Abstract
Induction of the Pho response in Bacillus subtilis occurs when the Pi concentrations in the growth medium fall below 0.1 mM, a condition which results in slowed cellular growth followed by entry into stationary phase. The phoPR promoter region contains three σA-responsive promoters; only promoter PA4 is PhoP autoregulated. Expression of the phoPR operon is postexponential, suggesting the possibility of a repressor role for a transition-state-regulatory protein(s). Expression of a phoPR promoter-lacZ fusion in a scoC loss-of-function mutant strain grown in low-phosphate defined medium was significantly higher than expression in the wild-type strain during exponential growth or stationary phase. Derepression in the scoC strain from a phoP promoter fusion containing a mutation in the CcpA binding site (cre1) was further elevated approximately 1.4-fold, indicating that the repressor effects of ScoC and CcpA on phoP expression were cumulative. DNase I footprinting showed protection of putative binding sites by ScoC, which included the −10 and/or −35 elements of five (PB1, PE2, PA3, PA4, and PA6) of the six promoters within the phoPR promoter region. PA6 was expressed in vivo from the phoP cre1 promoter fusion in both wild-type and scoC strains. Evidence for ScoC repression in vivo was shown by primer extension for PA4 and PA3 from the wild-type promoter and for PA4 and PE2 from the phoP cre1 promoter. The latter may reflect ScoC repression of sporulation that indirectly affects phoPR transcription. ScoC was shown to repress PA6, PA4, PE2, and PB1 in vitro.
The majority of Bacillus subtilis genes, which are induced in response to phosphate-limiting growth conditions, are controlled by one of two major global regulatory systems, the PhoP-PhoR two-component signal transduction (TCS) or SigB, a stress sigma factor that is activated in response to limiting Pi. An unknown regulatory system may also exist since a few genes identified as Pi starvation-induced do not depend on either PhoP-PhoR or SigB (3).
Phosphorylated PhoP (PhoP∼P) is required for activation or repression of Pho regulon genes. During activation, PhoP binds to a core binding region located between −20 and −60 (relative to the translation start site) on the coding strand, which consists of four repeats of a 6-bp consensus sequence, TT(A/C/T)A(C/T)A, separated by four to six nonconserved base pairs (11, 12). Activated promoters may have additional binding sites either 5′ (11, 12) of the core binding region or 3′ (25) within the coding region that are required for full promoter activity. Activated promoters have no −35 consensus and require PhoP∼P for activation (33). The exception is that during autoinduction of the phoPR promoter, PhoP∼P enhances activity of promoters that have low-level activity without PhoP∼P (27). At repressed promoters there are usually two consensus repeats on the noncoding strand upstream of the transcription start site, and PhoP oligomerizes along the DNA into the coding region (19).
The characterization of the B. subtilis phosphate deficiency response controlled by PhoPR has revealed that regulatory networks involving multiple two-component systems function in an interdependent manner to make the best use of environmental conditions at the time. The PhoP-PhoR TCS is part of a signal transduction network that consists of at least three TCSs (PhoP-PhoR, ResD-ResE, and Spo0A∼P) and a transition state regulator, AbrB (19, 41). The Pho response is positively activated upstream of PhoPR via two parallel pathways involving the ResD-ResE TCS and AbrB (41). An abrB mutation causes a slight reduction in the Pho regulon response (18, 21) while a deletion mutation in resD leads to an 80% reduction in Pho regulon gene expression (41). AbrB was shown to be essential for the 20% remaining Pho regulon expression in an resD mutant strain when an abrB resD double mutant (41) showed no Pho regulon gene induction.
Characterization of ResDE TCS regulon genes has shown that ResD plays an indirect role in Pho regulon induction via heme A synthesis required for terminal oxidases (aa3 and caa3) (27, 42) that oxidize reduced quinones. Reduced quinones were shown to inhibit autophosphorylation of the PhoR in vitro, suggesting that it was the ResD role in terminal oxidase production that positively modulates the PhoR signal (35) upstream of PhoPR. Consistent with this idea, resD mutants containing a spontaneous mutation in rex (formerly ydiH), a repressor of cydABCD encoding bd oxidase (34), allowed expression of cydABCD during Pho induction, which bypassed the requirement for ResD for full Pho induction (35). Together, these data indicate that the terminal oxidase bd, encoded by cydABCD, was sufficient to replace the loss of caa3 and aa3 in the resD mutant strain by restoring the terminal oxidase function of oxidation of reduced quinones that inhibit PhoR autophosphorylation. Spo0A∼P, produced by the Spo0A phosphorelay system (6), represses the Pho response by negatively regulating abrB expression (37) and resDE expression via the resA promoter (M. Hulett and G. Sun, unpublished data). Positive regulation of Pho regulon gene expression via AbrB is not well understood.
The complexity of the phoPR operon promoter region has become apparent over the past decade, revealing a very versatile promoter. Expression from the phoPR promoter represents the sum of the six promoters (Table 1), with each responding to specific growth phase and environmental signals (27, 32). Several forms of RNA polymerase (RNAP) holoenzymes are required for transcription of the phoPR operon: three EσA-responsive promoters (PA3, PA4, and PA6), one EσB promoter (PB1), and one EσE promoter (PE2) (Table 1). The form of RNAP required for the P5 promoter remains unknown. PA4 is largely responsible for low-level transcription from the phoPR promoter during exponential growth in low-phosphate defined medium (LPDM) before autoinduction. PE2 is expressed during stationary phase but not in a sigE mutant strain (27, 30). Autoinduction by PhoP∼P enhances transcription from PA4 and PE2 (27). P5 was induced only in a sigB mutant strain, perhaps in response to increased Pi deficiency stress caused by the absence of SigB-regulated phosphate starvation-induced (PSI) genes (19, 27, 30). PA6 was expressed in a ccpA mutant strain (32). PA3 is more strongly induced in vivo than in vitro, probably because it lacks an unknown activator required to compensate for its poor −35 consensus (27). To date, we have accumulated data that uncover layers of regulation placed on the Pho response by exploring the ResDE TCS, CcpA, and Spo0A∼P (19, 27, 32, 35), but how transition state regulators affect the Pho response in B. subtilis during transition from late exponential growth to the early stationary phase (when Pi concentration is <0.1 mM) (21) remains a question. AbrB binds extensively to the phoPR operon promoter region (M. A. Strauch and F. M. Hulett, unpublished data).
TABLE 1.
Characteristics of each promoter identified within the phoPR operon promoter region
| phoPR promoter | Positiona | σ Factorb | Activator | Repressor |
|---|---|---|---|---|
| PA6 | −175 | σA | CcpA | |
| P5 | −93 | Unknownc | Unknownd | Unknown |
| PA4 | −69 | σA | PhoP∼P | |
| PA3 | −48 to −49 | σA | Unknowne | |
| PE2 | −36 to −38 | σE | PhoP∼P | |
| PB1 | −23 | σB |
Relative to the A, position +1, of the translation start site, ATG.
Not EσA, EσB, EσE, or EσH responsive (Paul and Hulett, unpublished data).
The transition state of B. subtilis has been characterized by the expression of functions that are not expressed during exponential growth but initiate expression as cells enter the stationary growth phase. Transition phase functions include production of antibiotics, synthesis of flagella, development of competence for DNA uptake, motility, and production of degradative enzymes, including alkaline phosphatases, that have been shown to be regulated by transition state regulators (28, 29, 38-40). Among the best-studied B. subtilis transition state regulators are AbrB, ScoC, and Sin.
