Abstract
Human dental biofilm communities comprise several species, which can interact cooperatively or competitively. Bacterial interactions influence biofilm formation, metabolic changes, and physiological function of the community. Lactic acid, a common metabolite of oral bacteria, was measured in the flow cell effluent of one-, two- and three-species communities growing on saliva as the sole nutritional source. We investigated single-species and multispecies colonization by using known initial, early, middle, and late colonizers of enamel. Fluorescent-antibody staining and image analysis were used to quantify the biomass in saliva-fed flow cells. Of six species tested, only the initial colonizer Actinomyces oris exhibited significant growth. The initial colonizer Streptococcus oralis produced lactic acid but showed no significant growth. The early colonizer Veillonella sp. utilized lactic acid in two- and three-species biofilm communities. The biovolumes of all two-species biofilms increased when Veillonella sp. was present as one of the partners, indicating that this early colonizer promotes mutualistic community development. All three-species combinations exhibited enhanced growth except one, i.e., A. oris, Veillonella sp., and the middle colonizer Porphyromonas gingivalis, indicating specificity among three-species communities. Further specificity was seen when Fusobacterium nucleatum (a middle colonizer), Aggregatibacter actinomycetemcomitans (a late colonizer), and P. gingivalis did not grow with S. oralis in two-species biofilms, but inclusion of Veillonella sp. resulted in growth of all three-species combinations. We propose that commensal veillonellae use lactic acid for growth in saliva and that they communicate metabolically with initial, early, middle, and late colonizers to establish multispecies communities on enamel.
The human oral cavity contains a widely diverse community of resident bacteria composed of several hundred species (1, 18). They organize into multispecies communities through a recurrent sequence of colonization that occurs after each oral hygiene treatment; for example, dental plaque development on enamel starts with the initial colonizers streptococci and actinomyces (7, 15), which are followed by early-colonizing veillonellae (7, 11, 14), middle-colonizing porphyromonads (7) and fusobacteria (7, 10, 11), and late-colonizing aggregatibacters (9).
During the initial stage of biofilm formation, streptococci and actinomyces bind to host-derived receptors in the salivary pellicle coating of enamel. In turn, other species bind to already-adherent cells, a process called coadhesion (2). This process and coaggregation (10), defined as specific cell-to-cell recognition between genetically distinct cells, as well as growth of adherent cells contribute to dental plaque development. While it is known that pure cultures of oral bacteria metabolize dietary sugars to lactic acid, little is known about the importance of lactic acid to community growth on saliva as a sole nutrient source. Most pure cultures and many combinations of species are unable to grow on whole saliva, which is a complex nutritional source. Growth might, in fact, require spatial organization and mutualistic interactions among selected species that collectively possess a combination of metabolic properties that are capable of converting latent nutrition into usable nutrition. In succession, groups of other selected species with other combined metabolic capabilities can further process this complex nutritional source, with a resultant assembling and disassembling of constantly changing oral biofilm communities.
Streptococci make up 60 to 90% of the supragingival plaque biomass in the first 24 h of colonization (12, 15). They catabolize carbohydrates to short-chain organic acids, such as lactic acid and pyruvic acid (4). Veillonellae constitute as much as 5% of the initial plaque biomass but are unable to catabolize sugars. They rely on the fermentation of organic acids such as lactic acid (6) and thus set up a convenient metabolic food chain in dental plaque.
In vivo studies using gnotobiotic rats demonstrated that veillonellae were unable to establish monoinfections. Yet when a strain of Veillonella was inoculated into rats already monoinfected with a strain of Streptococcus mutans that coaggregates with that Veillonella strain, the number of veillonellae on the teeth of the coinfected animals was 1,000-fold higher than the number when a noncoaggregating Veillonella strain was used (13). Also, in gnotobiotic rats, lower caries and plaque scores were obtained for two-species biofilms than for single-species colonization by streptococci, and inclusion of veillonellae reduced caries activity and demineralization of the enamel by streptococci (13). Streptococcus-Veillonella communities containing coaggregation partners were micromanipulated from 8-h human dental plaque, providing additional evidence of the close association of these two species in vivo (3). Further, Veillonella spp. are juxtaposed with coaggregation receptor polysaccharide-bearing streptococci in early communities in vivo, and a rapid succession of veillonella phylotypes occurs in these communities (16). These reports offer broad-based evidence that veillonellae and streptococci are linked in oral biofilms.
