Abstract
SpoIID is a membrane-anchored enzyme that degrades peptidoglycan and is essential for engulfment and sporulation in Bacillus subtilis. SpoIID is targeted to the sporulation septum, where it interacts with two other proteins required for engulfment: SpoIIP and SpoIIM. We changed conserved amino acids in SpoIID to alanine to determine whether there was a correlation between the effect of each substitution on the in vivo and in vitro activities of SpoIID. We identified one amino acid substitution, E88A, that eliminated peptidoglycan degradation activity and one, D210A, that reduced it, as well as two substitutions that destabilized the protein in B. subtilis (R106A and K203A). Using these mutants, we show that the peptidoglycan degradation activity of SpoIID is required for the first step of engulfment (septal thinning), as well as throughout membrane migration, and we show that SpoIID levels are substantially above the minimum required for engulfment. The inactive mutant E88A shows increased septal localization compared to the wild type, suggesting that the degradation cycle of the SpoIID/SpoIIP complex is accompanied by the activity-dependent release of SpoIID from the complex and subsequent rebinding. This mutant is also capable of moving SpoIIP across the sporulation septum, suggesting that SpoIID binding, but not peptidoglycan degradation activity, is needed for relocalization of SpoIIP. Finally, the mutant with reduced activity (D210A) causes uneven engulfment and time-lapse microscopy indicates that the fastest-moving membrane arm has greater concentrations of SpoIIP than the slower-moving arm, demonstrating a correlation between SpoIIP protein levels and the rate of membrane migration.
Endospore formation is an evolutionarily conserved process that allows Bacillus subtilis and related Gram-positive bacteria to adapt to changes in the environment, such as nutrient depletion. Many dramatic morphological changes occur during sporulation, each requiring a multitude of specialized proteins (reviewed in references 13 and 17). First, a sporulation septum is formed near one of the cell poles, forming two separate compartments of unequal sizes and with differing fates (Fig. 1A). The smaller of the two, the forespore, will eventually become the spore, while the larger, the mother cell, will ultimately lyse. Next, the mother cell membranes move up and around the forespore in the poorly understood process of engulfment. Although this process is superficially similar to eukaryotic engulfment, it is complicated by the thick cell wall that surrounds and separates the two compartments. After engulfment, the migrating membranes pinch off from the mother cell membrane, thereby releasing the forespore into the cytoplasm of the mother cell, where it can be enveloped with protective coat proteins and eventually released into the environment as a mature spore. Sporulation provides an ideal, nonessential system for understanding how bacterial cells are capable of undergoing dramatic morphological changes.
FIG. 1.
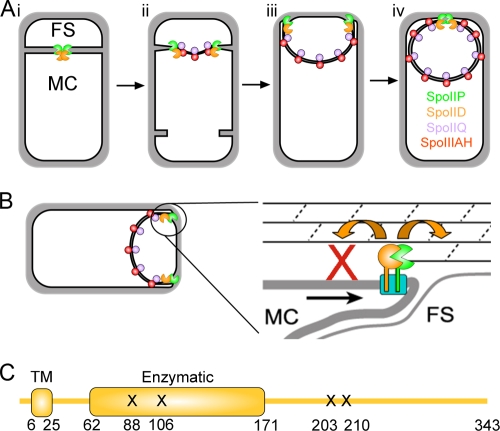
Engulfment in B. subtilis. (A) (i) Engulfment begins with formation of an asymmetric septum that divides the cell into the forespore (FS) and mother cell (MC). SpoIID (orange pacman) and SpoIIP (green pacman) peptidoglycan degradation enzymes localize to the center of the septum. (ii) SpoIID and SpoIIP thin the septal peptidoglycan, starting from the center and moving toward the cell edges. SpoIIQ (purple ball) and SpoIIIAH (red ball) form a zipper across the septum, assembling foci behind the leading edges. (iii) The peptidoglycan degradation enzymes localize to the leading edges during membrane migration, while additional SpoIIQ-SpoIIIAH complexes assemble around the forespore. (iv) Engulfment membrane fission occurs at the top of the forespore, releasing the forespore into the mother cell cytoplasm. (B) Burnt-bridge Brownian ratchet model for membrane migration, adapted from earlier studies (1, 7). (C) Schematic representation of the SpoIID domain structure. The transmembrane domain (TM) and putative enzymatic domain, as defined by Pfam (14), are indicated. Amino acid numbers are below the schematic, and mutations causing in vivo phenotypes are indicated by an “X”.
Engulfment involves dynamic protein localization and large-scale rearrangements of cellular membranes and peptidoglycan to accommodate internalization of the forespore. The physical basis for engulfment remains unclear, but two separate protein machineries that contribute to engulfment have been discovered. The first module involves the only three proteins known to be required for engulfment under all physiological conditions: SpoIID, SpoIIM, and SpoIIP (16, 24, 35). Zymography assays have demonstrated that both SpoIID and SpoIIP degrade peptidoglycan in vitro (1, 8), and this function is thought to be essential for engulfment in wild-type cells (1, 2, 8). SpoIID and SpoIIP are membrane-spanning proteins that directly interact both in vivo and in vitro, as demonstrated by coimmunoprecipitation and affinity chromatography techniques (2, 8). These studies failed to demonstrate an interaction between SpoIIM and either SpoIID or SpoIIP, perhaps because SpoIIM is an integral membrane protein. However, SpoIIM is required for localization of SpoIID and SpoIIP (2, 8), suggesting that all three proteins interact to form a peptidoglycan degradation module that is essential for engulfment.
The second system influencing membrane migration is the SpoIIQ/SpoIIIAH zipper, which is required for engulfment only under certain conditions (2, 7, 38). SpoIIQ is produced in the forespore (23) and SpoIIIAH is produced in the mother cell (19). SpoIIQ and SpoIIIAH interact both in vitro and in vivo via their extracellular domains (6, 7, 10). Because these two proteins are produced in separate compartments, the only possible place for an interaction is the intermembrane space between the mother cell and forespore, forming a protein-protein zipper between the two cells. This zipperlike interaction is necessary for septal localization of SpoIIIAH and other mother cell proteins (6, 10, 18) and is capable of holding the two cells together when peptidoglycan is removed with lysozyme (7). Surprisingly, digestion of the peptidoglycan with lysozyme also allows membrane migration in about half of treated cells, in a process requiring the SpoIIQ/SpoIIIAH zipper but not the SpoIIDMP peptidoglycan degradation module. The SpoIIQ-SpoIIIAH zipper also contributes to engulfment in living cells, since strains lacking SpoIIQ or SpoIIIAH complete engulfment more slowly than the wild type and have synergistic engulfment defects when certain secondary mutations are introduced (2, 7, 38). Together, these results strongly support a role for the SpoIIQ/SpoIIIAH module in engulfment, demonstrating that the zipper contributes to the efficiency of membrane migration even when the SpoIIDMP module is present and functional. They also suggest that the engulfment machinery displays functional redundancy and that the zipper module provides a backup machinery for membrane migration.
The precise role of the SpoIIDMP module during engulfment remains unclear. One model proposes that SpoIID and SpoIIP act as a burnt-bridge Brownian ratchet (Fig. 1B) (1, 7). This model asserts that as SpoIID and SpoIIP degrade peptidoglycan, they eliminate their own enzymatic targets, resulting in the absence of substrate in one direction and therefore, overall movement in the opposite direction. As the enzymes move forward toward new targets, the mother cell membranes are dragged along with them because they are anchored in the membrane. This hypothesis predicts that SpoIID and SpoIIP are processive enzymes and that the SpoIIDMP complex could function as a motor, moving along peptidoglycan as a track and pulling the membranes with it (1, 7). A second model predicts that peptidoglycan degradation could simply remove a steric hindrance to membrane migration (such as links between the forespore membrane and the cell wall) and that some other mechanism provides the force required for membrane migration. Although the SpoIIQ-SpoIIIAH module can contribute to membrane migration, these proteins are not always essential for engulfment in intact cells (7, 38), suggesting that another unidentified system must generate the force required for membrane movement if the DMP module does not act as a burnt-bridge Brownian ratchet. Recent evidence suggests that peptidoglycan biosynthesis, which is localized to the leading edge of the engulfing membrane and necessary for membrane migration in the absence of the SpoIIQ-SpoIIIAH proteins, might be this missing force generating mechanism (26).
Both models predict that the activities of SpoIID and SpoIIP are essential for membrane migration. This requirement has been demonstrated for SpoIIP (8) and, while this work was under review, for SpoIID. SpoIID shows no sequence similarity to any characterized enzyme that degrades peptidoglycan and thus constitutes the founding member of a new class of enzymes that remodel peptidoglycan (1, 27). However, SpoIID does show some similarity to B. subtilis LytB (24), a protein that enhances the activity of the amidase LytC (5, 20, 34), while SpoIIP is related to LytC (14). A recent study demonstrated that SpoIIP is both an amidase and endopeptidase and that SpoIID both activates SpoIIP and functions as a lytic transglycosylase, cleaving peptidoglycan between NAG and NAM (27). Together, these two enzymes degrade peptidoglycan into its smallest repeating subunits. However, it remains unclear which of the demonstrated or suggested biochemical functions of SpoIID are required for its various in vivo activities (interaction with SpoIIP, localization, septal thinning, and membrane migration), and it is unclear whether peptidoglycan degradation activity is required throughout engulfment or only for the initial stage of septal thinning.
We use site-directed mutagenesis to test the role of 56 conserved amino acids in SpoIID, focusing on hydrophilic amino acids that might be involved in protein-protein interactions and peptidoglycan degradation. We identified one mutation (E88A) that eliminates and three others (R106A, K203A, and D210A) that reduce peptidoglycan degradation activity and show that SpoIID activity is required for the earliest stage of engulfment (septal thinning), as well as throughout membrane migration. Our results confirm and extend those of Morlot et al. (27) and also demonstrate that SpoIID activity is required throughout engulfment. Furthermore, our data indicate that the enzymatically inactive mutant protein (E88A) shows increased septal localization compared to the wild-type protein, suggesting that peptidoglycan degradation contributes to the release of SpoIID from the septum. We propose a modified model for the enzymatic cycle of the SpoIID and SpoIIP complex.
