Abstract
Most effector proteins of bacterial type III secretion (T3S) systems require chaperone proteins for translocation into host cells. Such effectors are bound by chaperones in a conserved and characteristic manner, with the chaperone-binding (Cb) region of the effector wound around the chaperone in a highly extended conformation. This conformation has been suggested to serve as a translocation signal in promoting the association between the chaperone-effector complex and a bacterial component required for translocation. We sought to test a prediction of this model by identifying a potential association site for the Yersinia pseudotuberculosis chaperone-effector pair SycE-YopE. We identified a set of residues in the YopE Cb region that are required for translocation but are dispensable for expression, SycE binding, secretion into the extrabacterial milieu, and stability in mammalian cells. These residues form a solvent-exposed patch on the surface of the chaperone-bound Cb region, and thus their effect on translocation is consistent with the structure of the chaperone-bound Cb region serving as a signal for translocation.
The type III secretion (T3S) system is crucial to the virulence of many Gram-negative bacterial pathogens (14, 18). These pathogens use the T3S system to translocate effector proteins from the bacterial cytosol directly into the interior of host cells. Typically, effectors contribute to the virulence of the pathogen by modifying or interacting with specific host cell targets. Effector proteins are arranged in a modular fashion, with sequence elements required for translocation located within their N-terminal ∼100 to 150 residues and with domains that interact with host components following (20, 49). For most effectors, including the extensively characterized Yersinia effector YopE (23 kDa), two different N-terminal sequence elements are required for translocation into host cells. The first, termed signal 1 (S1), occurs at the very N terminus and spans ∼10 to 15 residues (Fig. 1A). The S1 element is highly degenerate in sequence (29, 36), and in YopE the S1 region has been shown to be structurally disordered (37). While the S1 element is sufficient for the nonphysiological process of secretion of effectors into the extrabacterial milieu, it is not sufficient for translocation into host cells (41, 44).
FIG. 1.
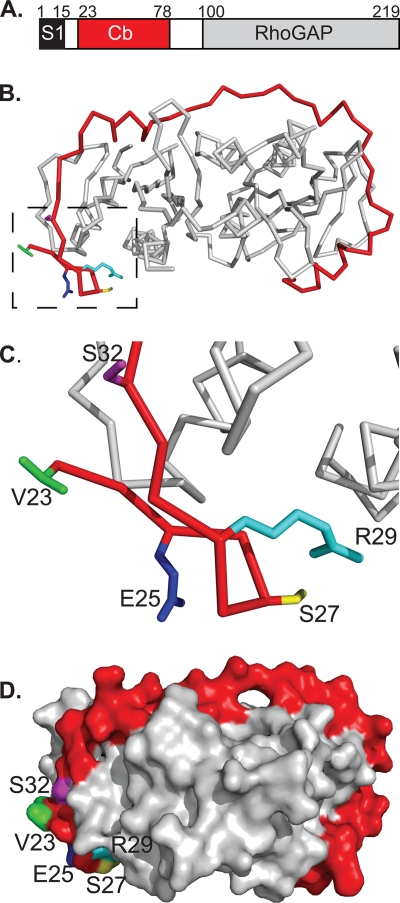
The SycE-YopE chaperone-effector complex. (A) Schematic of YopE domains. S1, signal 1; Cb, chaperone-binding region; RhoGAP, Rho GTPase activating protein domain. Residue numbers for domain boundaries are indicated. (B) Structure of the SycE-YopE(Cb) complex. The Cb region of YopE (red) and the SycE dimer (gray) are shown in C-α stick representation. Side chains that were mutated in the YopE Cb region are depicted for the following residues: V23 (green), E25 (blue), S27 (yellow), R29 (cyan), and S32 (magenta). Molecular figures were made with PyMol (http://pymol.sourceforge.net). (C) Enlarged view of boxed region in panel B. (D) Molecular surface representation of the SycE-YopE(Cb) complex. The YopE(Cb) surface is shown in red, except for surfaces formed by V23 (green), E25 (blue), S27 (yellow), R29 (cyan), and S32 (magenta). The SycE homodimer is shown in gray.
