Abstract
The bacterial phosphoenolpyruvate phosphotransferase system (PTS) is a highly conserved phosphotransfer cascade that participates in the transport and phosphorylation of selected carbohydrates and modulates many cellular functions in response to carbohydrate availability. It plays a role in the virulence of many bacterial pathogens. Components of the carbohydrate-specific PTS include the general cytoplasmic components enzyme I (EI) and histidine protein (HPr), the sugar-specific cytoplasmic components enzymes IIA (EIIA) and IIB (EIIB), and the sugar-specific membrane-associated multisubunit components enzymes IIC (EIIC) and IID (EIID). Many bacterial genomes also encode a parallel PTS pathway that includes the EI homolog EINtr, the HPr homolog NPr, and the EIIA homolog EIIANtr. This pathway is thought to be nitrogen specific because of the proximity of the genes encoding this pathway to the genes encoding the nitrogen-specific σ factor σ54. We previously reported that phosphorylation of HPr and FPr by EI represses Vibrio cholerae biofilm formation in minimal medium supplemented with glucose or pyruvate. Here we report two additional PTS-based biofilm regulatory pathways that are active in LB broth but not in minimal medium. These pathways involve the glucose-specific enzyme EIIA (EIIAGlc) and two nitrogen-specific EIIA homologs, EIIANtr1 and EIIANtr2. The presence of multiple, independent biofilm regulatory circuits in the PTS supports the hypothesis that the PTS and PTS-dependent substrates have a central role in sensing environments suitable for a surface-associated existence.
Attachment of a free-swimming bacterium to a surface, which is termed biofilm formation, is the result of a complex decision tree that occurs when a bacterium encounters a surface (19). Environmental signals dictate the decisions made at each branch point. The advantages afforded to a bacterium by surface attachment depend on the environmental niche of the bacterium being studied, and the environmental signals that induce biofilm formation reflect this.
Formation of a multilayer bacterial biofilm often requires synthesis of an extracellular matrix composed of biological polymers that enhance interbacterium attachment. These extracellular polymers may be proteins, polysaccharides, and/or DNA (7). Synthesis of the biofilm matrix is often tightly regulated. The environmental activators of many signaling pathways that modulate multilayer biofilm accumulation have been identified. These activators include specific carbohydrates, quorum-sensing molecules, nucleic acids and their precursors, and polyamines (1, 6, 9, 13, 15, 17, 18, 26, 31, 33, 35, 41, 45, 53). However, there are many known biofilm regulatory pathways for which no environmental activator has been identified yet.
The phosphoenolpyruvate phosphotransferase system (PTS) is a multicomponent phosphotransfer cascade that mediates transport and phosphorylation of selected sugars, such as glucose, sucrose, mannose, and N-acetylglucosamine (10). In addition, it has been implicated in the formation of biofilms by diverse organisms (1, 2, 17, 31, 42). Phosphate enters the PTS through transfer from phosphoenolpyruvate to the first PTS component, the phosphoenolpyruvate-protein phosphotransferase or enzyme I (EI). EI in turn transfers the phosphate group to another component of the PTS, histidine protein (HPr). Many bacterial genomes also encode a protein homologous to HPr termed FPr, which is preferred for transport of fructose through the PTS. HPr and FPr transfer phosphate to a number of enzymes II, which are multisubunit, membrane-associated complexes that carry out transport and phosphorylation of specific PTS substrates. Because transport of PTS substrates rapidly depletes the PTS of phosphorylated intermediates, the phosphorylation states of PTS components serve as cytoplasmic reporters of environmental nutrient availability. These reporters then modulate cellular functions such as chemotaxis (60), uptake and catabolism of PTS-independent carbohydrates (1, 12, 39), and glycogen breakdown (54, 55).
The genomes of many Gram-negative organisms contain genes encoding another phosphotransfer cascade that is homologous to the carbohydrate-transporting PTS. Because these genes are close to rpoN, which encodes the sigma factor involved in transcription of many genes required for nitrogen assimilation, this phosphotransfer cascade is termed the nitrogen-related PTS or PTSNtr (22, 49, 50). The components of this phosphotransfer cascade include EINtr, NPr, and EIIANtr. Unlike the carbohydrate-transporting PTS, PTSNtr does not include membrane-associated components and, therefore, does not participate directly in transport of nutrients into the cell. Rather, its primary function in the cell is thought to be regulatory (22, 24, 25, 32, 44). While the breadth and overarching goal of the PTSNtr have not been defined, transfer of phosphate between the two PTS cascades may be one mechanism by which the PTSNtr influences cellular function (46).
Vibrio cholerae is a Gram-negative bacterium whose natural habitat includes environments with low and intermediate salinities, such as ponds and estuaries (8). There is some evidence that V. cholerae forms a multilayer, exopolysaccharide-based biofilm in freshwater environments (20). The exopolysaccharide is referred to as VPS (Vibrio polysaccharide), and the region of the V. cholerae genome containing many of the genes required to make the biofilm matrix is known as the vps island (64).
When pathogenic V. cholerae is ingested by humans in contaminated food or water, the diarrheal disease cholera results. While many studies of human and murine infection have suggested that proteins required for biofilm matrix synthesis are expressed in vivo (14, 30), no study has found that these proteins are essential for colonization of the mammalian intestine (16, 23, 63).
The V. cholerae genome encodes 25 PTS components, including two EI homologs, three HPr homologs, and nine EIIA homologs (Fig. 1). A role in carbohydrate transport has been established for a subset of these components (16). We recently discovered that supplementation of minimal medium (MM) with PTS sugars activates transcription of the vps genes and formation of a multilayer biofilm (40). Furthermore, we found that phosphorylated form of EI represses V. cholerae biofilm formation in minimal medium (17). Subsequent biofilm assays conducted with Luria-Bertani (LB) broth suggested that there are additional PTS-based regulatory pathways. Here we define three PTS-based biofilm regulatory pathways that are present when this medium is used. These studies illustrate the complexity of the regulation of V. cholerae biofilm accumulation by PTS components and underscore the importance of carbohydrates as signals in the decision of V. cholerae cells to join a biofilm.
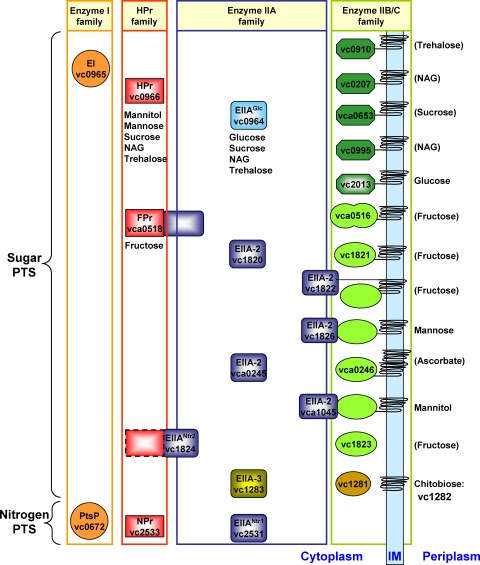
FIG. 1.
Schematic diagram of the 25 V. cholerae PTS components. Homologs of the EI, HPr, and EII proteins are indicated by the same colors. Furthermore, proteins belonging to the same EIIA superfamily are color coded (blue, glucose superfamily; violet, mannitol-fructose superfamily; yellow, lactose superfamily). Sugar specificities are indicated for PTS components if they are known (16). Parentheses indicate presumptive specificities. The VC1824 HPr-like domain is surrounded by a dashed line to indicate its low level of similarity to HPr. The sugar specificity of VC1281 is based on previously described data (3). NAG, N-acetylglucosamine.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. V. cholerae strain PW357 was used for biofilm assays. Bacteria were cultivated in Luria-Bertani broth or minimal medium supplemented with 0.5% (wt/vol) glucose or pyruvate (Sigma) (17). Where indicated below, the medium was also supplemented with streptomycin (100 μg/ml), ampicillin (50 or 100 μg/ml), or l-arabinose (0.04% wt/vol). Where specified below, a 0.1 M phosphate-buffered saline solution (PBS) (pH 7.0) was used.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype and/or phenotype | Reference |
|---|---|---|
| E. coli SM10λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mul pir R6K; Kmr | 38 |
| V. cholerae strains | ||
| PW357 | MO10 lacZ::vpsLp→lacZ; Smr | 15 |
| PW751 | MO10 lacZ::vpsLp→lacZ ΔPTS; Smr | 17 |
| PW961 | MO10 lacZ::vpsLp→lacZ ΔEI; Smr | 17 |
| PW964 | MO10 lacZ::vpsLp→lacZ ΔHPr; Smr | 17 |
| PW954 | MO10 lacZ::vpsLp→lacZ ΔFPr; Smr | 16 |
| PW965 | MO10 lacZ::vpsLp→lacZ ΔHPr ΔFPr; Smr | 16 |
| PW836 | MO10 lacZ::vpsLp→lacZ ΔEIIAGlc; Smr | 17 |
| PW989 | MO10 lacZ::vpsLp ΔEIIBCGlc; Smr | 17 |
| PW988 | MO10 lacZ::vpsLp ΔEIIAGlc ΔEIIBCGlc; Smr | This study |
| PW996 | MO10 lacZ::vpsLp ΔEIIBCGlc ΔMlc; Smr | This study |
| PW994 | MO10 lacZ::vpsLp ΔEIIAGlc ΔEIIBCGlc ΔMlc; Smr | This study |
| PW993 | MO10 lacZ::vpsLp ΔMlc | This study |
| PW992 | MO10 lacZ::vpsLp→lacZ ΔEIIANtr1; Smr | 16 |
| PW991 | MO10 lacZ::vpsLp→lacZ ΔEIIANtr2; Smr | 16 |
| PW1002 | MO10 lacZ::vpsLp→lacZ VC1283::pGP704; Smr Apr | 16 |
| PW1003 | MO10 lacZ::vpsLp→lacZ VC1820::pGP704; Smr Apr | 16 |
| PW1004 | MO10 lacZ::vpsLp→lacZ VC1822::pGP704; Smr Apr | 16 |
| PW1005 | MO10 lacZ::vpsLp→lacZ VC1826::pGP704; Smr Apr | 16 |
| PW1006 | MO10 lacZ::vpsLp→lacZ VCA0245::pGP704; Smr Apr | 16 |
| PW1007 | MO10 lacZ::vpsLp→lacZ VCA1045::pGP704; Smr Apr | 16 |
| Plasmids used in rescue experiments | ||
| pBAD-TOPO-EI | pBAD-TOPO carrying the coding sequence of VC0965 | 17 |
| pBAD-TOPO-EI(H189A) | pBAD-TOPO carrying the coding sequence of VC0965 with an H-to-A mutation at position 189 | 17 |
| pBAD-TOPO-EIIAGlc | pBAD-TOPO carrying the coding sequence of VC0964 | 17 |
| pBAD-TOPO-EIIAGlc(H91A) | pBAD-TOPO carrying the coding sequence of VC0964 with an H-to-A mutation at position 91 | This study |
| pBAD-TOPO-EIIAGlc(H91D) | pBAD-TOPO carrying the coding sequence of VC0964 with an H-to-D mutation at position 91 | This study |
| pBAD-TOPO-mlc | pBAD-TOPO carrying the coding sequence of VC2007 | This study |
| pBAD-TOPO-EIIANtr1 | pBAD-TOPO carrying the coding sequence of VC2531 | This study |
| pBAD-TOPO-EIIANtr1(H66A) | pBAD-TOPO carrying the coding sequence of VC2531 with an H-to-A mutation at position 66 | This study |
| pBAD-TOPO-EIIANtr1(H66D) | pBAD-TOPO carrying the coding sequence of VC2531 with an H-to-D mutation at position 66 | This study |
| pBAD-TOPO-EIIANtr2 | pBAD-TOPO carrying the coding sequence of VC1824 | This study |
| pBAD-TOPO-EIIANtr2(H172A) | pBAD-TOPO carrying the coding sequence of VC1824 with an H-to-A mutation at position 172 | This study |
| pBAD-TOPO-EIIANtr2(H172D) | pBAD-TOPO carrying the coding sequence of VC1824 with an H-to-D mutation at position 172 | This study |
Construction of mutants.