MATERIALS AND METHODS
Bacterial strains, genetic manipulations, and growth conditions.
Bacillus subtilis strains used in the present study are derivatives of the wild-type strain PY79 (39) and are shown in Table 2. Mutations and plasmids were introduced into PY79 by transformation (12). B. subtilis strains were grown and sporulated at 37°C unless otherwise indicated. Sporulation was induced by the modified resuspension method (36) using 25% LB as a growth medium (4) for all cell biological experiments. Heat-resistant spore assays were performed on cultures grown and sporulated in DSM broth (31) for 24 h at 37°C. Cultures were then heated at 80°C for 20 min, serially diluted, and plated on LB. Spore titers were calculated based on colony counts.
TABLE 2.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| B. subtilis | ||
| PY79 | Wild type | 39 |
| KP7 | spoIID298 | 24 |
| KP38 | spoIID298 amyE::spoIID38-kan | 1 |
| KP718 | spoIID298 amyE::spoIID-kan | 1 |
| KP719 | spoIIP::tet | 16 |
| KP1065 | spoIID298 amyE::spoIIDY65A-kan | This study |
| KP1066 | spoIID298 amyE::spoIIDR66A-kan | This study |
| KP1067 | spoIID298 amyE::spoIIDE73A-kan | This study |
| KP1068 | spoIID298 amyE::spoIIDE78A-kan | This study |
| KP1069 | spoIID298 amyE::spoIIDE79A-kan | This study |
| KP1070 | spoIID298 amyE::spoIIDY80A-kan | This study |
| KP1071 | spoIID298 amyE::spoIIDS87A-kan | This study |
| KP1072 | spoIID298 amyE::spoIIDE88A-kan | This study |
| KP1073 | spoIID298 amyE::spoIIDM89A-kan | This study |
| KP1074 | spoIID298 amyE::spoIIDP90A-kan | This study |
| KP1075 | spoIID298 amyE::spoIIDK94A-kan | This study |
| KP1076 | spoIID298 amyE::spoIIDE96A-kan | This study |
| KP1077 | spoIID298 amyE::spoIIDK99A-kan | This study |
| KP1078 | spoIID298 amyE::spoIIDQ101A-kan | This study |
| KP1079 | spoIID298 amyE::spoIIDR106A-kan | This study |
| KP1080 | spoIID298 amyE::spoIIDT107A-kan | This study |
| KP1081 | spoIID298 amyE::spoIIDF108A-kan | This study |
| KP1082 | spoIID298 amyE::spoIIDD128A-kan | This study |
| KP1083 | spoIID298 amyE::spoIIDD129A-kan | This study |
| KP1084 | spoIID298 amyE::spoIIDT130A-kan | This study |
| KP1085 | spoIID298 amyE::spoIIDQ134A-kan | This study |
| KP1086 | spoIID298 amyE::spoIIDY136A-kan | This study |
| KP1087 | spoIID298 amyE::spoIIDK137A-kan | This study |
| KP1088 | spoIID298 amyE::spoIIDT164A-kan | This study |
| KP1089 | spoIID298 amyE::spoIIDT170A-kan | This study |
| KP1090 | spoIID298 amyE::spoIIDY171A-kan | This study |
| KP1091 | spoIID298 amyE::spoIIDF181A-kan | This study |
| KP1092 | spoIID298 amyE::spoIIDS182A-kan | This study |
| KP1093 | spoIID298 amyE::spoIIDT183A-kan | This study |
| KP1095 | spoIID298 amyE::spoIIDT188A-kan | This study |
| KP1097 | spoIID298 amyE::spoIIDE192A-kan | This study |
| KP1098 | spoIID298 amyE::spoIIDY201A-kan | This study |
| KP1099 | spoIID298 amyE::spoIIDK203A-kan | This study |
| KP1100 | spoIID298 amyE::spoIIDS204A-kan | This study |
| KP1101 | spoIID298 amyE::spoIIDS207A-kan | This study |
| KP1102 | spoIID298 amyE::spoIIDD210A-kan | This study |
| KP1103 | spoIID298 amyE::spoIIDS213A-kan | This study |
| KP1104 | spoIID298 amyE::spoIIDK215A-kan | This study |
| KP1105 | spoIID298 amyE::spoIIDT219A-kan | This study |
| KP1106 | spoIID298 amyE::spoIIDK220A-kan | This study |
| KP1107 | spoIID298 amyE::spoIIDT221A-kan | This study |
| KP1108 | spoIID298 amyE::spoIIDT223A-kan | This study |
| KP1109 | spoIID298 amyE::spoIIDE226A-kan | This study |
| KP1110 | spoIID298 amyE::spoIIDQ228A-kan | This study |
| KP1111 | spoIID298 amyE::spoIIDQ229A-kan | This study |
| KP1112 | spoIID298 amyE::spoIIDK230A-kan | This study |
| KP1113 | spoIID298 amyE::spoIIDK234A-kan | This study |
| KP1114 | spoIID298 amyE::spoIIDS239A-kan | This study |
| KP1115 | spoIID298 amyE::spoIIDT262A-kan | This study |
| KP1116 | spoIID298 amyE::spoIIDK282A-kan | This study |
| KP1117 | spoIID298 amyE::spoIIDR283A-kan | This study |
| KP1118 | spoIID298 amyE::spoIIDD286A-kan | This study |
| KP1119 | spoIID298 amyE::spoIIDF295A-kan | This study |
| KP1120 | spoIID298 amyE::spoIIDH297A-kan | This study |
| KP1121 | spoIID298 amyE::spoIIDV299A-kan | This study |
| KP1122 | spoIID298 amyE::spoIIDM301A-kan | This study |
| KP1123 | spoIID298 amyE::spoIIDQ303A-kan | This study |
| KP1124 | spoIID::cat::tet amyE::gfp-spoIID | This study |
| KP1125 | spoIID::cat::tet amyE::gfp-spoIIDE88A | This study |
| KP1142 | spoIID::cat::tet amyE::gfp-spoIIDR106A | This study |
| KP1126 | spoIID::cat::tet amyE::gfp-spoIIDK203A | This study |
| KP1127 | spoIID::cat::tet amyE::gfp-spoIIDD210A | This study |
| KP1128 | spoIIP::tet amyE::gfp-spoIID-cm | This study |
| KP1129 | spoIID298 amyE::spoIID-kan ΔspoIIPtetΩPIIPgfp-spoIIP-erm | This study |
| KP1137 | spoIID298 ΔspoIIPtetΩPIIPgfp-spoIIP-erm | This study |
| KP1130 | spoIID298 amyE::spoIIDA-kan ΔspoIIPtetΩPIIPgfp-spoIIP-erm | This study |
| KP1141 | spoIID298 amyE::spoIIDR106A-kan ΔspoIIPtetΩPIIPgfp-spoIIP-erm | This study |
| KP1131 | spoIID298 amyE::spoIIDK203A-kan ΔspoIIPtetΩPIIPgfp-spoIIP-erm | This study |
| KP1132 | spoIID298 amyE::spoIIDD210A-kan ΔspoIIPtetΩPIIPgfp-spoIIP-erm | This study |
| E. coli | ||
| KP1133 | BL21; pSA12 (His6-SpoIID27-343) | This study |
| KP1134 | BL21; pSA12 (His6-SpoIID27-343 E88A) | This study |
| KP1147 | BL21; pSA12 (His6-SpoIID27-343 R106A) | This study |
| KP1135 | BL21; pSA12 (His6-SpoIID27-343 K203A) | This study |
| KP1136 | BL21; pSA12 (His6-SpoIID27-343 D210A) | This study |
Construction of His6-SpoIID.
The His6-SpoIID overexpression plasmid pSA12 was constructed in pET30a (Novagen). The spoIID region encoding amino acids 27 to 343 was PCR amplified from PY79 chromosomal DNA using the primers: 5′-GAGCCGCATTAGCACCATCATCATCATCATCAGCATAATAAGGAAGCGGGG and 5′-GCGGCGGTCGACTTACTTTTTCGCCATATATTTATT (the NdeI and SalI restriction sites are underlined). pET30a was used as the vector with the N-terminal His6 tag encoded on the primer. PCR fragments were digested with NdeI and SalI (New England Biolabs) and ligated into NdeI- and SalI-digested pET30a. The ligation mix was transformed into the Escherichia coli strain DH5α and transformants selected for on LB-kanamycin (50 μg/ml). All constructs were confirmed to be correct by DNA sequencing.
Targeted mutagenesis of spoIID.
Mutations in spoIID were introduced using a site-directed mutagenesis protocol adapted from an earlier study (30). Plasmid pKP01 (1), an amyE-integrating vector that includes spoIID, was used as the template for initial QuikChange PCRs. Primers used in the PCRs are listed in Table SA2 in the supplemental material and generally included 12 bp upstream and 12 bp downstream of the mutation that substituted the native codon with a GCA alanine codon. Plasmids were transformed into DH5α or TOP10 cells (Invitrogen). The entire spoIID coding region of the mutagenized plasmids was sequenced by EtonBio (San Diego, CA) or Genewiz (San Diego, CA), using the primers JGP2dp or JGPD358 (see Table SA2 in the supplemental material). After transformation into B. subtilis, the resultant colonies were purified, patched to DSM, and incubated at 30°C to assess the ability of each construct to support sporulation. Mutations with a sporulation phenotype were subsequently introduced into pRSE15 and pSA12, the latter encoding His6-tagged SpoIID in an E. coli expression construct (Novagen) (construction described above). The GFP-spoIID plasmid pRSE15 was constructed from pAAD9 (1), an amyE inserting vector containing spoIID in frame and downstream of the fusion of green fluorescent protein (GFP) to the spoIID promoter, translational initiation sequences, and the first five codons encoded in pMDS14 (33). This plasmid had three mutations in the spoIID coding sequence, which were converted to wild type by using QuikChange mutagenesis and primers RMSP12 and RMSP13 to give pRSE10 and then using RMSP10 and RMSP11 (see Table SA2 in the supplemental material) to yield pRSE15.
