR-PA2662 operon in PAO1, leading to positive control of the psl operon via PpyR, which is a small protein (85 amino acids) predicted to be located in the cytoplasmic membrane (3; www.pseudomonas.com). Consistent with this hypothesis, expressing ppyR in PAOU by using the pMMB207-PA2663 plasmid (3) allowed the strain to form flow-resistant biofilms in 24 h, even though the biovolume and average thickness values remained modest compared to those of induced PAOU PBAD::psl and PAOU(pBAUC) biofilms (Table 3). This prompted us to use another method: we evaluated the stability of PAOU(pMMB207-PA2663) biofilms grown in a microtiter plate under static conditions (17). The crystal violet turbidity values were 1.7 ± 0.1 and 0.74 ± 0.16 after gentle and harsh washing, respectively. These values are similar to those obtained with the wild-type PAO1 strain (Fig. 1B), confirming that expressing ppyR in PAOU increased the biofilm stability.
DISCUSSION
The present study revealed that the ECF sigma factor AlgU is required for efficient attachment to abiotic surfaces and for development of robust, shear-resistant biofilms by the nonmucoid P. aeruginosa PAO1 strain. Such a strong biofilm formation defect of the algU mutant strain was unexpected and is likely multifactorial. Likewise, we identified the psl operon and the lecA and lecB genes as being underexpressed in the algU mutant. This led us to propose three mechanisms by which AlgU could contribute to biofilm formation: Psl polysaccharide matrix synthesis, production of LecA and LecB lectins, and type IV pilus biogenesis. It is likely that additional AlgU-dependent genes remain to be identified, but these three mechanisms may be sufficient to explain the attachment and biofilm stability defects of the algU mutant. The critical roles of Psl, LecA, LecB, and type IV pili in adhesion and biofilm formation are indeed extremely well documented (3, 8, 14, 19, 29, 30, 31, 35, 36, 38, 41, 42, 62).
Control of psl expression in P. aeruginosa appears to be complex. Prior studies showed that the psl genes are transcribed as an operon, which is constitutively expressed in planktonic cells but tightly regulated in biofilm-grown bacteria (38, 42). Several recent studies have reported the regulation of psl by the secondary messenger molecule c-di-GMP (26, 58) as well as by the GacS-GacA-rsmZ-RsmA signal transduction pathway (20). Furthermore, the involvement of PpyR in P. aeruginosa PAO1 biofilm formation (4) was explained by a dramatic reduction in psl gene expression in the ppyR mutant (3). Likewise, the lecA gene is the object of multiple regulation levels: lecA transcription requires the Rhl QS system, the stationary-phase sigma factor RpoS, and the Pseudomonas quinolone signal (PQS), while it is repressed by MvaT (12, 13, 66). As with psl, reports indicate that the posttranscriptional GacS-GacA-rsmZ-RsmA regulatory pathway and other signaling proteins feeding into this pathway control lecA expression, but these controls could occur indirectly through the QS network (20, 44, 51). Little is known regarding lecB regulation, except that similar to that of lecA, lecB transcription requires the Rhl QS system and RpoS (66). Notably, expression of psl, lecA, and lecB was not previously identified as AlgU dependent. This discovery adds another level of complexity to the regulation of these genes. As a sigma factor, AlgU could be directly responsible for their transcription. Analysis of the promoters of these genes, however, indicated that this is probably not the case, since the transcription initiation sites are not preceded by an AlgU-like promoter sequence with appropriate spacing (Fig. 4) (42, 66). AlgU is therefore likely to act indirectly, via the transcription of genes encoding regulators acting on psl, lecA, and/or lecB. We observed that AlgU contributes to expression of ppyR, whose product positively controls psl expression via an unknown mechanism (3). The control of the fhp-ppyR-PA2662 operon by AlgU is likely indirect, since the fhp promoter is dependent on the alternative sigma factor RpoN (σ54) and on the activator FhpR (1). The fhp and fhpR genes are adjacent, sharing a bidirectional promoter region. Proteins encoded by PA0779 and PA3697, which both bind the fhp-fhpR intergenic region, also activate transcription of fhp and fhpR (32). We did not observe a clear downregulation of the PA0779 and PA3697 genes in PAOU (not shown). The control of the psl operon by PpyR is also likely indirect, since the latter is a putative membrane protein, suggesting that it plays the role of sensor rather than transcriptional regulator (3, 4). The complete pathway from AlgU to psl thus remains largely unknown. The control exerted by QS on lecA and lecB suggested that the absence of AlgU could somehow affect the QS network, but this hypothesis was invalidated by the following lines of evidence. Production of the Las and Rhl autoinducer molecules and of PQS was not significantly altered by algU inactivation under our conditions (not shown). Consistently, PAOU was able to produce rhamnolipids at a level close to that of PAO1 (not shown), although this process is directly dependent on RhlR and C4-HSL and indirectly dependent on LasR and PQS (39, 43). Therefore, future work will be required to unravel the mechanism(s) by which AlgU contributes to regulate psl, lecA, and lecB genes.