The plasmid used to make the Δmlc mutant, pKEKΔmlc, was generously provided by J. Reidl (3). All other plasmids used in the construction of insertion and in-frame deletion mutants were available in our laboratory (16, 17). Suicide plasmids were used to generate insertion and in-frame deletion mutants by single and double homologous recombination, respectively.
Construction of rescue plasmids.
Plasmids used for rescue experiments, which are listed in Table 1, were constructed as previously described (17, 61) using the primers listed in Table 2. Briefly, the native sequence of the targeted gene was amplified by PCR and cloned into pBAD-TOPO (Invitrogen) using the manufacturer's protocol. Point mutation sequences were generated by amplification of two gene fragments using internal primers with overlapping sequences containing the desired mutation. The two fragments were joined using the splicing by overlap extension (SOE) technique and cloned into pBAD-TOPO (Invitrogen) using the manufacturer's protocol. The sequence of the inserted fragment was confirmed by amplification and sequence analysis. The rescue plasmids were then introduced into V. cholerae strains by electroporation, and transformants were selected on LB agar supplemented with ampicillin. A pBAD-TOPO vector containing an antisense fragment of the coding sequence for EIIAGlc(H91A) was used as a control plasmid for rescue experiments. Expression of the cloned protein was evaluated by Western blot analysis as previously described (16).
TABLE 2.
Primers used in this study
| Protein | Primer sequences |
|---|---|
| Primers used for construction of expression plasmids | |
| Primers for wild-type alleles | |
| Mlc | TAAGAGGAATAATAAATGTACATGGCTCAA CCCGG, TCCTTCGACCACTTTCATTAA GAG |
| PtsN | CAATTGAGCGAAGTACTTTCATTGGACTG, TTGGTTAACCATGATGTTGTACAGC |
| VC1824 | GCGTGACATCGCGTACATCAG, CATGAATTCCAAGTCACCTTCTTGG |
| Internal primers for construction of mutant alleles | |
| EIIAGlc(H91A) | GCGTTGAGCTGTTTGTTGCCTTCGGTATCG, CGATACCGAAGGCAACAAACAGCTCAACGC |
| EIIAGlc(H91D) | GCGTTGAGCTGTTTGTTGACTTCGGTATCG, CGATACCGAAGTCAACAAACAGCTCAACGC |
| PtsN(H66A) | ATAGCCATCCCAGCCGCACGCATGT, ACATGCGTGCGGCTGGGATGGCTAT |
| PtsN(H66D) | ATAGCCATCCCAGACGCACGCATGT, ACATGCGTGCGTCTGGGATGGCTAT |
| VC1824(H172A) | GGGATTGCTATTCCCGCTGTGATGTTTGC, GCAAACATCACAGCGGGAATAGCAATCCC |
| VC1824(H172D) | GGGATTGCTATTCCCGATGTGATGTTTGC, GCAAACATCACATCGGGAATAGCAATCCC |
| Primers used in quantitative RT-PCR | |
| EI | ACCGTTATCGAAGAGCAAGCCACT, TCTGCGCAGTTTCAGAAGGCGTTA |
| EIIAGlc | GCGATCAAGCCTGCTGGTAACAAA, AAGCCTTCACCTTTCAGCTCAACC |
| HPr | TGTTCAAGCTGCAAACGCTAGGTC, GCAACTAGGTGCTCAACAGCTTCT |
| EIIANtr1 | GCTATTGCAGTGTGAAGAGCCGAT, CTGCGTTACGAAGCTGTTTGAGGA |
| EIIANtr2 | TCGGTATTGATGCTGAACTCGCCT, GGGCTTTGGCATAGTGCCATTTGA |
| ClpX | AGAGTTCATTGGTCGTCTGCCTGT, AACAACGCCGCATACTGTTTGGTC |
Quantitative analysis of total growth and biofilm formation.
Quantification of surface association was performed as described previously (20). Briefly, the strains were grown overnight on LB agar plates at 27°C. The following morning, the resulting colonies were used to inoculate borosilicate tubes filled with 300 μl of LB broth or MM supplemented with glucose or pyruvate. After incubation for 18 to 24 h at 27°C, each planktonic cell suspension was collected, and the planktonic cell density was estimated by measuring the optical density at 655 nm (OD655) using a Benchmark Plus microplate spectrophotometer (Bio-Rad). To quantify the surface-attached cells, 300 μl of PBS and a small number of 1-mm glass beads were added to the surface-attached cells remaining in the borosilicate tube, and the cells were dispersed by vortexing. The OD655 of the resulting cell suspension was measured. Each total growth value represented the sum of the OD655 recorded for the planktonic and surface-associated cell suspensions. All values are the means of at least three experimental replicates, and standard deviations were determined. Statistical significance was calculated using a two-tailed t test.
Quantification of PTS component transcripts.
Wild-type V. cholerae was cultured for 18 h in LB broth or MM supplemented with pyruvate (0.5%, wt/vol). The cells were then pelleted by centrifugation, and total RNA was isolated using an RNeasy kit (Qiagen), followed by RNase-free DNase I treatment to remove contaminating DNA. Reverse transcription-PCR (RT-PCR) was performed using 1 to 2 ng of total RNA with a SuperScriptIII first-strand kit (Invitrogen). Subsequently, 15 ng of the resulting cDNA was used as the template for quantitative PCR using iTaq SYBR green Supermix with ROX (Bio-Rad) and 5 pmol of the relevant primers (Table 2) in a 20-μl reaction mixture. The level of the clpX (VC1921) transcript was used to normalize all measurements. Template-free reaction mixtures were included as controls to confirm the absence of contaminating DNA or RNA. The experiments were conducted with a StepOnePlus PCR system (Applied Biosystems) using the following steps: (i) 95°C for 10 min, (ii) 40 cycles of denaturation for 15 s at 95°C and annealing and extension for 1 min at 60°C, and (iii) dissociation curve analysis using temperatures from 60°C to 90°C. Data were analyzed using the StepOne V2.0 software (Applied Biosystems). Measurements were obtained in triplicate.
Measurement of vpsL transcription using a β-galactosidase reporter.
Strains were inoculated into LB broth to obtain an initial OD655 of approximately 0.05. Cultures were incubated at 27°C for 4 h with shaking until the OD655 was approximately 0.5. The OD655 was determined and subsequently used for normalization of measurements of β-galactosidase activity. Fifty or 100 μl of each culture was moved to a white 96-well plate (Nunc). Three freeze-thaw cycles were performed, and an equal volume of the β-Glo luminescent substrate (Promega) was added to each well. After incubation in the dark at room temperature for 30 min, luminescence was measured with an Infinite 200 spectrophotometer (Tecan). Three experimental replicates were included each time that the experiment was performed, the experiment was repeated three times, and similar results were obtained in the three experiments. Values for a representative experiment are reported below and are the means of three experimental replicates; standard deviations were also determined, and statistical significance was calculated using a two-tailed t test.
RESULTS
The biofilm phenotypes of ΔEI and ΔEIIAGlc mutants in LB broth are additive.
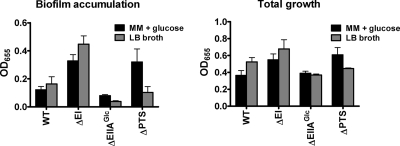
We first compared biofilm formation in LB broth and minimal medium supplemented with glucose by a ΔEI mutant, a ΔEIIAGlc mutant, and a ΔPTS mutant lacking the genes encoding EI, HPr, and EIIAGlc (Fig. 2). Similar to what was observed in minimal medium, in LB broth a ΔEI mutant exhibited increased surface accumulation compared with wild-type V. cholerae, while a ΔEIIAGlc mutant exhibited decreased surface accumulation. As previously noted (17), surface accumulation by a ΔPTS mutant lacking EI, HPr, and EIIAGlc was increased in minimal medium supplemented with glucose. In contrast, the surface accumulation by the ΔPTS mutant was similar to that by wild-type V. cholerae in LB broth. This suggested that the biofilm phenotype of the ΔPTS mutant in LB broth might reflect the additive effects of two opposing regulatory pathways.