Microscopy and image analysis.
To visualize the GFP, samples from sporulating cultures were taken at the indicated times, stained with a final concentration of 5 μg of FM 4-64/ml and 2 μg of DAPI/ml, and applied to poly-l-lysine-coated coverslips (32). For membrane fusion assays, the cells were stained with a final concentration of 5 μg of FM 4-64/ml and 4 μg of MitoTracker Green (MTG)/ml (32). Images were collected by using an Applied Precision Spectris optical sectioning microscope equipped with a Photometrix CoolsnapHQ charge-coupled device camera. Images were deconvolved by using SoftWoRx software (Applied Precision, Inc.).
Time-lapse microscopy.
Time-lapse microscopy was performed as previously described (4). Briefly, cells were grown at 37°C in 25% LB to an optical density at 600 nm (OD600) of 0.5 and resuspended in A+B sporulation salts (36). A 2-ml aliquot of each resuspended culture was shifted to a small culture tube, followed by incubation in a roller at 30°C with FM 4-64 added to a final concentration of 0.5 μg/ml at the time of resuspension, while the remainder of the culture was grown without dye. The unstained culture was spun down 20 min before t2 and the supernatant used to make a 1.2% agarose/A+B pad containing 0.5 μg of FM 4-64/ml. The stained culture was spread on this pad, covered with a glass coverslip, and equilibrated to 30°C for 10 min prior to viewing. Cells were grown and imaged on the pad in a climate controlled chamber (Precision Control Weather Station) set to 30°C.
His6-SpoIID overexpression and purification.
For purification of His6-SpoIID, a 250-ml culture of BL21 containing pSA12, or else pSA12 with the indicated single amino acid substitution, was incubated at 37°C until reaching an OD600 of 0.6 to 0.8. Cultures were then induced for 3 h with 2 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cells were harvested by centrifugation, and the pellet was frozen at −80°C. The pellet was resuspended in buffer J (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole [pH 8.0]) including 2 mM phenylmethylsulfonyl fluoride (PMSF) and sonicated to lyse the cells. Cell debris was removed by centrifugation, and the supernatant was added to equilibrated His-Select Ni/NTA resin (Sigma) and shaken at 4°C overnight. The slurry was added to a column, washed with 10 column volumes of buffer J and 5 column volumes of buffer J plus 40 mM imidazole, and eluted with buffer J plus 250 mM imidazole. His6-SpoIID protein was further purified by ion-exchange chromatography (except for His6-SpoIIDR106A). Protein was dialyzed into buffer A (100 mM NaH2PO4, 10 mM TrisHCl, 150 mM NaCl [pH 8]) and run over a column of Q-Sepharose beads. The His-tagged protein was eluted by running buffer A, with stepwise increases in NaCl concentration, over the column. Fractions of 1-ml volume were collected and analyzed for protein content by using a spectrophotometer to monitor the A280. Fractions containing protein were combined and dialyzed into activity buffer (20 mM Tris-HCl, 150 mM NaCl [pH 8.0]). Protein was stored at −80°C until use with an approximate concentration of 0.5 mg/ml.
Renaturing gel electrophoresis assay for peptidoglycan degradation activity.
Zymography was performed as previously described (15, 37) using 0.1% Micrococcus luteus cells (Sigma) as a substrate. Gels were rocked in renaturing solution (25 mM Tris-HCl, 1% Triton X-100 [pH 7.2]) at 37°C overnight, imaged using an Olympus E-410 Digital SLR camera, stained with fresh 0.01% methylene blue in 0.01% KOH for 3 h at room temperature, and then rinsed. Stained gels were imaged using a Typhoon scanner (GE Healthcare) without an emission filter.
Western blotting.
Samples for Western blots were prepared as described in reference 9. Briefly, 1.0 ml of cell culture was harvested at the indicated time points and precipitated by the addition of 110 μl of 50% trichloroacetic acid and frozen overnight. Samples were spun on high for 5 min, washed with Tris buffer (pH 8), and resuspended in 54 μl of Tris sucrose buffer (33 mM Tris [pH 8], 40% sucrose, 1 mM EDTA, 300 μg of PMSF/ml). Lysozyme was added to a final concentration of 1 μg/ml, and the samples were incubated at 37°C for 10 min. An equal volume of 2× sodium dodecyl sulfate (SDS) loading buffer was added to each sample, and the samples were heated to 42°C. Samples were applied to a SDS-10% PAGE gel, transferred to a polyvinylidene difluoride membrane, and probed with previously prepared antibodies to SpoIID at a 1:3,000 dilution (9). Secondary, anti-rabbit antibody conjugated to horseradish peroxidase (GE Healthcare) was added at 1:3,000 and developed using an ECL-Plus kit (GE Healthcare).
In-gel GFP fluorescence.
The levels of GFP-SpoIID proteins were assessed using an in-gel GFP fluorescence protocol adapted from Dubnau and Davidoff-Abelson (11). Cultures were sporulated by resuspension. At the indicated times after initiation of sporulation, 1.5-ml samples were collected and resuspended in 45 μl of phosphate-buffered saline plus 300 μg of PMSF/ml. Lysozyme was added to a final concentration of 1 mg/ml, and the samples were incubated at 37°C for 10 min. Then, 50 μl of fresh solubilization buffer (200 mM Tris-HCl [pH 8.8], 20% glycerol, 5 mM EDTA, 0.02% bromophenol blue, 4% SDS, 50 mM dithiothreitol) was added, and the samples were again heated at 37°C for 10 min. A needle and syringe were used to break up DNA and reduce sample viscosity. A 20-μl portion of the sample was applied to a SDS-10% PAGE gel and run at 60 V. GFP fluorescence was visualized by using a Typhoon scanner with 520 BP 40 emission filter and a blue (488-nm) laser.
RESULTS
Site-directed mutagenesis of spoIID.
SpoIID is a 343-amino-acid, membrane-spanning protein with a short cytoplasmic N terminus, a single transmembrane domain, and a large extracytoplasmic C-terminal domain (Fig. 1C). The SpoIID extracytoplasmic domain is conserved throughout the endospore forming bacteria, with more distant relatives found in many other bacterial phyla (14). Our results indicated that this domain was responsible for the peptidoglycan degradation activity of purified SpoIID in zymography gels (see below), but until recently (27), none of the proteins that are homologous to SpoIID have had characterized activities.
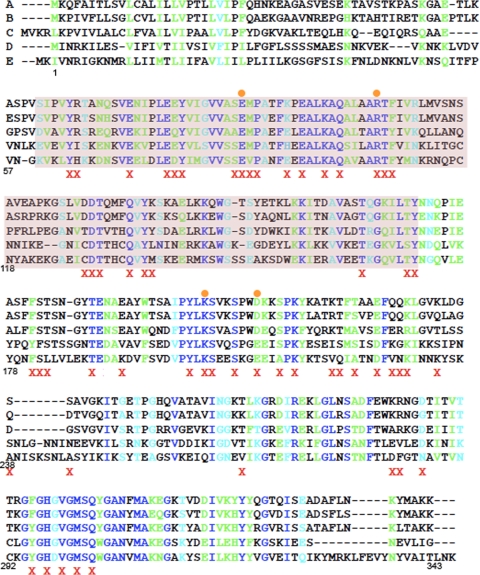
To further investigate the mechanism by which SpoIID cleaves peptidoglycan, we identified conserved amino acids in SpoIID using the CLUSTAL W program (21), which performs multiple HMM alignments of SpoIID homologues (Fig. 2). We selected 56 conserved hydrophilic amino acids for site-directed mutagenesis to alanine, including all conserved amino acids identified in the putative enzymatic domain (Pfam 08486, Fig. 2, purple highlight). The entire spoIID coding and promoter region was sequenced, and the mutations were screened for the ability to complement a spoIID-null strain and support sporulation both by examining pigmentation on DSM plates and by measuring the ability to produce heat resistant spores. This strategy identifies only amino acids that are essential for sporulation. We found three amino acid substitutions within SpoIID that cause severe sporulation blocks: E88A and R106A, conserved residues within the putative enzymatic domain, and K203A, which is on the C-terminal side of this domain and within the extracellular domain (Fig. 1C). Each mutation decreased the viable spore counts 10,000-fold compared to that of the wild type (Table 1). We also identified four amino acid substitutions that cause an intermediate phenotype, ranging from a 10- to a 1,000-fold decrease in spores (Table 1). Of these four, we chose to characterize D210A as a representative of the group. Importantly, all 13 codons that were mutagenized both here and in the study of Morlot et al. (27) had similar effects on sporulation in both studies.
FIG. 2.
Amino acids selected for mutagenesis of SpoIID. Alignments of spoIID were performed by using CLUSTAL W (21). The species presented here are as follows: A, Bacillus subtilis; B, Bacillus amyloliquefaciens; C, Geobacillus thermodinitrificans; D, Clostridium perfringens; E, Clostridium saccharobutylicum. Amino acid numbers below the alignment refer to the B. subtilis sequence for easier identification of mutations. A red X indicates an amino acid changed to alanine and an orange circle indicates a mutant characterized in the present study. Sequence homology is represented by color, with completely conserved amino acids indicated by royal blue, highly conserved amino acids indicated by green and weakly conserved amino acids indicated by cyan. The putative peptidoglycan degradation domain is highlighted in purple. Recently, an alignment and compilation of all 667 SpoIID family members has become available on Pfam (14). This alignment was used to confirm our earlier alignments and verify that all residues showing significant conservation were mutated during our study. In this more complete alignment, only six amino acids are completely conserved, and just two of these, E88 and R106, resulted in an in vivo phenotype when changed to alanine.
TABLE 1.
