Abstract
The Escherichia coli RNA degradosome proteins are organized into a helical cytoskeletal-like structure within the cell. Here we describe the ATP-dependent assembly of the RhlB component of the degradosome into polymeric filamentous structures in vitro, which suggests that extended polymers of RhlB are likely to comprise a basic core element of the degradosome cytoskeletal structures.
The RNA degradosome plays an essential role in normal RNA processing and degradation. Within the cell, the degradosome proteins (RNA helicase B [RhlB], RNase E, polynucleotide phosphorylase [PNPase], and enolase) (4, 13, 15, 16) are organized into coiled structures that resemble the pole-to-pole helical structures of the MreB and MinCDE bacterial cytoskeletal systems (4, 12, 13). However, the degradosomal structures are also present in cells that lack the MreB and MinCDE cytoskeletal elements, suggesting that the degradosomal structures may be part of an independent class of prokaryotic cytoskeletal elements (19-21).
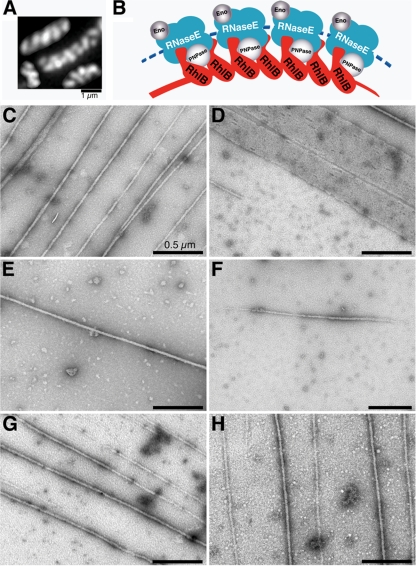
One of the degradosomal proteins, RhlB, is organized into similar helical cellular structures in cells that lack the other degradosome proteins (Fig. 1 A). In addition, RhlB recruits PNPase to the helical framework in the absence of other degradosome proteins, suggesting that the RhlB structures are core elements of the degradosomal cytoskeletal-like elements of the cell (Fig. 1B) (20). The cellular RhlB structures could be generated in two ways: (i) individual RhlB molecules may bind to an as-yet-undefined underlying track, or (ii) RhlB may polymerize to form the filamentous helical structures independent of any underlying template.
FIG. 1.
The RhlB filamentous cytoskeletal-like structures. (A) Cellular organization of RhlB based on immunofluorescence microscopy using purified anti-RhlB antibody in the absence of RNase E filamentous elements in AT8 cells (rne1-417), which fail to generate RNase E coiled structures because of the absence of the RNase E cytoskeletal localization domain (20). (B) Proposed model for the cytoskeletal-like organization of the RNA degradosome (modified from reference 20). Arcs depict the RNase E (blue) and RhlB (red) helical strands. It is not known whether the RNase E helical strand is formed by RNase E polymerization or by the association of RNase E with an unknown underlying cytoskeletal structure. Enolase (Eno) and PNPase are shown in gray. Molecular dimensions and stoichiometry of the proteins were arbitrarily chosen to simplify the figure. (C to H) Electron micrographs of uranyl acetate-stained RhlB filaments (C, E, and H) and RhlB sheets (D). Unless otherwise indicated, the sample contained 9 μM RhlB, 2 mM ATP, 5 mM MgCl2, and 5 mM CaCl2. (E) Calcium was omitted. (F) ATP was omitted. (G and H) ATP was replaced by ATPγS (G) or AMP-PNP (H). Samples were loaded on glow-discharged 300-mesh carbon-coated copper grids and then stained. Images were taken with a JEOL 100CX transmission electron microscope. Magnification, ×10,000 to 50,000.
Here we report that RhlB can self-assemble into extended polymeric structures in vitro in a process that requires ATP binding but not ATP hydrolysis. It is likely that extended RhlB polymers such as those described here are the basic components of the RhlB filamentous helical elements that comprise the core of the degradosomal cytoskeletal structures of the Escherichia coli cell.