ScoC was first identified as a sporulation control locus (4) and has also been referred to as hpr (17) or cat (15). ScoC is a negative regulator of extracellular proteases (nprE and aprE), sinI, and both oligopeptide permeases, app and opp, either of which is sufficient to supply the essential oligopeptide permease function for sporulation initiation (23). ScoC is a MarR-type regulator whose transcription is activated by AbrB and has been reported to repress alkaline phosphatases (4, 7). A consensus DNA binding sequence, RATANTATY, was shown by footprint analysis to lie upstream of the nprE, aprE, and sinI genes (22). More recently, it was shown that in the presence of 2% glucose, scoC does not require functional AbrB for its expression (36). These observations suggest that AbrB and ScoC might independently regulate the phoPR operon by affecting the expression of one or more transcription start sites.
Several observations led us to explore the possibility of a role for ScoC in phoPR expression: (i) the presence of three σA transcription start sites in the phoPR promoter region although most phoPR expression is postexponential, (ii) the presence of two putative consensus ScoC binding sites within the phoPR promoter region that contain an 8/9-bp match to the ScoC binding consensus (RATANTATY) (22), and (iii) the occurrence of Pho induction in the medium when the Pi concentrations fall below 0.1 mM during the transition state of the cells.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains, plasmids, and primers used in this study are described in Tables 2 and 3. To construct MH7415, JH642 was transformed with linearized plasmid pJM2501 (28), and the transformants were selected for chloramphenicol (Cm) resistance (5 μg/ml) on tryptose blood agar base (Difco), containing 0.5% glucose. To construct MH7416, MH7415 (Cmr) was transformed with linearized pJL62 (1.1-kb spectinomycin cassette cloned into NcoI site of pJH101) (14), and transformants were selected for spectinomycin (Spc) resistance (100 μg/ml) and screened for Cm sensitivity. To construct MH7400, an isogenic scoC mutant strain containing a phoPR-lacZ fusion in the amy locus, MH5562, was transformed with chromosomal DNA from MH7416, and transformants were selected for Spc resistance (100 μg/ml). MH7417 was constructed via a single crossover through homologous recombination of the phoP cre1-lacZ fusion in pMUTIN2 (pAT14 [32]) into the scoC mutant, MH7416, selecting for erythromycin resistance (5 μg/ml).
TABLE 2.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | Lab stock | |
| BL21(DE3) pLysS | Novagen | |
| B. subtilis strains | ||
| JH642 | pheA1 trpC2 | J. A. Hoch |
| MH5562 | pheA1 trpC2 amy::pho-lacZ Cmr | 27 |
| MH7415 | pheA1 trpC2 scoC::(pJM2501) Cmr | This work |
| MH7416 | pheA1 trpC2 scoC::(pJM2501) Cms::pJL62 Spcr | This work |
| MH7400 | pheA1 trpC2 scoC::(pJM2501) Cms::pJL62 Spcramy::phoPR-lac Z Cmr | This work |
| MH7417 | pheA1 trpC2 scoC::(pJM2501) Cms::pJL62 Spcr pAT14Ω phoP cre1-lacZ Ermr | This work |
| MH6040 | pheA1 trpC2 pAT14Ω phoP cre1-lacZ Ermr | 32 |
| MH6024 | pheA1 trpC2 pAT3Ω phoP-lacZ Ermr | 32 |
| Plasmids | ||
| pJM2501 | A pUC19 plasmid carrying a 900-bp PvuII fragment containing the scoC gene with cat gene insertion mutation at PvuI | 28 |
| pJL62 | 1.1-Kb spectinomycin resistance cassette cloned into NcoI site of pJH101; Ampr Spcr Tetr | I. Smith |
| PCR2.1 | A linearized vector having single 3′ deoxythymidine (T) residues; Ampr Kanr | Invitrogen |
| pHT4-phoPR | pHT315::Sau3A1 fragment of B. subtilis chromosome containing 3′ region of mdh, phoP, and phoR and 5′ polA | 8; T. Masdek |
| pBK1 | 1.364-Kb SmaI-Pau I insert from pHT4-phoPR cloned into pUC18 | 26; this work |
| pBK6.1 | PCR-amplified scoC gene (636 bp) from JH642 chromosomal DNA ligated into PCR2.1 vector | This work |
| pBK6 | Vector for overexpression of ScoC; XhoI-BamHI fragment from pBK6.1 encoding scoC gene cloned into pET16b at the same restriction sites | This work |
| pAT3 | Full-length phoPR promoter from pES2 in pMUTIN2; bp −705 to +92 relative to phoP translation start site | 32 |
| pAT14 | phoP cre1 promoter from pAT12 in pMUTIN2; Ermr | 32 |
| pAT12 | phoP cre1 promoter in pCR2.1 | 32 |
| pSB5 | phoPR promoter in PCR2.1; Ampr Kanr; bp −301 to +92 relative to phoP translation start site | 27 |
| pSB40 | 396-bp BamHI/EcoRI fragment from pSB5 subcloned into pDH32; Ampr Cmr | 27 |
TABLE 3.
Primers
| Primer | Sequence (restriction enzyme)a | Function |
|---|---|---|
| FMH1011 | 5′-GACTCGAGATGAATCGAGTGGAACCGCCC-3′ (XhoI) | Forward primer for the amplification of scoC gene |
| FMH1012 | 5′-GCGGATCCCATCATGAAGCATTTTGATTA-3′ (BamHI) | Reverse primer for the amplification of scoC gene |
| FMH880 | 5′-GAATTC−326GTAGGCGGCAACGG−313-3′ (EcoRI) | Forward primer for the amplification of the 5′ phoPR promoter region |
| FMH881 | 5′-−100CGACAATTCGCCTTTTACA−118-3′ | Reverse primer for the amplification of the 5′ promoter region |
| FMH1018 | 5′-GCGGCCGC−120GATGTAAAAGGCGAATTGTCGG−99-3′ (NotI) | Forward primer for the amplification of the 3′ phoPR promoter region |
| FMH1019 | 5′-CATATG+24CACAACTAAAATTTTCTTGTTC+3-3′ (NdeI) | Reverse primer for the amplification of the 3′ phoPR promoter region |
| FMH1025 | 5′-ATATAAAAGCATTAGTGTATCAATTCAAGC-3′ | Primer within lacZ fusions used for primer extension analysis |
Superscript base pair numbering is relative to the A of the ATG translation start site of phoP. Restriction sites are underlined.
Growth media and enzyme assays.
For the expression of phoPR-lacZ fusions in the wild-type (WT) and scoC mutant strains, the cells were cultured in LPDM containing 2% glucose (LPDMG) as the carbon source as described previously (20). Isogenic scoC mutant strains exhibited a very severe growth defect when grown for expression of the phoPR-lacZ fusions in LPDMG. Because microarray studies (7) indicated that scoC mutations affect the synthesis of isoleucine, arginine, and valine, addition of these amino acids (0.5 mg/ml) along with routinely added amino acids in LPDMG overcame the growth defect and allowed us to study the effect of scoC mutations on phoPR expression under phosphate starvation conditions. β-Galactosidase specific activity (U/mg protein) was determined as described previously (13). The assay was performed at 37°C. The activity unit was defined as 0.33 nmol of ortho-nitrophenol produced per minute.
Overexpression and purification of ScoC.