FIG. 2.
PTS mutant displays distinct biofilm phenotypes in LB broth and minimal medium (MM) supplemented with glucose. Total growth and biofilm accumulation were determined for wild-type V. cholerae (WT), a ΔEI mutant, a ΔEIIAGlc mutant, and a ΔPTS mutant in minimal medium supplemented with 0.5% (wt/vol) glucose (MM + glucose) and LB broth (LB). The values are the averages of four experimental replicates, and the error bars indicate standard deviations. There was a statistically significant difference (P = 0.0062)between biofilm formation by wild-type V. cholerae and biofilm formation by the ΔPTS mutant in minimal medium supplemented with glucose (indicated by an asterisk) but not in LB broth (P = 0.116).
Evidence for repression of biofilm accumulation by EI-P and HPr-P in LB broth (regulatory pathway 1).
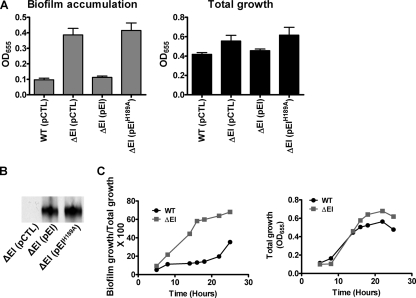
In both minimal medium and LB broth, EI represses biofilm formation (17). To determine if the phosphorylated form of EI is required for this repression, we measured biofilm formation by a ΔEI mutant rescued with a control plasmid, with a plasmid carrying a wild-type EI allele, or with a plasmid carrying a mutant EI allele in which phosphorylated histidine 189 had been changed to alanine. As shown in Fig. 3A, while the biofilm phenotype of a ΔEI mutant was rescued by a wild-type EI allele provided in trans, the EIH189A allele was unable to rescue the biofilm phenotype despite adequate expression of the protein (Fig. 3B). This suggests that, similar to what was observed in minimal medium (17), EI-P represses V. cholerae biofilm formation in LB broth. We previously reported that in minimal medium supplemented with glucose, repression of biofilm accumulation by EI becomes apparent only during the stationary phase of growth. This corresponds to the time at which the glucose in the growth medium is depleted (17) and phosphorylated PTS intermediates begin to accumulate. Because PTS substrates are less abundant in LB broth (20), we predicted that during growth in LB broth, repression of biofilm accumulation by EI-P would be observed much earlier in the growth cycle. As predicted, the difference in biofilm accumulation between wild-type V. cholerae and the ΔEI mutant occurred during the exponential phase of growth and increased in early stationary phase (Fig. 3C).
FIG. 3.
EI-P represses biofilm formation and vps gene transcription in LB broth (biofilm regulatory pathway 1). (A) Total growth and biofilm accumulation in LB broth by wild-type V. cholerae (WT) or a ΔEI mutant rescued with either a control pBAD plasmid (pCTL), a pBAD plasmid expressing a wild-type EI allele (pEI), or a pBAD plasmid expressing an EI allele encoding an H-to-A mutation at position 189 (pEIH189A). Protein expression was induced by addition of 0.04% l-arabinose. The values are the averages of four experimental replicates, and the error bars indicate standard deviations. Biofilm formation by the ΔEI mutant rescued with a wild-type EI allele was not statistically different from biofilm formation by wild-type V. cholerae (P = 0.290), while biofilm formation by a ΔEI mutant rescued with the EIH189A allele was statistically different (P = 0.0007). (B) Western blot analysis demonstrating that the wild-type EI and EIH189A alleles are well expressed in the ΔEI genetic background. (C) Quantification of total growth and biofilm formation over time for wild-type V. cholerae and a ΔEI mutant. The values are the means of two experimental replicates. The ΔEI mutant demonstrated more biofilm accumulation than wild-type V. cholerae throughout the course of the experiment.
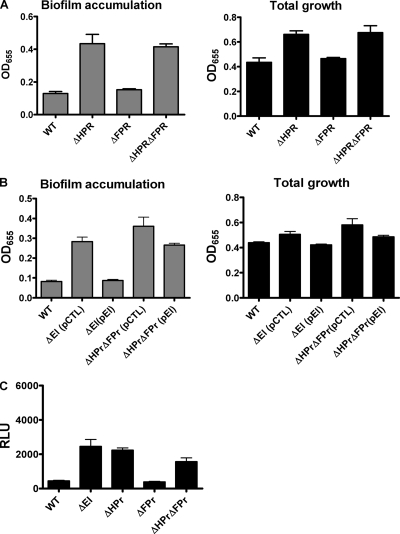
In minimal medium, repression of biofilm formation by EI requires phosphotransfer to either HPr or FPr (16). Furthermore, this repression occurs at the level of transcription. To determine if phosphotransfer from EI to HPr or FPr contributed to repression of biofilm accumulation in LB broth, we compared biofilm accumulation by wild-type V. cholerae and biofilm accumulation by ΔHPr, ΔFPr, and ΔHPr ΔFPr mutants. As shown in Fig. 4A, biofilm accumulation by a ΔHPr mutant was approximately 2-fold greater than that by wild-type V. cholerae, while biofilm formation by a ΔFPr mutant was not significantly different. To demonstrate that EI is upstream of HPr/FPr in the signaling pathway, we documented that a wild-type EI allele provided in trans repressed biofilm formation by a ΔEI mutant but was unable to repress biofilm formation in the absence of HPr and FPr (Fig. 4B). Lastly, we measured activation of the vps genes in wild-type V. cholerae and ΔEI, ΔHPr, ΔFPr, and ΔHPr ΔFPr mutants using a chromosomal lacZ reporter fusion to the vpsL promoter. As shown in Fig. 4C, deletion of EI and HPr but not FPr increased transcription of the vps genes. Thus, the regulatory pathway leading to repression of biofilm formation and vps gene transcription through phosphorylation of EI and HPr is present in both minimal medium and LB broth. This pathway is referred to in this paper as pathway 1 (see Fig. 14).
FIG. 4.
EI and HPr are in the same biofilm regulatory pathway. (A) Total growth and biofilm accumulation by wild-type V. cholerae (WT), a ΔHPr mutant, a ΔFPr mutant, and a ΔHPr ΔFPr double mutant. Biofilm accumulation by a ΔHPr mutant is significantly different from that by wild-type V. cholerae (P = 0.002), while biofilm accumulation by a ΔHPr ΔFPr is not significantly different from that by ΔHPr (P = 0.758). (B) Total growth and biofilm accumulation by wild-type V. cholerae (WT), as well as ΔEI and ΔHPr ΔFPr mutants rescued either with a control pBAD plasmid (pCTL) or a pBAD plasmid expressing a wild-type EI allele (pEI). Protein expression was induced by addition of 0.04% l-arabinose. Deletion of HPr and FPr blocked the ability of EI to repress biofilm formation. (C) Measurement of vps gene transcription in wild-type V. cholerae and various PTS mutants harboring a chromosomal vpsL-lacZ fusion. vpsL transcription in the ΔEI (P = 0.0002), ΔHPr (P < 0.0001), and ΔHPr ΔFPr (P = 0.0001) mutants is significantly different from that in wild-type V. cholerae. The error bars indicate standard deviations. RLU, relative light units.
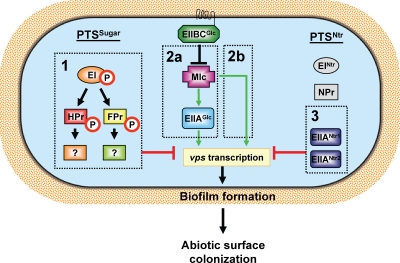
FIG. 14.
Schematic diagram of three pathways in the PTS network that regulate biofilm formation. Pathway 1 requires phosphorylation of the PTS components EI and HPr or FPr to repress transcription of the vps genes. Pathway 2a is comprised of the components EIIBCGlc, Mlc, and EIIAGlc. EIIAGlc activates vps transcription in a phosphorylation-independent manner. Mlc potentiates the action of EIIAGlc but may also activate vps gene transcription through an EIIAGlc-independent route (pathway 2b). EIIBCGlc interferes with the action of Mlc. Pathway 3 includes EIIANtr1and EIIANtr2, whose functions display some redundancy in repression of vps transcription.
Three EIIA homologs regulate biofilm accumulation.
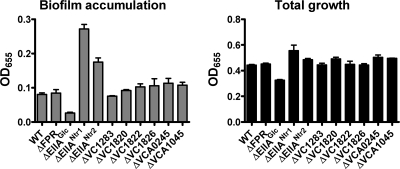
To further define regulation of biofilm formation by the PTS, we compared biofilm accumulation in LB broth by wild-type V. cholerae and biofilm accumulation in LB broth by strains having mutations in the FPr gene, which encodes both EIIA and HPr domains, as well as in each of the nine genes in the V. cholerae genome encoding EIIA homologs (Fig. 1). As shown in Fig. 5, we identified three EIIA mutants with altered biofilm phenotypes compared with wild-type V. cholerae. First, approximately 3-fold fewer ΔEIIAGlc mutant cells accumulated in a biofilm than wild-type V. cholerae cells. Second, mutations in VC2531 and VC1824 led to increases in biofilm accumulation. VC2531 (EIIANtr1) and VC1824 (EIIANtr2) both contain EIIA domains homologous to EIIANtr, and VC1824 also includes an N-terminal domain which is homologous to HPr but is not as closely related as the other V. cholerae HPr homologs (Fig. 1). Our observations suggested that at least two independent PTS pathways control biofilm formation at the level of EIIA in LB broth.
FIG. 5.