For translocation, a second sequence element, termed the chaperone-binding (Cb) region, is required (Fig. 1A) (44). The Cb region, which consists of ∼50 to 100 residues downstream of S1, promotes translocation only when bound by a member of the effector-dedicated chaperone protein family (49). The chaperone protein is dissociated from the effector by a T3S ATPase (2), as shown in Salmonella, and remains in the bacterial cytosol upon transport of the effector (17). There are a large number of such effector-dedicated chaperones, as each individual chaperone protein binds just a single effector, or in some cases a few effectors. These chaperones are divergent in sequence (≤20% identity) but have similar protein folds, dimeric oligomerization states, and effector-binding modes (4, 5, 10, 28, 30, 31, 35, 42, 43, 46, 47). In the binding mode, the Cb region of the effector winds around the surface of the dimeric chaperone in a highly extended fashion (Fig. 1B and C). The chaperone-bound Cb region lacks independent tertiary structure but has short α-helical, β-strand, and random coil stretches that contact the chaperone. This mode has been observed for the Cb region of YopE (residues 23 to 78) bound to its homodimeric chaperone, SycE (29 kDa per dimer) (5), as well as for a number of other chaperone-effector complexes (28, 35, 42, 46). The structural conservation among these chaperone-effector pairs is especially striking because the various effectors have no obvious sequence relationship to one another in their Cb regions. However, recent work showed that the chaperone-Cb-region binding mode can be predicted from de novo models (22).
The functional significance of the conserved chaperone-Cb-region structure is not yet clear, but this structure has been suggested to constitute a translocation signal (5). It has been proposed that the conformation of the chaperone-bound Cb region promotes association between the chaperone-effector complex and a bacterial component required for translocation, i.e., a receptor that recognizes this three-dimensional translocation signal. Consistent with this hypothesis, recent evidence indicated that the chaperone SycE brings about the structuring of an otherwise unstructured Cb region in YopE (37). Nuclear magnetic resonance studies demonstrated that the Cb region of YopE in its free state is unstructured and flexible and that it undergoes a pronounced disorder-to-order transition upon SycE binding. The effect of SycE was strictly localized to the Cb region, while other portions of YopE, including the S1 region and the C-terminal RhoGAP domain, were impervious to SycE binding. Additional lines of evidence also support the translocation signal model of chaperone action (8, 13, 50).
We sought here to test a prediction of the translocation signal model. This is the prediction that the surface of the chaperone-Cb-region complex ought to provide a receptor-binding site. We surmised that mutation of such a solvent-exposed site in the Cb region of YopE would affect translocation but not chaperone binding. Mutations of residues in the Cb region that contact the chaperone have already been shown to reduce translocation (5, 28, 41). These are simple to explain, as they diminish the affinity of the effector for the chaperone. In contrast, residues in the Cb region that are exposed in the chaperone-Cb-region complex, and hence available to form a receptor-binding site, have not yet been shown to be important for translocation. We now report the identification of such residues in the YopE Cb region. These residues are crucial for translocation but not for other aspects of YopE function, including steady-state expression, binding to SycE, secretion, and stability in mammalian cells. These results are consistent with a translocation signal model of action for the chaperone-bound Cb region and identify a potential receptor-binding site.
MATERIALS AND METHODS
Expression and purification of SycEΔ122-YopE(Cb) constructs.
SycEΔ122 (residues 1 to 121) and wild-type, 3Ala (V23A/E25A/S32A mutant), and 5Ala (V23A/E25A/S27A/R29A/S32A mutant) YopE(Cb) (residues 1 to 80) constructs were expressed in inducible fashion in Escherichia coli BL21(DE3) by use of a bicistronic vector derived from pET28b (Novagen). Bacteria were induced for protein expression at the mid-log growth phase by use of 1 mM isopropyl β-d-thiogalactopyranoside (IPTG) and were grown further at room temperature for 3 h. Bacteria were harvested by centrifugation, resuspended in 150 mM NaCl, 50 mM phosphate buffer (pH 8.0), and 10 mM β-mercaptoethanol (βME), and lysed by sonication. Polyethyleneimine was added to the lysate to a final concentration of 0.5%, and the solution was centrifuged. The supernatant, which contained SycEΔ122-YopE(Cb), was then further precipitated with 50% saturated (NH4)2SO4. The resulting pellet, which contained SycEΔ122-YopE(Cb), was resuspended and dialyzed in 20 mM NaCl, 30 mM Tris, pH 8.0, and 10 mM βME. SycEΔ122-YopE(Cb) was purified by anion-exchange (Poros HQ/M), cation-exchange (Poros HS/M), and size exclusion (Superdex 75) chromatography.