Escherichia coli DH5α was used as a host for plasmid construction. E. coli BL21(DE3) pLysS (Novagen) served as host for overexpressing the ScoC protein. The scoC gene was amplified from B. subtilis JH642 chromosomal DNA by PCR using primers FMH1011 at the 5′ end of the gene and FMH1012 at the 3′ end of the gene. The PCR product was cloned into pCR2.1 (Invitrogen) to construct pBK6.1. The scoC gene was then released from pBK6.1 by XhoI and BamHI digestion and subcloned into the XhoI and BamHI sites of pET16b (Novagen), generating pBK6. The scoC gene was confirmed by sequencing. The pBK6 plasmid contains a T7 lac promoter, the codons for 10 histidine residues, and an engineered factor Xa site upstream of the scoC gene. BL21(DE3) pLysS cells containing pBK6 were grown in Luria Bertani medium (100 ml) containing carbenicillin (50 μg/ml) and ampicillin (100 μg/ml) at 37°C overnight and were inoculated into 2 liters of the same medium at a ratio of 1 to 100. The cells were grown at 30°C until the optical density at 540 nm (OD540) was 0.6; isopropyl-β-d-thiogalactopyranoside (IPTG; 1 mM) was added to the culture, and the cells were collected by centrifugation at 8,000 × g for 45 min after a 3-h incubation period. The pellet was resuspended in 30 ml of sonication buffer (50 mM Tris-HCl [pH 8.0], 500 mM NaCl, 5 mM MgCl2, and 20% glycerol); after 1 mM phenylmethylsulfonyl fluoride (PMSF) was added, the cells were sonicated immediately and centrifuged at 16,000 × g for 1 h at 4°C. The supernatant fraction was filtered through 0.45-μm-pore-size membrane and applied to a 2.5-ml nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen) affinity column (the Ni-NTA resin was previously equilibrated with sonication buffer in a 1.0-cm by 1.0-cm Econo-column [Bio-Rad]). This column was incubated at 4°C on a rotoshaker for 1.5 h. The column was sequentially washed with the sonication buffer (20 times, 2.5 ml each time) until the OD280 of the elute was less than 0.03; it was then washed with 30 mM imidazole in sonication buffer (two times, 2.5 ml each time) at 4°C. The bound protein was eluted with a four-step imidazole gradient from 100 to 400 mM (2.5 ml each of 100 mM, 200 mM, 300 mM, and 400 mM imidazole in the sonication buffer at 4°C). The eluted protein (300 mM and 400 mM imidazole) was dialyzed overnight against sonication buffer at 4°C to remove the imidazole. The protein concentration was determined using a Bio-Rad protein assay (Bio-Rad Laboratories), with bovine serum albumin (BSA) as the standard. The protein was aliquoted and stored at −70°C.
DNase I footprinting.
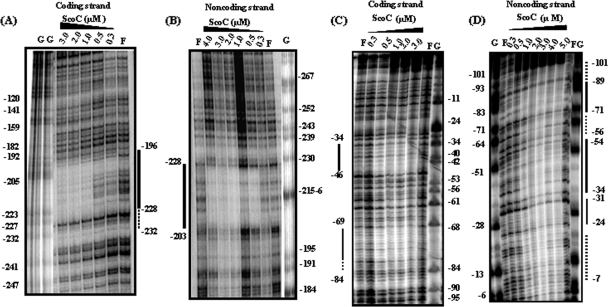
DNase I footprinting experiments were performed as described previously (24). The phoPR promoter was amplified from pBK1 in two different reactions to make two fragments, each containing a putative ScoC binding site(s). To assess DNase I protection by ScoC on the 5′ promoter region containing a single putative binding site, 5′-−227GAAAGTATT−219-3′ (see Fig. 1), on the coding strand, a 232-bp-long probe for the coding strand was prepared by amplifying the phoPR promoter from pBK1 using radiolabeled primer FMH880 and nonradiolabeled primer FMH881. For the noncoding strand preparation, a PCR was set up using nonradiolabeled primer FMH880 and radiolabeled primer FMH881. PCR products were gel extracted using a Qiagen gel extraction kit. To assess the DNase I protection by ScoC on the 3′ promoter region containing a single putative binding site, −78AATAAAATC−71 (see Fig. 1), on the coding strand, a 156-bp-long probe for the coding strand was prepared by amplifying the phoPR promoter from pBK1 using radiolabeled primer FMH1018 and nonradiolabeled primer FMH1019. For the noncoding strand, a PCR was set up using nonradiolabeled primer FMH1018 and radiolabeled primer FMH1019.
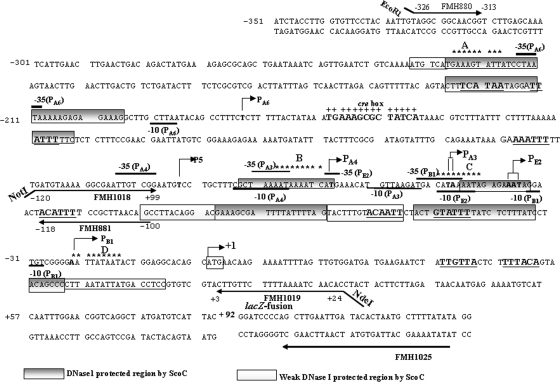
FIG. 1.
The phoPR promoter sequence and 5′ PhoP coding sequence showing six transcription start sites along with the putative ScoC binding sites. Transcription start sites for PB1, PE2, PA3, PA4, P5, and PA6 are indicated by boldface sequence and are identified by a bent arrow followed by the promoter number and a letter identifying the form of RNAP (where known) required for the transcription. The −10 region is marked below, and the −35 is marked above the sequence for each promoter based on published consensus sequences (16). The region of +1 to +92 is the 5′ PhoP coding sequence followed by the lacZ fusion. The translation start site, ATG, is boxed and identified by a bent arrow marked +1. Sequence numbering is relative to the A of ATG as +1. A single putative ScoC binding site in the 5′ region of the phoPR promoter, 5′-−227GAAAGTATT−219-3′ (site A), is located on the coding strand. Three conserved putative ScoC binding sites in the 3′ region of the phoPR promoter are 5′-78AATAAAATC−71-3′ (site B), 5′-−51CATAAAATA−43-3′ (site C), and 5′-−23AATTATAAT−15-3′ (site D) located on the coding strand. The ScoC binding consensus sequence is RATANTATY, where R is A or G, Y is C or T, and N is A, G, C, or T. The putative ScoC binding sites are indicated with nine stars above the sequences. The CcpA binding site (cre box) is shown in bold, marked above with plus signs. Primers used are shown as underlined or overlined sequences with an arrow in the direction of synthesis. FMH1025 is the primer used for primer extension analysis specific to the lacZ fusion. The ScoC DNase I-protected regions are shown as gray boxes on both strands of the phoPR promoter. The white boxes show the weaker ScoC protection. The consensus repeats for PhoP dimer binding, TT(A/C/T)A(C/T)A, are underlined, and the sequence is in boldface.
In each reaction mixture, the ScoC protein at increasing concentrations of 0.0, 0.3, 0.5, 1.0, 2.0, 3.0, 4.0, and 5.0 μM was incubated with the probe at a 50 nM concentration in the binding buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA [pH 7.5], 50 mM KCl, 10% glycerol, and 1 mM dithiothreitol) at room temperature and digested with 2 μl of DNase I (1 U/μl) for 1 min for protein-containing samples and for 30 s for protein-free samples. The reaction was stopped, and the DNA fragments were purified by phenol-chloroform extraction, followed by ethanol precipitation. The DNA fragments were run on a 6% polyacrylamide gel containing 7 M Urea and detected by using a Storm 860 PhosphorImager (Molecular Dynamics) or X-ray film.
RNA preparation and primer extension analysis.
