Abstract
The bacterial pathogen Brucella abortus was recently demonstrated to recruit the essential cytoplasmic histidine kinase PdhS to its old pole. Here, we report identification of the fumarase FumC as a specific partner for the N-terminal “sensing” domain of PdhS, using an ORFeome-based yeast two-hybrid screen. We observed that FumC and PdhS colocalize at the old pole of B. abortus, while the other fumarase FumA is not polarly localized. FumC is not required for PdhS localization, and polar FumC localization is not FumA dependent. FumC homologs are not polarly localized in Sinorhizobium meliloti and Caulobacter crescentus, suggesting that polar recruitment of FumC by PdhS is evolutionarily recent.
It was previously reported that morphological asymmetry, here defined by the production of two sibling cells different in length at each cell cycle, is a common feature shared by at least four members of the alphaproteobacteria with diversified lifestyles, namely, the free-living model bacterium Caulobacter crescentus, the plant pathogen Agrobacterium tumefaciens, the plant symbiont Sinorhizobium meliloti, and the facultative intracellular pathogen Brucella abortus (5). In B. abortus, an essential cytoplasmic histidine kinase named PdhS is anchored to the old pole of the bacterium (6). PdhS interacts with the response regulator DivK (6).
PdhS is a rather atypical histidine kinase. It is cytoplasmic and contains an N-terminal “sensing” domain with no predicted function, except for a PAS domain that remains to be characterized. Nevertheless, the N-terminal part preceding the PAS domain of PdhS is sufficient for polar localization in B. abortus, as well as in the alphaproteobacteria S. meliloti and C. crescentus (6). This suggests that a molecular mechanism and/or polar structure required for PdhS targeting is conserved in these alphaproteobacteria.
FumC is identified as a PdhS binding protein in a yeast two-hybrid screen.
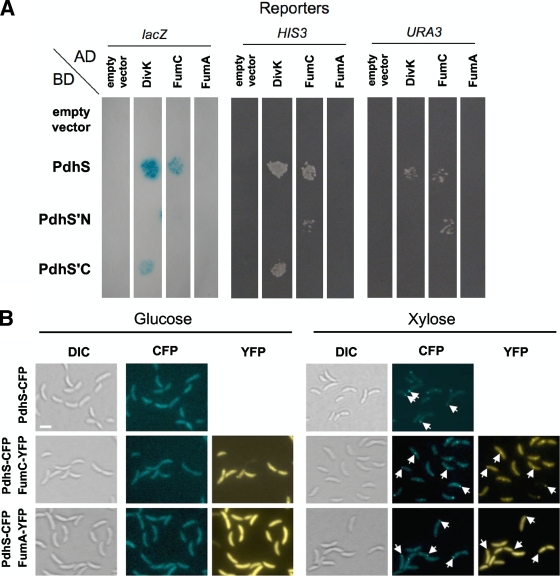
Given that PdhS contains a long, soluble (predicted cytoplasmic) N-terminal domain, we hypothesized that this part of the protein could be involved in protein-protein interactions. We therefore carried out a yeast two-hybrid (Y2H) screen between the full-length version of PdhS and the majority (96.7%) of the 3,198 proteins predicted to be encoded in the B. melitensis 16M genome, whose coding sequences (CDS) are cloned in the ORFeome (2). A detailed description of the protocols used is provided in the supplemental material; all strains and plasmids are described in Table S1, and primers are reported in Table S2. Four candidate clones were positive for at least two Y2H reporters. One of them was DivK (data not shown). We also identified two predicted hypothetical proteins (data not shown) and a predicted 463-residue protein (BAB2_0186; NCBI accession number YP_418415), renamed FumC here, that was positive for the three reporter genes used in the Y2H assay. We performed a second Y2H assay in which the activation domain (AD) and DNA binding domain (BD) fusions were switched, in the sense that AD-PdhS was tested against BD fusions to FumC or other proteins (Fig. 1 A). The AD fusion with PdhS, as well as with its N-terminal (PdhS′N, amino acids [aa] 1 to 613) and C-terminal (PdhS′C, aa 611 to 1035) parts, was tested. As expected, BD-DivK was able to interact with AD-PdhS′C and AD-PdhS (Fig. 1A), and the interaction between BD-FumC and AD-PdhS was confirmed. In addition, an interaction between BD-FumC and AD-PdhS′N was observed, suggesting that the N-terminal domain of PdhS is sufficient to recruit FumC. The PdhS-FumC interaction discovered in the Y2H screen was further supported in this study by use of a distinct technical approach based on the particular localization of B. abortus PdhS when it is produced in C. crescentus (Fig. 1B). PdhS-CFP (cyan fluorescent protein) is able to form foci in C. crescentus, and when FumC-YFP (yellow fluorescent protein) is produced in the same strain, it systematically colocalizes with PdhS-CFP. A detailed description of these assays is provided in the supplemental material.