SycE-His (residues 1 to 130), which contains a C-terminal His tag, was expressed in E. coli BL21(DE3) by use of pET28b (Novagen) as described above and was purified as described previously for His-tagged SycE-YopE complexes (37).
Guanidine denaturation.
Wild-type, 3Ala, and 5Ala SycEΔ122-YopE(Cb) constructs were concentrated to 1.55 μM in 100 mM NaCl and 10 mM phosphate buffer, pH 8.0. Likewise, SycE-His was concentrated to 0.775 μM (dimers) in the same buffer. Proteins were denatured through stepwise, automated titration (Hamilton Microlab 500 titrator) with a solution consisting of 5.7 M guanidine, 100 mM NaCl, 10 mM phosphate buffer, pH 8.0, and either 1.55 μM SycEΔ122-YopE(Cb) (in the case of denaturation of this complex) or 0.775 μM SycE-His (in the case of SycE-His denaturation) in order to maintain a constant protein concentration. The concentration of guanidine was checked by refractometry. Protein samples were stirred for 10 min between titration steps (to reach equilibrium, as determined empirically), and the circular dichroism signal at 222 nm was monitored for 15 s on an Aviv 202 circular dichroism spectrometer. Denaturation data were fitted to a two-state folding model, with the pre- and posttransition baselines treated to have linear dependence on the concentration of denaturant, as described previously (16).
Strain and plasmid construction.
The serogroup III Yersinia pseudotuberculosis strain 126 (a gift from James Bliska) has been described previously (6), as have Y. pseudotuberculosis 126 ΔyopB (34) and Y. pseudotuberculosis 71 (7). A strain derived from Y. pseudotuberculosis 126 in which the coding sequence for yopE was precisely replaced by the coding sequence for aminoglycoside 3′-phosphotransferase (Kanr) was constructed by standard allelic exchange methods, using the suicide vector pSB890 (34), and bacterial mating with E. coli S17-1 λpir. The site of integration was verified by DNA sequencing.
Expression of wild-type YopE, YopE-3Ala, and YopE-5Ala in Y. pseudotuberculosis 126 ΔyopE was achieved with a 2.9-kb low-copy-number plasmid derived from pKK223-3 which carries the ampicillin resistance gene and the tac promoter (9).
For expression of wild-type YopE, YopE-3Ala, and YopE-5Ala in HeLa cells, pcDNA3.1+ (Invitrogen) was used.
Y. pseudotuberculosis expression of YopE.
Strains of Y. pseudotuberculosis were grown overnight at 26°C in Luria broth (LB) or brain heart infusion (BHI) medium with appropriate antibiotics (30 μg/ml kanamycin or 30 μg/ml kanamycin plus 30 μg/ml carbenicillin). To examine expression of YopE under nonsecreting conditions, cultures were diluted in LB or BHI medium containing 2.5 mM CaCl2 and appropriate antibiotics to a final A600 of 0.1 and grown for 1 h at 26°C, at which time IPTG was added to a final concentration of 1 mM; cultures were then grown for a further 2 h at 37°C. After this time, the quantities of cells in the samples were set to equivalent levels by diluting the cultures to a final A600 of 0.3, and bacterial cells were harvested by centrifugation (10 min, 4°C, 4,000 × g). Cell pellets were lysed by the addition of 5 mg/ml lysozyme and 5 mg/ml DNase and by being resuspended and boiled in SDS-PAGE sample buffer. Samples were analyzed by Western blotting as described below.
