Abstract
We identified a pathway in Bacillus subtilis that is used for recovery of N-acetylglucosamine (GlcNAc)-N-acetylmuramic acid (MurNAc) peptides (muropeptides) derived from the peptidoglycan of the cell wall. This pathway is encoded by a cluster of six genes, the first three of which are orthologs of Escherichia coli genes involved in N-acetylmuramic acid dissimilation and encode a MurNAc-6-phosphate etherase (MurQ), a MurNAc-6-phosphate-specific transcriptional regulator (MurR), and a MurNAc-specific phosphotransferase system (MurP). Here we characterized two other genes of this cluster. The first gene was shown to encode a cell wall-associated β-N-acetylglucosaminidase (NagZ, formerly YbbD) that cleaves the terminal nonreducing N-acetylglucosamine of muropeptides and also accepts chromogenic or fluorogenic β-N-acetylglucosaminides. The second gene was shown to encode an amidase (AmiE, formerly YbbE) that hydrolyzes the N-acetylmuramyl-l-Ala bond of MurNAc peptides but not this bond of muropeptides. Hence, AmiE requires NagZ, and in conjunction these enzymes liberate MurNAc by sequential hydrolysis of muropeptides. NagZ expression was induced at late exponential phase, and it was 6-fold higher in stationary phase. NagZ is noncovalently associated with lysozyme-degradable particulate material and can be released from it with salt. A nagZ mutant accumulates muropeptides in the spent medium and displays a lytic phenotype in late stationary phase. The evidence for a muropeptide catabolic pathway presented here is the first evidence for cell wall recovery in a Gram-positive organism, and this pathway is distinct from the cell wall recycling pathway of E. coli and other Gram-negative bacteria.
Bacteria are covered by an exoskeleton-like cell wall that protects the fragile membrane-enclosed cell, the protoplast, and withstands the high internal pressure of the cell, the turgor pressure (27). Despite its stabilizing function, the cell wall is not rigid and static but is highly flexible. It undergoes continuous resynthesis, remodeling, and degradation (turnover) in which a substantial amount of the murein (peptidoglycan), the stabilizing component of the bacterial cell wall, is released during logarithmic growth (5, 47). The Gram-positive model organism Bacillus subtilis was shown to release about 50% of its murein into the medium in one generation during growth (9, 42, 43). Gram-negative bacteria have an outer membrane that keeps most of the cell wall turnover products in the periplasmic space, but Gram-positive bacteria lack such a membrane barrier and therefore cannot retain their turnover products. So far, more than 30 peptidoglycan hydrolases of B. subtilis have been characterized or identified as candidate autolysins on the basis of amino acid sequence identity (50), but the nature of the turnover products remains ambiguous. Moreover, a cell wall recycling-recovery pathway has not been identified in this organism or in any other Gram-positive bacterium so far.
In contrast, cell wall turnover and recycling in the Gram-negative model bacterium Escherichia coli have been well studied (for a recent review, see reference 47). E. coli also degrades about 50% of the murein of the cell wall in each generation during growth, and the turnover products are efficiently reutilized (i.e., recycled) (23). The principal degradation products are N-acetylglucosamine (GlcNAc)-1,6-anhydro-N-acetylmuramic acid (anhMurNAc) peptides (anhydromuropeptides), which are released from the cell wall during cell growth by the action of lytic transglycosylases, endopeptidases, and carboxypeptidases, yielding primarily GlcNAc--anhMurNAc-l-Ala-γ-d-Glu-meso-diaminopimelic acid (m-DAP)-d-Ala (for a review, see reference 27). Anhydromuropeptides are taken up by the AmpG secondary transport system (12) and are degraded further in the cytoplasm by NagZ, a β-N-acetylglucosaminidase (11, 59), AmpD, an anhMurNAc-l-Ala amidase (28, 34), and LdcA, an ld-carboxypeptidase (53), which together generate GlcNAc, anhMurNAc, d-Ala, and l-Ala-γ-d-Glu-m-DAP in the cytosol. The tripeptide is directly fed into the murein biosynthesis pathway by the muropeptide ligase Mpl, which transfers the tripeptide to UDP-N-acetylmuramic acid (MurNAc) (44). The tripeptide may also be degraded to its individual amino acids by γ-d-glutaminyl-m-DAP amidase (MpaA) and l-Ala-dl-Glu epimerase (YcjG) (49, 54). GlcNAc and anhMurNAc are also recycled (46). GlcNAc is converted to GlcNAc-6-phosphate by the GlcNAc kinase NagK (55), whereas anhMurNAc is first phosphorylated by the kinase AnmK, yielding MurNAc-6-phosphate, which is then converted to GlcNAc-6-phosphate by the etherase MurQ, which cleaves the lactyl ether substituent of MurNAc-6-phosphate (24, 35, 36, 56, 57). Besides recycling anhydromuropeptides, E. coli was shown to utilize MurNAc as a sole source of carbon (13). MurNAc is imported and phosphorylated by the MurP-specific phosphotransferase system, yielding cytoplasmic MurNAc-6-phosphate (13). MurQ, the same lactyl etherase that is required for recycling of anhMurNAc, is also essential for growth on MurNAc (35). Recently, the transcriptional regulator MurR was characterized, which, in conjunction with the catabolite activator protein (CAP), regulates the MurNAc etherase operon in E. coli (37).
In this work we identified a muropeptide rescue pathway in B. subtilis and characterized two of the proteins involved. The first protein, B. subtilis NagZ (BsNagZ) (formerly YbbD) is an ortholog of the cell wall recycling β-N-acetylglucosaminidases of E. coli (EcNagZ), but in contrast to the latter enzymes, it is secreted and has an additional C-terminal domain with an unknown function. BsNagZ occurs in the late exponential and stationary growth phases mainly in a particulate form in the supernatant. It cleaves cell wall-derived muropeptides, thereby generating a substrate for a second enzyme of the pathway, BsAmiE (formerly YbbE), an N-acetylmuramyl-l-alanine amidase. A nagZ deletion strain accumulates muropeptides in the medium, and the cells lyse at late stationary phase. We recognized that BsNagZ is identical to an enzyme that was characterized in the early 1970s and was proposed to be involved in recycling of the endogenous cell wall during growth (2, 45).
MATERIALS AND METHODS
Bacterial strains, plasmids, chemicals, and growth conditions.
E. coli and B. subtilis strains, as well as plasmids used in this work, are listed in Table 1. Bacteria were grown aerobically at 37°C in LB medium. When appropriate, the chromosomal drug resistance marker in B. subtilis was selected with erythromycin (1 μg/ml). E. coli cells carrying plasmids were selected with ampicillin (100 μg/ml). 4-Methylumbelliferyl-β-d-glucosaminide (4-Mu-β-GlcNH2) was synthesized (see the supplemental material). GlcNAc-MurNAc was obtained from Carbosynth Ltd., Berkshire, United Kingdom. All other reagents were obtained from Sigma-Aldrich unless otherwise stated.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference and/or source |
|---|---|---|
| E. coli strains | ||
| MG1655 | Sequenced E. coli K-12 strain | 4 |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | 52 |
| B. subtilis strains | ||
| 168 | trpC2; sequenced B. subtilis strain | Bacillus Genetic Stock Center |
| ybbD::pMUTIN4 | pMUTIN4 insertion in BsnagZ, Ermr | Bacillus Knockout Collection (http://bacillus.genome.jp/) (38) |
| Plasmids | ||
| pET16b | E. coli expression vector, PT7 AmproripBR322lacI, N-terminal His10 tag | Novagen |
| pASK-IBA32 | E. coli expression vector, Ptet Amprorif1ompA signal sequence, C-terminal His6 tag | IBA |
| pET16b-BsNagZ | IPTG-inducible cytoplasmic BsNagZ-His6 expression vector | This study |
| pASK-IBA32-BsAmiE | Anhydrotetracycline-inducible periplasmic BsAmiE His6 expression vector | This study |
Ampr, ampicillin resistant; Ermr, erythromycin resistant.
Cloning of nagZ and amiE.
DNA preparation, restriction enzyme digestion, ligation, and transformation were performed using standard techniques. BsnagZ was amplified by PCR without a signal sequence using 30 ng of chromosomal DNA of B. subtilis 168, 5 U Pwo DNA polymerase (Genaxxon Biosciences, Biberach, Germany), and the following primers: BsNagZ/FP (5′-AAA ACC ATG GGC CAT ATG TTT TTC GGG GCC AGA CAG AC-3′) and BsNagZ/RP (5′-T TTT CTC GAG TTA AAG CGG TCT TCC CGT TTT G-3′) (underlining indicates the recognition sites for restriction endonucleases NdeI and XhoI, respectively). BsamiE was amplified without a signal sequence using Phusion DNA polymerase (Finnzymes Oy, Espoo, Finland) and the following primers with recognition sites for restriction endonuclease BsaI (underlined): AmiE/FP (5′-ATC GTC GGT CTC AGG CCC AAA CAG CAG GCA ACT TGA TTG AG-3′) and AmiE/RP (5′-ATC GTC GGT CTC AGC GCT CTC CAT CGC TTC ATA AAT CGC G-3′). Thirty cycles of 15 s at 94°C, 30 s at 55°C, and 120 s at 72°C for nagZ and 35 cycles of 10 s at 98°C, 30 s at 50°C, and 30 s at 72°C for amiE were performed with a thermal cycler, and the results revealed single 1.9-kb (nagZ) and 1.3-kb (amiE) fragments as analyzed by agarose gel electrophoresis. BsnagZ was cloned into the vector pET16b under control of the T7 promoter (pET16b-BsNagZ), and its product was overexpressed in E. coli as a cytoplasmic N-terminal His10-tagged fusion protein. BsamiE was cloned into the vector pASK-IBA32 (pASK-IBA32-BsAmiE) under control of the tetracycline promoter, and its product was overexpressed in E. coli as a periplasmic C-terminal His6-tagged fusion protein.