Spore titers of SpoIID mutantsa
| Strain | SpoIID mutation | Avg spore titer | Avg sporulation (% of wild type) |
|---|---|---|---|
| KP7 | spoIID298 | 0 × 108 | 0.00 |
| PY79 | Wild type | 1.8 × 108 | 100.00 |
| KP1124 | GFP-SpoIID | 2.1 × 108 | 116.67 |
| KP1065 | Y65A | 1.8 × 108 | 100.00 |
| KP1066 | R66A | 2.6 × 108 | 144.44 |
| KP1067 | E73A | 2.6 × 108 | 144.44 |
| KP1068 | E78A | 2.0 × 108 | 111.11 |
| KP1069 | E79A | 5.5 × 108 | 305.56 |
| KP1070 | Y80A | 8.4 × 107 | 46.67 |
| KP1071 | S87A | 2.5 × 108 | 138.89 |
| KP1072 | E88A | 3.5 × 104 | 0.02 |
| KP1125 | GFP-SpoIIDE88A | 1.5 × 104 | 0.01 |
| KP1073 | M89A | 2.6 × 108 | 144.44 |
| KP1074 | P90A | 2.6 × 108 | 127.78 |
| KP1075 | K94A | 1.1 × 108 | 61.11 |
| KP1076 | E96A | 3.1 × 108 | 172.22 |
| KP1077 | K99A | 5.0 × 108 | 277.78 |
| KP1078 | Q101A | 1.8 × 108 | 100.00 |
| KP1079 | R106A | 6.5M × 104 | 0.04 |
| KP1142 | GFP-SpoIIDR106A | 1.1 × 106 | 0.61 |
| KP1080 | T107A | 1.0 × 108 | 55.56 |
| KP1081 | F108A | 2.2 × 108 | 122.22 |
| KP1082 | D128A | 1.6 × 108 | 88.89 |
| KP1083 | D129A | 1.6 × 108 | 88.89 |
| KP1084 | T130A | 2.0 × 108 | 111.11 |
| KP1085 | Q134A | 1.8 × 108 | 100.00 |
| KP1086 | Y136A | 3.0 × 108 | 166.67 |
| KP1087 | K137A | 1.5 × 108 | 83.33 |
| KP1088 | T164A | 1.3 × 108 | 72.22 |
| KP1089 | T170A | 1.8 × 108 | 100.00 |
| KP1090 | Y171A | 1.2 × 108 | 66.67 |
| KP1091 | F181A | 2.7 × 108 | 150.00 |
| KP1092 | S182A | 1.9 × 108 | 105.56 |
| KP1093 | T183A | 4.0 × 108 | 222.22 |
| KP1095 | T188A | 1.0 × 107 | 5.56 |
| KP1097 | E192A | 2.3 × 108 | 127.78 |
| KP1098 | Y201A | 1.3 × 108 | 72.22 |
| KP1099 | K203A | 1.0 × 104 | 0.01 |
| KP1126 | GFP-SpoIIDK203A | 2.2 × 108 | 122.22 |
| KP1100 | S204A | 1.6 × 108 | 88.89 |
| KP1101 | S207A | 2.5 × 108 | 138.89 |
| KP1102 | D210A | 2.0 × 107 | 11.11 |
| KP1127 | GFP-SpoIIDD210A | 1.3 × 105 | 0.07 |
| KP1103 | S213A | 2.4 × 108 | 133.33 |
| KP1104 | K215A | 3.8 × 108 | 211.11 |
| KP1105 | T219A | 3.3 × 108 | 183.33 |
| KP1106 | K220A | 1.3 × 108 | 72.22 |
| KP1107 | T221A | 3.5 × 108 | 194.44 |
| KP1108 | T223A | 2.6 × 108 | 144.44 |
| KP1109 | E226A | 2.6 × 108 | 144.44 |
| KP1110 | Q228A | 3.6 × 108 | 200.00 |
| KP1111 | Q229A | 3.7 × 108 | 205.56 |
| KP1112 | K230A | 2.6 × 108 | 144.44 |
| KP1113 | K234A | 1.5 × 108 | 83.33 |
| KP1114 | S239A | 1.3 × 108 | 72.22 |
| KP1115 | T262A | 1.2 × 108 | 66.67 |
| KP1116 | K282A | 2.0 × 108 | 111.11 |
| KP1117 | R283A | 2.0 × 108 | 111.11 |
| KP1118 | D286A | 1.5 × 108 | 83.33 |
| KP1119 | F295A | 2.3 × 108 | 127.78 |
| KP1120 | H297A | 1.4 × 105 | 0.08 |
| KP1121 | V299A | 1.7 × 108 | 94.44 |
| KP1122 | M301A | 2.9 × 107 | 16.11 |
| KP1123 | Q303A | 3.1 × 108 | 172.22 |
Heat kill assays were performed (31) on strains containing each spoIID mutant. Three mutations (E88A, R106A, and K203A) drastically lowered spore titers, while four other mutations lowered spore titers to intermediate levels (T188A, D210A, H297A, and M301A). For this study, we focused on those mutations causing a severe sporulation block and one mutation with an intermediate spore titer as a representative of the intermediate group. Strains further characterized in this study are indicated in boldface.
Characterization of engulfment in the mutant strains.
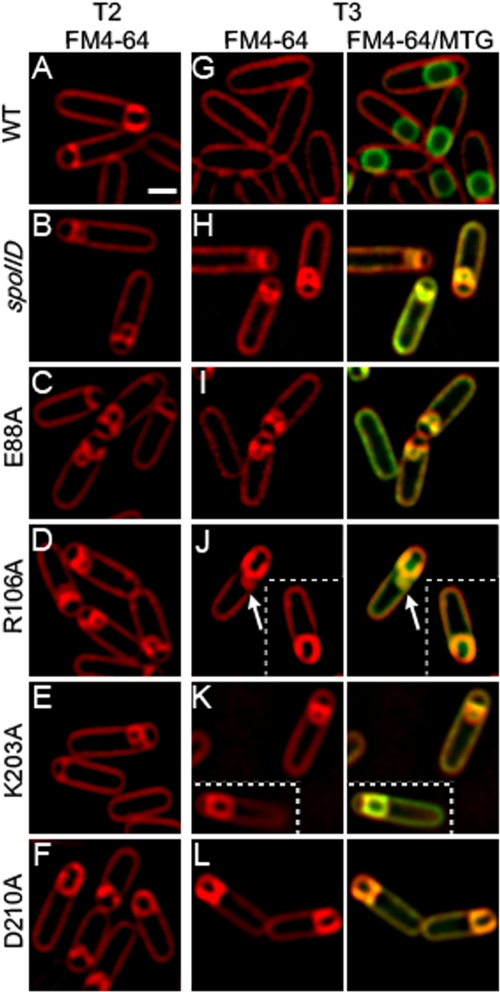
Strains expressing the four mutant proteins SpoIIDE88A, SpoIIDR106A, SpoIIDK203A, and SpoIIDD210A were further characterized by using FM 4-64 membrane staining and fluorescence microscopy to determine the stage at which sporulation is arrested. SpoIIDE88A, SpoIIDR106A, and SpoIIDK203A exhibited flat septa and septal bulges with no membrane migration at early time points (Fig. 3C to E), a finding similar to that seen with a spoIID-null strain (Fig. 3B). Thus, these mutations block the very earliest stage of engulfment: septal thinning. SpoIIDD210A, however, displayed some septal bulging at early time points but also showed significant membrane migration (Fig. 3F), suggesting that this mutant is slow to complete septal thinning but able to initiate membrane migration.
FIG. 3.
Mutant SpoIID proteins show engulfment defects. Sporulation was induced by resuspension at 37°C and samples were collected at indicated times of sporulation (in hours). Scale bar, 1 μm. (A to E) Membranes were stained with FM 4-64 in strains expressing a single copy of either wild-type or mutant SpoIID at the amyE locus. A, wild type (KP718); B, spoIID null (KP7); C, SpoIIDE88A (KP1072); D, SpoIIDR106A (KP1079); E, SpoIIDK203A (KP1099); F, SpoIIDD210A (KP1102). Membranes in the wild type (A) show membrane migration and no septal bulges, while membranes in spoIID (B), SpoIIDE88A (C), and SpoIIDK203A (E) show flat or slightly curved septa with bulges. SpoIIDD210A (F) shows some open bulges but also membrane migration. Migration is often strikingly asymmetric in this strain. SpoIIDR106A (D) shows an intermediate phenotype with slow migration and persistent bulges. (G to L) Fusion assays. Membranes were stained with FM 4-64 and Mitotracker Green (MTG) to examine the completion of engulfment. Forespores that stain green but not red have completed engulfment membrane fission. Wild-type (G) cells have completed engulfment. The spoIID null (H), SpoIIDE88A (I), and SpoIIDK203A (K) show no migration and no fusion but retain septal bulges. SpoIIDR106A (J) shows some migration but very little fusion; an arrow illustrates septal bulge persistence even when some migration is present. SpoIIDD210A (L) shows significant membrane migration but little fusion (no fusion is shown in this panel, although 3% of cells have fused at t3; see Table SA1 in the supplemental material).
We next used a membrane fusion assay (32) to determine whether strains expressing the mutant SpoIID proteins were able to complete engulfment. This assay uses two membrane stains, MTG, which is membrane permeable, and FM 4-64, which is membrane impermeable and therefore unable to stain the forespore membranes after engulfment. The completion of engulfment is indicated by the staining of the forespore membranes with the membrane permeable dye MTG but not with FM 4-64. In wild-type B. subtilis, our observations agree with published data (1, 22, 29) that ca. 65% of cells have completed engulfment by 3 h after the initiation of sporulation by resuspension (t3) at 37°C (Fig. 3G and see Table SA1 in the supplemental material). In contrast, strains expressing SpoIIDE88A (Fig. 3I) or SpoIIDK203A (Fig. 3K and Table SA1 in the supplemental material) failed to complete engulfment by t3, t4, or t5. In a strain expressing SpoIIDD210A, ca. 4 and 9% of sporangia complete engulfment by t3 and t4, respectively (Fig. 3L and see Table SA1 in the supplemental material). This is consistent with spore titer data showing that this strain produces ca. 10% the level of heat-resistant spores as the wild type (Table 1). A strain expressing SpoIIDR106A also shows bulges at early time points, but, although migration sometimes initiates, forespores often maintain indents at the original septum site and show evidence of bulge persistence (Fig. 3J, arrow), indicating that dissolution of the septum is less effective than in a strain expressing SpoIIDD210A, but more than in a null mutant. Completion of engulfment is very rarely seen in strains expressing SpoIIDR106A, reflecting the 10,000-fold reduction in heat-resistant spores compared to the wild type (Table 1). Thus, R106A has a phenotype that is subtly different from spoIID-null or spoIIDE88A strains.