The AlgU functions and the mechanisms leading to its hyperactivity were studied specifically in the context of mucoid P. aeruginosa strains (16, 21, 46, 48, 61). Relying on the inhibition of AlgU by its anti-sigma factor MucA (21, 48), the notion that AlgU activity was negligible in nonmucoid strains likely prevented investigation of AlgU functions in such strains. Although AlgU activity is certainly low in nonmucoid cells, our data showed that it is very far from being negligible. Consistently, whereas 67% of the AlgU pool in PAO1 was held associated with the inner membrane, likely in an inactive state, the remaining 33% was in the cytosol (50), which could lead to a basal AlgU activity. Since the expression of psl, lecA, and lecB genes was partially AlgU dependent in PAO1 (low AlgU activity), we expected that these genes would be identified as strongly induced in transcriptomic studies of mucoid strains (high AlgU activity) and after AlgU activation by stress conditions. This, however, was not the case (16, 60, 67, 68). This could be due to the fact that these genes are only indirectly regulated by AlgU and that other necessary conditions were not met in mucoid cells and/or under the experimental conditions used. Alternatively, it could reflect differential AlgU functions in mucoid and nonmucoid cells. A transcriptome study of the AlgU regulon in nonmucoid cells in the absence of stress would be of great interest to further address this question.
AlgU has been studied primarily for its essential role in mucoidy and therefore in the maintenance of chronic infection in the lungs of CF patients, and it is furthermore known to be involved in stress responses (2, 45, 67, 68). Here we identified new AlgU functions in P. aeruginosa ecology and pathology. Since AlgU is required for attachment of P. aeruginosa to surfaces and for development of robust biofilms, it plays an important role in colonization of abiotic environments by nonmucoid P. aeruginosa strains, providing a source of bacteria for subsequent infections (23). In addition, AlgU contributes to the production of virulence factors by nonmucoid cells, including LecA, LecB, and probably type IV pilus and protease IV as a consequence of a low LecB level (9, 56, 63). Finally, in the process leading to chronic infection of CF patients, it is believed that the lung is initially colonized by nonmucoid strains, which develop biofilms before conversion to mucoidy (48). For this conversion, Wozniak et al. (69) pointed out the importance of a switch from unknown polysaccharides (later identified as Pel and Psl [51]) to alginate in the primary polysaccharide content of the extracellular polymeric substances. Although the involvement of AlgU in the formation of nonmucoid biofilms on biotic surfaces, especially mucin-covered epithelial cells, remains to be investigated, our data suggest that AlgU could play a key role in the initial infection steps (adhesion and development of nonmucoid biofilm) and in the polysaccharide production switch.
Acknowledgments
We are grateful to G. A. O'Toole, B. Polack, and T. K. Wood for the kind gifts of plasmids. We thank O. Maillot for technical assistance.
This work was supported by the Region Bretagne, France (doctoral fellowship to G.H. [Bioclean program]), by European FEDER funds, by the Ministère de la Recherche et de la Technologie, France (RITMER grant, doctoral fellowship to A.B., and postdoctoral fellowship to K.S.), and by Public Health Service grants AI061396 and HL058334 (D.J.W.).
Footnotes
Published ahead of print on 26 March 2010.