Overexpression and purification of NagZ and AmiE.
E. coli strain BL21(DE3) harboring pET16b-BsNagZ and E. coli strain BL21(DE3) harboring pASK-IBA32-BsAmiE were grown at 37°C in LB medium supplemented with ampicillin (100 μg/ml) with vigorous shaking. Expression of BsNagZ was induced at log phase (optical density at 578 nm [OD578], 0.5 to 0.6) by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 1 mM; expression of BsAmiE was induced at an OD578 of 1.5 by addition of 200 μg liter−1 anhydrotetracycline. Then the cultures were incubated for another 3 h. Cells were harvested by centrifugation (4,000 × g for 30 min at 4°C), resuspended in ice-cold sodium phosphate buffer (20 mM Na2HPO4·2H2O, 500 mM NaCl; pH 7.5), and disrupted by passage through a French pressure cell three times. Debris and unbroken cells were removed by centrifugation at 100,000 × g for 1 h at 4°C. The His-tagged proteins in the supernatant were purified by Ni2+ affinity chromatography with a 5-ml HiTrap HP column (Amersham-Pharmacia, Freiburg, Germany) preequilibrated with 20 mM sodium phosphate buffer (pH 7.5) and were eluted from the column with elution buffer (sodium phosphate buffer supplemented with 500 mM imidazole, pH 7.5). The purity of the enzymes was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Peak fractions containing pure protein were concentrated and desalted by dialysis at 4°C against sodium phosphate buffer for enzymatic assays. The protein concentrations were determined by UV absorption at 280 nm (the calculated molar extinction coefficients [ɛ280] for NagZ and AmiE are 36,330 M−1 cm−1 and 50,770 M−1 cm−1, respectively) and by the Bradford method with bovine serum albumin as the standard (6).
Sedimentation of NagZ and extraction with salt.
Eight-milliliter portions of 12-h cultures of B. subtilis grown in 50 ml of LB medium in 300-ml Erlenmeyer flasks at 37°C with vigorous shaking were fractionated by centrifugation at 1,200 × g for 10 min, at 30,000 × g for 30 min, or at 100,000 × g for 30 min. The pellets were resuspended in 1 ml Clark and Lubs buffer (0.1 M KH2PO4, 0.1 M NaOH; pH 5.8), and the supernatants were concentrated to 1 ml using Amicon Ultra-15 centrifugal filter devices (Millipore). The amounts of the enzyme present in each supernatant and in each pellet after centrifugation were determined as described below.
Two-milliliter portions of the 12-h cultures were mixed with 6 ml of aqueous solutions of NaCl at different concentrations in 0.05 M Tris-HCl buffer (pH 8.0). The cell suspensions were incubated at room temperature for 20 min and then centrifuged at 30,000 × g for 30 min, using a previously described protocol (45). The sedimented cells were resuspended in 1 ml Clark and Lubs buffer (pH 5.8), and the amounts of NagZ remaining in the pellets were determined as described below.
Expression of NagZ during growth.
NagZ expression was assayed in B. subtilis strain 168 and the ybbD::pMUTIN4 strain; both strains were grown in 1 liter LB medium at 37°C in 5-liter shaking flasks. Growth was monitored by measuring the optical density at 600 nm using an Ultrospec 3000 spectrophotometer (Pharmacia Biotech). A volume that was equivalent to an OD600 of 10 per ml was collected at different time points, and cells were sedimented by centrifugation at 30,000 × g for 30 min. The cell sediment was suspended in 1 ml of Clark and Lubs buffer (pH 5.8) (see above), and the enzyme activity was determined as described below.
Enzyme assays.
NagZ activity was determined using 300-μl reaction mixtures containing equal volumes of an enzyme solution (cell suspension or supernatant), Clark and Lubs buffer (pH 5.8) (see above), and substrate (either 3 mM 4-methylumbelliferyl-β-N-acetyl-d-glucosaminide [4-Mu-β-GlcNAc] or 4-nitrophenyl-β-N-acetyl-d-glucosaminide [pNP-β-GlcNAc]) that were incubated at 37°C in 96-well plates (Greiner Bio-one) for 5 to 30 min. The reaction was initiated by addition of preheated substrate. The release of 4-methylumbelliferone was determined by measurement of fluorescence with a Spectramax M2 microplate reader (Molecular Devices) with an excitation wavelength of 362 nm and an emission wavelength of 448 nm. The release of 4-nitrophenol was determined with the same device by measuring the absorption at 400 nm. One unit was defined as the amount of enzyme that hydrolyzed 1 μmol of substrate per min at pH 5.8 and 37°C. The extinction coefficient was determined by calibration using 4-methylumbelliferone and 4-nitrophenol standards. Kinetic parameters were determined using different concentrations of 4-Mu-β-GlcNAc in Clark and Lubs buffer (pH 5.8) and were evaluated by nonlinear regression of the reaction curve using the program Prism4 (GraphPad). The pH activity profile of NagZ was determined by using 0.1 M citric acid-0.2 M disodium phosphate buffer (McIlvaine) having pHs ranging from 4.0 to 8.0, 0.2 M sodium acetate acetic acid buffer having pHs ranging from 4.0 to 5.6, and Clark and Lubs buffer having pHs ranging from 5.8 to 8.0. The 300-μl enzyme reaction mixtures containing 1.5 mM 4-Mu-β-GlcNAc and 1.5 × 10−3 mg ml−1 NagZ were incubated for 5 to 30 min at 37°C, and the reactions were stopped by adding 9 volumes of a 0.2 M Na2CO3 solution (pH 10.0).
Preparation of peptidoglycan from E. coli and B. subtilis.
Peptidoglycan was isolated from B. subtilis 168 and E. coli MG1655 cells as described previously (20, 25, 61). In brief, bacteria were grown at 37°C and harvested at an OD578 of 0.5 to 0.7. Cells were rapidly cooled to 4°C, which was followed by centrifugation at 4,000 × g for 30 min at 4°C. Cell pellets were resuspended in ice-cold distilled water (0.2 g/ml) and added dropwise with vigorous stirring to an equal volume of boiling 8% sodium dodecyl sulfate (SDS). When all of the cells had been added, the suspensions were boiled for 30 min to solubilize the membranes and degrade the DNA. The lysate was then cooled to room temperature (RT), and the SDS-insoluble material was collected by centrifugation (100,000 × g for 60 min at RT). The pellet was resuspended in H2O and dialyzed against H2O until most of the SDS was removed. The SDS content in the supernatant was controlled by using the methylene blue assay as described previously (26). Then the peptidoglycan preparation was pelleted again by ultracentrifugation and resuspended in a minimal volume of H2O. Contaminating high-molecular-weight glycogen trapped in the peptidoglycan preparation (39) was degraded by adding 100 μg/ml α-amylase (Bacillus licheniformis type XII-A in 10 mM Tris-HCl [pH 7.0], 10 mM NaCl) and incubating the preparation for 2 h at 37°C. To inhibit lysozyme contamination in the α-amylase preparation, 0.32 M imidazole (in 10 mM Tris-HCl [pH 7.0], 10 mM NaCl) was added. After this, lipoproteins covalently bound to E. coli peptidoglycan (7, 8) were degraded by incubation with 200 μg/ml pronase (in 10 mM Tris-HCl [pH 7.0], 10 mM NaCl) for 1.5 h at 60°C. Pronase (type XXV from Streptomyces griseus) was pretreated by incubation for 2 h at 60°C. The sample was then added to an equal volume of boiling 8% SDS and boiled for 15 min. Then the purified peptidoglycan was ultracentrifuged, and the SDS was removed by dialysis as described above.
Preparation of muropeptides.
Muropeptides were prepared by digestion of peptidoglycan at 37°C overnight with 1 μg/40 μl mutanolysin (from Streptomyces globisporus; 1 mg/ml in 20 mM sodium phosphate buffer, pH 5.5). The enzyme reaction was stopped by boiling the mixture for 5 min, insoluble contaminants were removed by centrifugation, and the released muropeptides in the supernatant were stored at −20°C. After digestion with mutanolysin, the muropeptides released from the peptidoglycan contained MurNAc as the terminal reducing sugar, which could be labeled with 8-aminonaphthalene-1,3,6-trisulfonic acid (ANTS) and separated by polyacrylamide gel electrophoresis as described below (61).