Bulge formation at the sporulation septum is due to a defect in septal thinning, which allows the growing forespore to push through the septal midpoint and into the mother cell (16, 24, 35). Bulges are apparent in all four mutant strains at early time points, indicating that SpoIIDE88A, SpoIIDR106A, SpoIIDK203A, and SpoIIDD210A all have a septal thinning defect. However, SpoIIDE88A and SpoIIDK203A show no membrane migration and significant lysis is observed, exactly mimicking spoIID-null strains. Thus, SpoIIDE88A and SpoIIDK203A have severe early engulfment defects. In contrast, SpoIIDD210A shows septal bulges at early time points, indicating decreased septal thinning, but the strain often initiates, and sometimes completes, membrane migration. This phenotype is somewhat similar to that of a spoIIB-null strain, which also initially shows septal bulges, followed by the initiation of membrane migration (25, 28). In a spoIIB deletion, localization of SpoIIDMP is delayed and is facilitated by SpoIVFA (2). The similarity of the SpoIIDD210A and spoIIB phenotypes could indicate that the interaction between SpoIIDD210A and SpoIIP is impaired and localization of both proteins is delayed or transient, resulting in slow migration. SpoIIDR106A has a unique presentation combining elements of both the spoIID-null and SpoIIDD210A phenotypes with persistent septal bulges, indicating very poor septal thinning even though some membrane migration is evident.
Peptidoglycan degradation activity of the mutant proteins.
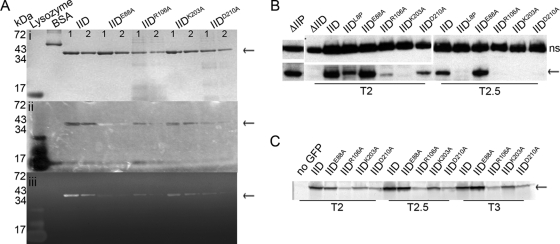
We used a renaturing polyacrylamide gel assay (zymography) to test whether the proteins could degrade bacterial cell walls incorporated into a gel (1, 15). Briefly, the wild type and the four mutant SpoIID proteins with in vivo phenotypes were purified from E. coli, and 2- and 1-μg portions of each protein were run on two SDS-PAGE gels, one with M. luteus cells suspended in the gel and one without. Prior experiments indicated that M. luteus cells are a better substrate for examining SpoIID activity than are purified B. subtilis cell walls (1). The recent finding indicating that SpoIID cleaves only glycan strands that have been denuded by SpoIIP (27) suggests that this substrate shows clearing because it consists of a nonuniform mixture of peptidoglycan targets with cells in various stages of division and growth that might have been cleaved by endogenous amidases. After renaturation, the M. luteus gel was photographed against a dark background to directly visualize lysis of the M. luteus cells as zones of clearing at the positions of the lysozyme control band and SpoIID (Fig. 4A, panel ii), thereby excluding the possibility that an apparent cleavage was caused by exclusion of the methylene blue stain normally used in such assays (as suggested by Morlot et al. [27]). The gel was then stained with methylene blue, rinsed, and then imaged again to more clearly visualize clearing in the gel caused by loss of peptidoglycan and, therefore, peptidoglycan degradation activity (Fig. 4Aiii). As seen in Fig. 4A, the mutant SpoIID proteins display a range of degradation activity. SpoIIDE88A showed no clearing on zymography gels, suggesting that it is inactive. Interestingly, SpoIIDE88A forms an opaque band on zymography gels (Fig. 4Aii) that is visible prior to staining of the gel, while the other mutants that retain some activity form clear areas. This suggests that SpoIIDE88A may be modifying peptidoglycan, but in a way that does not cause cell lysis or support engulfment. This result agrees with the findings of Morlot et al. (27) and reinforces the conclusion that E88 is involved in peptidoglycan degradation.
FIG. 4.
Biochemical tests of SpoIID. SpoIID is indicated with arrows. (A) Renaturing gel electrophoresis assay for cell wall degradation (zymography). Purified SpoIID and mutant SpoIID proteins were run on two SDS-PAGE gels. Band size is indicated on the left and amount of protein loaded is at the top of each column: lane 1 for each protein contains 2 μg, and lane 2 contains 1 μg. (i) Gel stained with Coomassie blue to visualize protein purity and concentration. (ii) Photograph of gel containing M. luteus cells as substrate and incubated overnight in renaturing solution. The gel was imaged without stain and over black paper, with clearing evidenced by the appearance of the black background through the opacity of the gel. SpoIIDE88A appears as an opaque band. (iii) Same gel as in panel ii, with the peptidoglycan stained with methylene blue before imaging on a Typhoon scanner. Clearing indicates peptidoglycan degradation. Lysozyme is a positive control for peptidoglycan degradation and bovine serum albumin (BSA) is a negative control. (B) Western blot of SpoIID protein levels. Mutant constructs are inserted at amyE and indicated above each column; time of sample collection (after sporulation initiation) is below. The upper panels show a nonspecific (ns) band bound by the SpoIID antibody and used as a loading control. Lower panels show SpoIID. (C) In-gel GFP fluorescence of GFP-SpoIID levels. Cell extracts were collected from cultures containing the GFP-SpoIID fusion protein indicated above each column at the time indicated below. Extracts were run on SDS-PAGE gels, and GFP was visualized directly using a Typhoon imager.
SpoIIDD210A shows minimal but reproducible clearing, suggesting that it has a severe reduction in peptidoglycan degradation activity. The ability of SpoIIDD210A to complete engulfment in ∼10% of sporangia suggests that the septal thinning system in B. subtilis is robust, since enzyme activity levels well below wild-type levels can support membrane migration. SpoIIDR106A and SpoIIDK203A show intermediate levels of clearing, with more clearing than SpoIIDD210A but less than the wild type. This suggests that the reductions in the peptidoglycan degradation activities of these mutant proteins are not sufficient to explain their severe engulfment defect. Although Morlot et al. characterized SpoIIDR106A as an inactive mutant based on their mass spectrometry assay, the protein has clear activity on zymography gels. The different results could be due to different buffer conditions or to different substrates present in the M. luteus preparation used here compared to purified peptidoglycan.
Variable accumulation of the mutant SpoIID proteins in vivo.
To determine whether the mutant SpoIID proteins are stably expressed in vivo, we used Western blot analysis and SpoIID-specific antibodies to probe the steady-state levels of the proteins throughout sporulation. These experiments showed that levels of SpoIIDE88A were greater than wild-type SpoIID (Fig. 4B), either because the inability of this strain to complete engulfment prolonged σE activity and SpoIID synthesis or because the mutation increased the proteolytic stability. In contrast, SpoIIDK203A was undetectable at all time points tested (Fig. 4B) despite its wild-type expression signals, suggesting that this mutant protein is unstable and quickly degraded. The instability of SpoIIDK203A explains the observation that although this mutant protein shows significant peptidoglycan degradation activity, the strain expressing this protein has a phenotype similar to that of the spoIID-null strain, with no septal thinning or membrane migration. SpoIIDD210A was also detected at levels below that of the wild type (Fig. 4B). However, low protein levels do not explain the low spore titers, because SpoIIDL8P (1) accumulates to a level equivalent to that of SpoIIDD210A (Fig. 4B), but SpoIIDL8P supports ca. 40% wild-type spore production (1), whereas SpoIIDD210A produces just 10% of the level of spores compared to the wild type (Table 1). This suggests that SpoIIDD210A has an additional defect that reduces spore production, likely its reduced peptidoglycan degradation activity. SpoIIDR106A was also detected at levels well below that of the wild type, suggesting that instability of this protein in vivo explains the absence of membrane fusion in cells despite retention of peptidoglycan degradation activity in vitro. The intermediate level of SpoIIDR106A, between that of SpoIIDK203A and SpoIIDD210A, coupled with its intermediate membrane migration phenotype, suggests that the amount of SpoIIDR106A protein available is just below the threshold necessary to complete membrane migration in vivo.
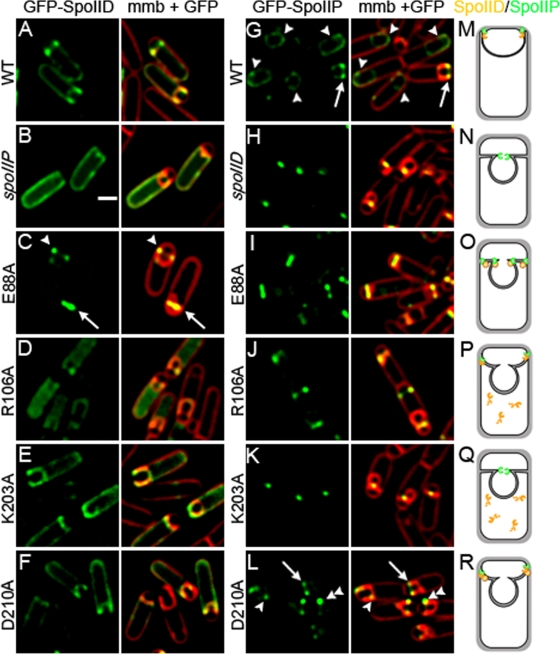
Localization of GFP-SpoIID.