FIG. 1.
B. abortus PdhS interacts with FumC. (A) Yeast two-hybrid assay for the PdhS-FumC interaction. The fusions to either the AD or the BD of Gal4p are indicated next to the pictures. Three reporters were assayed for activation. PdhS′N and PdhS′C correspond to the N-terminal (613 aa) and C-terminal (425 aa) regions of PdhS, respectively. The PdhS-FumC interaction is positive for lacZ, HIS3, and URA3 reporters. The PdhS′N-FumC interaction is positive for HIS3 and URA3 reporters, suggesting that FumC interacts with the N-terminal part of PdhS. The divK and fumA CDS were used as positive and negative controls for the PdhS-FumC interaction, respectively. Another set of negative controls corresponded to the plasmids (“empty vector”) coding for either the Gal4p AD or the Gal4p BD, without insert. (B) Reconstruction of FumC recruitment by PdhS in C. crescentus. Three strains of C. crescentus carrying a pdhS-cfp fusion (XDB1166) under the control of the xylX promoter were constructed. In two of them, an additional fusion, either fumC-yfp (pJM080) or fumA-yfp (pJM095; negative control), is carried on a replicative plasmid. The cells were grown overnight in PYE (peptone-yeast extract) with glucose to reach a starting optical density at 600 nm (OD600) of 0.05. The medium was then supplemented with glucose (no induction) or xylose (induction) for 5 h, and the cells were subsequently immobilized on agarose pad slides and visualized by differential interference contrast (DIC) microscopy and in CFP and/or YFP channels. The fusions produced by each strain are indicated on the left. The type of observation, either DIC microscopy (Normarski) or CFP or YFP typical fluorescence, is also indicated for each culture medium. White arrows indicate the positions of some PdhS-CFP foci. All pictures are shown with the same magnification. Bar, 2 μm.
The B. abortus genome encodes two fumarases, FumC and FumA.
BAB2_0186 is the homolog of Escherichia coli FumC (see Fig. S1 in the supplemental material), which belongs to the family of class II fumarate hydratases (also called fumarases) and is involved in the tricarboxylic acid (TCA) cycle, in which it reversibly converts fumarate into l-malate (7). The E. coli genome also contains two genes sharing a high percentage of identity (89%) and named fumA and fumB, encoding, respectively, FumA and FumB, belonging to the family of class I fumarate hydratases (8). In B. abortus, the protein BAB1_0977 (NCBI accession number YP_414395) shares 66 and 65% identities (80% and 79% similarities) with E. coli FumA and FumB, respectively. BAB1_0977 was therefore named FumA. FumA is not able to interact with PdhS in a Y2H assay (Fig. 1). A demonstration of fumarase activity by recombinant FumA and FumC is provided in the supplemental material.
We constructed an in-frame deletion of fumA or fumC CDS in the B. abortus 544 Nalr strain (named, respectively, XDB1160 or XDB1157) by using an allelic-replacement procedure. The two single deletions did not alter overall cell morphology or cell growth compared to that for the wild-type strain (data not shown). However, fumA and fumC deletions were synthetic lethal (see the supplemental material). From these results, we concluded that fumA and fumC are functionally redundant, at least under the culture conditions used here, and that translational fusions of fumC and fumA to yfp are at least partially functional, because they rescued the lethality of the fumA fumC double deletion.
FumC localizes at one pole in B. abortus.