REFERENCES
- 1.Arai, H., M. Hayashi, A. Kuroi, M. Ishii, and Y. Igarashi. 2005. Transcriptional regulation of the flavohemoglobin gene for aerobic nitric oxide detoxification by the second nitric oxide-responsive regulator of Pseudomonas aeruginosa. J. Bacteriol. 187:3960-3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aspedon, A., K. Palmer, and M. Whiteley. 2006. Microarray analysis of the osmotic stress response in Pseudomonas aeruginosa. J. Bacteriol. 188:2721-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Attila, C., A. Ueda, and T. K. Wood. 2008. PA2663 (PpyR) increases biofilm formation in Pseudomonas aeruginosa PAO1 through the psl operon and stimulates virulence and quorum-sensing phenotypes. Appl. Microbiol. Biotechnol. 78:293-307. [DOI] [PubMed] [Google Scholar]
- 4.Attila, C., A. Ueda, S. L. G. Cirillo, J. D. Cirillo, W. Chen, and T. K. Wood. 2008. Pseudomonas aeruginosa PAO1 virulence factors and poplar tree response in the rhizosphere. Microb. Biotechnol. 1:17-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bazire, A., F. Diab, L. Taupin, S. Rodrigues, M. Jebbar, and A. Dufour. 2009. Effects of osmotic stress on rhamnolipid synthesis and time-course production of cell-to-cell signal molecules by Pseudomonas aeruginosa. Open Microbiol. J. 3:128-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bloemberg, G. V., G. A. O'Toole, B. J. J. Lugtenberg, and R. Kolter. 1997. Green fluorescent protein as a marker for Pseudomonas spp. Appl. Environ. Microbiol. 63:4543-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrd, M. S., I. Sadovskaya, E. Vinogradov, H. Lu, A. B. Sprinkle, S. H. Richardson, L. Ma, B. Ralston, M. R. Parsek, E. M. Anderson, J. S. Lam, and D. J. Wozniak. 2009. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol. Microbiol. 73:622-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campisano, A., C. Schroeder, M. Schemionek, J. Overhage, and B. H. A. Rehm. 2006. PslD is a secreted protein required for biofilm formation by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 72:3066-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chemani, C., A. Imberty, S. de Bentzmann, M. Pierre, M. Wimmerová, B. P. Guery, and K. Faure. 2009. Role of LecA and LecB lectins in Pseudomonas aeruginosa-induced lung injury and effect of carbohydrate ligands. Infect. Immun. 77:2065-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corbella, M. E., and A. Puyet. 2003. Real-time reverse transcription-PCR analysis of expression of halobenzoate and salicylate catabolism-associated operons in two strains of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 69:2269-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davey, M. E., N. C. Caiazza, and G. A. O'Toole. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:1027-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diggle, S. P., K. Winzer, A. Lazdunski, P. Williams, and M. Cámara. 2002. Advancing the quorum in Pseudomonas aeruginosa: MvaT and the regulation of N-acylhomoserine lactone production and virulence gene expression. J. Bacteriol. 184:2576-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diggle, S. P., K. Winzer, S. R. Chhabra, K. E. Worrall, M. Cámara, and P. Williams. 2003. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol. Microbiol. 50:29-43. [DOI] [PubMed] [Google Scholar]
- 14.Diggle, S. P., R. E. Stacey, C. Dodd, M. Cámara, P. Williams, and K. Winzer. 2006. The galactophilic lectin, LecA, contributes to biofilm development in Pseudomonas aeruginosa. Environ. Microbiol. 8:1095-1104. [DOI] [PubMed] [Google Scholar]
- 15.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Firoved, A. M., and V. Deretic. 2003. Microarray analysis of global gene expression in mucoid Pseudomonas aeruginosa. J. Bacteriol. 185:1071-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman, L., and R. Kolter. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675-690. [DOI] [PubMed] [Google Scholar]
- 18.Friedman, L., and R. Kolter. 2004. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giltner, C. L., E. J. van Schaik, G. F. Audette, D. Kao, R. S. Hodges, D. J. Hassett, and R. T. Irvin. 2006. The Pseudomonas aeruginosa type IV pilin receptor binding domain functions as an adhesin for both biotic and abiotic surfaces. Mol. Microbiol. 59:1083-1096. [DOI] [PubMed] [Google Scholar]
- 20.Goodman, A. L., B. Kulasekara, A. Rietsch, D. Boyd, R. S. Smith, and S. Lory. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7:745-754. [DOI] [PubMed] [Google Scholar]
- 21.Govan, J. R. W., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guyard-Nicodème, M., A. Bazire, G. Hémery, T. Meylheuc, D. Mollé, N. Orange, L. Fito-Boncompte, M. Feuilloley, D. Haras, A. Dufour, and S. Chevalier. 2008. Outer membrane modifications of Pseudomonas fluorescens MF37 in response to hyperosmolarity. J. Proteome Res. 7:1218-1225. [DOI] [PubMed] [Google Scholar]