EIIA homologs EIIAGlc, EIIANtr1, and EIIANtr2 regulate biofilm formation: quantification of total growth and biofilm accumulation by wild-type V. cholerae, as well as strains having mutations in each of the 10 genes encoding EIIA domains. The biofilm accumulation by the ΔEIIAGlc (P < 0.0001), ΔEIIANtr (P < 0.0001), and ΔVC1824 (P = 0.0004) mutants was significantly different from that by wild-type V. cholerae (WT). The error bars indicate standard deviations.
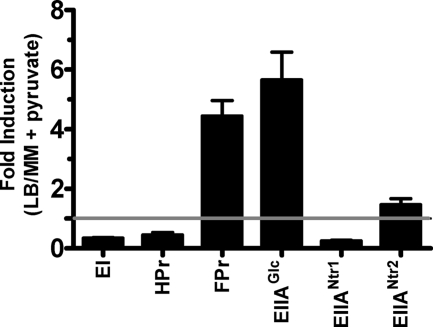
Because independent control of biofilm formation by an EIIA component was not observed in minimal medium (16), we hypothesized that transcription of EIIAGlc, EIIANtr1, and EIIANtr2 might be activated in LB broth compared to their transcription in minimal medium. To test this hypothesis, we used quantitative RT-PCR to compare the transcript levels of EI, HPr, FPr, EIIAGlc, EIIANtr1, and EIIANtr2 in LB broth and minimal medium supplemented with pyruvate (Fig. 6). Pathway 1 was present in both minimal medium and LB broth. However, the transcript levels of the pathway 1 components EI and HPr were lower in LB broth. The transcription of FPr, which does not appear to play a role in regulation of biofilm accumulation in LB broth, was elevated in LB broth compared with minimal medium. The transcription of EIIAGlc was approximately 6-fold greater in LB broth, suggesting that activation of biofilm formation by EIIAGlc in LB broth may be due at least in part to increased transcription of EIIAGlc. Transcription of the PTS components EIIANtr1 and EIIANtr2 was not increased in LB broth. Therefore, differential transcription of these components does not underlie the observation that they regulate biofilm formation in LB broth but not in minimal medium.
FIG. 6.
Differential transcription of PTS components is not solely responsible for the patterns of biofilm regulation observed in LB broth compared with those observed in minimal medium. The EI, HPr, FPr, EIIAGlc, EIIANtr1, and ΔEIIANtr2 transcript levels in wild-type V. cholerae grown in LB broth were compared to the transcript levels in MM supplemented with pyruvate. The data were analyzed in triplicate using the ΔΔCt method. clpX was used as a standard. The error bars indicate standard deviations.
Activation of biofilm formation by EIIAGlc does not require phosphorylation of the conserved histidine at position 91 (regulatory pathway 2).
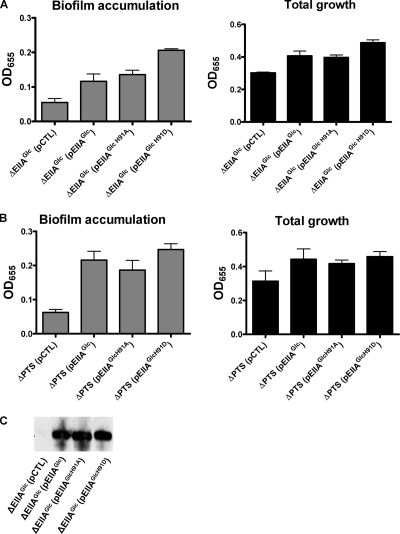
Phosphotransfer through EI and HPr is predicted to lead to EIIAGlc phosphorylation at histidine 91. To determine whether regulation of biofilm formation by EIIAGlc requires phosphorylation of this residue, we studied biofilm accumulation by a ΔEIIAGlc mutant containing an expression plasmid carrying either a control sequence, a wild-type EIIAGlc allele, an EIIAGlc allele with a histidine-to-alanine mutation at position 91, or an EIIAGlc allele with a histidine-to-aspartate mutation at position 91. As shown in Fig. 7A, while these EIIAGlc alleles did not affect the total growth, all of them activated biofilm accumulation in the ΔEIIAGlc mutant background. To further establish that this rescue was not dependent on either EI or HPr and, therefore, was not part of regulatory pathway 1, we measured activation of biofilm accumulation by the same EIIAGlc alleles in a ΔPTS mutant background, which lacks EI, HPr, and EIIAGlc (Fig. 7B). If EIIAGlc were dependent on EI or HPr for activation of biofilm formation, an EIIAGlc allele would be unable to activate biofilm accumulation in the absence of EI or HPr. However, we observed that rescue of biofilm accumulation by the ΔPTS mutant was possible with each of the three EIIAGlc alleles but not with the control plasmid. Lastly, we demonstrated by Western analysis that all rescue constructs resulted in expression of a full-length protein in the ΔEIIAGlc mutant background (Fig. 7C). Therefore, we concluded that activation of biofilm accumulation by EIIAGlc does not require phosphorylation of the conserved histidine at position 91 and that it represents a second, independent PTS-based biofilm regulatory pathway (regulatory pathway 2).
FIG. 7.
Activation of biofilm formation by EIIAGlc does not require phosphorylation and is independent of EI and HPr (biofilm regulatory pathway 2). (A) Quantification of total growth and biofilm accumulation by a ΔEIIAGlc mutant rescued with a pBAD plasmid carrying a control sequence (pCTL) and pBAD plasmids with a wild-type EIIAGlc allele (pEIIAGlc) and a variety of mutant EIIAGlc alleles. Protein expression was induced with 0.04% arabinose. Biofilm accumulation by the ΔEIIAGlc mutant rescued with a wild-type EIIAGlc glucose allele (P = 0.0024), an EIIAGlc allele encoding a histidine-to-alanine substitution at position 191 (pEIIAGlcH91A) (P < 0.0001), or an EIIAGlc allele encoding a histidine-to-aspartate substitution at position 191 (pEIIAGlcH91D) (P < 0.0001) was significantly increased compared with biofilm accumulation by the ΔEIIAGlc mutant rescued with a control sequence. (B) Quantification of total growth and biofilm accumulation by a ΔPTS mutant (lacking EI, HPr, and EIIAGlc) rescued with a pBAD plasmid carrying a control sequence (pCTL), a wild-type EIIAGlc allele (pEIIAGlc), an EIIAGlc allele encoding a histidine-to-alanine substitution at position 191 (pEIIAGlcH91A), or an EIIAGlc allele encoding a histidine-to-aspartate substitution at position 191 (pEIIAGlcH91D). Protein expression was induced with 0.04% arabinose. Biofilm accumulation by the ΔPTS mutant rescued with any of the EIIAGlc alleles was significantly increased compared with biofilm accumulation by the ΔPTS mutant rescued with a control sequence (P ≤ 0.0002 for all comparisons). The error bars indicate standard deviations. (C) Western blot demonstrating that the wild-type and mutant EIIAGlc alleles used in these experiments are well expressed and produce full-length proteins.
Evidence that EIIBCGlc and the transcription factor Mlc participate in regulatory pathway 2.
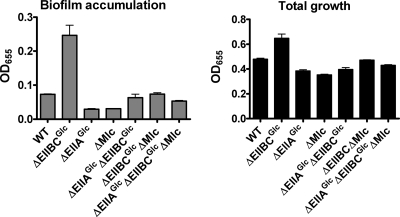
Several proteins are known to interact with EIIAGlc. Two of these proteins are EIIBCGlc and Mlc. EIIBCGlc is downstream of EIIAGlc in the glucose-specific PTS phosphotransfer cascade. Furthermore, in Escherichia coli, the DNA-binding protein Mlc has been shown to repress transcription of EIIBCGlc when PTS substrates are scarce (21). Unphosphorylated EIIBCGlc interacts directly with Mlc to relieve this repression (27, 43). The V. cholerae Mlc and EIIBCGlc proteins are very similar to the corresponding proteins of E. coli. We therefore asked whether EIIBCGlc and Mlc might play a role in regulation of V. cholerae biofilm formation by EIIAGlc. We first measured biofilm accumulation by wild-type V. cholerae, a ΔEIIBCGlc mutant, a ΔMlc mutant, and a ΔEIIAGlc mutant, as well as by strains with combinations of the mutations. As shown in Fig. 8, the total levels of growth were comparable for all of the strains studied. However, biofilm formation by the ΔMlc and ΔEIIAGlc mutants was significantly decreased, while biofilm accumulation by the ΔEIIBCGlc mutant was increased compared with wild-type V. cholerae biofilm accumulation. Furthermore, deletion of either Mlc or EIIAGlc in a ΔEIIBCGlc mutant background was sufficient to decrease the biofilm accumulation almost to the levels observed for the ΔEIIAGlc mutant. This led us to hypothesize that the biofilm regulatory pathway shown in Fig. 9A is present. In this signal transduction pathway, EIIBCGlc inhibits activation of EIIAGlc by Mlc. EIIAGlc, in turn, activates transcription of the vps genes by an as-yet-unidentified mechanism.
FIG. 8.
Evidence for involvement of Mlc and EIIBCGlc in pathway 2: quantification of total growth and biofilm accumulation by wild-type V. cholerae and various pathway 2 mutants. Biofilm accumulation by the ΔEI (P = 0.0008) or ΔEIIBCGlc (P = 0.0001) mutant was significantly greater than that by wild-type V. cholerae (WT). Biofilm accumulation by the ΔEIIAGlc or ΔMlc mutants was significantly less than that by wild-type V. cholerae (P < 0.0001). Biofilm accumulation by the ΔEIIAGlc ΔEIIBCGlc mutant (P = 0.04) or the ΔEIIAGlc ΔEIIBCGlc ΔMlc mutant (P = 0.001) was significantly greater than that by the ΔEIIAGlc mutant. Biofilm accumulation by the ΔEIIBCGlc ΔMlc mutant (P = 0.0006) or the ΔEIIAGlc ΔEIIBCGlc ΔMlc mutant (P = 0.0005) was significantly greater than that by the ΔMlc mutant. The error bars indicate standard deviations.