Evidence that RhlB can self-assemble into filamentous polymeric structures came from electron microscopic studies of purified His-tagged RhlB negatively stained with 2% uranyl acetate. This staining showed large numbers of long uniform filamentous structures when the purified protein was incubated in the presence of ATP and Ca2+ (Fig. 1C). The filaments were 25 ± 1.8 nm wide (n = 91; mean ± standard deviation) and were generally more than 10 μm long. Some wider sheets were also observed (Fig. 1D). Optimal assembly of the RhlB filamentous structures required ATP and Ca2+, as shown by the observation that only occasional single structures were present when the polymerization reaction was carried out in the absence of Ca2+ (Fig. 1E) or ATP (Fig. 1F). The polymeric RhlB-His structures were observed with approximately similar frequencies when ATP was replaced by the nonhydrolyzable ATP analog adenosine 5′-(γ-thiotriphosphate) (ATPγS) or AMP-PNP (Fig. 1G and H).
Cellular localization studies showed that the presence of the His tag did not interfere with the ability of RhlB to form the helical cellular structures. Thus, RhlB-His was present in extended helical filamentous structures that were indistinguishable from those formed by untagged RhlB (20, 21). Similarly, the RhlB-His structures recruited PNPase to the helical framework in a manner similar to untagged RhlB (20) (see Fig. S1 in the supplemental material).
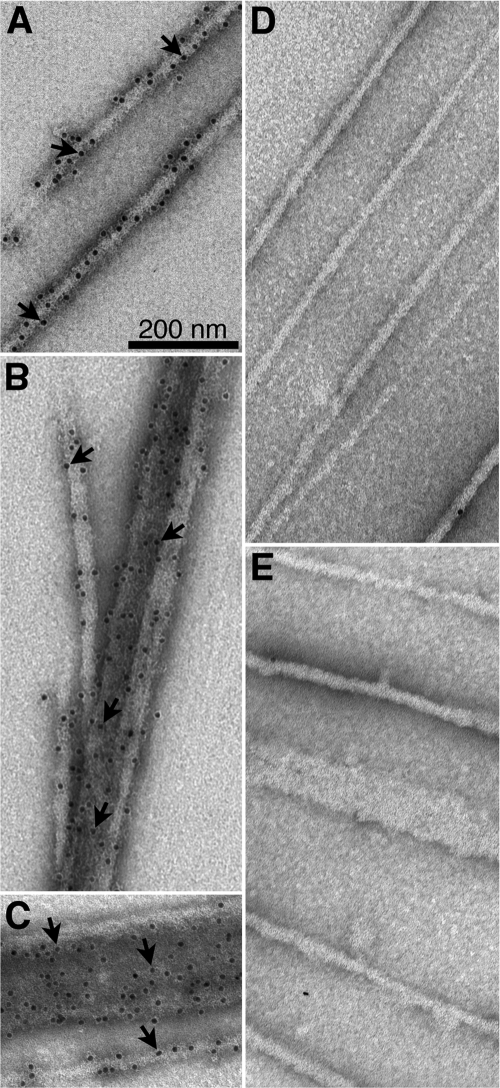
Immunogold staining showed that the filaments and sheets were decorated with gold particles when stained with mouse anti-His tag antibody and gold-labeled secondary antibody (Fig. 2 A to C), confirming that the structures were composed of RhlB. In contrast, the structures were not decorated with gold particles in the absence of the primary antibody or when mouse anti-His tag antibody was replaced by nonimmune mouse IgG (Fig. 2D and E). The polymeric structures were observed with C-terminally His-tagged RhlB, which is functional in terms of helicase activity (8), but not when the tag was present at the amino terminus of the protein, where the His tag may interfere with RhlB self-assembly.
FIG. 2.
Immunogold electron microscopy of RhlB structures. Samples were prepared as describe for Fig. 1C, except that the grids were stained with 2% uranyl acetate after exposure to primary and/or secondary antibodies as indicated. (A to C) RhlB structures decorated with 10-nm gold particles in samples stained with mouse anti-His tag monoclonal antibody and gold-labeled secondary antibody. (A) single filaments; (B) clustered filaments; (C) a single RhlB filament and RhlB sheet. Arrows indicate gold particles. (D) The primary mouse anti-His tag antibody was replaced by mouse IgG. (E) The primary antibody was omitted.