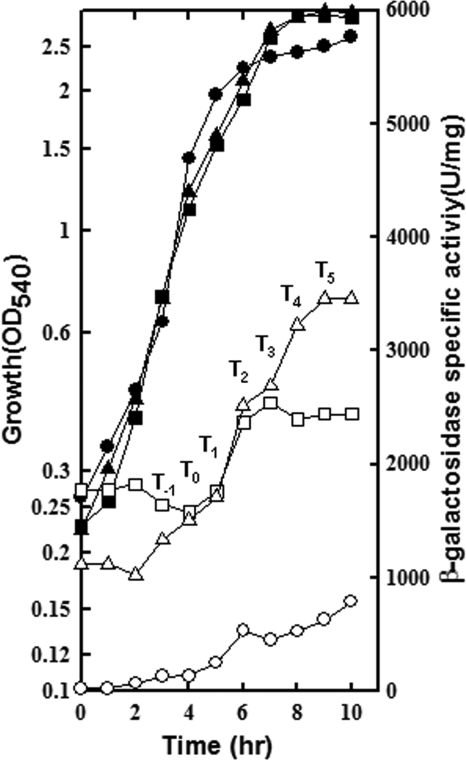
Total RNA was isolated, by using an RNeasy Midi Kit (Qiagen), from MH5562, MH7400, MH6040, and MH7417 strains grown in LPDMG at various time intervals (exponential stage, transition stage, and postexponential growth) during a 12-h growth period. Two volumes of the RNA Later stabilization reagent (Qiagen) were added to 1 volume of the cell culture; they were mixed by vortexing and kept at room temperature for 10 min. Cells were immediately collected by centrifugation at 5,000 rpm at 4°C for 10 min. This pellet was then used to isolate total RNA using the kit according to the manufacturer's instructions. Purified RNA was stored at −80°C until use. A primer specific for lacZ was used for primer extension analysis of RNA. FMH1025 was end labeled for 30 min at 37°C in a 50-μl 1× T4 polynucleotide kinase (PNK) forward reaction buffer (MBI Fermentas) containing 150 μCi [γ-32P]ATP (6,000 Ci/mmol or 10 mCi/ml; Perkin-Elmer) and 50 units of PNK (10 U/μl). The labeling reaction was stopped by heating the mixture at 90°C for 10 min. A total of 50 μg of total RNΑ was used in each primer extension reaction mixture. A primer extension procedure previously described (9) was used with some modifications. The end-labeled primer was annealed to 50 μg of total RNA by mixing and precipitating it together with 0.1 volume of 3 M sodium acetate and 2.5 volumes of 100% ethanol; the sample was chilled on dry ice for 30 min and centrifuged at 12,000 rpm for 10 min; the pellet was air dried, suspended in 25 μl of 1× reverse transcriptase buffer (RTB) (MBI Fermentas), and incubated in a preheated water bath at 80°C. The water bath was allowed to cool at room temperature until it reached 37°C. Two microliters of RNase inhibitor (MBI Fermentas) was added, and incubation continued at 37°C for 16 h. To each reaction mixture, 25 μl of the primer extension mix (1× RTB, 10 mM each deoxynucleoside triphosphate [dNTP], 1 U of avian myeloblastosis virus [AMV] reverse transcriptase, 0.3 units of RNase inhibitor [MBI Fermentas]) was added, and samples were incubated in a water bath at 42°C for 1 h. Reactions were stopped by incubation at 90°C for 15 min. Primer extension products were precipitated with 0.1 volume of 3 M sodium acetate and 2.5 volumes of 100% ethanol at −80°C for 30 min; they were collected by centrifugation (12,000 rpm) at 4°C, and the pellets were air dried. The pellets were suspended in 8 μl of Tris-EDTA (TE) buffer, pH 8.0, and 4 μl of a sequencing dye (98% formamide, 10 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue). Samples were heated at 85°C for 5 min and run on a preheated 5% sequencing gel. A sequencing ladder was made by annealing the same end-labeled primer, FMH1025, to pSB40 or pAT14 for experiments shown in Fig. 4 and 6, respectively, using a Sequenase, version 2.0, sequencing kit (U.S. Biochemicals Corp.) according to the manufacturer's instructions.
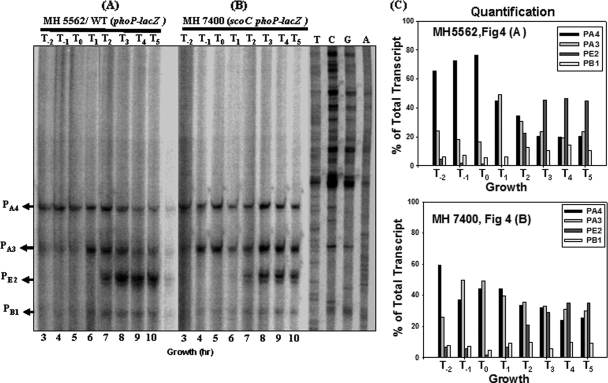
FIG. 4.
Primer extension analysis of the phoPR promoter region from the total RNA isolated from a WT (MH5562) or a scoC strain (MH7400) grown in LPDMG. The end-labeled primer FMH1025 was annealed to RNA isolated from exponential phase, transition stage, or postexponential phase cultures. (A) Lanes 3 to 10 show the primer extension reactions of RNA samples taken from a WT strain (MH5562) growing in LPDMG at the times indicated. The expression from four transcriptional start sites (PB1, PE2, PA3, and PA4) is indicated with arrows. T0 is the time of Pho induction and T1, T2, T3, T4, and T5 are 1, 2, 3, 4, and 5 h of growth, respectively, in LPDMG after Pho induction. T−2 and T−1 samples were taken at 2 h and 1 h before Pho induction, respectively. (B) Lanes 3 to 10 show the primer extension of RNA samples taken from the scoC (MH7400) strain at the times indicated. (C) Quantification of individual transcripts. Radioactivity was determined in arbitrary units for each transcript by phosphorimaging. The sum of all transcripts from a reaction was considered 100%. The percent contribution of each transcript to the total transcription from each primer extension reaction was calculated and plotted in a set of bar graphs. The time of RNA sampling was indicated relative to Pho induction, T0. Bars correspond to PA4, PA3, PE2, and PB1, respectively.
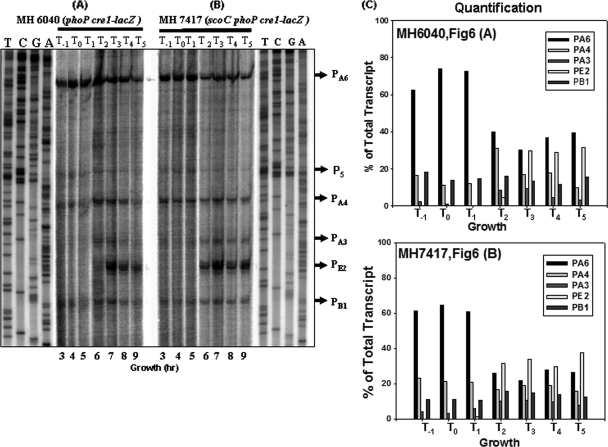
FIG. 6.
Primer extension analysis of the phoPR promoter region from the total RNA isolated from the phoP cre1-lacZ (MH6040) strain or scoC phoP cre1-lacZ (MH7417) strain grown in LPDMG. The end-labeled primer FMH1025 was annealed to RNA isolated from exponential phase, transition stage, and postexponential phase cultures. (A) Lanes 3 to 9 show the primer extension reactions of RNA samples taken at 3 to 9 h from the phoP cre1-lacZ (MH6040) strain grown in LPDMG (Fig. 5) at the times indicated. T0 is the time of Pho induction, and T1, T2, T3, T4, and T5 are 1, 2, 3, 4, and 5 h of growth, respectively, after Pho induction in LPDMG. T−1 is 1 h before Pho induction (growth hour 3). Promoter expression from six transcriptional start sites, PB1, PE2, PA3, PA4, P5, and PA6, is indicated with arrows. (B) Lanes 3 to 9 show the primer extension of RNA samples taken at the indicated times from the scoC phoP cre1-lacZ (MH7417) strain. (C) Quantification of individual transcripts as described for Fig. 4. Bars correspond to PA6, PA4, PA3, PE2, and PB1, respectively.
RESULTS
ScoC represses the transcription of the phoPR promoter.
The phoPR operon promoter region contains three vegetative σA promoters, which raised the question of why the expression of this operon is so low during exponential growth and induced principally postexponentially. Part of the answer is that PhoP∼P, which is present only after a culture experiences growth-limiting Pi concentrations (0.1 mM), enhances expression of one of the three σA promoters, PA4. Transition state regulators, which silence promoters during exponential growth that would otherwise be active, are also candidates for phoPR repression.