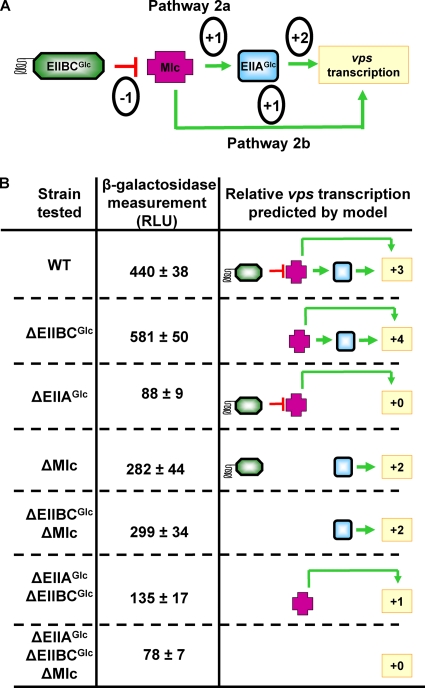
FIG. 9.
Pathway 2 regulates biofilm accumulation at the transcriptional level. (A) Schematic diagram of pathways 2a and 2b. In this model, Mlc activates vps gene transcription through both EIIAGlc-dependent and EIIAGlc-independent pathways. EIIBCGlc interferes with the action of Mlc. The numbers in circles indicate the predicted relative effect of each component in the regulatory cascade on vps gene transcription. (B) All the strains tested have vpsL-lacZ promoter fusions in the V. cholerae lacZ locus. The measurements of β-galactosidase activity reflect vpsL transcription. The measurements for the ΔEIIAGlc (P < 0.0001), ΔEIIBCGlc (P = 0.04), and ΔMlc (P = 0.0154) mutants were significantly different from that for wild-type V. cholerae. The vpsL transcription in the ΔEIIBCGlc ΔMlc mutant was significantly different from that in the ΔEIIBCGlc mutant (P = 0.0003) but not from that in the ΔMlc mutant (P = 0.766). The vps transcription in the ΔEIIAGlc ΔEIIBCGlc mutant was significantly different from that in the ΔEIIAGlc mutant (P = 0.032) or the ΔEIIBCGlc mutant (P < 0.0001). The vps transcription in the ΔEIIAGlc ΔEIIBCGlc ΔMlc mutant was significantly different both from that in the ΔEIIAGlc ΔEIIBCGlc mutant (P = 0.0086) or the ΔEIIBCGlc ΔMlc mutant (P < 0.0001) but not from that in the ΔEIIAGlc mutant (P = 0.415). The models on the right show the predicted impact of each genetic background on vps gene transcription. The relative contributions of the regulatory components to vps gene transcription as shown in panel A were added to obtain the predicted impact of the genetic background on vps gene transcription. WT, wild type; RLU, relative light units.
To further evaluate the presence of this signal transduction pathway, we measured vpsL transcription in wild-type V. cholerae and ΔEIIBCGlc, ΔMlc, and ΔEIIAGlc mutants, as well as in strains with combinations of these mutations. The results of this experiment are shown in Fig. 9B along with the components of the signal transduction pathway present in each genetic background and the relative amount of vps transcription expected based on functional pathway components. The data support a model in which Mlc activates ΔEIIAGlc and is repressed by EIIBCGlc. Furthermore, the level of vps transcription in a ΔEIIAGlc ΔEIIBCGlc mutant is higher than that in a ΔEIIAGlc ΔEIIBCGlc ΔMlc mutant, suggesting that Mlc activates vps transcription even in the absence of EIIAGlc. Therefore, an additional pathway must be postulated, in which Mlc activates vpsL transcription independent of EIIAGlc. These pathways are referred to in Fig. 9A as pathways 2a and 2b, respectively.
In general, the measurements of biofilm accumulation correlated well with measurements of vpsL transcription. However, differences were found. First, there was no difference in biofilm accumulation between ΔEIIAGlc and ΔMlc mutants. Second, biofilm formation by strains having mutations in EIIBCGlc as well as in EIIAGlc and/or Mlc all formed biofilms that were slightly but significantly and reproducibly larger than those formed by ΔEIIAGlc and ΔMlc mutants. One possible explanation for these observations is that EIIBCGlc also represses biofilm accumulation at the posttranscriptional level. The complex effect of EIIBCGlc on biofilm formation is currently under investigation.
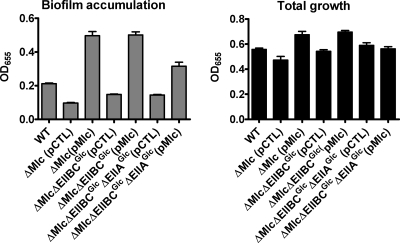
We hypothesized that if Mlc were partially dependent on EIIAGlc for its effect on biofilm formation, the absence of EIIAGlc would compromise the ability of an mlc allele provided in trans to rescue an Mlc mutation. To test this hypothesis, the following epistasis experiment was performed. We introduced a wild-type allele of mlc into ΔMlc, ΔEIIBCGlc ΔMlc, and ΔEIIAGlc ΔEIIBCGlc ΔMlc mutants. As shown in Fig. 10, while a control vector had no effect on biofilm accumulation by any of these mutants, introduction of the mlc allele into the ΔMlc and ΔEIIBCGlc ΔMlc mutants increased biofilm accumulation approximately 4-fold. In contrast, introduction of the mlc allele into a ΔEIIAGlc ΔEIIBCGlc ΔMlc mutant increased biofilm accumulation only 2-fold. This result supports our model for pathway 2 in which Mlc activates biofilm accumulation through EIIAGlc-dependent and -independent pathways.
FIG. 10.
Activation of biofilm accumulation by Mlc is partially dependent on EIIAGlc: quantification of biofilm accumulation and total growth by wild-type V. cholerae (WT) and ΔMlc, ΔEIIBCGlc ΔMlc, and ΔEIIAGlc ΔEIIBCGlc ΔMlc mutants rescued with either a control vector (pCTL) or a plasmid carrying a wild-type mlc allele (pMlc). Biofilm formation by ΔMlc (P < 0.0001), ΔEIIBCGlc ΔMlc (P < 0.0001), and ΔEIIAGlc ΔEIIBCGlc ΔMlc (P = 0.002) mutants rescued with the plasmid carrying the wild-type mlc allele was significantly greater than that by mutants carrying a control vector. The error bars indicate standard deviations.
EIIANtr1 and EIIANtr2 repress biofilm accumulation (regulatory pathway 3).
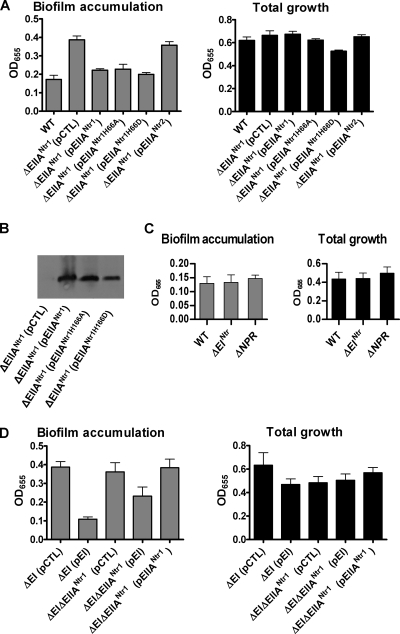
The gene encoding EIIANtr, which is located close to rpoN, has been found in many bacterial genomes (36, 48, 51). Mutation of the two V. cholerae EIIANtr homologs, EIIANtr1 (60% identity and 75% similarity to E. coli EIIANtr) and EIIANtr2 (26% identity and 42% similarity to E. coli EIIANtr), resulted in increased biofilm accumulation (Fig. 5). The conserved histidine residues at position 66 of EIIANtr1 and position 172 of EIIANtr2 are predicted to be phosphorylated by phosphotransfer from the HPr homolog NPr, which in turn receives a phosphate from EINtr. We first determined whether phosphorylation of the conserved histidine was required for the function of EIIANtr1 by comparing biofilm accumulation data for ΔEIIANtr1 mutants carrying either a plasmid encoding a control sequence, a wild-type allele of EIIANtr1, a mutant allele having a histidine-to-alanine substitution at residue 66, or a mutant allele having a histidine-to-aspartate substitution at residue 66. Rescue of the biofilm phenotype of a ΔEIIANtr1 mutant was observed with both mutant alleles, as well as with the wild-type allele of EIIANtr1 (Fig. 11A). Furthermore, all alleles of EIIANtr1 were well expressed as full-length transcripts (Fig. 11B). This result suggests that phosphorylation of the conserved histidine at position 66 is not required for repression of biofilm accumulation by EIIANtr1. Based on this observation, we also predicted that the upstream components of the PTSNtr would not be required for repression of biofilm accumulation by EIIANtr1. To test this hypothesis, we constructed strains having in-frame deletions in EINtr and NPr and compared the biofilm accumulation by these strains to that by wild-type V. cholerae. As expected, we found that biofilm accumulation by ΔEINtr and ΔNPr mutants was not significantly different from that by wild-type V. cholerae (Fig. 11C).
FIG. 11.
Repression of biofilm formation by EIIANtr1 does not require phosphorylation of the conserved histidine at residue 66 and is independent of pathway 1 (biofilm regulatory pathway 3). (A) Quantification of total growth and biofilm accumulation by wild-type V. cholerae or a ΔEIIANtr1 mutant rescued with a control plasmid (pCTL), a plasmid carrying a wild-type EIIANtr1 allele (pEIIANtr1), a plasmid carrying an EIIANtr1 allele resulting in an alanine substitution for the histidine at position 66 (pEIIANtr1H66A), a plasmid carrying an EIIANtr1 allele resulting in an aspartate substitution for the histidine at position 66 (pEIIANtr1H66D), and a plasmid carrying a wild-type EIIANtr2 allele (pEIIANtr2). Only biofilm accumulation by the ΔEIIANtr1 (pCTL) (P = 0.0003) and ΔEIIANtr1 (pEIIANtr2) (P = 0.0004) mutants was significantly different from that by wild-type V. cholerae (WT). The error bars indicate standard deviations. (B) Western blot demonstrating that the wild-type and mutant EIIANtr1 alleles used in these experiments are well expressed and produce full-length proteins. (C) Quantification of total growth and biofilm accumulation by wild-type V. cholerae and strains having mutations in the other two components of the PTSNtr, EINtr, and NPr. Biofilm accumulation by the two mutants was not significantly different from that by wild-type V. cholerae. (D) Quantification of total growth and biofilm formation by a ΔEI mutant or a ΔEI ΔEIIANtr1 mutant rescued with a pBAD plasmid carrying a control sequence (pCTL), a wild-type EI allele (pEI), or a wild-type EIIANtr1 allele (pEIIANtr1). Biofilm accumulation by the ΔEI ΔEIIANtr1 mutant rescued with a wild-type EI allele was significantly different from that by the ΔEI ΔEIIANtr1 mutant carrying a control sequence (P = 0.0102).