The focus of the current investigation was to explore Veillonella-based mixed-species communities in saliva-fed flow cells. The concentration of lactic acid in the effluent of flow cells containing biofilm communities was determined. We hypothesize that spatiotemporal metabolic interactions and coaggregation of Veillonella sp. with Streptococcus oralis and early, middle, and late colonizers allow these organisms to form three-species biofilm communities. We show high specificity of community partnerships among the six species examined, suggesting that successions of species in naturally recurring dental plaque in vivo are centered on metabolic and physical interactions of the community participants which support the nonrandom sequential appearance of species in the development of oral biofilms.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
S. oralis 34 and Actinomyces oris ATCC 43146 were routinely cultured in Todd-Hewitt broth (THB) (Difco Laboratories, Detroit, MI) or on THB agar. Veillonella sp. strain PK1910 was grown in THB supplemented with 0.6% lactic acid (THBL). A. actinomycetemcomitans JP2 (a kind gift from D. Demuth, University of Louisville, Louisville, KY) was grown in brain heart infusion medium (BHI) (Difco) supplemented with 0.04% sodium bicarbonate and 2% yeast extract. Fusobacterium nucleatum ATCC 10953 was grown in BHI supplemented with 0.25% l-glutamic acid. Porphyromonas gingivalis ATCC 33277 (a kind gift from R. J. Lamont, University of Florida, Gainesville, FL) was grown in BHI supplemented with 0.1% yeast extract, 0.5% hemin, and 0.001% menadione. All species were grown in a Bactron anaerobic (N2-CO2-H2, 90:5:5) environmental chamber (Sheldon Manufacturing Inc., Cornelius, OR) at 37°C.
Saliva preparation.
Saliva from 6 to 10 healthy individuals was collected on ice, pooled on ice, and treated with 2.5 mM dithiothreitol for 10 min with stirring to reduce salivary protein aggregation. The saliva was then centrifuged and processed as previously described (17). Briefly, the supernatant was diluted with distilled water to produce 25% saliva and then was filtered through a 0.22-μm-pore-size surfactant-free cellulose acetate (SFCA) low-protein-binding filter (Nalge Nunc International, Rochester, NY) and stored at −20°C. Prior to use, saliva was thawed and centrifuged to remove any precipitate that resulted from freezing and thawing.
Lactate assay.
Lactate concentrations (μM) were calculated based on standard curves obtained by using 2-fold serial dilutions of lactate. The lactic acid concentrations in filter-sterile saliva in the reservoir and the effluent saliva collected from the flow cell outlet at 4 h and 18 h were quantified by using a lactic acid assay kit (BioVision, Mountain View, CA). The effluent saliva was filtered through a 0.22-μm filter prior to assay.
Flow cell preparation.
Two tracks (each track was 40 mm long, 3 mm wide, and 2 mm deep) were milled into a high-density polyethylene block, resulting in two chambers, each with a 240-μl volume. A glass coverslip, which serves as the attachment substratum for the growing biofilm, was secured to the reusable flow cells with a silicone adhesive. The flow cells were cleaned overnight with 0.1 M HCl and rinsed with 5 ml of distilled water, followed by injecting 70% ethanol into the flow cells and incubating for 20 min. The flow cells were then treated with 25% sterile human saliva for 15 min at 37°C in an anaerobic chamber to condition the glass surface with salivary components.
Biofilm growth conditions in flow cells.
Overnight bacterial cultures were harvested by centrifugation and washed twice with 25% sterile human saliva, and the optical density at 600 nm was adjusted by using saliva to 0.1, which was equivalent to about 1 × 107 to 3 × 107 cells/ml. Flow cells were inoculated with one, two, or three species. Two-species and three-species inoculations were first coaggregated (by mixing 0.1-ml portions of appropriate combinations of Veillonella sp., S. oralis, A. oris, F. nucleatum, P. gingivalis, and A. actinomycetemcomitans [equivalent to about 1 × 106 to 3 × 106 cells of each species]), which was followed by incubation of the flow cells in the anaerobic chamber to provide an environment favorable to Veillonella sp., P. gingivalis, and F. nucleatum, which are strict anaerobes. The sole nutritional source was sterile 25% saliva supplied at a flow rate of 0.2 ml/min, which approximated unstimulated salivary flow in the mouth (5).