Wild-type and mutant SpoIID proteins were fused to green fluorescent protein (GFP) in order to directly visualize localization of the mutant proteins. As previously noted (1), GFP-SpoIID does not localize exclusively to the septum (Fig. 5A) but rather shows some enrichment at the leading edge of the engulfing membrane and diffuse fluorescence throughout the mother cell membrane. This localization is dependent on SpoIIP (Fig. 5B) (2, 8). The only GFP-SpoIID construct that exhibited strong septal localization was GFP-SpoIIDE88A (Fig. 5C), which localized almost exclusively to the septal membranes. This suggests that GFP-SpoIIDE88A is tightly bound at the septum (Fig. 5C, arrow), either to peptidoglycan or to another protein, supporting a model in which SpoIID enzymatic activity is required for movement of the protein around the forespore and also suggesting that this activity releases the protein from the septum. However, GFP-SpoIIDE88A was capable of relocalizing to the edge of the septum in ca. 16% of sporulating cells (Fig. 5C, arrowhead), demonstrating that SpoIID peptidoglycan degradation activity is not always required to move SpoIID across the septum.
FIG. 5.
Localization of GFP fusion proteins. All cells were grown at 37°C and images taken at t3. Membranes were stained with FM 4-64. Scale bar, 1 μm. (A to E) GFP-SpoIID localization in: A, wild type (KP1124); B, spoIIP null (KP1128); C, GFP-SpoIIDE88A (KP1125); D, GFP-SpoIIDR106A (KP1142); E, GFP-SpoIIDK203A (KP1126); and F, GFP-SpoIIDD210A (KP1127). GFP-SpoIID shows some enrichment at the leading edge together with diffuse fluorescence throughout the mother cell membranes. The only strain showing significantly enhanced localization is GFP-SpoIIDE88A (C), which is almost exclusively localized to the septum (C, arrow) or at the edges of the septum (C, arrowhead), although no membrane migration is seen. Signal is also more diffuse in the spoIIP-null strain (B) than in other strains, suggesting that, as previously reported, SpoIID localization depends on SpoIIP (8). GFP-SpoIIDR106A (D) is diffuse in the cytoplasm, suggesting protein degradation. (G to L) GFP-SpoIIP localization in: G, wild type (KP1129); H, spoIID null (KP1137); I, SpoIIDE88A (KP1130); J, SpoIIDR106A (KP1141); K, SpoIIDK203A (KP1131); and L, SpoIIDD210A (KP1132). GFP-SpoIIP localizes to the leading edges of migrating membranes in the wild type (G, arrow), as a diffuse signal after fusion (G, arrowheads) and as a sharp central focus in the absence of SpoIID (H). GFP-SpoIIP localizes across the septum and to the edges of the septal disk in SpoIIDE88A (I), as a central focus in SpoIIDR106A (J) and SpoIIDK203A (K), and similarly to the wild type in SpoIIDD210A (L, arrow), although GFP-SpoIIP signal is sometimes uneven (L, double arrowhead) or lagging behind the leading edge in SpoIIDD210A (L, arrowhead). (M to R) Diagrams representing SpoIIP (green) and SpoIID (orange) localization at t3 in: M, wild type; N, spoIID null; O, SpoIIDE88A; P, SpoIIDR106A; Q, SpoIIDK203A; and R, SpoIIDD210A. Panel Q summarizes the results in the strain expressing untagged SpoIIDK203A (which fails to accumulate) and not GFP-SpoIIDK203A.
Both GFP-SpoIIDK203A and GFP-SpoIIDD210A localize in a manner similar to that of the wild type, with some enrichment at the leading edge of the engulfing membrane and diffuse fluorescence throughout the mother cell membrane (Fig. 5E and F). Interestingly, fusion of GFP to SpoIIDK203A abolished the engulfment defect, allowing membrane migration and restoring spore titers to wild-type levels (Table 1), whereas fusion of GFP to SpoIIDD210A enhanced the sporulation phenotype, blocking membrane migration and dropping spore titers to essentially zero (Table 1). GFP-SpoIIDR106A, although occasionally seen at the edges of the septal disk, was typically present in the mother cell cytoplasm (Fig. 5D), supporting the conclusion that this SpoIID variant is unstable.
We next examined whether GFP stabilized the mutant proteins using in-gel GFP fluorescence (11). The GFP tag did not change the relative accumulation of SpoIID, SpoIIDE88A, SpoIIDR106A, or SpoIIDD210A compared to Western blots of the untagged protein, since in each case SpoIIDE88A accumulated at higher levels than wild-type SpoIID, which accumulated at higher levels than SpoIIDD210A, which was higher than SpoIIDR106 (Fig. 4C). However, GFP-SpoIIDK203A accumulated to significantly higher levels than expected, since the untagged protein could not be detected on Western blots (Fig. 4C compared to Fig. 4B). This suggests that the N-terminal GFP tag stabilizes the SpoIIDK203A mutant protein, as has been observed for some other proteins (3). The stabilization of SpoIIDK203A by the GFP tag likely explains the increased ability of the GFP fusion protein to support engulfment.
Localization of GFP-SpoIIP in the mutant strains.
We next investigated the ability of GFP-SpoIIP to localize in strains expressing the mutant SpoIID proteins. SpoIIP and SpoIID interact both in vitro (8) and in vivo (2) and the absence of either protein alters localization of the other (Fig. 5B and H) (2, 8). GFP-SpoIIP clearly localizes to the leading edges of the migrating membranes during engulfment (Fig. 5G, arrow), becoming more diffusely localized around the forespore membranes after engulfment (Fig. 5G, arrowheads). In a spoIID-null strain, GFP-SpoIIP remains as a sharp focus at the center of the septum, without moving the edge of the septal disk (Fig. 5H), suggesting that SpoIID is required to move SpoIIP to the leading edge of the engulfing membrane. These two clearly distinct localization patterns allow minor variations to be distinguished in the mutant SpoIID proteins. In a SpoIIDE88A background, GFP-SpoIIP spreads across the septum and sometimes localizes to the edges of the septal disk (Fig. 5I), a phenotype clearly distinct from the tight focus observed in the spoIID-null strain (Fig. 5H). Thus, SpoIIDE88A is localized at the septum and likely capable of interacting with SpoIIP to mediate its movement across the septum, since SpoIIP localization is distinct from that seen in the absence of SpoIID. However, no membrane migration occurs, suggesting that the activity of both proteins is required for membrane migration.
In the engulfment defective SpoIIDR106A and SpoIIDK203A strains, GFP-SpoIIP forms a tight focus at the septum center (Fig. 5J and K), identical to the spoIID-null strain. This is consistent with Western blot data indicating that SpoIIDK203A fails to accumulate in the cell and SpoIIDR106A accumulates at very low levels. In the SpoIIDD210A strain, which is slow for both septal thinning and membrane migration, GFP-SpoIIP localizes to the leading edges of the migrating membranes in a manner similar to the wild type (Fig. 5L, arrow), except that some cells appear to have an asymmetric distribution of GFP-SpoIIP at the leading edges (Fig. 5L, double arrowhead) and in others GFP-SpoIIP foci appear to lag behind the leading edge of the engulfing membrane (Fig. 5L, arrowhead). This suggests that the interaction between SpoIIP and SpoIIDD210A may be weak or transient, leading to poor GFP-SpoIIP localization.
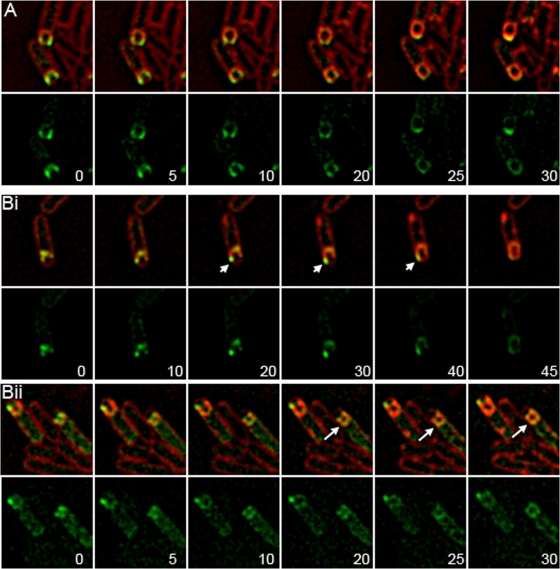
Time-lapse microscopy of GFP-SpoIIP.
If the rate of membrane migration depends on SpoIID and SpoIIP activity, then an increased localization of GFP-SpoIIP on one leading edge in SpoIIDD210A cells should correlate with an increased rate of membrane migration on that side of the sporangium. We used time-lapse fluorescence microscopy to test this hypothesis, visualizing GFP-SpoIIP localization and the engulfing membranes in wild-type and SpoIIDD210A strains (Fig. 6). In wild-type cells, membrane migration occurs either evenly, with membranes moving around the forespore at similar rates, or in a inch-worm fashion, with membranes on the right side of the forespore moving slightly forward and then membranes on the left side moving slightly forward until meeting at the top middle. GFP-SpoIIP localizes to the leading edges as a focus or with enrichment at the leading edge together with a gradient along the side of the forespore. Ultimately, GFP-SpoIIP localizes as a cap at the top of the forespore (Fig. 6A and see Movie SA1 in the supplemental material). In a SpoIIDD210A strain with GFP-SpoIIP, we observed membrane migration in only 17% of cells, with most cells forming large blebs. These blebs are distinct from the septal bulges seen in spoIID-null strains as septal thinning appears to be complete, evidenced by deeply curved septa. However, it appears that membrane synthesis at the edges of the septal disk continues without advancement, leading to membranes blebbing inward that effectively reform a “septum,” although there might not be peptidoglycan present in these structures. In such cells, GFP-SpoIIP is localized either in faint foci at the edges of the bleb or diffusely throughout the bleb (Fig. 6Bii, arrows). When membrane migration occurred in this strain, it was often strikingly asymmetric, with one arm advancing far ahead of the other. In these cells, GFP-SpoIIP is localized unevenly, with a brighter focus at the faster moving region of the membrane (Fig. 6Bi, arrowheads; see also Movie SA2 in the supplemental material). This suggests that higher concentrations of the SpoIID/SpoIIP complex are correlated with increased rates of membrane migration.