Because biofilm formation is also repressed by EI, we asked whether EI and EIIANtr1 function independently. As shown in Fig. 11D, return of a wild-type EI allele to a ΔEI ΔEIIANtr1 mutant partially rescued the increased biofilm phenotype, suggesting that EI regulates biofilm formation independent of EIIANtr1. Return of a wild-type EIIANtr1 allele did not reduce biofilm formation by a ΔEI ΔEIIANtr1 mutant. This suggests that the ΔEI mutation is dominant in the ΔEI ΔEIIANtr1 mutant. While we have not ruled out a model in which one function of EI is to act downstream of EIIANtr1 in a common signaling pathway, we maintain that pathway 1 is independent of pathway 3 because it is present in minimal medium (16), while pathway 3 is not.
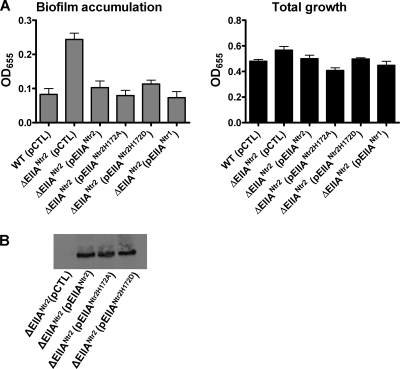
We then asked whether phosphorylation of the conserved histidine at position 172 was required for repression of biofilm accumulation by EIIANtr2. Biofilm accumulation was measured for ΔEIIANtr2 mutants harboring plasmids containing either a control sequence, a wild-type allele of EIIANtr2, an allele of EIIANtr2 resulting in substitution of the conserved histidine for alanine at position 172, or an allele of EIIANtr2 resulting in substitution of the conserved histidine for aspartate at position 172. As shown in Fig. 12A, the biofilm phenotype of a ΔEIIANtr2 mutant was rescued by the wild-type EIIANtr2 allele, as well as by both nonphosphorylatable mutant alleles. Furthermore, all rescue constructs were expressed as full-length proteins (Fig. 12B).
FIG. 12.
Repression of biofilm formation by EIIANtr2 does not require phosphorylation at histidine 172 and is rescued by EIIANtr1 (biofilm regulatory pathway 3) (A) Quantification of total growth and biofilm accumulation by wild-type V. cholerae or a ΔEIIANtr2 mutant rescued with a pBAD plasmid carrying a control sequence (pCTL), a wild-type ΔEIIANtr2 allele (pEIIANtr2), a ΔEIIANtr2 allele encoding an H-to-A point mutation at conserved residue 172 (pEIIANtr2H172A), a ΔEIIANtr2 allele encoding an H-to-D point mutation at conserved residue 172 (pEIIANtr2H172D), or a wild-type EIIANtr1 allele (pEIIANtr1). Protein expression was induced with 0.04% arabinose. Biofilm accumulation by a ΔEIIANtr2 mutant rescued with wild-type EIIANtr2 (P = 0.263), EIIANtr2H172A (P = 0.776), EIIANtr2H172D (P = 0.126), or EIIANtr1 (P = 0.514) was not significantly different from that by wild-type V. cholerae (WT). The error bars indicate standard deviations. (B) Western blot demonstrating that the wild-type and mutant EIIANtr2 alleles used in these experiments are well expressed and produce full-length proteins.
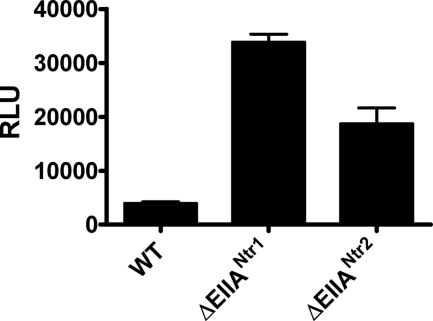
In other systems, EIIANtr has been shown to operate at the transcriptional level (25). We used β-galactosidase assays to measure the effects of the EIIANtr homologs on vpsL gene transcription. As shown in Fig. 13, while both EIIANtr1 and EIIANtr2 repress biofilm formation at the transcriptional level, EIIANtr1, which is more similar to the EIIANtr of E. coli, has a more pronounced effect.
FIG. 13.
EIIANtr1 and EIIANtr2 operate at the level of transcription: measurements of vps gene transcription in wild-type V. cholerae, a ΔEIIANtr1 mutant, and a ΔEIIANtr2 mutant harboring a chromosomal vpsL-lacZ fusion. The vpsL transcription in the ΔEIIANtr1 and ΔEIIANtr2 mutants was significantly different from that in wild-type V. cholerae (WT) (P < 0.0001). The error bars indicate standard deviations. RLU, relative light units.
Lastly, we examined whether EIIANtr1 and EIIANtr2 have redundant roles in repression of biofilm formation by attempting to rescue the biofilm phenotype of a ΔEIIANtr1 mutant with a wild-type EIIANtr2 allele and vice versa. A wild-type EIIANtr1 allele provided in trans did rescue the biofilm phenotype of the ΔEIIANtr2 mutant, supporting our conclusion that these two EIIANtr homologs function in the same biofilm regulatory pathway (Fig. 12A). EIIANtr2 provided in trans was not able to rescue the ΔEIIANtr1 mutant phenotype (Fig. 11A). We attribute the inability of EIIANtr2 to rescue the phenotype of the ΔEIIANtr1 mutant to its smaller effect on vps gene transcription and biofilm formation.
DISCUSSION
We previously described a PTS-dependent biofilm regulatory pathway present in minimal medium. This pathway requires phosphotransfer from EI to HPr or FPr for action (pathway 1) (16, 17). Here, we examined regulation of biofilm formation by the PTS in LB broth, a complex medium. Our studies established the action of pathway 1 in V. cholerae biofilms formed in LB broth and also identified two novel PTS-dependent pathways, one of which (pathway 2) leads to activation of biofilm formation by EIIAGlc and one of which (pathway 3) results in repression of biofilm formation by EIIANtr (Fig. 14). All three of these pathways regulate transcription of the vps genes, which encode the biofilm matrix biosynthetic machinery. The regulatory cascade comprising pathway 2 suggests that there are differences from the homologous PTS-dependent regulatory pathway in E. coli, while pathway 3 defines a new role for EIIANtr in regulation of bacterial biofilm formation.
In minimal medium, pathway 1 proceeds through EI-P to HPr-P and FPr-P, which have redundant functions. As a result, only a ΔHPr ΔFPr mutant forms a biofilm that is distinguishable from that formed by wild-type V. cholerae. In contrast, in LB broth, HPr is the dominant protein in the signal transduction cascade, resulting in increased biofilm accumulation by a ΔHPr mutant compared with wild-type V. cholerae. This is not due to increased transcription of HPr in LB broth compared with minimal medium (Fig. 6). Rather, we hypothesize that phosphotransfer through HPr is preferred in LB broth. Alternatively, if pathway 1 remains divergent downstream of HPr and FPr, subsequent components may be responsible for the differences observed in LB broth.
Signaling through pathway 2 results in activation of vps gene transcription. This activation requires EIIAGlc and is observed in the absence of phosphorylation of the conserved histidine residue at position 91. In E. coli, EIIAGlc has two important regulatory functions. EIIAGlc-P potentiates the action of adenylate cyclase to increase intracellular concentrations of cyclic AMP (cAMP), a cofactor of the global regulator, the cAMP receptor protein (CRP) (56). The unphosphorylated form of EIIAGlc directly blocks transport of non-PTS substrates (52). Here we discovered a phosphorylation-independent regulatory role for EIIAGlc in activation of the vps genes and biofilm formation in V. cholerae. It is unlikely that the regulation is due to activation of adenylate cyclase as multiple studies have shown that cAMP represses rather than activates vps transcription and biofilm formation in V. cholerae (11, 17, 28).
Regulation of biofilm formation by EIIAGlc does not require phosphorylation of the conserved histidine at position 91. Therefore, the mechanism by which this function of EIIAGlc is controlled in response to environmental cues is unclear. Because transcription of V. cholerae EIIAGlc is 6-fold greater in LB broth than in minimal medium and 10-fold greater in response to addition of a PTS substrate to minimal medium (17), one possibility is that the activity of EIIAGlc is regulated by its intracellular abundance.
The results of epistasis experiments and measurements of vpsL transcription are consistent with a model in which Mlc activates biofilm accumulation through EIIAGlc-dependent and EIIAGlc-independent pathways. EIIBCGlc, in turn, inhibits the action of Mlc. Because Mlc is a transcription factor, we hypothesize that it acts at the transcriptional level to increase EIIAGlc activity either directly or indirectly. E. coli Mlc functions as a transcriptional repressor of many PTS components (47, 59). However, it does not regulate transcription of EIIAGlc. E. coli EIIBCGlc, an integral membrane protein, blocks the action of Mlc by sequestering it at the inner membrane (27, 58). We hypothesize that a similar interaction between Mlc and EIIBCGlc may block activation of EIIAGlc by Mlc (pathway 2a) (Fig. 14). Our data also suggest that a small portion of the observed activation of vps gene transcription by Mlc may proceed through an EIIAGlc-independent pathway (pathway 2b) (Fig. 14).