Biofilm staining.
Staining of cells varied in the different experiments. When strains were grown as monocultures, they were visualized by primary immunofluorescence with each Alexa Fluor 488 (Invitrogen)-conjugated immunoglobulin G of a polyclonal antiserum to S. oralis, A. oris, and Veillonella sp. When multiple species were inoculated, visualization of species was by primary immunofluorescence with Alexa Fluor 488-, Alexa Fluor 633-, Alexa Fluor 488-, Alexa Fluor 546-, Alexa Fluor 488-, and Alexa Fluor 546-conjugated immunoglobulin G of a polyclonal antiserum to S. oralis, Veillonella sp., A. oris, F. nucleatum, A. actinomycetemcomitans, and P. gingivalis, respectively. Immunofluorescence was performed by injecting the antibody (5 μg/ml in filter-sterilized phosphate-buffered saline [PBS] containing 1% wt/vol bovine serum albumin) into the appropriate flow cell track and incubating for 20 min. A final wash with PBS preceded confocal laser scanning microscopy. No cross-reactivity with nonhomologous strains was observed based on staining by antibodies.
Image and statistical analysis.
A TCS-SP2 confocal microscope (Leica, Exton, PA) with a 40×, 1.25-numerical-aperture (NA) oil immersion lens was used to record confocal image stacks in five random locations near the center of the flow cells, after which biofilm biovolumes were determined by volumetric analyses (IMARIS version 5.71; Bitplane AG, Zurich, Switzerland). Fluorescence intensity thresholds were set manually for red, green, and blue pixels, with cubic voxels used for the biovolume determination. Five confocal data sets were analyzed for each time point, and the mean and standard deviation were calculated. A one-way analysis of variance at the 95% confidence level with a nonparametric Tukey pairwise comparison test was used to determine if means were statistically significantly different between the 4-h and 18-h time points for each condition (one, two, and three species) in the flow cell saliva. All images presented are maximum projections of the entire confocal image stack as produced by the Leica TCS software (Leica).
RESULTS
Lactic acid assay.
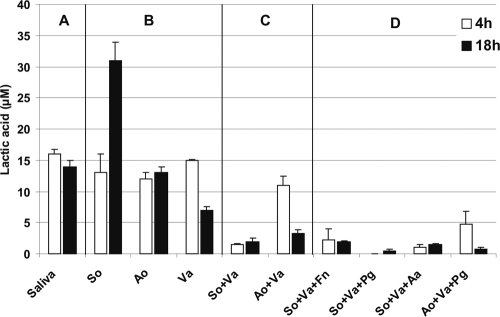
Lactic acid was quantified from uninoculated saliva and from flow cell effluent saliva of one-, two-, and three-species-inoculated flow cells at 4 h and 18 h of incubation. The lactic acid content of saliva was 16 μM (Fig. 1 A). Among the one-species inocula, only S. oralis produced lactic acid from saliva, and only Veillonella sp. used lactic acid; A. oris appeared to be unable to use or produce lactic acid (Fig. 1B). Lactic acid was not produced or consumed by F. nucleatum, A. actinomycetemcomitans, or P. gingivalis (data not shown). Veillonella sp. in partnership with S. oralis or A. oris utilized lactic acid to levels of below 5 μM (Fig. 1C). Compared to its ability to use lactic acid by itself (Fig. 1B), Veillonella sp. clearly was able to reduce the lactic acid concentration to a much lower level when in two-species biofilms with streptococci or actinomyces (Fig. 1C), suggesting that veillonellae grow well with these two initial colonizers. In three-species biofilms (Fig. 1D), lactic acid was used quickly (in 4 h) by veillonellae, except with A. oris and P. gingivalis, where lactic acid was used more slowly.
FIG. 1.