Muropeptides were also prepared from spent medium. B. subtilis wild-type and nagZ mutant cells were grown for 2 days in minimal salt medium supplemented with 0.2% succinate as the carbon source. Ten milliliters of spent medium was dried with a SpeedVac, the dried material was dissolved in 0.5 ml water, and 20 μl was subjected to high-performance liquid chromatography (HPLC) analysis.
Muropeptide degradation by NagZ and AmiE.
Muropeptides, as well as intact peptidoglycan from E. coli or B. subtilis, were incubated with purified NagZ and AmiE (5 μg/40 μl) at 37°C for 2 h, and each reaction was monitored by using reversed-phase HPLC (RP-HPLC) and fluorophore-assisted carbohydrate electrophoresis (FACE). An RP-C18 column (Gemini 5 μ C18; 150 by 4.60 mm; Phenomenex) was used to separate the muropeptides and other cell wall components, which were monitored at multiple wavelengths (205, 235, and 280 nm) using a UV detector (UltiMate 3000 photodiode array detector). Samples (25 μl) were applied to the HPLC column, and elution was performed for 5 min with buffer A (0.05% trifluoroacetic acid) and then for 50 min with a linear gradient from 0 to 100% buffer B (0.035% trifluoroacetic acid and 10% CH3CN) at a flow rate of 0.5 ml/min (as described previously [11]). The peaks obtained were collected and lyophilized. One part of the samples was tagged with ANTS and separated by FACE, as described below, and another part was analyzed by matrix-assisted laser desorption ionization (MALDI)-time of flight (TOF) mass spectrometry. For analysis by the MALDI-TOF method the samples were dissolved in acetonitrile, and 0.7 μl of a sample solution was mixed with 0.7 μl of a matrix solution (a saturated solution of cyano-4-hydroxycinnamic acid in acetonitrile) directly on the MALDI target. MALDI-TOF mass spectra were recorded with a Bruker Biflex III spectrometer in positive reflectron mode with a delayed-extraction MALDI source and a pulsed nitrogen laser (337 nm).
Fluorescent labeling of carbohydrate standards and muropeptides.
Reducing carbohydrates and muropeptides were tagged at their terminal semiacetal with the charged, fluorescent molecule 8-aminonaphthalene-1,3,6-trisulfonic acid (ANTS) as described previously (32, 51). Carbohydrate stock solutions (10 mM) were prepared from d-glucose, maltose, maltotriose, maltotetraose, maltopentaose, maltohexaose, maltoheptaose, GlcNAc, MurNAc, and GlcNAc-MurNAc in water. One-micromole aliquots of the carbohydrate solutions and 40-μl aliquots of the muropeptides and cell wall fragments were dried with a Speed Vac (31). The dried carbohydrate and muropeptide samples were suspended in 5.0 μl of 0.2 M ANTS dissolved in acetic acid-water (3:17, vol/vol) and 5.0 μl of 1.0 M sodium cyanoborohydride (NaCNBH3) dissolved in dimethyl sulfoxide (DMSO). After incubation for 16 h (overnight) at 37°C, the samples were dried again with a Speed Vac at 45°C for 4 h. The carbohydrate standards were suspended at a concentration of 100 pmol/μl in loading buffer (62.5 mM Tris-HCl [pH 6.8], 20% glycerol) and stored at −80°C. The cell wall samples were dissolved in 50 μl loading buffer and diluted subsequently in loading buffer as required, and samples were also stored at −80°C.
Electrophoresis and visualization of ANTS-labeled muropeptides.
FACE was performed as described by Goins and Cutler (21). Gels that were 8.3 cm wide, 7.2 cm high, and approximately 0.75 mm thick were prepared. The resolving gel contained 32% acrylamide and 2.4% bisacrylamide (21). For 10 ml of resolving gel (two acrylamide gels), 5 μl of N,N,N′,N′-tetramethylethylenediamine (TEMED) and 50 μl of 10% ammonium persulfate (APS) were added. The stacking gel consisted of 8% acrylamide and 0.6% bisacrylamide containing 5 μl TEMED and 50 μl APS for 5 ml of stacking gel. The gel buffer was 0.42 M Tris-HCl (pH 8.5), and the running buffer contained 0.025 M Tris base and 0.192 M glycine (pH 8.4) (21). Eight-hundred-picomole portions of the carbohydrate standards and muropeptides were loaded on the acrylamide gel. Trace amounts of bromphenol blue in loading buffer were loaded in a single well as a visual reference. Electrophoresis was performed at 30 mA for 3 h using a cooled system. Fluorescently labeled carbohydrates and muropeptides were visualized by exposing the gel to 312-nm UV light, and images were obtained with a gel-imaging system.
RESULTS
Identification of a pathway for muropeptide rescue in B. subtilis.
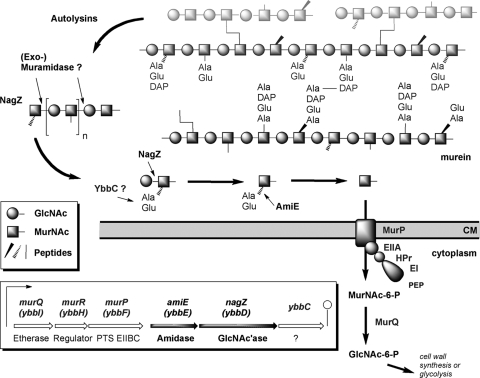
We identified a putative operon (ybbIHFEDC) on the chromosome of B. subtilis whose genes exhibit high levels of sequence similarity with genes involved in MurNAc dissimilation and cell wall recycling in E. coli (Fig. 1). The first three genes, ybbI, ybbH, and ybbF, are orthologs of the genes encoding the MurNAc-6-phosphate etherase (murQ), the MurNAc-6-phosphate-specific transcriptional repressor (murR), and the MurNAc-specific phosphotransferase system (murP) of E. coli, respectively. The levels of amino acid sequence identity of the Bacillus proteins and the E. coli orthologs are 47%, 26%, and 38%, respectively. The growth of ybbI (BsmurQ) and ybbF (BsmurP) mutants on MurNAc was impaired (S. Litzinger, unpublished results), and a function in the utilization of MurNAc like that in E. coli is proposed for the B. subtilis genes (13, 35-37). The functions of the three other genes of the Bacillus gene cluster, ybbE, ybbD, and ybbC, have not been determined previously, but ybbD encodes an ortholog of the cell wall-recycling N-acetylglucosaminidase NagZ of E. coli and was therefore renamed BsNagZ. In contrast to EcNagZ, the Bacillus ortholog is secreted and has a signal sequence with a putative signal peptidase cleavage site (MRPVFPLILSAVLFLSCFFGARQTEASA-SK [the putative signal peptidase cleavage site is indicated by bold type]). In addition, BsNagZ differs from the E. coli ortholog by having a C-terminal domain with an unknown function.
FIG. 1.
Diagram of the proposed muropeptide rescue and recovery pathway of B. subtilis and organization of the corresponding gene cluster. Autolysins release approximately 50% of the endogenous cell wall in one generation during growth (50). During this process long-chain peptidoglycan fragments are generated, to which NagZ is noncovalently bound and which are processed further by an unidentified muramidase. Soluble muropeptides generated in this way are processed by NagZ, which cleaves the β-1,4-glycosidic bond, and subsequently by AmiE, which cleaves the muramyl-l-Ala amide bond, releasing GlcNAc, MurNAc, and peptides. Recovery of MurNAc then proceeds like MurNAc dissimilation in E. coli; this process involves a MurNAc-specific phosphotransferase system (YbbF/MurP), by which MurNAc is taken up and simultaneously phosphorylated, yielding MurNAc-6-phosphate. In the cytoplasm, the etherase YbbI (MurQ) converts MurNAc-6-phosphate to GlcNAc-6-phosphate. YbbH (MurR) is a putative MurNAc-6-phosphate-specific transcriptional regulator, and the function of YbbC is unknown. CM, cell membrane; EIIA, enzyme IIA; EI, enzyme I; PEP, phosphoenolpyruvate; PTS, phosphotransferase system.
nagZ encodes an exo-GlcNAcase.