We identified two EIIANtr homologs that repress V. cholerae biofilm formation and vps gene transcription in LB broth but not in minimal medium (pathway 3) (Fig. 14). Transcription of the EIIANtr homologs in these two media is similar. Therefore, this regulatory pathway is not controlled at the transcriptional level. The mechanism by which this pathway is activated remains to be identified.
We have not ruled out the possibility that there is an interaction between pathways 2 and 3. However, mutation of EIIANtr1 or EIIANtr2 in a ΔEIIAGlc background activates biofilm formation (data not shown), suggesting that EIIANtr1 and EIIANtr2 act independent of EIIAGlc.
While EIIANtr homologs have not been associated with biofilm formation in other organisms, they have been implicated in regulation of diverse metabolic functions, some of which have a clear role in regulating the balance of carbon and nitrogen in the cell. Two examples of this are repression of polyhydroxyalkanoate synthesis in Pseudomonas putida and repression of β-hydroxybutyrate synthesis in Azotobacter vinelandii by EIIANtr homologs (44, 62). Both of these carbohydrate polymers are intracellular carbon storage compounds that are accumulated when the environmental carbohydrate supply exceeds the bacterial requirement. VPS, the extracellular polysaccharide which is a component of the V. cholerae biofilm matrix, may also play a role in carbohydrate balance. VPS synthesis is activated when PTS substrates are abundant and is repressed when PTS substrates become scarce (17). Thus, in repressing VPS synthesis, V. cholerae EIIANtr may have a function parallel to that of its homologs in P. putida and A. vinelandii.
The mechanism by which EIIANtr represses polysaccharide synthesis in P. putida and A. vinelandii has not been discovered yet. However, the mechanism of action of EIIANtr in transcriptional derepression of the ilvBN operon, which encodes acetohydroxy acid synthase I, a common enzyme in biosynthesis of branched-chain amino acids, has been established (25). By an unknown mechanism, transcription of this operon is inhibited by high intracellular levels of K+. The unphosphorylated form of EIIANtr inhibits the action of the K+ transporter TrkA, which keeps intracellular K+ levels low and preserves transcription of the ilvBN operon (24). In our experiments, phosphorylation of neither EIIANtr1 nor EIIANtr2 was required for activation of vpsL gene transcription. One possibility is that regulation of gene transcription by EIIANtr in V. cholerae is governed by intracellular K+ levels in a phosphorylation state-independent manner. Alternatively, EIIANtr may be involved in a regulatory interaction with a different protein.
There are several examples of V. cholerae functions that are governed by complex regulatory networks with multiple inputs. These functions include production of virulence factors (34), quorum sensing (37), chemotaxis, and regulation of biofilm formation by cyclic-di-GMP (4, 5, 29, 57). In each case, the mechanism by which various environmental signals are integrated and result in the observed phenotype is unknown. Regulation of biofilm formation in V. cholerae by the PTS also appears to be quite complex. Immediate goals include identification of the environmental signals to which pathways 2 and 3 respond, identification of downstream components of these pathways, and integration of the three pathways in regulation of biofilm formation by the PTS. In particular, it seems likely that these pathways eventually interface with members of the large family of V. cholerae proteins that modulate intracellular levels of c-di-GMP and whose activators remain largely unidentified.
The V. cholerae PTS participates in colonization of both environmental surfaces and the mammalian intestine (16). We previously demonstrated that the influence of the PTS on VPS-dependent biofilm formation does not play a role in colonization of the mammalian intestine. The regulatory pathways outlined here are hypothesized to participate in colonization of the environment. In particular, a close environmental relationship between V. cholerae and zooplankton has been demonstrated. V. cholerae is thought to degrade the chitinaceous exoskeletons of these organisms to N-acetylglucosamine, a sugar transported exclusively by the PTS (16). Therefore, accumulation of V. cholerae on the exoskeletons of zooplankton is likely to be regulated by these PTS-dependent pathways. We envision that after formation of a monolayer on a surface, augmentation of the biofilm through cell division coupled with VPS synthesis is finely tuned to the nutritive potential of the surface itself and the environment in which the surface occurs. The signaling pathways outlined here contribute to bacterial sensing of surfaces and their environments and highlight the importance of PTS substrates in this process.
Acknowledgments
We thank Joachim Reidl for generously providing plasmids for mutant construction.
This work was supported by NIH grant AI50082 to P.I.W.
Footnotes
Published ahead of print on 16 April 2010.
REFERENCES
- 1.Abranches, J., M. M. Candella, Z. T. Wen, H. V. Baker, and R. A. Burne. 2006. Different roles of EIIABMan and EIIGlc in regulation of energy metabolism, biofilm development, and competence in Streptococcus mutans. J. Bacteriol. 188:3748-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barriere, C., M. Veiga-da-Cunha, N. Pons, E. Guedon, S. A. van Hijum, J. Kok, O. P. Kuipers, D. S. Ehrlich, and P. Renault. 2005. Fructose utilization in Lactococcus lactis as a model for low-GC gram-positive bacteria: its regulator, signal, and DNA-binding site. J. Bacteriol. 187:3752-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg, T., S. Schild, and J. Reidl. 2007. Regulation of the chitobiose-phosphotransferase system in Vibrio cholerae. Arch. Microbiol. 187:433-439. [DOI] [PubMed] [Google Scholar]
- 4.Beyhan, S., and F. H. Yildiz. 2007. Smooth to rugose phase variation in Vibrio cholerae can be mediated by a single nucleotide change that targets c-di-GMP signalling pathway. Mol. Microbiol. 63:995-1007. [DOI] [PubMed] [Google Scholar]
- 5.Bomchil, N., P. Watnick, and R. Kolter. 2003. Identification and characterization of a Vibrio cholerae gene, mbaA, involved in maintenance of biofilm architecture. J. Bacteriol. 185:1384-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonet, R., M. D. Simon-Pujol, and F. Congregado. 1993. Effects of nutrients on exopolysaccharide production and surface properties of Aeromonas salmonicida. Appl. Environ. Microbiol. 59:2437-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Branda, S. S., S. Vik, L. Friedman, and R. Kolter. 2005. Biofilms: the matrix revisited. Trends Microbiol. 13:20-26. [DOI] [PubMed] [Google Scholar]
- 8.Colwell, R. R., and A. Huq. 1994. Environmental reservoir of Vibrio cholerae. The causative agent of cholera. Ann. N. Y. Acad. Sci. 740:44-54. [DOI] [PubMed] [Google Scholar]
- 9.De Kievit, T. R., R. Gillis, S. Marx, C. Brown, and B. H. Iglewski. 2001. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl. Environ. Microbiol. 67:1865-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deutscher, J., C. Francke, and P. W. Postma. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70:939-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fong, J. C., and F. H. Yildiz. 2008. Interplay between cyclic AMP-cyclic AMP receptor protein and cyclic di-GMP signaling in Vibrio cholerae biofilm formation. J. Bacteriol. 190:6646-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gershanovitch, V. N., N. V. Yourovitskaya, L. V. Komissarova, T. N. Bolshakova, R. S. Erlagaeva, and G. I. Bourd. 1975. Catabolite repression in Escherichia coli K12 mutants defective in glucose transport. Mol. Gen. Genet. 140:81-90. [DOI] [PubMed] [Google Scholar]
- 13.Hammer, B. K., and B. L. Bassler. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50:101-104. [DOI] [PubMed] [Google Scholar]
- 14.Hang, L., M. John, M. Asaduzzaman, E. A. Bridges, C. Vanderspurt, T. J. Kirn, R. K. Taylor, J. D. Hillman, A. Progulske-Fox, M. Handfield, E. T. Ryan, and S. B. Calderwood. 2003. Use of in vivo-induced antigen technology (IVIAT) to identify genes uniquely expressed during human infection with Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 100:8508-8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haugo, A. J., and P. I. Watnick. 2002. Vibrio cholerae CytR is a repressor of biofilm development. Mol. Microbiol. 45:471-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Houot, L., S. Chang, C. Absalon, and P. I. Watnick. 2010. Vibrio cholerae phosphoenolpyruvate phosphotransferase system control of carbohydrate transport, biofilm formation, and colonization of the germfree mouse intestine. Infect. Immun. 78:1482-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Houot, L., and P. I. Watnick. 2008. A novel role for enzyme I of the Vibrio cholerae phosphoenolpyruvate phosphotransferase system in regulation of growth in a biofilm. J. Bacteriol. 190:311-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karatan, E., T. R. Duncan, and P. I. Watnick. 2005. NspS, a predicted polyamine sensor, mediates activation of Vibrio cholerae biofilm formation by norspermidine. J. Bacteriol. 187:7434-7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karatan, E., and P. Watnick. 2009. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 73:310-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kierek, K., and P. I. Watnick. 2003. Environmental determinants of Vibrio cholerae biofilm development. Appl. Environ. Microbiol. 69:5079-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, S. Y., T. W. Nam, D. Shin, B. M. Koo, Y. J. Seok, and S. Ryu. 1999. Purification of Mlc and analysis of its effects on the pts expression in Escherichia coli. J. Biol. Chem. 274:25398-25402. [DOI] [PubMed] [Google Scholar]
- 22.King, N. D., and M. R. O'Brian. 2001. Evidence for direct interaction between enzyme I(Ntr) and aspartokinase to regulate bacterial oligopeptide transport. J. Biol. Chem. 276:21311-21316. [DOI] [PubMed] [Google Scholar]