The phoPR promoter contains two putative consensus DNA binding sites for ScoC with eight of nine residues matched to the ScoC DNA binding consensus sequence RATANTATY (22) shown in Fig. 1. The 5′ promoter region contains a putative binding site, −227GAAAGTATT−219 (Fig. 1, site A) on the coding strand. Similarly, the 3′ promoter region contains one putative binding site, −78AATAAAATC−71 (Fig. 1, site B) on the coding strand, with eight of nine residues matched, and two less-conserved ScoC binding sites, −51CATAAAATA−43 and −23AATTATAAT−15 (Fig. 1, sites C and D, respectively).
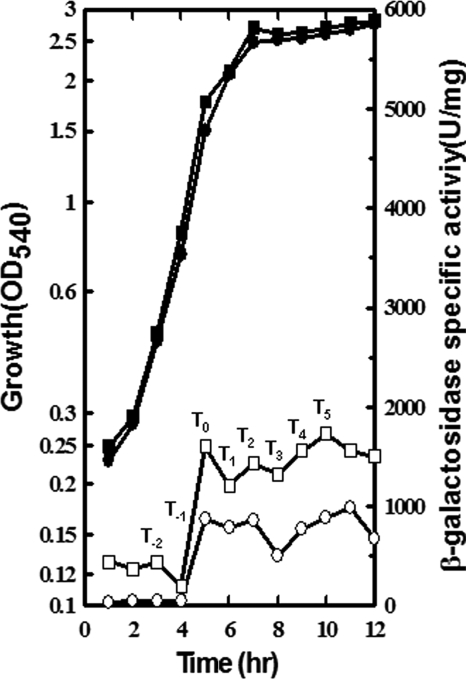
To determine if ScoC had a role in phoPR promoter regulation, we grew strain MH5562, which contained a single copy of the phoPR-lacZ promoter fusion at the amyE locus in JH642, and an isogenic strain, MH7400 (MH5562 with a scoC mutation), in LPDMG plus isoleucine, arginine, and valine in addition to routinely added amino acids, over a period of 12 h (Fig. 2). The WT strain, MH5562, exhibited low-level transcription from the phoPR promoter fusion during Pi-replete exponential growth (0 to 4 h). As the concentration of Pi decreased (5 h to 12 h) below 0.1 mM (2, 21), there was induction of the transcription of the phoPR operon in the WT strain, which is normally observed in JH642 grown in LPDMG (27). The isogenic scoC mutant strain, MH7400, had a higher level of phoPR transcription (5- to 10-fold) during exponential growth (at 0 to 4 h) than the WT strain (MH5562). After induction in both the strains, the scoC mutant maintained phoPR expression approximately 2-fold higher than the WT strain. These data suggested that ScoC represses, directly or indirectly, phoPR expression throughout growth, albeit to a lesser extent after transition into stationary phase (Fig. 2).
FIG. 2.
Effect of scoC mutation on phoPR transcription throughout growth. Cells were grown in LPDMG over a period of 12 h to monitor the growth and level of phoPR transcription. The first hour when APase activity was induced as a reporter for Pho induction is identified as T0. Solid symbols represent the growth, and open symbols represent the β-galactosidase specific activity of the full-length phoPR-lacZ fusion in each strain. Circle, WT (MH5562); square, scoC (MH7400).
ScoC binds directly to the phoPR promoter.
The above study revealed higher transcriptional levels in the scoC mutant than in the WT strain, leading us to ask if the repressor function of ScoC might be due to the binding of the ScoC protein at the observed putative binding sequences on the phoPR promoter. To test this hypothesis, we preformed gel retardation studies and observed evidence that purified ScoC protein could interact with DNA sequences located in the promoter region of the phoPR operon (data not shown). To determine the regions of the phoPR promoter protected by binding of ScoC protein, DNase I footprinting experiments were performed using purified ScoC and each of two phoPR promoter fragments. One fragment was a 226-bp-long probe (bp −326 to bp −100), called the 5′ promoter region, that contained the PA6 transcription start site, and the second was a 144-bp-long probe (bp −120 to bp +24), called the 3′ promoter region, that contained five transcription start sites (PB1, PE2, PA3, PA4, and P5).
Using the 5′ region of the phoPR promoter as a probe, ScoC protected the promoter region from −228 to −196 on the coding strand (Fig. 3A) and from −203 to −228 on the noncoding strand (Fig. 3B). The distinct protection by ScoC on the 5′ promoter region shown by footprint analysis encompasses the putative ScoC binding site on both strands (Fig. 1, site A). Using the 3′ phoPR promoter region as a probe, ScoC protected the promoter region from −34 to −46 and from −69 to −84 on the coding strand (Fig. 3C). The protected regions on the noncoding strand were from −24 to −31, −34 to −54, and −71 to −89 (Fig. 3D). Weaker ScoC protection on the noncoding strand could be seen to extend from −7 to −24, −56 to −71, and −90 to −101. The location of the DNase I-protected regions on the 3′ phoPR promoter region suggested that the second specific ScoC binding sequence (Fig. 1, site B) was an ScoC binding site on the phoPR promoter.
FIG. 3.
DNase I footprint analysis of ScoC binding to the phoPR promoter. The plasmid pBK1 was used as a template for a PCR probe. The ScoC concentration (μM) is shown at the top of each lane. F, free of ScoC; G, Maxam-Gilbert G sequencing reaction, used as a marker. Base pairs are numbered on coding and noncoding strands relative to the translation start site (as +1). Solid lines identify the DNase I protection. Dotted lines identify the weaker protection by ScoC. (A) DNase I protection by on the coding strand in the 5′ region of the phoPR promoter. End-labeled FMH880 and nonlabeled FMH881 were used to create the PCR probe. (B) DNase I protection on the noncoding strand in the 5′ region of the phoPR promoter. End-labeled FMH881 and nonlabeled FMH880 were used to create the PCR probe. (C) DNase I protection on the coding strand of the 3′ promoter region. End-labeled FMH1018 and nonlabeled FMH1019 were used to create the PCR probe. (D) DNase I protection on the noncoding strand of the 3′ promoter region. End-labeled FMH1019 and nonlabeled FMH1018 were used to create the PCR probe.
Closer inspection of the sequences protected by ScoC binding on the 3′ phoPR promoter region identified two other consensus binding sites, albeit less conserved (Fig. 1, sites C and D). The binding activity of the ScoC protein to the phoPR promoter in the present study protected the consensus binding sequence RATANTATY, which was shown by footprint analysis to lie upstream of the nprE, aprE, and sinI genes (22). The footprint analysis of the phoPR promoter also revealed that ScoC binding protects the −10 elements of the existing transcription start sites PB1, PE2, and PA4 and the −35 elements of the PB1, PE2, PA3, and PA6 promoters. These results suggest that the putative ScoC binding sites are important for the interaction of ScoC on the phoPR promoter to exert its negative effect.
Increased expression of PA3 prior to Pho induction and prolonged expression of PA4 and PA3 during stationary phase appeared to account for increased phoPR expression in an scoC strain compared to the WT.
Primer extension was preformed to determine which phoPR promoter(s) accounts for the increased phoPR expression in an scoC strain (MH7400) compared to the WT strain (MH5562) shown in Fig. 2. Figure 4 shows the results of primer extension analysis using RNA from MH5562 and MH7400 cultures grown in LPDMG that was isolated hourly, starting 2 h prior to (time T−2) and continuing until 5 h after Pho induction (defined as T0), as determined by APase assays. Transcription of PA3 was observed earlier in the scoC mutant strain (Fig. 4B T−1) than in the WT strain (Fig. 4A, T1) suggesting that PA3 contributes to the increased levels of total phoPR transcript during exponential growth (Fig. 2). Levels of both PA4 and PA3 transcripts appeared to decrease during stationary phase in the WT strain but not in the scoC mutant strain. Levels of PE2 appeared higher in the WT than in the scoC strain. Together, the last two observations suggest that the sustained expression of PA3 and PA4 during stationary phase contributed to the higher total phoPR expression in the scoC strain during that time.