- 23.Hall-Stoodley, L., and P. Stoodley. 2005. Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 13:7-10. [DOI] [PubMed] [Google Scholar]
- 24.Hentzer, M., G. M. Teitzel, G. J. Balzer, A. Heydorn, S. Molin, M. Givskov, and M. R. Parsek. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183:5395-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 26.Hickman, J. W., D. F. Tifrea, and C. S. Harwood. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. U. S. A. 102:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 28.Holloway, B. W., V. Krishnapillai, and A. F. Morgan. 1979. Chromosomal genetics of Pseudomonas. Microbiol. Rev. 43:73-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson, K. D., M. Starkey, S. Kremer, M. R. Parsek, and D. J. Wozniak. 2004. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J. Bacteriol. 186:4466-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klausen, M., A. Heydorn, P. Ragas, L. Lambertsen, A. Aaes-Jørgensen, S. Molin, and T. Tolker-Nielsen. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48:1511-1524. [DOI] [PubMed] [Google Scholar]
- 31.Klausen, M., A. Aaes-Jørgensen, S. Molin, and T. Tolker-Nielsen. 2003. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 50: 61-68. [DOI] [PubMed] [Google Scholar]
- 32.Koskenkorva, T., N. Aro-Kärkkäinen, D. Bachmann, H. Arai, A. D. Frey, and P. T. Kallio. 2008. Transcriptional activity of Pseudomonas aeruginosa fhp promoter is dependent on two regulators in addition to FhpR. Arch. Microbiol. 189:385-396. [DOI] [PubMed] [Google Scholar]
- 33.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 34.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 35.Ma, L., M. Conover, H. Lu, M. R. Parsek, K. Bayles, and D. J. Wozniak. 2009. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 5:e1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma, L., K. D. Jackson, R. M. Landry, M. R. Parsek, and D. J. Wozniak. 2006. Analysis of Pseudomonas aeruginosa conditional Psl variants reveals roles for the Psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J. Bacteriol. 188:8213-8221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marx, C. J., and M. E. Lidstrom. 2002. Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. Biotechniques 33:1062-1067. [DOI] [PubMed] [Google Scholar]
- 38.Matsukawa, M., and E. P. Greenberg. 2004. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J. Bacteriol. 186:4449-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKnight, S. L., B. H. Iglewski, and E. C. Pesci. 2000. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 182:2702-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neut, D., J. G. E. Hendriks, J. R. van Horn, H. C. van der Mei, and H. J. Busscher. 2005. Pseudomonas aeruginosa biofilm formation and slime excretion on antibiotic-loaded bone cement. Acta Orthop. 76:109-114. [DOI] [PubMed] [Google Scholar]
- 41.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 42.Overhage, J., M. Schemionek, J. S. Webb, and B. H. A. Rehm. 2005. Expression of the psl operon in Pseudomonas aeruginosa PAO1 biofilms: pslA performs an essential function in biofilm formation. Appl. Environ. Microbiol. 71:4407-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pearson, J. P., E. C. Pesci, and B. H. Iglewski. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179:5756-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pessi, G., F. Williams, Z. Hindle, K. Heurlier, M. T. G. Holden, M. Cámara, D. Haas, and P. Williams. 2001. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol. 183:6676-6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Potvin, E., F. Sanschagrin, and R. C. Levesque. 2007. Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 32:38-55. [DOI] [PubMed] [Google Scholar]
- 46.Qiu, D., V. M. Eisinger, D. W. Rowen, and H. D. Yu. 2007. Regulated proteolysis controls mucoid conversion in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 104:8107-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quenée, L., D. Lamotte, and B. Polack. 2005. Combined sacB-based negative selection and cre-lox antibiotic marker recycling for efficient gene deletion in Pseudomonas aeruginosa. Biotechniques 38:63-67. [DOI] [PubMed] [Google Scholar]
- 48.Ramsey, D. M., and D. J. Wozniak. 2005. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospect for management of chronic infections in cystic fibrosis. Mol. Microbiol. 56:309-322. [DOI] [PubMed] [Google Scholar]