BsNagZ was overexpressed as a cytoplasmic construct in E. coli and was purified by Ni2+ affinity chromatography to apparent homogeneity (see Fig. S1 in the supplemental material). This protein had a molecular mass of about 70 to 75 kDa, as analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and displayed N-acetylglucosaminidase activity. Kinetic parameters for NagZ were determined with the chromogenic and fluorogenic substrates pNP-β-GlcNAc and 4-Mu-β-GlcNAc, respectively (see Table S1 in the supplemental material). Notably, this enzyme did not show activity with 4-Mu-β-GlcNH2 (see the supplemental material for a description of synthesis and characterization of this compound). This suggests that acetylation is important for the enzyme function. In another paper (S. Litzinger, S. Fischer, P. Polzer, K. Diederichs, W. Welte, and C. Mayer, submitted for publication) we describe the crystal structure and mechanistic studies of NagZ, which revealed that the N-acetamido group of the substrate is important for binding to the enzyme and hence specificity but is not directly involved in the hydrolysis mechanism, in contrast to the properties of other N-acetylglucosaminidases (41, 60) but similar to Nag3A of Cellulomonas fimi (43a). The purified protein was stable for several months at 4°C at pH 4.0 to 8.0. We also studied the pH dependence of purified NagZ using a discontinuous assay with 4-Mu-β-GlcNAc as the substrate, as described in Materials and Methods. Plotting kcat against pH resulted in a bell-shaped curve with an optimum pH in the range from 5.8 to 6.2 (see Fig. S2 in the supplemental material).
NagZ is associated with particulate material.
In B. subtilis N-acetylglucosaminidase activity was found to be associated primarily with particulate material in the culture supernatant (Table 2). About 20% of the activity was associated with cells (1,200-×-g sediment), and 25% of the activity was found in the soluble fraction. The majority of the enzyme activity, however, was in a sedimentable form. The percentages varied with culture age, but about two-thirds of the N-acetylglucosaminidase activity was found in the 30,000-×-g sediment of a 12-h culture. Most N-acetylglucosaminidase activity was related to NagZ since the N-acetylglucosaminidase activity in the nagZ mutant was less than 5% of the activity in wild-type Bacillus cells. NagZ was released from particulate material by treatment with NaCl. With 750 mM NaCl about 75% of the enzyme was released from sedimented material. Treatment of the sedimentable form of NagZ with lysozyme made it soluble, which indicates that this material contained murein. Notably, a significant amount of N-acetylglucosaminidase activity (about 20% of the total activity) remained cell wall associated (Table 2) and could not be removed with salt.
TABLE 2.
Sedimentation of β-N-acetylglucosaminidase activity from cultures of B. subtilisa
| Treatment | % of enzyme remaining in supernatant |
|
|---|---|---|
| This study | Study of Ortiz et al. | |
| 1,200 × g for 10 min | 81 | 87 |
| 30,000 × g for 30 min | 37 | 31 |
| 100,000 × g for 30 min | 25 | 26 |
Twelve-hour cultures (8 ml) of B. subtilis wild-type strain 168 were harvested by centrifugation as indicated. The amounts of enzyme present in the pellets and the supernatants remaining after centrifugation were determined as described in Materials and Methods. The values obtained were compared with data for a partially purified N-acetylglucosaminidase previously described by Ortiz et al. (45). No activity was detected in the NagZ mutant.
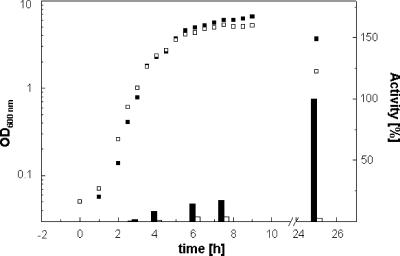
NagZ is induced during late exponential phase and stationary phase, and deletion of NagZ is associated with a lytic phenotype.
We also studied the expression of NagZ during growth of B. subtilis by measuring the activity with 4-Mu-β-GlcNAc in wild-type cells and a deletion strain (ybbD::pMUTIN4). The GlcNAcase activity appeared during the late exponential or early stationary phase of wild-type B. subtilis cells growing vegetatively (Fig. 2). However, the level of expression was 6-fold higher after 25 h, when the cells entered the autolysis phase. In this growth phase the GlcNAcase activity of the deletion strain was marginal (less than 5% of the wild-type activity). Intriguingly, there was a substantial decrease in the optical density of the nagZ mutant culture during late stationary phase that was associated with cell autolysis. Upon prolonged incubation both wild-type Bacillus and NagZ mutant cells lysed; however, the mutant lysed significantly earlier.
FIG. 2.
Occurrence of NagZ during growth of wild-type B. subtilis and a nagZ mutant. Filled squares and bars, wild-type strain; open squares and bars, nagZ mutant (ybbD::pMUTIN4). The symbols indicate the optical densities of the cultures (left y axis), and the bars indicate the sedimentable (30,000 × g) N-acetylglucosaminidase activity (right y axis). The soluble N-acetylglucosaminidase activity was about 25% or less of the total activity (Table 2). The N-acetylglucosaminidase activity appeared in the exponential phase during growth of wild-type B. subtilis cells at 37°C in LB medium, increased in stationary phase (filled squares), and was highest in the late stationary or decay phase (25 h). The activity in the late stationary or decay phase was defined as 100% and was 7.4 × 10−5 U/OD600 unit as determined with the fluorescent substrate 4-Mu-β-GlcNAc. Note that the N-acetylglucosaminidase activity in the nagZ mutant (ybbD::pMUTIN4) was less than 30% of that in wild-type B. subtilis during early stationary phase and less than 5% of that in wild-type B. subtilis in the late stationary or decay phase. The nagZ mutant had a lytic phenotype after prolonged cultivation (25 h).
NagZ is identical to a β-N-acetylglucosaminidase reported in the 1970s.
The results described above indicate that NagZ is identical to a β-N-acetylglucosaminidase reported in the early 1970s that was produced toward the end of growth and was found mainly in a sedimentable form in the growth medium (45). This enzyme was partially purified and characterized by Berkeley and colleagues (2) and had kinetic parameters similar to those that we determined for the recombinant NagZ. A comparison of the characteristics of the protein described by Berkeley et al. (2) and our results for BsNagZ is shown in Table S1 in the supplemental material, and on the basis of these characteristics it was concluded that the two enzymes are identical.
NagZ cleaves muropeptides but not peptidoglycan.
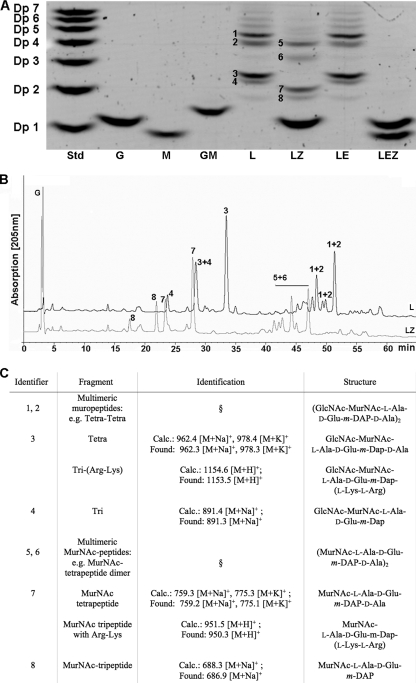
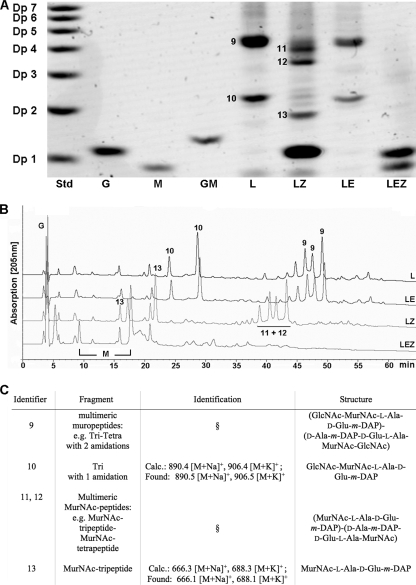
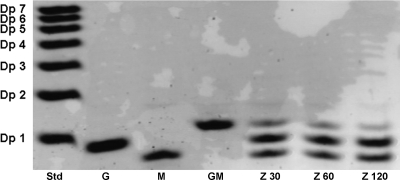
To determine the activity of NagZ with natural substrates, muropeptides were released from peptidoglycan of E. coli and B. subtilis by mutanolysin treatment. The degradation products were treated with isolated NagZ, and differences between the muropeptide composition before digestion and the muropeptide composition after digestion were analyzed by using two independent methods, fluorophore-assisted carbohydrate electrophoresis (FACE) and reversed-phase HPLC (RP-HPLC) (see Materials and Methods). For FACE the products were labeled with the fluorescent dye ANTS at their reducing ends and then separated by polyacrylamide gel electrophoresis and visualized by using UV light. Figures 3A and 4A show the results for the NagZ activity with muropeptides generated from E. coli (Fig. 3A) and B. subtilis (Fig. 4A) cell walls as determined by the FACE method. The muropeptides were identified by comparison with previously described data for muropeptide FACE analyses (40, 61) and by mass spectrometry (the composition of the muropeptides is summarized in Fig. 3C). The major muropeptides obtained from E. coli cell walls by mutanolysin treatment were identified as multimeric muropeptides, like disaccharide-tetrapeptide dimer Tetra-Tetra (identifiers 1 and 2) (Fig. 3A and C), as well as monomeric muropeptides, like Tetra (identifier 3) and disaccharide-tripeptide monomer (Tri) (identifier 4). In B. subtilis two prominent bands were visualized after mutanolysin treatment of isolated peptidoglycan (Fig. 4A). These muropeptides were identified by comparison with the vegetative cell wall peptidoglycans of B. subtilis described by Atrih et al. (1) and by mass spectrometry (Fig. 3C) as Tri-Tetra with two amidations (identifier 9), which accounted for 27.7% of the muropeptides, and Tri with one amidation (identifier 10), which accounted for 20.5% of the total cell wall fragments (1). A gel with a lower polyacrylamide concentration revealed that the Tri-Tetra (identifier 9) band consisted of at least two fragments, probably Tri-Tetra and other multimeric muropeptides (data not shown). NagZ was able to hydrolyze muropeptides generated from both E. coli and B. subtilis peptidoglycan, releasing GlcNAc (Fig. 3A and 4A). Multimeric muropeptides of E. coli containing Tetra-Tetra (identifiers 1 and 2) (Fig. 3A), as well as the monomeric muropeptides Tetra (identifier 3) and Tri (identifier 4), were converted to the following corresponding products lacking the terminal GlcNAc residues: a dimer of two MurNAc-tetrapeptide monomers (identifiers 5 and 6), a MurNAc-tetrapeptide (identifier 7), and a MurNAc-tripeptide (identifier 8). MALDI-TOF analysis of Tetra (identifier 3) (Fig. 3A) and the MurNAc-tetrapeptide (identifier 7) (Fig. 3A) revealed additional masses that matched those of Tri-Arg-Lys and MurNAc-tripeptide-Arg-Lys. These peptidoglycan fragments originated from peptidoglycan that had been covalently bound to Braun's lipoprotein (8). In B. subtilis multimeric muropeptides containing Tri-Tetra (identifier 9) and amidated Tri (identifier 10) were converted to multimeric MurNAc-peptides, including MurNAc-tripeptide-MurNAc-tetrapeptide (identifiers 11 and 12) and MurNAc-tripeptide (identifier 13). Some muropeptides of B. subtilis were amidated (identifier 10) (Fig. 3C), most likely at the m-DAP residue, which is a modification that was recognized previously (58). NagZ also released GlcNAc from muropeptides prepared from Micrococcus luteus peptidoglycan (data not shown) and cleaved the disaccharide GlcNAc-MurNAc (Fig. 5). This indicates that NagZ is not restricted by the peptide composition of the muropeptide substrate and that it does not depend on the peptides for activity. However, no GlcNAc was released from insoluble peptidoglycan (from any source) by NagZ.