Quantification of lactic acid in reservoir saliva (A) and effluent saliva (B to D) collected from flow cells after 4 h and 18 h of biofilm growth. (A) Sterile reservoir saliva; (B) single-species biofilms of S. oralis (So), A. oris (Ao), and Veillonella sp. (Va); (C) Two-species biofilms of S. oralis plus Veillonella sp. and A. oris plus Veillonella sp.; (D) three-species biofilms of S. oralis plus Veillonella sp. plus F. nucleatum (Fn), S. oralis plus Veillonella sp. plus P. gingivalis (Pg), S. oralis plus Veillonella sp. plus A. actinomycetemcomitans (Aa), and A. oris plus Veillonella sp. plus P. gingivalis. Error bars indicate standard deviations.
Flow cells inoculated with one species.
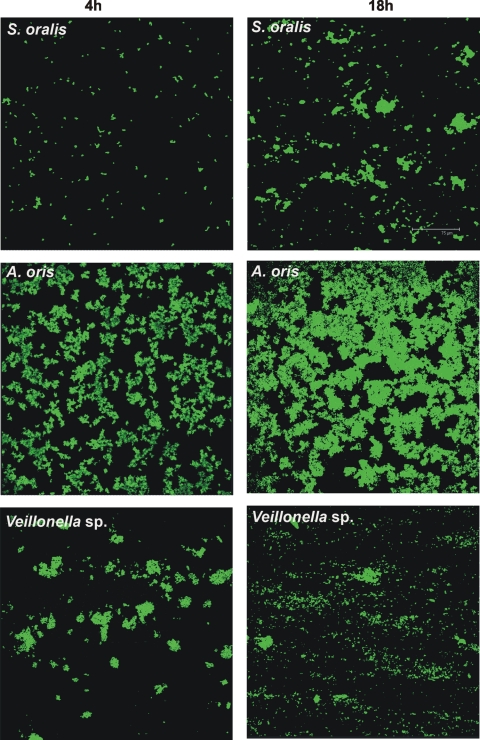
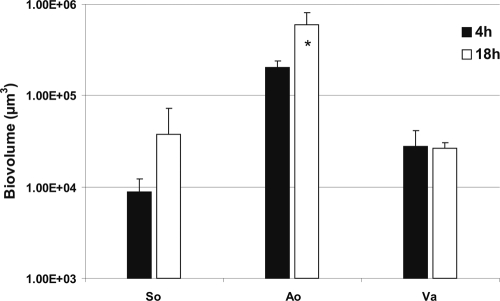
S. oralis, A. oris, and Veillonella sp. were inoculated into separate flow cells. Images of the biofilm formed were obtained after 4 h and 18 h of incubation and used to quantify biofilm cells as biovolume. Each species showed attachment to the saliva-conditioned surface at 4 h (Fig. 2). After 18 h of incubation, the numbers of S. oralis organisms appeared to increase (Fig. 2, top panels), but the increases were not significant (Fig. 3). Growth of A. oris increased 3-fold, but Veillonella sp. did not grow (Fig. 3). We reported earlier that P. gingivalis, F. nucleatum, and A. actinomycetemcomitans did not grow as single species (19-21).
FIG. 2.
Confocal micrographs of biofilms formed in flow cells inoculated with one species after 4 h (left column) and 18 h (right column) of growth on 25% saliva. Top panels, S. oralis; middle panels, A. oris; bottom panels, Veillonella sp. Colonization (4 h) and growth (18 h) of each species were monitored. Bacterial cells are stained with species-specific fluorophore-conjugated immunoglobulin G.
FIG. 3.
Time-resolved changes in biovolumes (μm3 per field of view) of S. oralis (So), A. oris (Ao), and Veillonella sp. (Va) following 4 h and 18 h of incubation in 25% saliva-fed flow cells. The biovolume values are for the single-species-inoculated flow cells (see Fig. 2). An asterisk indicates statistically significant increases (P < 0.05) in bacterial growth. Error bars indicate standard deviations.
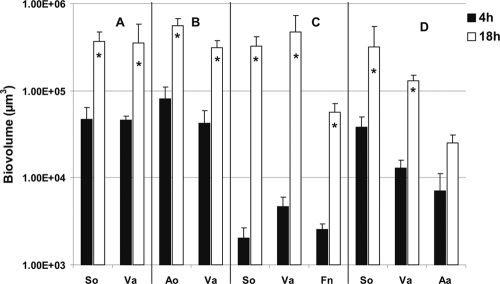
Flow cells inoculated with two species.