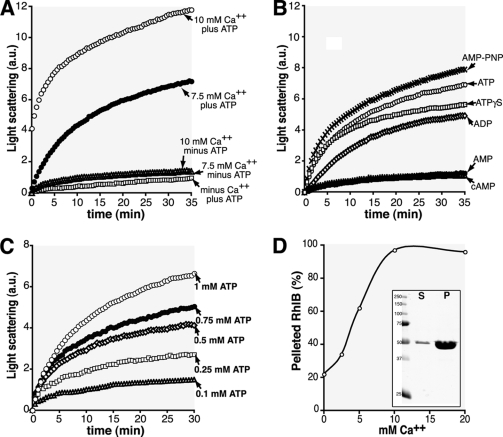
Changes in light scattering were used to follow the course of polymerization and to compare polymerization conditions in a more quantitative way than is possible by electron microscopy. The initial rate of increase in scattering was used to estimate polymerization rate (see Table S1 in the supplemental material). Significant rates of polymerization were observed in the presence of ATP and Ca2+, whereas there was very little increase in light scattering in the absence of nucleotide and/or Ca2+ (Fig. 3 A). ADP was less effective than ATP, whereas AMP and cyclic AMP (cAMP) were inactive (Fig. 3B). In the presence of Ca2+, the extent and rate of RhlB polymerization varied as a function of ATP concentration (Fig. 3C). Millimolar concentrations of Ca2+ were required to produce a measurable rate of polymerization in the light scattering assay (Fig. 3A). It is not known how these relatively high concentrations of Ca2+ promote the in vitro polymerization of RhlB and other cytoskeletal proteins, such as MreB and FtsZ (1, 11, 12, 14, 23).
FIG. 3.
RhlB polymerization. (A and B) RhlB polymerization as shown by 90° light scattering is indicated in arbitrary units (a.u.). RhlB polymerization was followed at room temperature in a 1-cm light path quartz cuvette using a Hitachi fluorometer (FL-2500) set to 400 V with excitation and emission wavelengths set at 455 nm and a slit width of 10 nm. The reaction (100 μl volume) was performed in a polymerization buffer (50 mM Tris, 50 mM KCl, 5 mM MgCl2; pH 8) as indicated. (A) The sample contained 9 μM RhlB-His, 1 mM ATP, and either 10 mM CaCl2, 7.5 mM CaCl2, or no CaCl2 (squares). In the lower three curves the samples lacked ATP or CaCl2, as indicated. (B) The sample contained 9 μM RhlB-His, 7.5 mM CaCl2, and 1 mM adenosine nucleotides: ATP, ADP, AMP, cAMP, AMP-PNP, and ATPγS. (C) The sample contained 9 μM RhlB-His, 7.5 mM CaCl2, and ATP as indicated (1 mM, 0.75 mM, 0.5 mM, 0.25 mM, or 0.1 mM ATP). (D) Effects of Ca2+ concentration on RhlB sedimentation in the presence of 2 mM ATP. RhlB in the pellet, expressed as a percentage of total RhlB present in the polymerization reaction mixture, was plotted against calcium concentration. The insert shows an example of a Coomassie blue-stained gel of supernatant (S) and pellet (P) fractions from the sedimentation assay in the presence of ATP and Ca2+ (see results for ATP in Table 1).
The nonhydrolyzable ATP analogs ATPγS and AMP-PNP were approximately equivalent to ATP in promoting polymerization as monitored by the light scattering assay (Fig. 3B) as well as in the electron microscopic studies. This suggests that nucleotide binding, but not hydrolysis, is required to promote RhlB polymerization. In this regard, RhlB resembles a number of other proteins, including F-actin, MreB, and MinD, where polymerization is induced by nucleotide binding (2, 5, 6, 18, 22). In these systems, subsequent ATP hydrolysis induces depolymerization, providing the basis for the dynamic behavior of the polymers within the cell. RhlB is an RNA-dependent ATPase (7), but it is not yet known whether ATP hydrolysis is associated with depolymerization in the RhlB system.
Similar results were obtained when the extent of polymerization was monitored by a sedimentation assay, measuring the proportion of RhlB in the pellet fraction after centrifugation at 278,000 × g for 10 min (Table 1; Fig. 3D). Essentially all of the protein was sedimentable at pH 8 in the presence of Ca2+ and ATP. RhlB sedimentation returned to background levels when EGTA or EDTA was added to the reaction mixture (Table 1), confirming the Ca2+ requirement for RhlB polymerization in the electron microscopic and light scattering analyses. ATPγS was equivalent to ATP in the sedimentation assay, confirming the results described above. The relatively high background of RhlB sedimentation was not affected by prespinning the samples prior to addition of nucleotides and/or Ca2+.