FIG. 3.
Fluorophore-assisted carbohydrate electrophoresis (FACE), HPLC, and mass spectrometry of muropeptides of E. coli degraded with NagZ and/or AmiE. (A) Muropeptides and amino sugars derived from peptidoglycan of E. coli by mutanolysin digestion were treated with NagZ and/or AmiE, labeled with the fluorophore ANTS, and separated by electrophoresis in a 32% polyacrylamide gel. The numbers on the left indicate the degrees of polymerization of a maltodextrin standard mixture (lane Std) (Dp 1, glucose; Dp 2, maltose; Dp 3, maltotriose; Dp 4, maltotetraose; Dp 5, maltopentaose; Dp 6, maltohexaose; Dp 7, maltoheptaose). The other lanes show the results for GlcNAc (lane G), MurNAc (lane M), GlcNAc-MurNAc (lane GM), mutanolysin-treated cell walls from E. coli (lane L), the sample in lane L also digested with NagZ (lane LZ), the sample in lane L also digested with AmiE (lane LE), and the sample in lane L also digested with NagZ and AmiE (lane LEZ). (B and C) Muropeptides were identified by HPLC separation and UV detection (B) and by mass spectrometry (C), as well as by comparison with previously described data (40, 61). The numbers next to the bands in panel A and above the HPLC peaks in panel B are identifiers for muropeptide fragments (C). Tri, disaccharide-tripeptide monomer; Tetra, disaccharide-tetrapeptide monomer; Tetra-Tetra, dimer of two Tetra monomers. Tri-(Arg-Lys) and MurNAc-tripeptide-(Arg-Lys) resulted from cell wall fragments that were covalently linked to lipoprotein via the C-terminal Lys residue and cleaved by pronase before Arg. A section sign indicates that fragments were identified on the basis of retention in FACE analyses as described previously (40, 61).
FIG. 4.
Fluorophore-assisted carbohydrate electrophoresis (FACE), HPLC, and mass spectrometry of muropeptides of B. subtilis degraded with NagZ and/or AmiE. (A) Muropeptides and amino sugars derived from peptidoglycan of B. subtilis by mutanolysin digestion were treated with NagZ and/or AmiE, labeled with the fluorophore ANTS, and separated by electrophoresis in a 32% polyacrylamide gel. The lanes contained a maltodextrin standard mixture (lane Std; for details see the legend to Fig. 3), GlcNAc (lane G), MurNAc (lane M), GlcNAc-MurNAc (lane GM), mutanolysin-treated cell walls from E. coli (lane L), the sample in lane L also digested with NagZ (lane LZ), the sample in lane L also digested with AmiE (lane LE), and the sample in lane L also digested with NagZ and AmiE (lane LEZ). (B and C) Muropeptides were identified by HPLC separation and UV detection (B) and by mass spectrometry (C), as well as by comparison with previously described data (1, 61). The numbers next to the bands in panel A and above the HPLC peaks in panel B are identifiers for muropeptide fragments (C). Tri, disaccharide-tripeptide monomer; Tetra, disaccharide-tetrapeptide monomer; Tri-Tetra, dimer of a Tri monomer and a Tetra monomer. A section sign indicates that fragments were identified on the basis of retention in FACE analyses as described previously (40, 61).
FIG. 5.
Fluorophore-assisted carbohydrate electrophoresis (FACE) of the disaccharide GlcNAc-MurNAc degraded with NagZ. The lanes contained a maltodextrin standard mixture (lane Std; for details see the legend to Fig. 3), GlcNAc (lane G), MurNAc (lane M), GlcNAc-MurNAc (lane GM), and GlcNAc-MurNAc digested with NagZ for 30, 60, and 120 min (lanes Z 30, Z 60, and Z 120, respectively).
Using a second approach, the muropeptide specificity of NagZ was analyzed by RP-HPLC. Under the conditions used muropeptides and cell wall sugars or sugar peptides were detected as two peaks related to their α- and β-anomeric forms. Figures 3B and 4B show the RP-HPLC elution patterns of muropeptides derived from mutanolysin-digested peptidoglycan of E. coli (Fig. 3B, line L) and B. subtilis (Fig. 4B, line L). NagZ-catalyzed hydrolysis of muropeptides of E. coli (Fig. 3B, line LZ) and B. subtilis (Fig. 4B, line LZ) was shown by the decrease in the peak heights or by the complete loss of several peaks, as well as by the appearance of new compounds, including GlcNAc. The main peaks before and after hydrolysis by NagZ were collected and analyzed by FACE, and the release of GlcNAc and the conversion of muropeptides into MurNAc-peptides as shown in Fig. 3C and 4C were confirmed. Although not all of the peaks in the RP-HPLC analysis could be identified, the specificity of NagZ cleavage of the β-1,4-glycosidic bond between GlcNAc and MurNAc and the broad specificity for peptide substitutions at MurNAc were shown by the two autonomous methods.
AmiE functions as an N-acetylmuramyl-l-alanine amidase.
Muropeptide mixtures were not affected by incubation with AmiE, as shown by FACE analysis (Fig. 3A and 4A, lanes LE). Consistently, the RP-HPLC elution pattern of muropeptides of B. subtilis after treatment with AmiE had only minor differences when they were compared to the muropeptide pattern obtained after mutanolysin treatment alone (Fig. 4B, line LE). This indicates that AmiE was not able to hydrolyze muropeptides containing GlcNAc at the nonreducing end. In contrast, this enzyme was active after previous digestion of the muropeptides with NagZ (Fig. 3A and 4A, lanes LE; Fig. 4B, line LEZ). The release of MurNAc from muropeptides by digestion with both NagZ and AmiE showed that AmiE functions as an N-acetylmuramyl-l-alanine amidase that hydrolyzes the amide bond between MurNAc and the l-alanine residue of the stem peptide. In contrast to NagZ, AmiE did not degrade muropeptides of M. luteus, although these compounds were substrates for NagZ (data not shown).
Muropeptides accumulate in the spent medium of a nagZ mutant.