- 23.Lauriano, C. M., C. Ghosh, N. E. Correa, and K. E. Klose. 2004. The sodium-driven flagellar motor controls exopolysaccharide expression in Vibrio cholerae. J. Bacteriol. 186:4864-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, C. R., S. H. Cho, M. J. Yoon, A. Peterkofsky, and Y. J. Seok. 2007. Escherichia coli enzyme IIANtr regulates the K+ transporter TrkA. Proc. Natl. Acad. Sci. U. S. A. 104:4124-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, C. R., B. M. Koo, S. H. Cho, Y. J. Kim, M. J. Yoon, A. Peterkofsky, and Y. J. Seok. 2005. Requirement of the dephospho-form of enzyme IIANtr for derepression of Escherichia coli K-12 ilvBN expression. Mol. Microbiol. 58:334-344. [DOI] [PubMed] [Google Scholar]
- 26.Lee, J., A. Jayaraman, and T. K. Wood. 2007. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 7:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, S. J., W. Boos, J. P. Bouche, and J. Plumbridge. 2000. Signal transduction between a membrane-bound transporter, PtsG, and a soluble transcription factor, Mlc, of Escherichia coli. EMBO J. 19:5353-5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang, W., A. J. Silva, and J. A. Benitez. 2007. The cyclic AMP receptor protein modulates colonial morphology in Vibrio cholerae. Appl. Environ. Microbiol. 73:7482-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim, B., S. Beyhan, and F. H. Yildiz. 2007. Regulation of Vibrio polysaccharide synthesis and virulence factor production by CdgC, a GGDEF-EAL domain protein, in Vibrio cholerae. J. Bacteriol. 189:717-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lombardo, M. J., J. Michalski, H. Martinez-Wilson, C. Morin, T. Hilton, C. G. Osorio, J. P. Nataro, C. O. Tacket, A. Camilli, and J. B. Kaper. 2007. An in vivo expression technology screen for Vibrio cholerae genes expressed in human volunteers. Proc. Natl. Acad. Sci. U. S. A. 104:18229-18234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loo, C. Y., K. Mitrakul, I. B. Voss, C. V. Hughes, and N. Ganeshkumar. 2003. Involvement of an inducible fructose phosphotransferase operon in Streptococcus gordonii biofilm formation. J. Bacteriol. 185:6241-6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao, X. J., Y. X. Huo, M. Buck, A. Kolb, and Y. P. Wang. 2007. Interplay between CRP-cAMP and PII-Ntr systems forms novel regulatory network between carbon metabolism and nitrogen assimilation in Escherichia coli. Nucleic Acids Res. 35:1432-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martino, P. D., R. Fursy, L. Bret, B. Sundararaju, and R. S. Phillips. 2003. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can. J. Microbiol. 49:443-449. [DOI] [PubMed] [Google Scholar]
- 34.Matson, J. S., J. H. Withey, and V. J. DiRita. 2007. Regulatory networks controlling Vibrio cholerae virulence gene expression. Infect. Immun. 75:5542-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGinnis, M. W., Z. M. Parker, N. E. Walter, A. C. Rutkovsky, C. Cartaya-Marin, and E. Karatan. 2009. Spermidine regulates Vibrio cholerae biofilm formation via transport and signaling pathways. FEMS Microbiol. Lett. [DOI] [PubMed]
- 36.Michiels, J., T. Van Soom, I. D'Hooghe, B. Dombrecht, T. Benhassine, P. de Wilde, and J. Vanderleyden. 1998. The Rhizobium etli rpoN locus: DNA sequence analysis and phenotypical characterization of rpoN, ptsN, and ptsA mutants. J. Bacteriol. 180:1729-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, M. B., K. Skorupski, D. H. Lenz, R. K. Taylor, and B. L. Bassler. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110:303-314. [DOI] [PubMed] [Google Scholar]
- 38.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitchell, W. J., T. P. Misko, and S. Roseman. 1982. Sugar transport by the bacterial phosphotransferase system. Regulation of other transport systems (lactose and melibiose). J. Biol. Chem. 257:14553-14564. [PubMed] [Google Scholar]
- 40.Moorthy, S., and P. I. Watnick. 2004. Genetic evidence that the Vibrio cholerae monolayer is a distinct stage in biofilm development. Mol. Microbiol. 52:573-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mueller, R. S., S. Beyhan, S. G. Saini, F. H. Yildiz, and D. H. Bartlett. 2009. Indole acts as an extracellular cue regulating gene expression in Vibrio cholerae. J. Bacteriol. 191:3504-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munoz-Elias, E. J., J. Marcano, and A. Camilli. 2008. Isolation of Streptococcus pneumoniae biofilm mutants and their characterization during nasopharyngeal colonization. Infect. Immun. 76:5049-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nam, T. W., H. I. Jung, Y. J. An, Y. H. Park, S. H. Lee, Y. J. Seok, and S. S. Cha. 2008. Analyses of Mlc-IIBGlc interaction and a plausible molecular mechanism of Mlc inactivation by membrane sequestration. Proc. Natl. Acad. Sci. U. S. A. 105:3751-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noguez, R., D. Segura, S. Moreno, A. Hernandez, K. Juarez, and G. Espin. 2008. Enzyme I NPr, NPr and IIA Ntr are involved in regulation of the poly-beta-hydroxybutyrate biosynthetic genes in Azotobacter vinelandii. J. Mol. Microbiol. Biotechnol. 15:244-254. [DOI] [PubMed] [Google Scholar]
- 45.Patel, C. N., B. W. Wortham, J. L. Lines, J. D. Fetherston, R. D. Perry, and M. A. Oliveira. 2006. Polyamines are essential for the formation of plague biofilm. J. Bacteriol. 188:2355-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfluger, K., and V. de Lorenzo. 2008. Evidence of in vivo cross talk between the nitrogen-related and fructose-related branches of the carbohydrate phosphotransferase system of Pseudomonas putida. J. Bacteriol. 190:3374-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plumbridge, J. 1998. Control of the expression of the manXYZ operon in Escherichia coli: Mlc is a negative regulator of the mannose PTS. Mol. Microbiol. 27:369-380. [DOI] [PubMed] [Google Scholar]
- 48.Powell, B. S., D. L. Court, T. Inada, Y. Nakamura, V. Michotey, X. Cui, A. Reizer, M. H. Saier, Jr., and J. Reizer. 1995. Novel proteins of the phosphotransferase system encoded within the rpoN operon of Escherichia coli. Enzyme IIANtr affects growth on organic nitrogen and the conditional lethality of an erats mutant. J. Biol. Chem. 270:4822-4839. [DOI] [PubMed] [Google Scholar]
- 49.Reizer, J., A. Reizer, M. J. Lagrou, K. R. Folger, C. K. Stover, and M. H. Saier, Jr. 1999. Novel phosphotransferase systems revealed by bacterial genome analysis: the complete repertoire of pts genes in Pseudomonas aeruginosa. J. Mol. Microbiol. Biotechnol. 1:289-293. [PubMed] [Google Scholar]
- 50.Reizer, J., A. Reizer, M. J. Merrick, G. Plunkett III, D. J. Rose, and M. H. Saier, Jr. 1996. Novel phosphotransferase-encoding genes revealed by analysis of the Escherichia coli genome: a chimeric gene encoding an enzyme I homologue that possesses a putative sensory transduction domain. Gene 181:103-108. [DOI] [PubMed] [Google Scholar]
- 51.Ren, J., S. Sainsbury, N. S. Berrow, D. Alderton, J. E. Nettleship, D. K. Stammers, N. J. Saunders, and R. J. Owens. 2005. Crystal structure of nitrogen regulatory protein IIANtr from Neisseria meningitidis. BMC Struct. Biol. 5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saier, M. H., Jr. 1989. Protein phosphorylation and allosteric control of inducer exclusion and catabolite repression by the bacterial phosphoenolpyruvate:sugar phosphotransferase system. Microbiol. Rev. 53:109-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seidl, K., C. Goerke, C. Wolz, D. Mack, B. Berger-Bachi, and M. Bischoff. 2008. Staphylococcus aureus CcpA affects biofilm formation. Infect. Immun. 76:2044-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seok, Y. J., B. M. Koo, M. Sondej, and A. Peterkofsky. 2001. Regulation of E. coli glycogen phosphorylase activity by HPr. J. Mol. Microbiol. Biotechnol. 3:385-393. [PubMed] [Google Scholar]
- 55.Seok, Y. J., M. Sondej, P. Badawi, M. S. Lewis, M. C. Briggs, H. Jaffe, and A. Peterkofsky. 1997. High affinity binding and allosteric regulation of Escherichia coli glycogen phosphorylase by the histidine phosphocarrier protein, HPr. J. Biol. Chem. 272:26511-26521. [DOI] [PubMed] [Google Scholar]
- 56.Stulke, J., and W. Hillen. 1998. Coupling physiology and gene regulation in bacteria: the phosphotransferase sugar uptake system delivers the signals. Naturwissenschaften 85:583-592. [DOI] [PubMed] [Google Scholar]
- 57.Tamayo, R., A. D. Tischler, and A. Camilli. 2005. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 280:33324-33330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tanaka, Y., K. Kimata, and H. Aiba. 2000. A novel regulatory role of glucose transporter of Escherichia coli: membrane sequestration of a global repressor Mlc. EMBO J. 19:5344-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanaka, Y., K. Kimata, T. Inada, H. Tagami, and H. Aiba. 1999. Negative regulation of the pts operon by Mlc: mechanism underlying glucose induction in Escherichia coli. Genes Cells 4:391-399. [DOI] [PubMed] [Google Scholar]
- 60.Titgemeyer, F. 1993. Signal transduction in chemotaxis mediated by the bacterial phosphotransferase system. J. Cell Biochem. 51:69-74. [DOI] [PubMed] [Google Scholar]
- 61.Van Dellen, K. L., L. Houot, and P. I. Watnick. 2008. Genetic analysis of Vibrio cholerae monolayer formation reveals a key role for ΔΨ 32 ιn the transition to permanent attachment. J. Bacteriol. 190:8185-8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Velazquez, F., K. Pfluger, I. Cases, L. I. De Eugenio, and V. de Lorenzo. 2007. The phosphotransferase system formed by PtsP, PtsO, and PtsN proteins controls production of polyhydroxyalkanoates in Pseudomonas putida. J. Bacteriol. 189:4529-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Watnick, P. I., C. M. Lauriano, K. E. Klose, L. Croal, and R. Kolter. 2001. Absence of a flagellum leads to altered colony morphology, biofilm development, and virulence in V. cholerae O139. Mol. Microbiol. 39:223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. U. S. A. 96:4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]