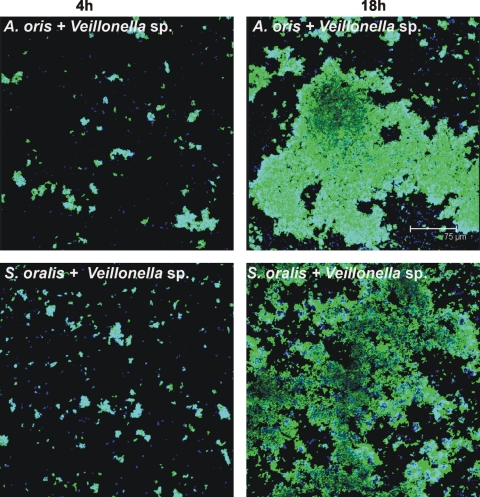
In flow cells inoculated with coaggregates of Veillonella sp. and S. oralis, both species attached to the saliva-conditioned surface, as shown by labeling with specific antibodies (Fig. 4, bottom panels). The biovolumes of Veillonella sp. and S. oralis each increased 8-fold between 4 h and 18 h (Fig. 5 A), indicating that both species grew on saliva. After inoculation of Veillonella sp. and A. oris, each increased 7-fold between 4 h and 18 h (Fig. 4, top panels, and 5B), indicating that both species grew on saliva.
FIG. 4.
Representative confocal micrographs of 4-h (left column) and 18-h (right column) biofilms showing growth in two-species-inoculated flow cells. Top panels, Veillonella sp. plus A. oris; bottom panels, Veillonella sp. plus S. oralis. Bacterial cells were stained with species-specific fluorophore-conjugated immunoglobulin G (blue, Veillonella sp.; green, A. oris and S. oralis) and show cell-cell contact.
FIG. 5.
Time-resolved changes in biovolumes (μm3 per field of view) of Veillonella sp. (Va), A. oris (Ao), S. oralis (So), F. nucleatum (Fn), and A. actinomycetemcomitans (Aa) following 4 h and 18 h of incubation in 25% saliva-fed flow cells. (A and B) Two-species-inoculated flow cells (see Fig. 4); (C and D) three-species-inoculated flow cells (see Fig. 6). An asterisk indicates statistically significant increases (P < 0.05) in bacterial growth. Error bars indicate standard deviations.
Flow cells inoculated with three species.
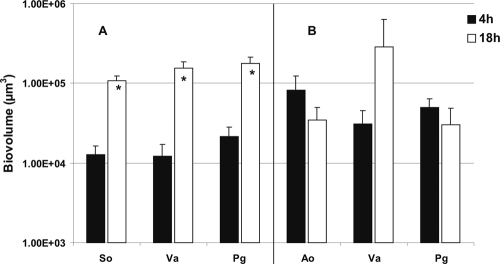
S. oralis grew when it was partnered with F. nucleatum (19), but it did not grow when partnered with A. actinomycetemcomitans (data not shown) or P. gingivalis (21). This specificity was explored further by including Veillonella sp. as the third species and examining the formation of resultant three-species communities in saliva-fed flow cells. Interestingly, S. oralis, Veillonella sp., and F. nucleatum biovolumes increased (Fig. 6, top panels) 163-, 100-, and 23-fold, respectively (Fig. 5C). With A. actinomycetemcomitans, the biovolumes of S. oralis and Veillonella sp. increased 9-fold and 13-fold, respectively; the biovolume of A. actinomycetemcomitans increased 4-fold (Fig. 6, bottom panels, and 5D). The biovolumes of S. oralis, Veillonella sp., and P. gingivalis increased significantly (9-, 13-, and 9-fold, respectively) (Fig. 7, top panels, and Fig. 8 A). In contrast, replacing S. oralis with A. oris resulted in lower biovolumes of A. oris and P. gingivalis (Fig. 8B), although initial binding to the saliva-coated substratum was excellent (Fig. 7, bottom panels). Only Veillonella sp. appeared to grow at 18 h (Fig. 8B). These data indicate high specificity of three-species communities for growth on saliva.