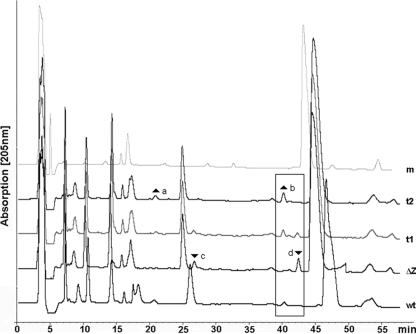
Differences in the muropeptide pattern for the spent medium of wild-type Bacillus and nagZ mutant cells were identified (Fig. 6). Two compounds (Fig. 6, peaks c and d) were present only in the spent medium of the nagZ mutant. After treatment with purified NagZ, these compounds disappeared and two other compounds appeared (Fig. 6, peaks a and b). For the peak c compound the mass determined by mass spectrometry was consistent with the theoretical mass of GlcNAc-MurNAc-Ala-Glu (calculated: 719.3 [M+Na]+, 757.3 [M+Na/K]+; found: 719.5 [M+Na]+, 757.5 [M+K]+). The peak d compound could not be unequivocally identified by mass spectrometry; however, it eluted with a retention time similar to that of Tri-Tetra muropeptides (Fig. 4). The peak a and b compounds, which were generated from the peak c and d compounds by treatment with NagZ, eluted with retention times typical of MurNAc-peptides and MurNAc-peptide dimers, respectively, consistent with the presumed identities of these compounds (MurNAc-Ala-Glu and MurNAc-tripeptide-MurNAc-tetrapeptide).
FIG. 6.
HPLC analysis of B. subtilis spent medium. The HPLC elution profile of spent medium of B. subtilis nagZ mutant cells (line ΔZ) has two minor peaks at retention times of 27 min (peak c) and 42 min (peak d) that are not present in the profile of the spent medium of wild-type cells (line wt). After treatment of the spent medium of nagZ cultures with recombinant NagZ protein for 5 min (line t1) and 60 min (line t2), these peaks disappeared and two peaks appeared at 20 min (peak a) and 40 min (peak b), which correspond to peaks that are found in the profile of spent medium of wild-type Bacillus cells (line wt) but not in the profile of the control containing minimal medium plus succinate and NagZ protein (line m).
DISCUSSION
Similar to findings for E. coli, the rate of cell wall turnover in some Gram-positive bacteria (e.g., B. subtilis W-23 [42] and Bacillus megaterium, Bacillus cereus, and Lactobacillus acidophilus [5, 9, 14]) is up to 50% of the cell wall material per generation. Curiously, however, the current view is that Gram-positive bacteria, in contrast to Gram-negative bacteria, do not recycle the peptidoglycan of the endogenous wall. This characteristic was assumed based on early studies showing that cell wall turnover products accumulate in the culture medium (42, 43). However, if the turnover products were not recovered, there would be a tremendous loss of resources, since peptidoglycan usually accounts for more than 20% of the total cell mass of Gram-positive bacteria. It is obvious that cell wall turnover products would be a fortuitous carbon and energy source, particularly for cells entering a starvation phase (e.g., stationary phase or beginning of sporulation).
In E. coli, anhydromuropeptides are the primary turnover products that are continuously generated during growth and transported into the cytoplasm by the permease AmpG, where further processing involves the recycling enzymes AmpD, the anhMurNAc amidase, and AnmK, the anhMurNAc kinase, among others (12, 28, 33, 34, 57). Although anhMurNAc has been recognized in Bacillus, anhydromuropeptides and the lytic transglycosylases which generate them play only a minor role in peptidoglycan turnover in this organism (1). Moreover, B. subtilis apparently lacks orthologs of the recycling proteins AmpG, AmpD, and AnmK. Peptidoglycan recycling in B. subtilis, therefore, must differ from peptidoglycan recycling in E. coli. At least 35 cell wall lytic enzymes (autolysins) have been identified in B. subtilis, but their precise specificities and functions, particularly those of muramidases and lytic transglycosylases, are for the most part unknown (50). There are three putative G-type lysozymes or lytic transglycosylases (family 23 glycosidases): YocA (BSU19130), YjbJ (BSU11570), and YomI (BSU21350). Furthermore two additional enzymes which belong to the SLT family have been annotated: YqbO (BSU26030, P45931) and XKDO (P54334). Recently, the putative lytic transglycosylase CwlT (YddH, BSU04970) was shown to act as a lysozyme-like muramidase rather than a lytic transglycosylase (19). CwlT is a bifunctional two-domain enzyme; the N-terminal domain functions as an N-acetylmuramidase, and the C-terminal domain has ld-endopeptidase activity and is capable of hydrolyzing d-γ-glutamyl-m-DAP linkages. This protein releases mostly peptidyl tetra-, hexa-, and octasaccharides from the peptidoglycan and only small amounts of the disaccharide GlcNAc-MurNAc-l-Ala-γ-d-Glu (19), the product that accumulates in the spent medium of the nagZ mutant. Clearly, the origin of the substrate of NagZ (GlcNAc-MurNAc peptides) needs to be investigated further. In addition to CwlT, a muramidase that in conjunction with NagZ would lead to complete degradation of peptidoglycan fragments is unknown. One candidate, an exomuramidase, was discovered in crude enzyme preparations and was able to hydrolyze the disaccharide MurNAc-GlcNAc (15), but so far neither the enzyme responsible nor the corresponding gene has been identified. Two of the most abundant autolysins in B. subtilis, the amidase LytC and the endo-N-acetylglucosaminidase LytD, as well as LytF, a γ-d-glutamate meso-diaminopimelate muropeptidase, are not involved in cell wall turnover but were shown recently to act as peptidoglycan-remodeling autolysins in flagellum synthesis (10).
Our group identified a pathway for the recovery of MurNAc consisting of the MurNAc-specific phosphotransferase system (MurP), the MurNAc-6-phosphate etherase (MurQ), and the MurNAc-6-phosphate-sensitive transcriptional repressor MurR (13, 35-37). Recently, Plumbridge showed that NagE, a GlcNAc-specific phosphotransferase system enzyme, is involved in murein recycling in E. coli (48). B. subtilis possesses a NagE ortholog, as well orthologs of the MurNAc-specific phosphotransferase system (ybbF/murP) and the MurNAc-6-phosphate etherase (murQ/ybbI). Therefore, recovery of the cell wall amino sugars GlcNAc and MurNAc in B. subtilis may proceed though transport and concomitant phosphorylation involving specific phosphotransferase systems (NagE and MurP), yielding the amino sugar 6-phosphate and resulting in further conversion of MurNAc-6-phosphate to GlcNAc-6-phosphate by an etherase (MurQ). GlcNAc-6-phosphate can then deacetylated by the NagA enzyme, yielding glucosamine-6-phosphate, which either is deaminated by NagB, yielding fructose-6-phosphate that enters glycolysis, or is converted to glucosamine-1-phosphate by the phosphoglucosamine mutase GlmM, which enters the amino sugar and cell wall biosynthesis pathway.
Interestingly, MurQ and MurP are encoded by genes in a putative operon in B. subtilis together with an ortholog of the NagZ β-N-acetylglucosaminidase of E. coli that was designated BsNagZ (formerly ybbD). This protein was annotated as a lipoprotein; however, a signal sequence for signal peptidase 1 was identified that potentially cleaves the N-terminal cysteine that would be required for covalent binding to phospholipids. Our results showed that BsNagZ occurs in the culture supernatant of B. subtilis mainly in a sedimentable form and can be solubilized with NaCl. Therefore, it is not a lipoprotein. In this study we characterized NagZ and AmiE (formerly YbbE), which together are able to release MurNAc from muropeptides. Notably, both of these proteins act outside the cell. NagZ cleaves GlcNAc residues from the nonreducing end and acts on muropeptides generated from B. subtilis, E. coli, or M. luteus peptidoglycan. It could also release GlcNAc from larger muropeptide fragments, such as that produced by CwlT; however, it was not able to degrade isolated peptidoglycan. As there may be some GlcNAc at the nonreducing ends of peptidoglycan strands, this indicates that peptidoglycan is not a substrate of NagZ. A small amount of GlcNAc might not be detectable by the FACE gel assay; however, FACE analysis can detect as little as 2 to 5 pmol muropeptide (40, 61), and hence it should be possible to detect even small amounts of GlcNAc released from the nonreducing end of peptidoglycan chains with this method.
NagZ is associated with the cell wall and cell wall-derived particulate material, likely via ionic interactions of the positively charged protein (calculated pI, 9.37) with the negatively charged Bacillus cell wall, and it can be released from the wall by high concentrations of salt. AmiE can cleave peptides from MurNAc-peptides of E. coli and B. subtilis but not from MurNAc-peptides of M. luteus, likely due to differences in the peptide composition. In M. luteus the peptide subunits have the sequence N-(l-alanyl-γ-[α-d-glutamylglycine])-l-lysyl-d-alanine, whereas in E. coli and B. subtilis the peptides consist of N-(l-alanyl-γ-d-glutamyl-m-DAP-d-alanine). Furthermore, in M. luteus the peptidoglycan is cross-linked by peptide bridges consisting of repeating tetrapeptide sequences.
NagZ is the major exo-β-N-acetylglucosaminidase of B. subtlis, since a NagZ mutant shows severely decreased activity with the substrate analogue 4-Mu-β-GlcNAc. Using stationary-phase cells, less than 5% of the wild-type B. subtilis N-acetylglucosaminidase activity was found in the nagZ mutant. During the mid-exponential growth phase the nagZ mutant had residual activity compared to the wild type, indicating that a second exo-acting enzyme was present (Fig. 2). This activity may be associated with the exo-acting N-acetylglucosaminidase LytG that is expressed mainly during this growth phase under control of the σA-dependent promoter (29). A lack of LytG resulted in appearance of GlcNAc at the nonreducing terminus, and hence LytG may have a role in modifying glycan strand length, since the glycan strands of mature peptidoglycan have a MurNAc at the nonreducing end in B. subtilis (Fig. 1).