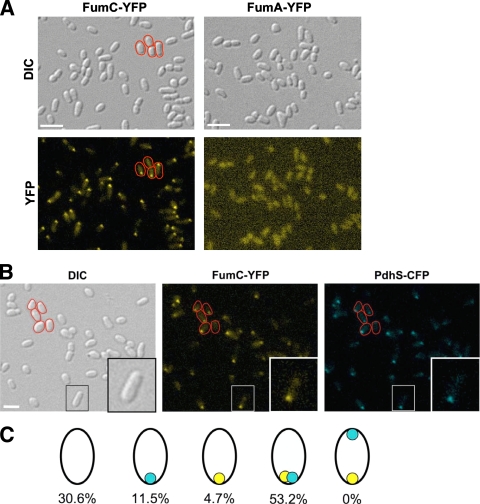
We generated strains carrying a chromosomal fusion of fumC CDS with yfp, cfp, and mCherry gene CDS (strains XDB1150, XDB1151, and XDB1152, respectively) in the B. abortus 544 Nalr background. The fumC CDS is not predicted to be cotranscribed with the downstream CDS (data not shown). By fluorescence microscopy, we observed that FumC-YFP accumulated at one pole in 74% (n = 431) of the XDB1150 cells, reminiscent of the localization pattern of PdhS (Fig. 2). Similar results were observed for FumC-CFP and FumC-mCherry fusions, suggesting that the nature of the fluorescent protein is not responsible for the polar localization. This localization pattern was also observed when a fumC-yfp fusion was expressed from the low-copy-number plasmid pJM063 introduced into either the wild-type or the ΔfumC strain (data not shown). An inactive version of FumC (with the H188N substitution [see the supplemental material]) was still able to be recruited to a pole when it was fused to YFP (data not shown). In contrast, FumA-YFP was homogeneously scattered throughout the cytoplasm whether it was produced from the chromosomal fumA locus (XDB1153 strain) (Fig. 2A) or from the low-copy-number plasmid pJM043 in B. abortus (data not shown). Western blot analysis performed on every strain indicated that the fusion proteins were stable (data not shown). We wondered whether the absence of one fumarase, either FumA or FumC, has an impact on the subcellular localization of the other. However, FumA-YFP and FumC-YFP fusions retained localization patterns in ΔfumC and ΔfumA mutants, respectively, that were similar to those found with the wild-type strain (see Fig. S2 in the supplemental material). These data suggested that FumC localization, at least under the conditions tested here, does not depend on the presence of FumA and that the FumA-YFP localization pattern is not altered when FumC is absent. We tested the localizations of other TCA cycle enzymes fused to the N or C terminus of the mCherry fluorescent reporter but did not detect polar localization (C. Van der Henst and M. Deghelt, unpublished data), suggesting that no TCA cycle enzyme is localized at the old pole in B. abortus.
FIG. 2.
FumC-YFP is localized at the old pole in B. abortus. (A) Localization of FumC-YFP and FumA-YFP in B. abortus. The XDB1150 and XDB1153 strains, producing, respectively, FumC-YFP and FumA-YFP from fusions integrated into the B. abortus 544 chromosome, were observed during the exponential growth phase by DIC microscopy (Normarski) and for the typical fluorescence spectrum of YFP. (B) Colocalization of PdhS-CFP with FumC-YFP in B. abortus. A fumC-yfp fusion was integrated by homologous recombination at the fumC locus in the chromosome of XDB1155, a B. abortus strain in which pdhS was replaced by a pdhS-cfp allele. Observations were made by DIC microscopy (Normarski) and for typical fluorescence of YFP and CFP, as indicated. Some bacteria are surrounded with a red line in the pictures to show that the FumC-YFP and PdhS-CFP signals are polar. Bar, 2 μm. (C) Schematic drawing illustrating the proportions of cells that exhibit a localization pattern for PdhS-CFP and/or FumC-YFP. A total of 425 bacteria were examined. The polar foci of PdhS-CFP and FumC-YFP are shown in blue and yellow, respectively. It is striking that all 226 bacteria (53.2%) for which a CFP focus and a YFP focus were detectable have colocalized signals and that none present foci labeling the opposite poles. A quite large fraction of the population (30.6%) does not display any focus, whether CFP or YFP. This could be due to a low expression level of the fusions, resulting in poor sensitivity for the fluorescent signal.
FumC and PdhS colocalize at the old pole of B. abortus.
As FumC and PdhS display similar unipolar localization patterns and physically interact in Y2H assays, we reasoned that they may colocalize in B. abortus. Therefore, a B. abortus 544 strain (XDB1155) in which the wild-type chromosomal pdhS allele was replaced by a pdhS-cfp fusion was constructed. PdhS-CFP, as the only copy of PdhS that supported viability, did not impair cell morphology and displayed a localization pattern consistent with that reported previously (6). A fumC-yfp fusion was then integrated at the fumC locus, as described for the XDB1150 strain. The FumC-YFP localization pattern in this pdhS-cfp background strain (XDB1156) was similar to those described above. As depicted in Fig. 2B and C, 92% (n = 246) of the cells for which a FumC-YFP polar focus was detectable exhibited a corresponding PdhS-CFP polar focus at the same localization. Moreover, although 16% of the entire bacterial cell population (n = 425) exhibited either a PdhS-CFP or a FumC-YFP polar focus, cells with two foci located at opposite poles were never observed. These data strongly argue in favor of a model in which FumC and PdhS colocalize at the old pole of B. abortus.
FumC is not required for PdhS polar localization.