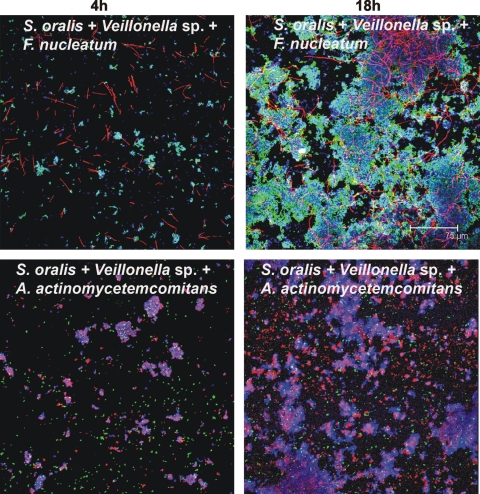
FIG. 6.
Representative confocal micrographs of biofilms from three-species-inoculated flow cells. Mixed-species communities at 4 h (left column) and at 18 h (right column) show intimate interaction of Veillonella sp. (blue) with S. oralis (green) and F. nucleatum (red) (top panels) and of S. oralis (blue) with A. actinomycetemcomitans (green) and Veillonella sp. (red) (bottom panels).
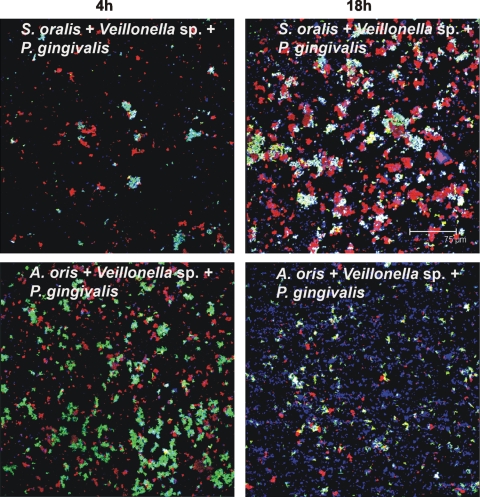
FIG. 7.
Representative confocal micrographs of biofilms from three-species-inoculated flow cells. Mixed-species communities at 4 h (left column) and at 18 h (right column) show intimate interaction of Veillonella sp. (blue) with S. oralis (green) and P. gingivalis (red) (top panels) and of Veillonella sp. (blue) with A. oris (green) and P. gingivalis (red) (bottom panels).
FIG. 8.
Time-resolved changes in biovolumes (μm3 per field of view) of Veillonella sp. (Va), A. oris (Ao), S. oralis (So), and P. gingivalis (Pg) following 4 h and 18 h of incubation in 25% saliva-fed flow cells (see Fig. 7). An asterisk indicates statistically significant increases (P < 0.05) in bacterial growth. Error bars indicate standard deviations.
DISCUSSION
Veillonellae and streptococci are metabolically linked through streptococcal fermentation of sugars to lactic acid, which is a carbon source for the nonsaccharolytic veillonellae. Also, the spatial relationship between these species influences gene regulation in vitro; diffusible signal exchange between the coaggregating partners Veillonella sp. strain PK1910 and Streptococcus gordonii V288 resulted in upregulation of an amylase gene (amyB) promoter in the streptococcal strain (8). Those data suggest a close metabolic association of the two species. In the present study we report that lactic acid is utilized in Veillonella sp.-S. oralis biofilms and in three-species biofilms with F. nucleatum, A. actinomycetemcomitans, or P. gingivalis. We show here that utilization of lactic acid from Veillonella sp.-S. oralis two-species biofilms occurs faster than it does with Veillonella sp.-A. oris two-species biofilms, suggesting that streptococci are preferred community partners over actinomyces in vivo (Fig. 1).
We show that the early colonizer Veillonella sp. adheres to saliva-coated glass but cannot grow without other species. The species most likely to be present early in vivo are the initial colonizing streptococci and actinomyces (7, 15). Microbial colonization of enamel is a repeatable and selective process involving a succession of species. By using molecular methods to characterize temporally the microbial diversity on a retrievable enamel model, Streptococcus, Actinomyces, and Veillonella were found to be among the dominant genera within the first 4 h of colonization (7). The genera Fusobacterium and Porphyromonas were present in only a low percentage of sequences analyzed and are thus considered middle colonizers; the genus Aggregatibacter was not detected and is considered a late colonizer. In our current investigation of two-species biofilm communities in vitro, Veillonella sp. is an active member when combined with S. oralis, A. oris, P. gingivalis, F. nucleatum, and A. actinomycetemcomitans, illustrating a central role of veillonellae in the selective process of colonization. Likewise, including Veillonella sp. as a third species with S. oralis and F. nucleatum, with S. oralis and A. actinomycetemcomitans, or with S. oralis and P. gingivalis resulted in excellent growth of all species. Interestingly, the biovolumes of S. oralis, Veillonella sp., and F. nucleatum significantly increased (163-, 100-, and 23-fold, respectively) between 4 h and 18 h, which demonstrates that Veillonella sp. plays an integral role in fostering robust community growth among coaggregates with S. oralis.