The regulation of the gene cluster of B. subtilis involved in MurNAc dissimilation was not examined in this study. However, we recognized a σA-dependent promoter in the region upstream of ybbI (murQ). In addition, the expression of this cluster likely is regulated by carbon catabolite repression, since a highly conserved catabolite response element (cre) is located upstream of the σA-dependent promoter. Glucose starvation should result in a switch to utilization of alternative carbon sources mediated mainly by the catabolite control protein CcpA. Furthermore, accumulation of MurNAc-6-phosphate might induce transcription of the operon, as is the case in E. coli (37). Eichenberger et al. have shown that ybbFEDC is also part of the σE regulon, which influences the transcription of more than 250 genes during sporulation in B. subtilis (17). NagZ is expressed during the transition to stationary growth, but the highest level of activity was found in the late stationary or decay phase, which can be attributed to σE at the onset of sporulation.
The nagZ mutant displayed normal cell morphology, and no change in the growth rate was observed during exponential growth in rich medium. However, a substantial decrease in the optical density was observed for the nagZ mutant compared to the wild type in stationary phase. The reason for this lytic effect is currently unclear but is being studied. One possibility is that the nagZ mutation affects sporulation. B. subtilis responds to nutritional limitation by entering the sporulation phase (16, 30). This is an energy- and time-intensive process, and therefore B. subtilis cells exhibit a cannibalistic tendency by killing their sister cells and feeding on the released nutrients to delay sporulation (18, 22). Recently, Bernhardt et al. showed that after a few hours of glucose starvation a few sporulation-specific proteins appeared (3), which indicated that at least a portion of the cell population initiated sporulation. The increased lysis of a nagZ null mutant during the late stationary phase compared with the wild type may have been a result of a connection, either direct or indirect, with sporulation, spore germination, cannibalism, or autolysis.
Supplementary Material
Acknowledgments
This work was supported by grant MA2436/4 from the Deutsche Forschungsgemeinschaft and Heisenberg stipend MA2436/3 to C.M.
Footnotes
Published ahead of print on 16 April 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Atrih, A., G. Bacher, G. Allmaier, M. P. Williamson, and S. J. Foster. 1999. Analysis of peptidoglycan structure from vegetative cells of Bacillus subtilis 168 and role of PBP 5 in peptidoglycan maturation. J. Bacteriol. 181:3956-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkeley, R. C., S. J. Brewer, J. M. Ortiz, and J. B. Gillespie. 1973. An exo-β-N-acetylglucosaminidase from Bacillus subtilis B; characterization. Biochim. Biophys. Acta 309:157-168. [DOI] [PubMed] [Google Scholar]
- 3.Bernhardt, J., J. Weibezahn, C. Scharf, and M. Hecker. 2003. Bacillus subtilis during feast and famine: visualization of the overall regulation of protein synthesis during glucose starvation by proteome analysis. Genome Res. 13:224-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Boothby, D., L. Daneo-Moore, M. L. Higgins, J. Coyette, and G. D. Shockman. 1973. Turnover of bacterial cell wall peptidoglycans. J. Biol. Chem. 248:2161-2169. [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Braun, V. 1975. Covalent lipoprotein from the outer membrane of Escherichia coli. Biochim. Biophys. Acta 415:335-377. [DOI] [PubMed] [Google Scholar]
- 8.Braun, V., and U. Sieglin. 1970. The covalent murein-lipoprotein structure of the Escherichia coli cell wall. The attachment site of the lipoprotein on the murein. Eur. J. Biochem. 13:336-346. [DOI] [PubMed] [Google Scholar]
- 9.Chaloupka, J., and P. Kreckova. 1971. Turnover of mucopeptide during the life cycle of Bacillus megaterium. Folia Microbiol. (Praha) 16:372-382. [DOI] [PubMed] [Google Scholar]
- 10.Chen, R., S. B. Guttenplan, K. M. Blair, and D. B. Kearns. 2009. Role of the sigmaD-dependent autolysins in Bacillus subtilis population heterogeneity. J. Bacteriol. 191:5775-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng, Q., H. Li, K. Merdek, and J. T. Park. 2000. Molecular characterization of the β-N-acetylglucosaminidase of Escherichia coli and its role in cell wall recycling. J. Bacteriol. 182:4836-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng, Q., and J. T. Park. 2002. Substrate specificity of the AmpG permease required for recycling of cell wall anhydro-muropeptides. J. Bacteriol. 184:6434-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahl, U., T. Jaeger, B. T. Nguyen, J. M. Sattler, and C. Mayer. 2004. Identification of a phosphotransferase system of Escherichia coli required for growth on N-acetylmuramic acid. J. Bacteriol. 186:2385-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daneo-Moore, L., J. Coyette, M. Sayare, D. Boothby, and G. D. Shockman. 1975. Turnover of the cell wall peptidoglycan of Lactobacillus acidophilus. The presence of a fraction immune to turnover. J. Biol. Chem. 250:1348-1353. [PubMed] [Google Scholar]
- 15.Del Rio, L. A., R. C. Berkeley, S. J. Brewer, and S. E. Roberts. 1973. An enzyme from Bacillus subtilis B with exo-β-N-acetylmuramidase activity. FEBS Lett. 37:7-9. [DOI] [PubMed] [Google Scholar]
- 16.Driks, A. 1999. Bacillus subtilis spore coat. Microbiol. Mol. Biol. Rev. 63:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eichenberger, P., S. T. Jensen, E. M. Conlon, C. van Ooij, J. Silvaggi, J. E. Gonzalez-Pastor, M. Fujita, S. Ben-Yehuda, P. Stragier, J. S. Liu, and R. Losick. 2003. The σE regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol. 327:945-972. [DOI] [PubMed] [Google Scholar]
- 18.Engelberg-Kulka, H., and R. Hazan. 2003. Cannibals defy starvation and avoid sporulation. Science 301:467-468. [DOI] [PubMed] [Google Scholar]
- 19.Fukushima, T., T. Kitajima, H. Yamaguchi, Q. Ouyang, K. Furuhata, H. Yamamoto, T. Shida, and J. Sekiguchi. 2008. Identification and characterization of novel cell wall hydrolase CwlT: a two-domain autolysin exhibiting N-acetylmuramidase and dl-endopeptidase activities. J. Biol. Chem. 283:11117-11125. [DOI] [PubMed] [Google Scholar]
- 20.Glauner, B. 1988. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal. Biochem. 172:451-464. [DOI] [PubMed] [Google Scholar]
- 21.Goins, T. L., and J. E. Cutler. 2000. Relative abundance of oligosaccharides in Candida species as determined by fluorophore-assisted carbohydrate electrophoresis. J. Clin. Microbiol. 38:2862-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez-Pastor, J. E., E. C. Hobbs, and R. Losick. 2003. Cannibalism by sporulating bacteria. Science 301:510-513. [DOI] [PubMed] [Google Scholar]
- 23.Goodell, E. W. 1985. Recycling of murein by Escherichia coli. J. Bacteriol. 163:305-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hadi, T., U. Dahl, C. Mayer, and M. E. Tanner. 2008. Mechanistic studies on N-acetylmuramic acid 6-phosphate hydrolase (MurQ): an etherase involved in peptidoglycan recycling. Biochemistry 47:11547-11558. [DOI] [PubMed] [Google Scholar]
- 25.Harz, H., K. Burgdorf, and J. V. Höltje. 1990. Isolation and separation of the glycan strands from murein of Escherichia coli by reversed-phase high-performance liquid chromatography. Anal. Biochem. 190:120-128. [DOI] [PubMed] [Google Scholar]
- 26.Hayashi, K. 1975. A rapid determination of sodium dodecyl sulfate with methylene blue. Anal. Biochem. 67:503-506. [DOI] [PubMed] [Google Scholar]
- 27.Höltje, J. V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Höltje, J. V., U. Kopp, A. Ursinus, and B. Wiedemann. 1994. The negative regulator of β-lactamase induction AmpD is a N-acetyl-anhydromuramyl-l-alanine amidase. FEMS Microbiol. Lett. 122:159-164. [DOI] [PubMed] [Google Scholar]
- 29.Horsburgh, G. J., A. Atrih, M. P. Williamson, and S. J. Foster. 2003. LytG of Bacillus subtilis is a novel peptidoglycan hydrolase: the major active glucosaminidase. Biochemistry 42:257-264. [DOI] [PubMed] [Google Scholar]
- 30.Itaya, M., and T. Tanaka. 1997. Experimental surgery to create subgenomes of Bacillus subtilis 168. Proc. Natl. Acad. Sci. U. S. A. 94:5378-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson, P. 1994. The analysis of fluorophore-labeled glycans by high-resolution polyacrylamide gel electrophoresis. Anal. Biochem. 216:243-252. [DOI] [PubMed] [Google Scholar]