For each primer extension reaction, the radioactivity from each promoter was detected using a PhosphorImager and was quantified using ImageQuant software. The sum of the activity was defined as 100%, and the percent contribution from each promoter was calculated. The data are shown in bar graphs. In the WT samples PA4 was the major transcript from T−2 to T0 (60 to 80% of total transcript), but in the scoC strain PA3 was a major contributor to the total transcript, i.e., nearly 50% of the total transcript by T−1. In the WT strain at T1, PA3 and PA4 were nearly equally expressed, with each accounting for 45 to 50% of the total transcript, but thereafter their transcripts decreased in unison between T2 and T5, at the same time that PE2 was increasing. PA3 and PA4 each represented less than 20% of total transcript while PE2 accounted for over 40% between T3 and T5. In the scoC strain there was no significant change in PA3 and PA4 transcripts (T3 to T5) such that as PE2 transcript increased, the three transcripts became close to 30% each.
PB1 transcripts were low throughout growth but increased slightly in both strains during stationary growth. Neither P5 nor PA6 was observed in either strain, as expected.
Maximum expression of the phoP cre1 promoter fusion in an scoC background was greater than in a wild-type strain.
A phoP promoter fusion in a WT strain which contained a mutation in the CcpA binding site was reported to express a phoPR-lacZ fusion at levels similar to those observed in a ccpA deletion strain. To ask how ScoC affected PA6 transcription, we used strain (MH7417) containing an scoC mutation and the cre1 mutation in the CcpA binding site (cre box) of the phoPR promoter fusion, which allowed the expression of the phoP cre1-lacZ fusion at the phoP locus and also retained an intact phoPR promoter for phoPR operon expression downstream of the plasmid insertion. Isogenic JH642 strains containing a WT phoPR-lacZ (MH6024) or a phoP cre1-lacZ (MH6040) promoter fusion at the phoPR locus were used as controls.
These three strains were grown in LPDMG and assayed for growth and promoter expression over a period of 10 h (Fig. 5). APase induction was measured to determine T0, the time of Pho regulon induction (data not shown). The strain with the WT phoPR-lacZ fusion (MH6024),which exhibited low-level phoPR expression under excess phosphate conditions (0 to 4 h), was induced after inorganic phosphate levels became limiting (at 5 to 12 h). Expression from the phoP cre1-lacZ fusion in the MH6040 (WT) or MH7417 (scoC) strain had considerable lacZ activity from the inoculum, as was observed from the same promoter fusion in a ccpA mutant that expressed PA6 (32). Induction in the scoC strain began between hours 2 and 3 (1 h prior to APase induction) and continued to increase until hour 10 h. Induction in the WT strain was delayed until hour 5 and continued until hour 7; thereafter, the specific activity remained stable. The maximum expression from the phoP cre1-lacZ fusion in the scoC strain was 1.4-fold higher than in the WT strain. The elevated expression of the phoPR promoter fusion in MH7417 (scoC phoP cre1-lacZ) may be attributed to the derepression of the transcription from the PA6 promoter as a result of the scoC mutation and the inability of CcpA to bind at the cre1 site and block the transcription.
FIG. 5.
Combined effect of scoC and phoP cre1 promoter mutation on phoPR transcription during growth and stationary phase. Cells were grown in LPDMG for 10 h to monitor growth and level of transcription. Solid symbols represent the growth, and open symbols represent the β-galactosidase specific activity of the full-length phoPR-lacZ fusion in each strain. Circle, WT phoPR-lacZ (MH6024); square, phoPR-lacZ cre1-lacZ strain (MH6040); triangle, scoC phoPR-lacZ cre1-lacZ strain (MH7417).
A cre1 mutation in the phoPR promoter is sufficient to allow expression of PA6.
Primer extension analysis was performed on RNA isolated at various times of growth before and after Pho induction from the MH7417 and MH6040 cultures used in the experiment shown in Fig. 5 to determine if PA6 were expressed and which of the six promoters were regulated by ScoC. The data shown in Fig. 6A confirmed the assumption that PA6 was expressed from the phoP cre1-lacZ fusion in MH6040 (32), which resulted in expression levels similar to those observed in a ccpA mutant strain. Also similar to the primer extensions studies using RNA isolated from a ccpA strain, weak transcripts from P5 were observed in either strain with the phoP cre1 promoter fusion (Fig. 6 A and B).
The absence of ScoC may affect phoPR promoter transcription directly and indirectly.
When individual promoter expression from the phoP cre1-lacZ fusion in the scoC strain (MH7417) was compared to that in the parent strain (MH6040), a number of changes were noted that may be due to the absence of ScoC (Fig. 6A and B). During the first 3 h (T−1 to T1), PA6 was the major transcript in both cultures (≈60% of total transcript) although PA4 showed a relative increase in the scoC mutant compared to the parent strain, which may be due to the absence of ScoC repression. Two interesting differences in expression were noted at T2. First PE2, which requires EσE for expression, was expressed in the scoC strain but not in the parent strain, indicating that sporulation initiation had occurred earlier in the scoC strain. Further, PA6 expression decreased more dramatically in the scoC strain than in the parent strain at T2, consistent with decreasing EσA concentrations after the initiation of sporulation. Sporulation control, for which scoC was named (4), results from ScoC repression of oligopeptide transport expression (opp operon) (23), thereby delaying sporulation initiation. The latter two observations discussed here may represent direct roles of ScoC that affect sporulation initiation, which indirectly affects phoPR transcription.
Of the three σA-responsive promoters, PA4 expression was the least affected by the decreasing concentrations of EσA associated with the initiation of sporulation. This is consistent with in vitro transcription studies which indicated that the concentration of EσA required was decreased with increasing concentrations of PhoP∼P at another PhoP∼P-enhanced promoter, the resA promoter (1).
DISCUSSION
ScoC represses several phoPR promoters.
Analysis of the data presented here indicated that ScoC downregulates phoPR expression during vegetative and postexponential growth. The activity of the phoPR-lacZ fusion in the scoC loss-of-function mutant strain was elevated 5- to 10-fold during Pi-replete exponential growth compared to activity in the WT strain, in which expression of the phoPR-lacZ fusion is barely detectable (Fig. 2). The phoPR-lacZ fusion induction, when the cultures entered the stationary phase due to limiting inorganic phosphate, was 2-fold higher in the scoC mutant strain than the in the WT strain. In vitro transcription studies showed ScoC repression of transcription from PA6, PA4, and PB1 transcription start sites using T0 RNAP and from PA4, PA6, and PE2 start sites using T3 RNAP (data not shown). Our knowledge of promoters P5 and PA3 (Table 1) is insufficient to evaluate them using in vitro transcription.
Previous studies have identified an alkaline phosphatase among ScoC-repressed proteins (4, 7). It seems unlikely that ScoC plays a negative role in alkaline phosphatase expression via the phoPR derepression reported here. First, increased expression of phoPR does not necessarily lead to corresponding changes in pho regulon gene expression (10, 31). Second, the conditions under which the scoC mutant strains were cultured in the microarray studies (7) were not phosphate limiting. Further, not all APase expression in B. subtilis is PhoP∼P activated. phoB (formerly phoAIII) encoding APase B (formerly APase III) (5, 9) is expressed from two differentially regulated promoters, PS (EσE-responsive promoter) and Pv (EσA-responsive promoter), which are PhoP∼P repressed and activated during PSI, respectively (2, 5, 9). The study by Caldwell et al. (7) found a 9.3-fold increase in phoB expression in an scoC mutant compared to the WT strain at the last time point assayed (310 min after inoculation), an increase which represented the greatest fold increase in a group of sporulation genes dependent on Spo0A and σF. This increase could have resulted indirectly from regulation via removal of the repressor role of ScoC in the sporulation process, leading to activation of σE and thereby the phoB Ps promoter, or directly from the absence of ScoC binding to several putative binding sites observed on the phoB promoter, which might account for direct ScoC repression of the upstream phoB PS promoter, or from both.