Although we have tested only six genera of oral bacteria in these flow cell studies, an image of saliva-fed community compatibility and specificity is emerging. Previously, we reported that F. nucleatum (a middle colonizer of enamel) requires A. oris (an initial colonizer) in order to grow in the presence of S. oralis (an initial colonizer), supporting the known succession of species in oral biofilms (19). Also, we reported previously that A. actinomycetemcomitans (a late colonizer) grows mutualistically with Veillonella sp. (an early colonizer) or F. nucleatum when paired in a two-species biofilm, but it failed to show significant growth when combined with these coaggregation partners in a three-species biofilm (20). In our current study, we substituted S. oralis for F. nucleatum, with the result that S. oralis and Veillonella sp. grew significantly but again A. actinomycetemcomitans did not (Fig. 5), suggesting that growth in a three-species biofilm requires different interspecies interactions than does growth in a two-species biofilm. A. actinomycetemcomitans cannot grow with S. oralis (data not shown); P. gingivalis (a middle colonizer) cannot grow with S. oralis (21); and in a P. gingivalis-S. oralis-A. actinomycetemcomitans three-species biofilm, none grow (21). Here we show that in a P. gingivalis-S. oralis-Veillonella sp. three-species biofilm, all three species grow (Fig. 8A); however, in a P. gingivalis-A. oris-Veillonella sp. three-species biofilm, none grow significantly (Fig. 8B). The emerging picture is that only certain combinations of species support growth and that mutualistic growth in two-species biofilms does not predict growth in three-species biofilms. For example, none of the species grow in the P. gingivalis-A. oris-Veillonella sp. three-species biofilm (Fig. 8B), although pairwise, all grow mutualistically (A. oris plus Veillonella sp. [Fig. 5B], P. gingivalis plus Veillonella sp. [21], and P. gingivalis plus A. oris [21]).
Clearly, Veillonella sp. is an integral participant in the mixed-species communities that form sequentially in developing early, middle, and late dental plaque. When mixed with S. oralis and F. nucleatum, Veillonella sp. exhibits the highest growth increase (Fig. 5C) we have observed to date, suggesting that this three-species community is well matched metabolically and supported by this community's rapid utilization of lactic acid (Fig. 1D). Further, our results indicate that Veillonella sp. pairs better with S. oralis than with A. oris when P. gingivalis is the third partner (compare Fig. 8A and B). This observation is corroborated by the more rapid utilization of lactic acid in the three-species S. oralis-Veillonella sp.-P. gingivalis biofilm than in the A. oris-Veillonella sp.-P. gingivalis biofilm (Fig. 1D). We propose that this central role of veillonellae in multispecies community formation, especially with streptococci, is a key element in facilitating the succession of species in developing dental plaque in vivo.
Acknowledgments
This research was supported in part by the Intramural Research Program of the National Institute of Dental and Craniofacial Research, National Institutes of Health, and in part by the Colgate-Palmolive Co. CRADA.
We thank R. J. Palmer for helpful comments on the manuscript.
Footnotes
Published ahead of print on 12 February 2010.