- 32.Jackson, P. 1990. The use of polyacrylamide-gel electrophoresis for the high-resolution separation of reducing saccharides labelled with the fluorophore 8-aminonaphthalene-1,3,6-trisulphonic acid. Detection of picomolar quantities by an imaging system based on a cooled charge-coupled device. Biochem. J. 270:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobs, C., L. J. Huang, E. Bartowsky, S. Normark, and J. T. Park. 1994. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for beta-lactamase induction. EMBO J. 13:4684-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobs, C., B. Joris, M. Jamin, K. Klarsov, J. Van Beeumen, D. Mengin-Lecreulx, J. van Heijenoort, J. T. Park, S. Normark, and J. M. Frere. 1995. AmpD, essential for both β-lactamase regulation and cell wall recycling, is a novel cytosolic N-acetylmuramyl-l-alanine amidase. Mol. Microbiol. 15:553-559. [DOI] [PubMed] [Google Scholar]
- 35.Jaeger, T., M. Arsic, and C. Mayer. 2005. Scission of the lactyl ether bond of N-acetylmuramic acid by Escherichia coli “etherase.” J. Biol. Chem. 280:30100-30106. [DOI] [PubMed] [Google Scholar]
- 36.Jaeger, T., and C. Mayer. 2008. N-acetylmuramic acid 6-phosphate lyases (MurNAc etherases): role in cell wall metabolism, distribution, structure, and mechanism. Cell. Mol. Life Sci. 65:928-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaeger, T., and C. Mayer. 2008. The transcriptional factors MurR and catabolite activator protein regulate N-acetylmuramic acid catabolism in Escherichia coli. J. Bacteriol. 190:6598-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S. C. Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A. Danchin, M. Debarbouille, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, D. Hosoya, M. F. Hullo, H. Kakeshita, D. Karamata, Y. Kasahara, F. Kawamura, K. Koga, P. Koski, R. Kuwana, D. Imamura, M. Ishimaru, S. Ishikawa, I. Ishio, D. Le Coq, A. Masson, C. Mauel, R. Meima, R. P. Mellado, A. Moir, S. Moriya, E. Nagakawa, H. Nanamiya, S. Nakai, P. Nygaard, M. Ogura, T. Ohanan, M. O'Reilly, M. O'Rourke, Z. Pragai, H. M. Pooley, G. Rapoport, J. P. Rawlins, L. A. Rivas, C. Rivolta, A. Sadaie, Y. Sadaie, M. Sarvas, T. Sato, H. H. Saxild, E. Scanlan, W. Schumann, J. F. Seegers, J. Sekiguchi, A. Sekowska, S. J. Seror, M. Simon, P. Stragier, R. Studer, H. Takamatsu, T. Tanaka, M. Takeuchi, H. B. Thomaides, V. Vagner, J. M. van Dijl, K. Watabe, A. Wipat, H. Yamamoto, M. Yamamoto, Y. Yamamoto, K. Yamane, K. Yata, K. Yoshida, H. Yoshikawa, U. Zuber, and N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. U. S. A. 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leutgeb, W., and W. Weidel. 1963. On a glycogen trapped in E. coli cell wall preparations. Z. Naturforsch. B 18:1060-1062. (In German.) [PubMed] [Google Scholar]
- 40.Li, S. Y., J. V. Holtje, and K. D. Young. 2004. Comparison of high-performance liquid chromatography and fluorophore-assisted carbohydrate electrophoresis methods for analyzing peptidoglycan composition of Escherichia coli. Anal. Biochem. 326:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mark, B. L., D. J. Vocadlo, S. Knapp, B. L. Triggs-Raine, S. G. Withers, and M. N. James. 2001. Crystallographic evidence for substrate-assisted catalysis in a bacterial beta-hexosaminidase. J. Biol. Chem. 276:10330-10337. [DOI] [PubMed] [Google Scholar]
- 42.Mauck, J., L. Chan, and L. Glaser. 1971. Turnover of the cell wall of Gram-positive bacteria. J. Biol. Chem. 246:1820-1827. [PubMed] [Google Scholar]
- 43.Mauck, J., and L. Glaser. 1970. Turnover of the cell wall of Bacillus subtilis W-23 during logarithmic growth. Biochem. Biophys. Res. Commun. 39:699-706. [DOI] [PubMed] [Google Scholar]
- 43a.Mayer, C., D. J. Vocadlo, M. Mah, K. Rupitz, D. Stoll, R. A. Warren, and S. G. Withers. 2006. Characterization of a β-N-acetylhexosaminidase and a β-N-acetylglucosaminidase/β-glucosidase from Cellulomonas fimi. FEBS J. 273:2929-2941. [DOI] [PubMed] [Google Scholar]
- 44.Mengin-Lecreulx, D., J. van Heijenoort, and J. T. Park. 1996. Identification of the mpl gene encoding UDP-N-acetylmuramate:l-alanyl-γ-d-glutamyl-meso-diaminopimelate ligase in Escherichia coli and its role in recycling of cell wall peptidoglycan. J. Bacteriol. 178:5347-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ortiz, J. M., J. B. Gillespie, and R. C. Berkeley. 1972. An exo-β-N-acetylglucosaminidase from Bacillus subtilis B; extraction and purification. Biochim. Biophys. Acta 289:174-186. [DOI] [PubMed] [Google Scholar]
- 46.Park, J. T. 2001. Identification of a dedicated recycling pathway for anhydro-N-acetylmuramic acid and N-acetylglucosamine derived from Escherichia coli cell wall murein. J. Bacteriol. 183:3842-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park, J. T., and T. Uehara. 2008. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol. Mol. Biol. Rev. 72:211-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plumbridge, J. 2009. An alternative route for recycling of N-acetylglucosamine from peptidoglycan involves the N-acetylglucosamine phosphotransferase system in Escherichia coli. J. Bacteriol. 191:5641-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmidt, D. M., B. K. Hubbard, and J. A. Gerlt. 2001. Evolution of enzymatic activities in the enolase superfamily: functional assignment of unknown proteins in Bacillus subtilis and Escherichia coli as l-Ala-d/l-Glu epimerases. Biochemistry 40:15707-15715. [DOI] [PubMed] [Google Scholar]
- 50.Smith, T. J., S. A. Blackman, and S. J. Foster. 2000. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146:249-262. [DOI] [PubMed] [Google Scholar]
- 51.Stack, R. J., and M. T. Sullivan. 1992. Electrophoretic resolution and fluorescence detection of N-linked glycoprotein oligosaccharides after reductive amination with 8-aminonaphthalene-1,3,6-trisulphonic acid. Glycobiology 2:85-92. [DOI] [PubMed] [Google Scholar]
- 52.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 53.Templin, M. F., A. Ursinus, and J. V. Höltje. 1999. A defect in cell wall recycling triggers autolysis during the stationary growth phase of Escherichia coli. EMBO J. 18:4108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uehara, T., and J. T. Park. 2003. Identification of MpaA, an amidase in Escherichia coli that hydrolyzes the γ-d-glutamyl-meso-diaminopimelate bond in murein peptides. J. Bacteriol. 185:679-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uehara, T., and J. T. Park. 2004. The N-acetyl-d-glucosamine kinase of Escherichia coli and its role in murein recycling. J. Bacteriol. 186:7273-7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uehara, T., K. Suefuji, T. Jaeger, C. Mayer, and J. T. Park. 2006. MurQ etherase is required by Escherichia coli in order to metabolize anhydro-N-acetylmuramic acid obtained either from the environment or from its own cell wall. J. Bacteriol. 188:1660-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uehara, T., K. Suefuji, N. Valbuena, B. Meehan, M. Donegan, and J. T. Park. 2005. Recycling of the anhydro-N-acetylmuramic acid derived from cell wall murein involves a two-step conversion to N-acetylglucosamine-phosphate. J. Bacteriol. 187:3643-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vollmer, W. 2008. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol. Rev. 32:287-306. [DOI] [PubMed] [Google Scholar]
- 59.Vötsch, W., and M. F. Templin. 2000. Characterization of a β-N-acetylglucosaminidase of Escherichia coli and elucidation of its role in muropeptide recycling and β-lactamase induction. J. Biol. Chem. 275:39032-39038. [DOI] [PubMed] [Google Scholar]
- 60.Williams, S. J., B. L. Mark, D. J. Vocadlo, M. N. James, and S. G. Withers. 2002. Aspartate 313 in the Streptomyces plicatus hexosaminidase plays a critical role in substrate-assisted catalysis by orienting the 2-acetamido group and stabilizing the transition state. J. Biol. Chem. 277:40055-40065. [DOI] [PubMed] [Google Scholar]
- 61.Young, K. D. 1996. A simple gel electrophoretic method for analyzing the muropeptide composition of bacterial peptidoglycan. J. Bacteriol. 178:3962-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.