TABLE 1.
Sedimentation assay for RhlB polymerization
| Nucleotide added | % Sedimented RhlBa |
|---|---|
| No additionb | 23 |
| No nucleotide | 28 |
| ATPc | 97 |
| ADP | 58 |
| ATPγS | 99 |
| ATP + EDTA | 22 |
| ATP + EGTA | 20 |
RhlB present in the pellet is expressed as the percentage of total RhlB present in the polymerization reaction mixture. The polymerization reaction mixture contained, unless otherwise indicated, 9 μM RhlB, 5 mM MgCl2, 7.5 mM CaCl2, 2 mM nucleotide and, when present, 7.5 mM EDTA or EGTA.
The reaction was performed in the absence of nucleotides, MgCl2, and CaCl2.
Similar results were obtained when the RhlB concentration was varied between 0.4 and 18 μM (data not shown).
The ability of RhlB to self-assemble in vitro into polymeric structures is of special interest because RhlB also assembles into extended filamentous structures in vivo, where the filaments are organized as pole-to-pole helical cytoskeletal-like elements. This occurs in the absence of RNase E helical structures (Fig. 1A) and in the absence of the other degradosomal proteins or of known cytoskeletal proteins that might provide nucleation sites or tracks for filament assembly (20). We suggest that extended RhlB filamentous homopolymers such as those seen in the in vitro system are likely to be fundamental elements of the filamentous degradosome cytoskeletal-like structures of the cell. RhlB-RhlB interactions that could participate in RhlB self-assembly into polymeric structures have been observed in bacterial two-hybrid and BIAcore surface plasmon resonance studies (9).
The number of RhlB molecules per E. coli cell (approximately 1,350 [see Fig. S1E in the supplemental material]) would be sufficient to form a single 8- to 10-μm-long polymer that winds around the length of a 2-μm cell as a three- to four-turn helical structure, assuming that the dimensions of the RhlB monomer are similar to those of the RNA helicase of Methanococcus jannaschii (≈70 Å) (17). Therefore, the cellular cytoskeletal-like structure conceivably could be composed of a single RhlB homopolymer. Alternatively, the cellular structure could be composed of a number of shorter parallel RhlB polymers, held together by a linking protein(s) or by direct side-to-side interactions near the ends of the polymeric protofilaments.
If the present interpretations are correct, RhlB plays two roles in the cell, acting both as an RNA unwinding enzyme and as an organizing element of the cytoskeletal-like degradosome structures. Within these structures RhlB cooperates with other degradosome proteins by catalyzing the unwinding of double-stranded RNA regions as part of the pathway of degradation of cellular RNAs by the RNase E and PNPase components of the RNA degradosome (reviewed in reference 3). Might there be any role for RhlB polymerization other than to help organize the degradosome structure? The mechanism of RNA duplex unwinding by RhlB is not known, but it is conceivable that dynamic RhlB polymerization between the RNA strands of a structured RNA substrate could provide the driving force for RNA unwinding. In this context, it has recently been reported that RNA unwinding by other DEAD box RNA helicases does not require ATP hydrolysis but does require ATP binding (10), thereby mimicking the requirements for RhlB polymerization reported here.
Supplementary Material
Acknowledgments
We thank L. Rothfield, M. Osborn, and A. Das for helpful discussions and A. Hand and J. Aghajanian for help with electron microscopy.
This work was supported by NIH grant R37 GM060632.