Negative regulation of the phoPR promoter by ScoC and CcpA is cumulative.
Another layer of transcriptional regulation on the phoP promoter via ScoC was revealed during this study. It was shown previously that CcpA caused repression of the phoPR promoter through the PA6 transcriptional start site which is positioned upstream of the cre box (catabolite response element or CcpA binding consensus sequence) (32). The role of ScoC protection in the 5′ promoter region of the phoPR promoter, upstream of the PA6 transcription start site, was assessed by exploring the transcription in an scoC mutant strain containing the phoP cre1-lacZ fusion (cre1 is a mutation in the cre box). Expression of the phoP cre1-lacZ fusion in a WT strain was previously reported (32) to be similar to the that of the phoPR-lacZ fusion in a ccpA mutant strain. These expression data were consistent with the severely reduced CcpA binding to the phoP cre1 promoter (32).
The difference in the extent of derepression from the phoP cre1-lacZ fusion in the wild-type strain compared to the scoC mutant strain suggested that the repression effect of ScoC and CcpA on the phoPR promoter was cumulative. Primer extension data from a scoC mutant or a WT strain containing the phoP cre1 promoter fusion showed PA6 expression during exponential and postexponential growth in addition to the normally expressed promoters (PB1, PE2, PA3, and PA4) in the WT strain (Fig. 6). The maximum phoP expression from the scoC strain during postexponential growth (Fig. 5) was the result of the following: (i) earlier and sustained elevated expression of PE2; (ii) sustained expression of PA4 and, to a lesser extent, PA3; and (iii) sustained PA6 expression, albeit reduced compared to the WT strain after initiation of sporulation. No PA6 promoter transcription start site was detected from the unmutated phoPR-lacZ promoter fusion in the scoC mutant background during growth in LPDMG (Fig. 4), indicating that CcpA could physically block any repression relief that the scoC mutation provided. How ScoC affects PA6 expression remains a question because no derepression was observed in vivo in the scoC strain compared to the WT strain (Fig. 6), but PA6 was repressed by ScoC in vitro (data not shown). Aside from EσA and CcpA, it is not clear what, or if, other regulators control PA6 expression. In addition to ScoC, two other proteins are known to bind to the phoPR promoter in the region of the PA6 promoter, PhoP∼P (27) and AbrB (F. M. Hulett and M. A. Strauch, unpublished data). If, or how, either of these proteins affects PA6 transcription is not known.
Complexities of the phoPR promoter regulation.
The major difference between the primer extension analyses of the WT phoPR-lacZ and the phoP cre1 promoter fusions was the dominating presence of the PA6 transcript among the phoP cre1 promoter fusion transcripts and the complete absence of PA6 expression of the phoPR-lacZ fusion transcripts. This is easily explained based on the published role of CcpA repression of PA6, the reduced affinity of CcpA binding to the promoter containing the cre1 mutation, and the dominant presence of PA6 transcript in a ccpA mutant background (32). More puzzling were the transcript differences from the two promoters, phoPR-lacZ (Fig. 4) and phoP cre1-lacZ (Fig. 6), in vivo. (i) The contribution of PA3 to the total transcript from the phoPR-lacZ promoter was major compared to its weak expression from the phoP cre1-lacZ fusion in both the WT and the scoC strains; PA3 promoter expression from the phoPR-lacZ fusion was dominant in the scoC strain during both exponential and postexponential growth (Fig. 4B). (ii) The apparent effect on sporulation initiation in the scoC strain, as judged by the early and increased expression of PE2 compared to that in a WT strain, was observed from the phoP cre1-lacZ promoter but not the phoPR-lacZ promoter fusion.
We considered several scenarios that may have contributed to these differences. Both promoter fusions share the four densely positioned overlapping promoters (PB1 to PA4) which may be exposed to multiple different proteins (different forms of RNAP, activators, repressors, and etc.) that compete for their overlapping binding sites, often at the same time. The RNAP machinery required for PA6 transcription must pass through the congested downstream region during transcription of the long untranslated region of the nascent mRNA. It appeared that PA3 was most negatively affected by these conditions, but the reason remains unknown. The question remains, Why does the apparent indirect ScoC regulation occur with the phoP cre1 promoter but not the phoP promoter?
Further studies will be required to determine if either the dynamics of regulatory protein-DNA interactions in the congested promoter regions or the locations of the promoter fusions are responsible for the differences in levels and timing of expression observed from the individual promoters of the two promoter fusions.
In summary, the in vivo phoPR-lacZ transcription data, ScoC footprinting data, primer extension data, and in vitro transcription data reported here together suggest that ScoC is a direct repressor of phoPR transcription. We know that a number of proteins are involved in direct regulation of the complex phoPR promoter via direct binding: PhoP, PhoP∼P, CcpA, AbrB, four different forms of RNAP, and now ScoC. We also have evidence that additional unknown proteins are involved: another RNAP form and possible activators and/or repressors for P5, an unknown activator protein for PA3, and a possible unknown activator for PA6. There is also preliminary evidence for direct binding and activation by ResD∼P of certain of the six promoters (S. Paul and F. M. Hulett, unpublished data). ScoC, Spo0A, and the ResDE TCS are also involved in indirect regulation.
The unknown regulators complicate the interpretation of in vitro transcription studies, making analysis P5 and PA3 incomplete. Further, the interpretation of the in vivo analysis by primer extension data would benefit from additional information concerning the unknown regulators.
Acknowledgments
This work was supported by Public Health Service grant GM-33471 from the National Institutes of Health.
We are thankful to Jae-Yong Park and W. Abdel-Fattah for helpful discussions.
Footnotes
Published ahead of print on 9 April 2010.
REFERENCES
- 1.Abdel-Fattah, W. R. 2007. Bacillus subtilis Pho∼P direct roles in Pho and Res regulation in response to Pi-stress. Ph.D. thesis. University of Illinois at Chicago, Chicago, IL.
- 2.Abdel-Fattah, W. R., Y. Chen, A. Eldakak, and F. M. Hulett. 2005. Bacillus subtilis phosphorylated PhoP: direct activation of the EσA and repression of the EσE responsive phoB-PS+V promoters during Pho response. J. Bacteriol. 187:5166-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antelmann, H., C. Scharf, and M. Hecker. 2000. Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis. J. Bacteriol. 182:4478-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balassa, G., B. Dod, V. Jeannoda, P. Milhaud, J. Zucca, J. C. Sousa, and M. T. Silva. 1978. Pleiotropic control mutations affecting the sporulation of Bacillus subtilis. Ann. Microbiol. (Paris) 129 B:537-549. [PubMed] [Google Scholar]
- 5.Bookstein, C., C. W. Edwards, N. V. Kapp, and F. M. Hulett. 1990. The Bacillus subtilis 168 alkaline phosphatase III gene: impact of a phoAIII mutation on total alkaline phosphatase synthesis. J. Bacteriol. 172:3730-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burbulys, D., K. A. Trach, and J. A. Hoch. 1991. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 64:545-552. [DOI] [PubMed] [Google Scholar]
- 7.Caldwell, R., R. Sapolsky, W. Weyler, R. R. Maile, S. C. Causey, and E. Ferrari. 2001. Correlation between Bacillus subtilis scoC phenotype and gene expression determined using microarrays for transcriptome analysis. J. Bacteriol. 183:7329-7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, Y., C. Birck, J. P. Samama, and F. M. Hulett. 2003. Residue R113 is essential for PhoP dimerization and function: a residue buried in the asymmetric PhoP dimer interface determined in the PhoPN three-dimensional crystal structure. J. Bacteriol. 185:262-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chesnut, R. S., C. Bookstein, and F. M. Hulett. 1991. Separate promoters direct expression of phoAIII, a member of the Bacillus subtilis alkaline phosphatase multigene family, during phosphate starvation and sporulation. Mol. Microbiol. 5:2181-2190. [DOI] [PubMed] [Google Scholar]
- 10.Choi, S. K., and M. H. Saier, Jr. 2005. Regulation of pho regulon gene expression by the carbon control protein A, CcpA, in Bacillus subtilis. J. Mol. Microbiol. Biotechnol. 10:40-50. [DOI] [PubMed] [Google Scholar]
- 11.Eder, S., W. Liu, and F. M. Hulett. 1999. Mutational analysis of the phoD promoter in Bacillus subtilis: implications for PhoP binding and promoter activation of Pho regulon promoters. J. Bacteriol. 181:2017-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eder, S. C. 1998. The mechanism of PhoP transcriptional activation of Bacillus subtilis phoD and other PHO regulon genes. M.S. thesis. University of Illinois at Chicago, Chicago, IL.