The discovery of a new PdhS partner, FumC, prompted us to explore the impact of the absence of FumC on PdhS subcellular distribution. A fumC deletion was therefore constructed in the B. abortus 544 pdhS-cfp strain (XDB1155) to generate the XDB1158 strain. The XDB1155 and XDB1158 strains displayed indistinguishable PdhS-CFP localization patterns, with, respectively, 76% and 75% of cells exhibiting a single polar focus, strongly suggesting that FumC alone is not required for PdhS polar targeting (see Fig. S3 in the supplemental material).
B. abortus FumC does not share its localization property with ccFumC and smFumC, its orthologs in C. crescentus and S. meliloti.
The sequence conservation among FumC homologs in alphaproteobacteria led us to investigate whether polar localization was a common trait of FumC homologs. To address this question, two alphaproteobacteria were selected: C. crescentus, which does not possess any close PdhS homolog, and S. meliloti, which possesses two close PdhS homologs (5), PdhS1 (or CbrA) (3, 4) and PdhS2. The localization of FumC homologs in these organisms (C. crescentus FumC [ccFumC] and S. meliloti FumC [smFumC]) was examined. In C. crescentus, the production of ccFumC-YFP from a chromosomal fusion under the control of the ccfumC promoter did not lead to the detection of foci but displayed a diffuse localization pattern (see Fig. S4 in the supplemental material). An smFumC-YFP fusion was produced from the low-copy-number plasmid pJM106 in S. meliloti. The smFumC-YFP fusion did not induce a major alteration of cell morphology and appeared to be distributed diffusely throughout the cytoplasm (see Fig. S4 in the supplemental material). Based on these data, we propose that FumC polar recruitment is not a common feature shared by all alphaproteobacteria.
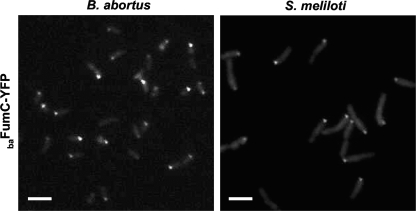
B. abortus FumC is polarly localized in S. meliloti but not in C. crescentus.
The PdhS property to localize at poles in S. meliloti and C. crescentus suggested the conservation of a polar structure among several alphaproteobacteria (6). We thus investigated whether B. abortus FumC (baFumC), ccFumC, and smFumC have the ability to be recruited at one pole in other alphaproteobacteria. For this, pJM063 was first introduced into C. crescentus and S. meliloti to monitor baFumC-YFP localization. The baFumC-YFP fusion was diffusely distributed throughout the C. crescentus cytoplasm (see Fig. S5 in the supplemental material), but when it was produced in S. meliloti, polar foci with a localization pattern similar to that observed for B. abortus were detected (Fig. 3). Then pJM105 (a low-copy-number plasmid carrying a ccfumC-yfp fusion) and pJM106 (a low-copy-number plasmid carrying an smfumC-yfp fusion) were separately introduced into the B. abortus ΔfumC (XDB1157) strain to ensure that the ccFumC-YFP and smFumC-YFP fusions, respectively, did not compete with endogenous baFumC. The ccFumC-YFP fusion was not recruited to a cell pole of B. abortus (see Fig. S5 in the supplemental material), whereas the control baFumC-YFP fusion was (see Fig. S6 in the supplemental material). Polar foci corresponding to the smFumC-YFP fusion were observed for only 7% of the B. abortus cell population (n = 1,330). ccFumC-YFP in S. meliloti and smFumC-YFP in C. crescentus did not display polar localization, although smFumC-YFP presented a patchy localization that could be due to its overproduction (see Fig. S5 in the supplemental material). Altogether, these results indicated that baFumC contains structural information allowing for its polar localization, a characteristic that is absent or altered in its ccFumC and smFumC homologs. Mapping of the substitutions between baFumC and smFumC sequences on the three-dimensional structure of E. coli FumC reveals that a belt of substitutions surrounds the FumC homotetramer (see Fig. S7 in the supplemental material). These substitutions do not directly affect the catalytic site (see Fig. S8 in the supplemental material) but may be involved in the generation of an interaction surface on the baFumC structure that is absent in smFumC. This baFumC interaction surface could be involved in binding to PdhS and possibly PdhS homologs.
FIG. 3.
Localization of baFumC in B. abortus and S. meliloti. For the two species, the localization of the B. abortus FumC-YFP fusion was observed in the exponential growth phase of the culture. The fumC-yfp fusion is carried on a low-copy-number plasmid (pJM063). Bar, 2 μm.