FIG. 6.
Time-lapse microscopy of GFP-SpoIIP. Sporulating cells were incubated on agarose pads at 30°C and membranes were stained with FM 4-64. Image sequences were initiated at t2.5 after the initiation of sporulation by resuspension and the first image was set to t = 0 min. Minutes after t = 0 is indicated in lower right of GFP image. The upper panels show FM 4-64 membranes merged with GFP-SpoIIP; the lower panels show GFP-SpoIIP alone. (A) GFP-SpoIIP in wild type (KP1129). GFP-SpoIIP is localized evenly to the leading edges throughout migration and becomes diffuse after the completion of membrane migration. (Bi and ii) Two examples of GFP-SpoIIP in a SpoIIDD210A background (KP1132). Localization corresponds with advancement of the membrane, and a faster membrane arm is indicated by arrowheads (i). GFP-SpoIIP is localized unevenly to the leading edges or diffuse throughout blebs; diffuse GFP-SpoIIP is indicated by arrows (ii).
DISCUSSION
SpoIID peptidoglycan degradation activity is essential for septal thinning and membrane migration.
SpoIID degrades peptidoglycan in vitro (1), and it was recently demonstrated to have a novel biochemical activity, since it is a lytic transglycosylase that cleaves peptidoglycan only after the peptide side chains have been removed by SpoIIP (27). We identify here an amino acid substitution within the SpoIID protein (SpoIIDE88A; also identified by Morlot et al. [27]) that abolishes the peptidoglycan degradation activity (Fig. 4A) and spore formation (Table 1). The membrane morphology of the spoIIDE88A mutant exactly matches that of the spoIID-null strain at t2 and t3 (Fig. 3C and I), showing flat septa and slightly curved septa with bulges. These bulges are thought to form due to the lack of septal thinning because in the absence of SpoIID, SpoIIP is trapped at the center of the septum (Fig. 5H) and does not move. This likely causes a weak spot at the center of the septum where SpoIIP is active, allowing membrane protrusion from the forespore and bulge formation. The identical structure of the membranes in a spoIID-null and spoIIDE88A strain strongly suggests that SpoIID-mediated peptidoglycan degradation is essential for septal thinning, the first step in the engulfment process.
We also identified a second mutant protein (SpoIIDD210A) that degrades peptidoglycan at a much lower level than wild-type SpoIID (Fig. 4A). This mutation is slow to complete septal thinning, showing transient septal bulges, and ultimately initiates membrane migration and completes engulfment at 10% the level of wild type (Table 1 and see Table SA1 in the supplemental material). By examining cells over a time course and by using time-lapse microscopy to visualize membrane migration in living cells, we found that the movement of the membranes around the forespore was much slower in the spoIIDD210A strain than in the wild type (Fig. 6B). This indicates that SpoIID activity is required throughout membrane migration. Together, these results show that SpoIID-mediated peptidoglycan degradation is critical throughout engulfment as predicted by both the burnt-bridge and steric barrier models (7).
We also noted that SpoIIP has an altered localization in a strain expressing SpoIIDD210A. Specifically, the GFP-SpoIIP signal is more mobile and less symmetrically localized to the leading edge than in the wild type, often showing diffuse localization throughout the blebbing membranes or foci behind the leading edge (Fig. 6). This suggests that SpoIIDD210A has a reduced ability to maintain the SpoIID/SpoIIP complex at the leading edge of the engulfing membrane. However, in some mutant cells, GFP-SpoIIP localizes in a manner similar to that of the wild type, and cells exhibiting this pattern show membrane migration, indicating that the correct assembly of the SpoIID/SpoIIP complex is important for rapid membrane migration. The SpoIIDD210A mutant strain also frequently shows uneven membrane migration, and in these sporangia, the brighter SpoIIP focus is associated with the most rapidly advancing leading edge, with the less intense focus on the lagging side (Fig. 6Bi, arrowheads). This suggests that, in this strain, peptidoglycan degradation by the SpoIID/SpoIIP complex is rate limiting for membrane migration.
Interaction with SpoIIP is not important for SpoIID stability.
In many complexes, the absence of one binding partner causes degradation of the other. Importantly, we show that SpoIID is readily detectable on Western blots in the absence of its binding partner SpoIIP (Fig. 4B). This strongly suggests that the altered levels of mutant SpoIID proteins are not simply due to a defect in SpoIIP interactions. In addition, the promoter region of each of the mutant proteins is identical, suggesting that transcription of the SpoIID region is the same for all proteins and that altered levels of protein are due either to translational differences caused by the specific mutations or to protein misfolding and degradation.
The SpoIIDMP module is robust.
Sporulation in B. subtilis is a robust process, with multiple redundant pathways contributing to its completion. For example, the SpoIIQ/SpoIIIAH module is normally dispensable for engulfment but is essential when engulfment is compromised by mutations that decrease the activity of the SpoIIDMP complex (7, 18, 38) or when peptidoglycan synthesis is inhibited with antibiotics (26). In addition, the initial localization of SpoIIDMP by the SpoIIB septal landmark protein can also be achieved by a second protein, SpoIVFA, when SpoIIB is absent (2). Finally, synergistic engulfment defects continue to be discovered that are not readily explained by our current models of sporulation (2, 7, 18), indicating as-yet-undiscovered layers of redundancy. It is not surprising, therefore, to discover that the SpoIID, SpoIIM, and SpoIIP proteins are robust as well. SpoIIDD210A degrades peptidoglycan at a significantly reduced level compared to the wild type (Fig. 4A), and it accumulates at levels below that of the wild type (Fig. 4B), and yet it still supports membrane migration in 40% of all cells and the completion of engulfment in 10% of cells (see Table SA1 in the supplemental material). In addition, a previously published mutant, SpoIIDL8P, accumulates at levels the same as or below that of SpoIIDD210A, and yet it supports wild-type levels of sporulation. Together, this indicates that the level of peptidoglycan degradation required for septal thinning and membrane migration is far below what is normally present in wild-type cells and that the peptidoglycan degradation module can withstand perturbations in both protein levels and protein activity.
Models for the role of SpoIID in engulfment.
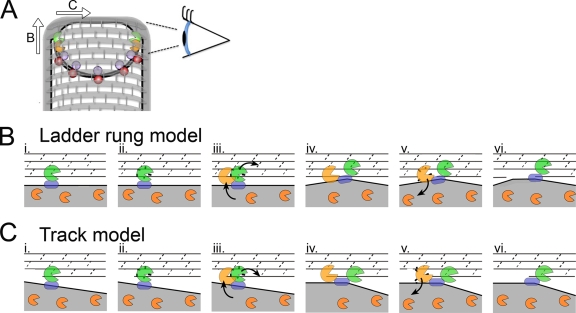
Our localization data suggest that SpoIID degradation activity causes the protein to be released from the SpoIID/SpoIIP complex, since the enzymatically inactive mutant SpoIIDE88A is localized exclusively to the septum, whereas only a fraction of the wild-type protein is localized. This is not the case for wild-type SpoIIP, which is well localized to the septum and leading edge of the engulfing membrane. This suggests that SpoIIP molecules are stably maintained at the leading edge during engulfment, whereas SpoIID moves between a localized pool where it interacts with SpoIIP and cleaves peptidoglycan and a delocalized pool that has little or no peptidoglycan degradation activity (because of the lack of SpoIIP or suitable peptidoglycan substrates). Our data further suggest that movement of SpoIIP across the septum depends on SpoIID and that the inactive SpoIIDE88A mutant protein is capable of moving SpoIIP across the septum. Based on these observations and the biochemical data of Morlot et al. (27), we propose a revised model for how the enzymatic activities of these proteins are coordinated and coupled to membrane migration (Fig. 7). First, we propose that SpoIIP binds to SpoIIM and the peptidoglycan at the septum (Fig. 7B, step 1) and cleaves the peptide chains (step 2), producing denuded glycan strands that are a SpoIID substrate. Second, we propose that SpoIID binds this denuded glycan strand (step 3), displacing SpoIIP to the next available substrate and moving the engulfing membrane forward (step 4). SpoIIP might move either along the same glycan strand, using it as a track (if degradation starts at one end of the strand) or by moving to the next glycan strand, using the glycan as a ladder (if degradation can start within a strand). Third, we propose that SpoIID then cleaves the glycan strand (step 5) and is released back into the delocalized membrane pool (step 6), allowing subsequent cycles of binding, degradation, and movement of the SpoIIP/SpoIID complex around the forespore. Our model also provides a potential mechanistic explanation of the previously observed activation of SpoIIP by SpoIID (27). If SpoIID is indeed required to move SpoIIP to a new peptidoglycan target after cleavage, without this push from SpoIID, SpoIIP would have to rely on slow release from the peptidoglycan and diffusion after cleavage to advance. However, if the two enzymes are added together, the rate at which SpoIIP advances should increase due to displacement of SpoIIP by SpoIID, thereby producing more cleavage products in the same amount of time.
FIG. 7.