- 13.Ferrari, E., S. Howard, and J. A. Hoch. 1986. Effect of stage 0 sporulation mutants on subtilisin expression. J. Bacteriol. 166:173-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrari, F. A., A. Nguyen, D. Lang, and J. A. Hoch. 1983. Construction and properties of an integrable plasmid for Bacillus subtilis. J. Bacteriol. 154:1513-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenleaf, A. L., and R. Losick. 1973. Appearance of a ribonucleic acid polymerase-binding protein in asporogenous mutants of Bacillus subtilis. J. Bacteriol. 116:290-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helmann, J. D., and C. P. Moran, Jr. 2002. RNA Polymerase and Sigma Factors, p. 289-312. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 17.Higerd, T. B., J. A. Hoch, and J. Spizizen. 1972. Hyperprotease-producing mutants of Bacillus subtilis. J. Bacteriol. 112:1026-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hulett, F., J. Lee, L. Shi, G. Sun, R. Chesnut, E. Sharkova, M. Duggan, and N. Kapp. 1994. Sequential action of two-component genetic switches regulates the PHO regulon in Bacillus subtilis. J. Bacteriol. 176:1348-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hulett, F. M. 2002. The Pho regulon, p. 193-203. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 20.Hulett, F. M., C. Bookstein, and K. Jensen. 1990. Evidence for two structural genes for alkaline phosphatase in Bacillus subtilis. J. Bacteriol. 172:735-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen, K. K., E. Sharkova, M. F. Duggan, Y. Qi, A. Koide, J. A. Hoch, and F. M. Hulett. 1993. Bacillus subtilis transcription regulator, Spo0A, decreases alkaline phosphatase levels induced by phosphate starvation. J. Bacteriol. 175:3749-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kallio, P. T., J. E. Fagelson, J. A. Hoch, and M. A. Strauch. 1991. The transition state regulator Hpr of Bacillus subtilis is a DNA-binding protein. J. Biol. Chem. 266:13411-13417. [PubMed] [Google Scholar]
- 23.Koide, A., M. Perego, and J. A. Hoch. 1999. ScoC regulates peptide transport and sporulation initiation in Bacillus subtilis J. Bacteriol. 181:4114-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, W., and F. M. Hulett. 1997. Bacillus subtilis PhoP binds to the phoB tandem promoter exclusively within the phosphate starvation-inducible promoter. J. Bacteriol. 179:6302-6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, W., Y. Qi, and F. M. Hulett. 1998. Sites internal to the coding regions of phoA and pstS bind PhoP and are required for full promoter activity. Mol. Microbiol. 28:119-130. [DOI] [PubMed] [Google Scholar]
- 26.Messing, J. 1983. New M13 vectors for cloning. Methods Enzymol. 101:20-78. [DOI] [PubMed] [Google Scholar]
- 27.Paul, S., S. Birkey, W. Liu, and F. M. Hulett. 2004. Autoinduction of Bacillus subtilis phoPR operon transcription results from enhanced transcription from EσA- and EσE-responsive promoters by phosphorylated PhoP. J. Bacteriol. 186:4262-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perego, M., and J. A. Hoch. 1988. Sequence analysis and regulation of the hpr locus, a regulatory gene for protease production and sporulation in Bacillus subtilis. J. Bacteriol. 170:2560-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perego, M., G. B. Spiegelman, and J. A. Hoch. 1988. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol. Microbiol. 2:689-699. [DOI] [PubMed] [Google Scholar]
- 30.Pragai, Z., N. E. Allenby, N. O'Connor, S. Dubrac, G. Rapoport, T. Msadek, and C. R. Harwood. 2004. Transcriptional regulation of the phoPR operon in Bacillus subtilis. J. Bacteriol. 186:1182-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puri-Taneja, A. 2007. Regulation of phoPR and cyd ABCD promoters of Bacillus subtilis by catabolite control protein A (CcpA). Ph.D. thesis. University of Illinois at Chicago, Chicago, IL.
- 32.Puri-Taneja, A., S. Paul, Y. Chen, and F. M. Hulett. 2006. CcpA causes repression of the phoPR promoter through a novel transcription start site, PA6. J. Bacteriol. 188:1266-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi, Y., and F. M. Hulett. 1998. PhoP∼P and RNA polymerase σA holoenzyme are sufficient for transcription of Pho regulon promoters in Bacillus subtilis: PhoP∼P activator sites within the coding region stimulate transcription in vitro. Mol. Microbiol. 28:1187-1197. [DOI] [PubMed] [Google Scholar]
- 34.Schau, M., Y. Chen, and F. M. Hulett. 2004. Bacillus subtilis YdiH is a direct negative regulator of the cydABCD operon. J. Bacteriol. 186:4585-4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schau, M., A. Eldakak, and F. M. Hulett. 2004. Terminal oxidases are essential to bypass the requirement for ResD for full Pho induction in Bacillus subtilis. J. Bacteriol. 186:8424-8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shafikhani, S. H., E. Nunez, and T. Leighton. 2003. ScoC mediates catabolite repression of sporulation in Bacillus subtilis. Curr. Microbiol. 47:327-336. [DOI] [PubMed] [Google Scholar]
- 37.Strauch, M., V. Webb, G. Spiegelman, and J. A. Hoch. 1990. The SpoOA protein of Bacillus subtilis is a repressor of the abrB gene. Proc. Natl. Acad. Sci. U. S. A. 87:1801-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strauch, M. A., and J. A. Hoch. 1992. Control of postexponential gene expression by transition state regulators. Biotechnology 22:105-121. [PubMed] [Google Scholar]
- 39.Strauch, M. A., and J. A. Hoch. 1993. Transition-state regulators: sentinels of Bacillus subtilis post-exponential gene expression. Mol. Microbiol. 7:337-342. [DOI] [PubMed] [Google Scholar]
- 40.Strauch, M. A., G. B. Spiegelman, M. Perego, W. C. Johnson, D. Burbulys, and J. A. Hoch. 1989. The transition state transcription regulator abrB of Bacillus subtilis is a DNA binding protein. EMBO J. 8:1615-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun, G., S. M. Birkey, and F. M. Hulett. 1996. Three two-component signal-transduction systems interact for Pho regulation in Bacillus subtilis. Mol. Microbiol. 19:941-948. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, X., and F. M. Hulett. 2000. ResD signal transduction regulator of aerobic respiration in Bacillus subtilis: ctaA promoter regulation. Mol. Microbiol. 37:1208-1219. [DOI] [PubMed] [Google Scholar]