Footnotes
Published ahead of print on 9 April 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bean, G. J., and K. J. Amann. 2008. Polymerization properties of the Thermotoga maritimactin MreB: roles of temperature, nucleotides, and ions. Biochemistry. 47:826-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlier, M. F. 1990. Actin polymerization and ATP hydrolysis. Adv. Biophys. 26:51-73. [DOI] [PubMed] [Google Scholar]
- 3.Carpousis, A. J. 2007. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 61:71-87. [DOI] [PubMed] [Google Scholar]
- 4.Carpousis, A. J., G. Van Houwe, C. Ehretsmann, and H. M. Krisch. 1994. Copurification of E. coli RNase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell 76:889-900. [DOI] [PubMed] [Google Scholar]
- 5.Esue, O., M. Cordero, D. Wirtz, and Y. Tseng. 2005. The assembly of MreB, a prokaryotic homolog of actin. J. Biol. Chem. 280:2628-2635. [DOI] [PubMed] [Google Scholar]
- 6.Hu, Z., E. P. Gogol, and J. Lutkenhaus. 2002. Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE. Proc. Natl. Acad. Sci. U. S. A. 99:6761-6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalman, M., H. Murphy, and M. Cashel. 1991. rhlB, a new Escherichia coli K-12 gene with an RNA helicase-like protein sequence motif, one of at least five such possible genes in a prokaryote. New Biol. 3:886-895. [PubMed] [Google Scholar]
- 8.Lin, P. H., and S. Lin-Chao. 2005. RhlB helicase rather than enolase is the beta-subunit of the Escherichia coli polynucleotide phosphorylase (PNPase)-exoribonucleolytic complex. Proc. Natl. Acad. Sci. U. S. A. 102:16590-16595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liou, G. G., H. Y. Chang, C. S. Lin, and S. Lin-Chao. 2002. DEAD box RhlB RNA helicase physically associates with exoribonuclease PNPase to degrade double-stranded RNA independent of the degradosome-assembling region of RNase E. J. Biol. Chem. 277:41157-41162. [DOI] [PubMed] [Google Scholar]
- 10.Liu, F., A. Putnam, and E. Jankowsky. 2008. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc. Natl. Acad. Sci. U. S. A. 105:20209-20214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Löwe, J., and L. A. Amos. 1999. Tubulin-like protofilaments in Ca2+-induced FtsZ sheets. EMBO J. 18:2364-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marrington, R., E. Small, A. Rodger, T. R. Dafforn, and S. G. Addinall. 2004. FtsZ fiber bundling is triggered by a conformational change in bound GTP. J. Biol. Chem. 279:48821-48829. [DOI] [PubMed] [Google Scholar]
- 13.Miczak, A., V. R. Kaberdin, C. L. Wei, and S. Lin-Chao. 1996. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc. Natl. Acad. Sci. U. S. A. 93:3865-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukherjee, A., and J. Lutkenhaus. 1999. Analysis of FtsZ assembly by light scattering and determination of the role of divalent metal cations. J. Bacteriol. 181:823-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Py, B., H. Causton, E. A. Mudd, and C. F. Higgins. 1994. A protein complex mediating mRNA degradation in Escherichia coli. Mol. Microbiol. 14:717-729. [DOI] [PubMed] [Google Scholar]
- 16.Py, B., C. F. Higgins, H. M. Krisch, and A. J. Carpousis. 1996. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381:169-172. [DOI] [PubMed] [Google Scholar]
- 17.Story, R. M., H. Li, and J. N. Abelson. 2001. Crystal structure of a DEAD box protein from the hyperthermophile Methanococcus jannaschii. Proc. Natl. Acad. Sci. U. S. A. 98:1465-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suefuji, K., R. Valluzzi, and D. RayChaudhuri. 2002. Dynamic assembly of MinD into filament bundles modulated by ATP, phospholipids, and MinE. Proc. Natl. Acad. Sci. U. S. A. 99:16776-16781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taghbalout, A., and L. Rothfield. 2008. New insights into the cellular organization of the RNA processing and degradation machinery of Escherichia coli. Mol. Microbiol. 70:780-782. [DOI] [PubMed] [Google Scholar]
- 20.Taghbalout, A., and L. Rothfield. 2008. RNase E and RNA helicase B play central roles in the cytoskeletal organization of the RNA degradosome. J. Biol. Chem. 283:13850-13855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taghbalout, A., and L. Rothfield. 2007. RNase E and the other constituents of the RNA degradosome are components of the bacterial cytoskeleton. Proc. Natl. Acad. Sci. U. S. A. 104:1667-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van den Ent, F., L. A. Amos, and J. Lowe. 2001. Prokaryotic origin of the actin cytoskeleton. Nature 413:39-44. [DOI] [PubMed] [Google Scholar]
- 23.Yu, X. C., and W. Margolin. 1997. Ca2+-mediated GTP-dependent dynamic assembly of bacterial cell division protein FtsZ into asters and polymer networks in vitro. EMBO J. 16:5455-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.