REFERENCES
- 1.Aas, J. A., B. J. Paster, L. N. Stokes, I. Olsen, and F. E. Dewhirst. 2005. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43:5721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bos, R., H. C. van der Mei, and H. J. Busscher. 1996. Co-adhesion of oral microbial pairs under flow in the presence of saliva and lactose. J. Dent. Res. 75:809-815. [DOI] [PubMed] [Google Scholar]
- 3.Chalmers, N. I., R. J. Palmer, Jr., J. O. Cisar, and P. E. Kolenbrander. 2008. Characterization of a Streptococcus sp.- Veillonella sp. community micromanipulated from dental plaque. J. Bacteriol. 190:8145-8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cotter, P. D., and C. Hill. 2003. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67:429-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawes, C., S. Watanabe, P. Biglow-Lecomte, and G. H. Dibdin. 1989. Estimation of the velocity of the salivary film at some different locations in the mouth. J. Dent. Res. 68:1479-1482. [DOI] [PubMed] [Google Scholar]
- 6.Delwiche, E. A., J. J. Pestka, and M. L. Tortorello. 1985. The veillonellae: gram-negative cocci with a unique physiology. Annu. Rev. Microbiol. 39:175-193. [DOI] [PubMed] [Google Scholar]
- 7.Diaz, P. I., N. I. Chalmers, A. H. Rickard, C. Kong, C. L. Milburn, R. J. Palmer, Jr., and P. E. Kolenbrander. 2006. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl. Environ. Microbiol. 72:2837-2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egland, P. G., R. J. Palmer, Jr., and P. E. Kolenbrander. 2004. Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc. Natl. Acad. Sci. U. S. A. 101:16917-16922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaplan, C. W., R. Lux, S. K. Haake, and W. Shi. 2009. The Fusobacterium nucleatum outer membrane protein RadD is an arginine-inhibitable adhesin required for inter-species adherence and the structured architecture of multispecies biofilm. Mol. Microbiol. 71:35-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolenbrander, P. E., R. N. Andersen, D. S. Blehert, P. G. Egland, J. S. Foster, and R. J. Palmer, Jr. 2002. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66:486-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li, J., E. J. Helmerhorst, C. W. Leone, R. F. Troxler, T. Yaskell, A. D. Haffajee, S. S. Socransky, and F. G. Oppenheim. 2004. Identification of early microbial colonizers in human dental biofilm. J. Appl. Microbiol. 97:1311-1318. [DOI] [PubMed] [Google Scholar]
- 12.Mager, D. L., L. A. Ximenez-Fyvie, A. D. Haffajee, and S. S. Socransky. 2003. Distribution of selected bacterial species on intraoral surfaces. J. Clin. Periodontol. 30:644-654. [DOI] [PubMed] [Google Scholar]
- 13.McBride, B. C., and J. S. van der Hoeven. 1981. Role of interbacterial adherence in colonization of the oral cavities of gnotobiotic rats infected with Streptococcus mutans and Veillonella alcalescens. Infect. Immun. 33:467-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nyvad, B., and M. Kilian. 1990. Comparison of the initial streptococcal microflora on dental enamel in caries-active and in caries-inactive individuals. Caries Res. 24:267-272. [DOI] [PubMed] [Google Scholar]
- 15.Nyvad, B., and M. Kilian. 1987. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand. J. Dent. Res. 95:369-380. [DOI] [PubMed] [Google Scholar]
- 16.Palmer, R. J., Jr., P. I. Diaz, and P. E. Kolenbrander. 2006. Rapid succession within the Veillonella population of a developing human oral biofilm in situ. J. Bacteriol. 188:4117-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmer, R. J., Jr., K. Kazmerzak, M. C. Hansen, and P. E. Kolenbrander. 2001. Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect. Immun. 69:5794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paster, B. J., I. Olsen, J. A. Aas, and F. E. Dewhirst. 2006. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontology 2000 42:80-87. [DOI] [PubMed] [Google Scholar]
- 19.Periasamy, S., N. I. Chalmers, L. Du-Thumm, and P. E. Kolenbrander. 2009. Fusobacterium nucleatum ATCC 10953 requires Actinomyces naeslundii ATCC 43146 for growth on saliva in a three-species community that includes Streptococcus oralis 34. Appl. Environ. Microbiol. 75:3250-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Periasamy, S., and P. E. Kolenbrander. 2009. Aggregatibacter actinomycetemcomitans builds mutualistic biofilm communities in saliva with Fusobacterium nucleatum and Veillonella sp. Infect. Immun. 77:3542-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Periasamy, S., and P. E. Kolenbrander. 2009. Mutualistic biofilm communities develop with Porphyromonas gingivalis and initial, early, and late colonizers of enamel. J. Bacteriol. 191:6804-6811. [DOI] [PMC free article] [PubMed] [Google Scholar]