- 49.Rashid, M. H., and A. Kornberg. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 97:4885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rowen, D. W., and V. Deretic. 2000. Membrane-to-cytosol redistribution of ECF sigma factor AlgU and conversion to mucoidy in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Mol. Microbiol. 36:314-327. [DOI] [PubMed] [Google Scholar]
- 51.Ryder, C., M. Byrd, and D. J. Wozniak. 2007. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol. 10:644-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schurr, M. J., H. Yu, J. C. Boucher, N. S. Hibler, and V. Deretic. 1995. Multiple promoters and induction by heat shock of the gene encoding the alternative sigma factor AlgU (σE) which controls mucoidy in cystic fibrosis isolates of Pseudomonas aeruginosa. J. Bacteriol. 177:5670-5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology (New York) 1:784-791. [Google Scholar]
- 54.Skerker, J. M., and H. C. Berg. 2001. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. U. S. A. 98:6901-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith, A. W., and B. H. Iglewski. 1989. Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 17:10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sonawane, A., J. Jyot, and R. Ramphal. 2006. Pseudomonas aeruginosa LecB is involved in pilus biogenesis and protease IV activity but not in adhesion to respiratory mucins. Infect. Immun. 74:7035-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stapper, A. P., G. Narasimhan, D. E. Ohman, J. Barakat, M. Hentzer, S. Molin, A. Kharazmi, N. Høiby, and K. Mathee. 2004. Alginate production affects Pseudomonas aeruginosa biofilm development and architecture, but is not essential for biofilm formation. J. Med. Microbiol. 53:679-690. [DOI] [PubMed] [Google Scholar]
- 58.Starkey, M., J. H. Hickman, L. Ma, N. Zhang, S. De Long, A. Hinz, S. Palacios, C. Manoil, M. J. Kirisits, T. D. Starner, D. J. Wozniak, C. S. Harwood, and M. R. Parsek. 2009. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J. Bacteriol. 191:3492-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sternberg, C., B. B. Christensen, T. Johansen, A. Toftgaard Nielsen, J. B. Andersen, M. Givskov, and S. Molin. 1999. Distribution of bacterial growth activity in flow-chamber biofilms. Appl. Environ. Microbiol. 65:4108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tart, A. H., M. C. Wolfgang, and D. J. Wozniak. 2005. The alternative sigma factor AlgT represses Pseudomonas aeruginosa flagellum biosynthesis by inhibiting expression of fleQ. J. Bacteriol. 187:7955-7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tart, A. H., M. J. Blanks, and D. J. Wozniak. 2006. The AlgT-dependent transcriptional regulator AmrZ (AlgZ) inhibits flagellum biosynthesis in mucoid, nonmotile Pseudomonas aeruginosa cystic fibrosis isolates. J. Bacteriol. 188:6483-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tielker, D., S. Hacker, R. Loris, M. Strathmann, J. Wingender, S. Wilhelm, F. Rosenau, and K.-E. Jaeger. 2005. Pseudomonas aeruginosa lectin LecB is located in the outer membrane and is involved in biofilm formation. Microbiology 151:1313-1323. [DOI] [PubMed] [Google Scholar]
- 63.van Delden, C. 2004. Virulence factors in Pseudomonas aeruginosa, p. 3-45. In J.-L. Ramos (ed.), Pseudomonas, vol. 2. Kluwer Academic/Plenum Publishers, New York, NY. [Google Scholar]
- 64.Whiteley, M., and E. P. Greenberg. 2001. Promoter specificity elements in Pseudomonas aeruginosa quorum-sensing-controlled genes. J. Bacteriol. 183:5529-5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wild, M., A. D. Caro, A. L. Hernández, R. M. Miller, and G. Soberón-Chávez. 1997. Selection and partial characterization of a Pseudomonas aeruginosa mono-rhamnolipid deficient mutant. FEMS Microbiol. Lett. 153:279-285. [DOI] [PubMed] [Google Scholar]
- 66.Winzer, K., C. Falconer, N. C. Garber, S. P. Diggle, M. Camara, and P. Williams. 2000. The Pseudomonas aeruginosa lectins PA-IL and PA-IIL are controlled by quorum sensing and by RpoS. J. Bacteriol. 182:6401-6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wood, L. F., and D. E. Ohman. 2009. Use of cell wall stress to characterize σ22 (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol. Microbiol. 72:183-201. [DOI] [PubMed] [Google Scholar]
- 68.Wood, L. F., A. J. Leech, and D. E. Ohman. 2006. Cell wall-inhibitory antibiotics activate the alginate biosynthesis operon in Pseudomonas aeruginosa: roles of σ22 (AlgT) and the AlgW and Prc proteases. Mol. Microbiol. 62:412-426. [DOI] [PubMed] [Google Scholar]
- 69.Wozniak, D. J., T. J. O. Wyckhoff, M. Starkey, R. Keyser, P. Azadi, G. A. O'Toole, and M. R. Parsek. 2003. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. U. S. A. 100:7907-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]