In conclusion, the data we report here strongly support that PdhS is able to directly recruit an important metabolic enzyme at the old pole in B. abortus, thereby suggesting that it could play multiple roles in the segregation of different functions between sibling cells. Moreover, baFumC localization suggests that the old pole could be a distinctive site of recruitment for various proteins involved, for example, in signaling or some aspects of metabolism. This is particularly relevant for B. abortus, since this facultative intracellular pathogen is able to inhibit its replication before reaching an endoplasmic reticulum-like compartment (1), and it is suspected that metabolic rearrangements are crucial for Brucella species adaptation and survival inside mammalian cells (1a). The links between metabolism, asymmetric division, and intracellular survival thus deserve further study.
Supplementary Material
Acknowledgments
We are very grateful to R.-M. Genicot for her generous technical assistance during the cloning procedures and to M. de Barsy for her help with the fumarase activity assays. We thank C. Michaux for her help in analysis of the fumarase three-dimensional structure and R. Hallez, J.-Y. Matroule, and C. Jacobs-Wagner for their critical readings of the first version of the manuscript.
This work was supported by the FRFC (Fonds de la Recherche Fondamentale Collective, agreements 2.4521.04 and 2.4541.08) and by the University of Namur. J. Mignolet held a Ph.D. fellowship from the FRS-FNRS (Fonds de la Recherche Scientifique-Fonds National de la Recherche Scientifique). C. Van der Henst and D. Dotreppe held Ph.D. fellowships from the FRIA (Fonds pour la Formation à la Recherche dans l'Industrie et dans l'Agriculture). C. Nicolas was supported by a fellowship from ARC (Actions de Recherche Concertée, agreements 04/09-325 and 08/13-015, French-Speaking Community of Belgium).
Footnotes
Published ahead of print on 9 April 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Celli, J., and J. P. Gorvel. 2004. Organelle robbery: Brucella interactions with the endoplasmic reticulum. Curr. Opin. Microbiol. 7:93-97. [DOI] [PubMed] [Google Scholar]
- 1a.Dozot, M., R. A. Boigegrain, R. M. Delrue, R. Hallez, S. Ouahrani-Bettache, I. Danese, J.-J. Letesson, X. De Bolle, and S. Köhler. 2006. The stringent response mediator Rsh is required for Brucella melitensis and Brucella suis virulence, and for expression of the type IV secretion system virB. Cell. Microbiol. 8:1791-1802. [DOI] [PubMed] [Google Scholar]
- 2.Dricot, A., J. F. Rual, P. Lamesch, N. Bertin, D. Dupuy, T. Hao, C. Lambert, R. Hallez, J. M. Delroisse, J. Vandenhaute, I. Lopez-Goni, I. Moriyon, J. M. Garcia-Lobo, F. J. Sangari, A. P. Macmillan, S. J. Cutler, A. M. Whatmore, S. Bozak, R. Sequerra, L. Doucette-Stamm, M. Vidal, D. E. Hill, J. J. Letesson, and X. De Bolle. 2004. Generation of the Brucella melitensis ORFeome version 1.1. Genome Res. 14:2201-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibson, K. E., M. J. Barnett, C. J. Toman, S. R. Long, and G. C. Walker. 2007. The symbiosis regulator CbrA modulates a complex regulatory network affecting the flagellar apparatus and cell envelope proteins. J. Bacteriol. 189:3591-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibson, K. E., G. R. Campbell, J. Lloret, and G. C. Walker. 2006. CbrA is a stationary-phase regulator of cell surface physiology and legume symbiosis in Sinorhizobium meliloti. J. Bacteriol. 188:4508-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hallez, R., A. F. Bellefontaine, J. J. Letesson, and X. De Bolle. 2004. Morphological and functional asymmetry in alpha-proteobacteria. Trends Microbiol. 12:361-365. [DOI] [PubMed] [Google Scholar]
- 6.Hallez, R., J. Mignolet, V. Van Mullem, M. Wery, J. Vandenhaute, J. J. Letesson, C. Jacobs-Wagner, and X. De Bolle. 2007. The asymmetric distribution of the essential histidine kinase PdhS indicates a differentiation event in Brucella abortus. EMBO J. 26:1444-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woods, S. A., J. S. Miles, R. E. Roberts, and J. R. Guest. 1986. Structural and functional relationships between fumarase and aspartase. Nucleotide sequences of the fumarase (fumC) and aspartase (aspA) genes of Escherichia coli K12. Biochem. J. 237:547-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woods, S. A., S. D. Schwartzbach, and J. R. Guest. 1988. Two biochemically distinct classes of fumarase in Escherichia coli. Biochim. Biophys. Acta 954:14-26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.