Model for SpoIIDMP function in vivo. (A) An engulfing B. subtilis cell with SpoIID (orange), SpoIIP (green), SpoIIQ (purple), and SpoIIIAH (red) indicated. Peptidoglycan is drawn with gray hoops representing glycan strands and short gray rods representing peptide cross-links. The eye orients the reader to the perspective used in panels B and C. As indicated by the arrows, the DMP protein complex can move in one of two directions relative to the long axis of the cell. If the proteins move from one glycan chain to the next (as shown in panel B and proposed by Morlot et al. [27]), each complex would move along the long axis of the cell. If the proteins travel along one glycan chain (as shown in panel C and proposed by Abanes-De Mello et al. [1]), then each complex would move across the short axis of the cell. (B and C) Top-down views of the leading edge of the engulfing mother cell membrane (gray fill) advancing over the forespore peptidoglycan, drawn with gray lines representing the glycan chains and dashed lines representing the peptide cross-links. (B) Ladder rung model, in which SpoIIP moves from one glycan chain to the next. (i) SpoIIP (green) is bound in the membrane by SpoIIM (blue) and to peptidoglycan. SpoIID (orange) is free in the membrane. (ii) SpoIIP cleaves peptide cross-links. (iii) SpoIID is recruited to the recently denuded glycan strands where it displaces SpoIIP to a new target on the next strand, laddering up the peptidoglycan. (iv) Both enzymes are in complex and bound to the peptidoglycan. (v) SpoIID cleaves the glycan strand, loses affinity for either SpoIIP or the peptidoglycan, and exits the complex. (vi) Same as panel i, but membranes have advanced. (C) Track model, similar to panel B, but depicting SpoIIDMP moving along the glycan chain, such that at step iii, SpoIID displaces SpoIIP to the next peptide cross-link on the same glycan strand.
This modified model of SpoIIDMP complex organization suggests that the complex could operate as a burnt-bridge Brownian ratchet, thus providing force for membrane migration (1, 27). However, two additional mechanisms also contribute to membrane migration, the SpoIIQ-SpoIIIAH zipper, which is essential for engulfment in protoplasts and in intact cells when SpoIIDMP activity is reduced (7), and peptidoglycan biosynthesis, which is essential for engulfment in the absence of SpoIIQ (26). The mechanisms by which these three protein complexes contribute to the directionality of engulfment and generate force for membrane movement are important topics for future research.
Supplementary Material
Acknowledgments
This research was supported by grants from the National Institutes of Health (R01-GM57045 and F32-GM087864).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the National Institute of General Medical Sciences.
We thank Richard Losick for strains with GFP-SpoIIP at the native locus that were used to create some of the strains for this study, Stefan Aung and Jon Shum for the creation of some expression plasmids, and James Gregory for the creation of some mutagenesis plasmids.
Footnotes
Published ahead of print on 9 April 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abanes-De Mello, A., Y. L. Sun, S. Aung, and K. Pogliano. 2002. A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes Dev. 16:3253-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aung, S., J. Shum, A. Abanes-De Mello, D. H. Broder, J. Fredlund-Gutierrez, S. Chiba, and K. Pogliano. 2007. Dual localization pathways for the engulfment proteins during Bacillus subtilis sporulation. Mol. Microbiol. 65:1534-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baens, M., H. Noels, V. Broeckx, S. Hagens, S. Fevery, A. D. Billiau, H. Vankelecom, and P. Marynen. 2006. The dark side of EGFP: defective polyubiquitination. PLoS One 1:e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, E. C., and K. Pogliano. 2007. Cell-specific SpoIIIE assembly and DNA translocation polarity are dictated by chromosome orientation. Mol. Microbiol. 66:1066-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackman, S. A., T. J. Smith, and S. J. Foster. 1998. The role of autolysins during vegetative growth of Bacillus subtilis 168. Microbiology 144 (Pt. 1):73-82. [DOI] [PubMed] [Google Scholar]
- 6.Blaylock, B., X. Jiang, A. Rubio, C. P. Moran, Jr., and K. Pogliano. 2004. Zipper-like interaction between proteins in adjacent daughter cells mediates protein localization. Genes Dev. 18:2916-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broder, D. H., and K. Pogliano. 2006. Forespore engulfment mediated by a ratchet-like mechanism. Cell 126:917-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chastanet, A., and R. Losick. 2007. Engulfment during sporulation in Bacillus subtilis is governed by a multi-protein complex containing tandemly acting autolysins. Mol. Microbiol. 64:139-152. [DOI] [PubMed] [Google Scholar]
- 9.Chiba, S., K. Coleman, and K. Pogliano. 2007. Impact of membrane fusion and proteolysis on SpoIIQ dynamics and interaction with SpoIIIAH. J. Biol. Chem. 282:2576-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doan, T., K. A. Marquis, and D. Z. Rudner. 2005. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol. Microbiol. 55:1767-1781. [DOI] [PubMed] [Google Scholar]
- 11.Drew, D., M. Lerch, E. Kunji, D. J. Slotboom, and J. W. de Gier. 2006. Optimization of membrane protein overexpression and purification using GFP fusions. Nat. Methods 3:303-313. [DOI] [PubMed] [Google Scholar]
- 12.Dubnau, D., and R. Davidoff-Abelson. 1971. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J. Mol. Biol. 56:209-221. [DOI] [PubMed] [Google Scholar]
- 13.Errington, J. 2003. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 1:117-126. [DOI] [PubMed] [Google Scholar]
- 14.Finn, R. D., J. Tate, J. Mistry, P. C. Coggill, S. J. Sammut, H. R. Hotz, G. Ceric, K. Forslund, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2008. The Pfam protein families database. Nucleic Acids Res. 36:D281-D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster, S. J. 1992. Analysis of the autolysins of Bacillus subtilis 168 during vegetative growth and differentiation by using renaturing polyacrylamide gel electrophoresis. J. Bacteriol. 174:464-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frandsen, N., and P. Stragier. 1995. Identification and characterization of the Bacillus subtilis spoIIP locus. J. Bacteriol. 177:716-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hilbert, D. W., and P. J. Piggot. 2004. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol. Mol. Biol. Rev. 68:234-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang, X., A. Rubio, S. Chiba, and K. Pogliano. 2005. Engulfment-regulated proteolysis of SpoIIQ: evidence that dual checkpoints control σK activity. Mol. Microbiol. 58:102-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kellner, E. M., A. Decatur, and C. P. Moran, Jr. 1996. Two-stage regulation of an anti-sigma factor determines developmental fate during bacterial endospore formation. Mol. Microbiol. 21:913-924. [DOI] [PubMed] [Google Scholar]
- 20.Kuroda, A., M. H. Rashid, and J. Sekiguchi. 1992. Molecular cloning and sequencing of the upstream region of the major Bacillus subtilis autolysin gene: a modifier protein exhibiting sequence homology to the major autolysin and the spoIID product. J. Gen. Microbiol. 138(Pt. 6):1067-1076. [DOI] [PubMed] [Google Scholar]
- 21.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. CLUSTAL W and CLUSTAL X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 22.Liu, N. J., R. J. Dutton, and K. Pogliano. 2006. Evidence that the SpoIIIE DNA translocase participates in membrane fusion during cytokinesis and engulfment. Mol. Microbiol. 59:1097-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Londono-Vallejo, J. A., C. Frehel, and P. Stragier. 1997. SpoIIQ, a forespore-expressed gene required for engulfment in Bacillus subtilis. Mol. Microbiol. 24:29-39. [DOI] [PubMed] [Google Scholar]
- 24.Lopez-Diaz, I., S. Clarke, and J. Mandelstam. 1986. spoIID operon of Bacillus subtilis: cloning and sequence. J. Gen. Microbiol. 132(Pt. 2):341-354. [DOI] [PubMed] [Google Scholar]
- 25.Margolis, P. S., A. Driks, and R. Losick. 1993. Sporulation gene spoIIB from Bacillus subtilis. J. Bacteriol. 175:528-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer, P., J. Gutierrez, K. Pogliano, and J. Dworkin. 1 April 2010. Cell wall synthesis is necessary for membrane dynamics during sporulation of Bacillus subtilis. Mol. Microbiol. [Epub ahead of print.] doi: 10.1111/j.1365-2958.2010.07155.x. [DOI] [PMC free article] [PubMed]
- 27.Morlot, C., T. Uehara, K. A. Marquis, T. G. Bernhardt, and D. Z. Rudner. 2010. A highly coordinated cell wall degradation machine governs spore morphogenesis in Bacillus subtilis. Genes Dev. 24:411-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez, A. R., A. Abanes-De Mello, and K. Pogliano. 2000. SpoIIB localizes to active sites of septal biogenesis and spatially regulates septal thinning during engulfment in Bacillus subtilis. J. Bacteriol. 182:1096-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez, A. R., A. Abanes-De Mello, and K. Pogliano. 2006. Suppression of engulfment defects in Bacillus subtilis by elevated expression of the motility regulon. J. Bacteriol. 188:1159-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawano, A., and A. Miyawaki. 2000. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 28:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaeffer, P., J. Millet, and J. Aubert. 1965. Catabolite repression of bacterial sporulation. Proc. Natl. Acad. Sci. U. S. A. 54:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharp, M. D., and K. Pogliano. 1999. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc. Natl. Acad. Sci. U. S. A. 96:14553-14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharp, M. D., and K. Pogliano. 2002. Role of cell-specific SpoIIIE assembly in polarity of DNA transfer. Science 295:137-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shida, T., H. Hattori, F. Ise, and J. Sekiguchi. 2001. Mutational analysis of catalytic sites of the cell wall lytic N-acetylmuramoyl-l-alanine amidases CwlC and CwlV. J. Biol. Chem. 276:28140-28146. [DOI] [PubMed] [Google Scholar]
- 35.Smith, K., M. E. Bayer, and P. Youngman. 1993. Physical and functional characterization of the Bacillus subtilis spoIIM gene. J. Bacteriol. 175:3607-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sterlini, J. M., and J. Mandelstam. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 113:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugai, M., T. Akiyama, H. Komatsuzawa, Y. Miyake, and H. Suginaka. 1990. Characterization of sodium dodecyl sulfate-stable Staphylococcus aureus bacteriolytic enzymes by polyacrylamide gel electrophoresis. J. Bacteriol. 172:6494-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun, Y. L., M. D. Sharp, and K. Pogliano. 2000. A dispensable role for forespore-specific gene expression in engulfment of the forespore during sporulation of Bacillus subtilis. J. Bacteriol. 182:2919-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Youngman, P., J. B. Perkins, and R. Losick. 1984. A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertions. Mol. Gen. Genet. 195:424-433. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.