Y. pseudotuberculosis secretion of YopE.
Strains of Y. pseudotuberculosis were cultured by shaking overnight at 26°C in 3 ml LB or BHI medium with appropriate antibiotics. Bacterial cultures were diluted in LB or BHI medium containing 10 mM EGTA, 10 mM MgCl2, and appropriate antibiotics to a final A600 of 0.1. Cultures were grown with shaking for 1 h at 26°C. IPTG was added to a final concentration of 1 mM, and cultures were grown with shaking for 2 h at 37°C or for a further hour at 26°C followed by 3 h at 37°C. Both protocols produced similar results. Following this period, cells were separated from the culture medium by centrifugation (10 min, 4°C, 4,000 × g), and the culture medium was boiled with SDS-PAGE sample buffer for Western blot analysis. The cell pellets were lysed as described above and were resuspended and boiled in SDS-PAGE sample buffer for Western blot analysis as well. As a negative control, bacterial strains were diluted in BHI medium containing 2.5 mM CaCl2 after overnight growth.
Western blotting.
Samples were resolved by 12% SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane (Amersham). Membranes were blocked with 5% milk in TBS (50 mM Tris, pH 8.0, 150 mM NaCl) or TBS containing 0.1% Tween (TBST) for 1 h at 26°C or 12 h at 4°C. Primary antibody (anti-YopE polyclonal antibodies were a gift from J. Bliska, anti-SycE polyclonal antibodies were described previously [37], anti-DnaK monoclonal antibodies were from Abcam, and anti-α-tubulin monoclonal antibodies were from Santa Cruz Biotechnology) diluted in 5% bovine serum albumin (BSA) (in TBS) or 5% milk (in TBST) was added and incubated for 50 min at 26°C or for 12 h at 4°C. Membranes were washed 4 to 8 times with 20 to 30 ml TBS or TBST (3 to 5 min per wash), and 20 to 25 ml of secondary antibody (horseradish peroxidase [HRP]-conjugated rabbit anti-mouse; Santa Cruz) was added and incubated for 30 to 45 min at 26°C. Membranes were washed 4 to 8 times with 20 to 30 ml TBS or TBST (3 to 5 min per wash), and immunodetection was carried out on X-ray film with ECL Plus (Amersham) according to the manufacturer's instructions. Densitometry of X-ray film was performed using ImageJ. Quantifications were ascertained to be within the linear range of measurement.
In cases in which membranes were stripped and reprobed, a stripping solution consisting of 67% guanidine hydrochloride, 50 μM EDTA, 50 mM glycine, pH 10.8, 2.5 mM KCl, and 1.4 mM βME was incubated with the membrane for 10 min. The membrane was then washed once with H2O and then with TBS for 5 min. Primary and secondary antibodies were added as described above.
Translocation assay.
HeLa cells were routinely cultured in Dulbecco's modified Eagle's medium (DMEM; Cellgro) supplemented with 10% fetal bovine serum (FBS), glutamine, streptomycin, and penicillin in a 5% CO2 humidified incubator at 37°C. HeLa cells were split 18 h prior to infection and were plated at a density of 2 × 106 cells per 10-cm dish. Two hours prior to infection, adherent cells were washed twice with phosphate-buffered saline (PBS) at 37°C and incubated in unsupplemented DMEM (i.e., DMEM without FBS, penicillin, streptomycin, or glutamine). The proteasome inhibitor MG-132 (Calbiochem) and the actin polymerization inhibitor cytochalasin D (Biomol) were added to final concentrations of 5 μM and 1 μg/ml, respectively. HeLa cells were then incubated for 2 h at 37°C and 5% CO2.
Strains of Y. pseudotuberculosis were grown overnight with shaking at 26°C in LB supplemented with appropriate antibiotics and then diluted to an A600 of 0.1 in LB containing 2.5 mM CaCl2 and appropriate antibiotics. After 1 h at 26°C, IPTG was added to a final concentration of 1 mM, and the cultures were then grown with shaking for an additional 2 h at 37°C. One milliliter of bacterial culture was centrifuged (1 min, 6,000 × g), and the pellet was resuspended to an A600 of 1.0 in DMEM supplemented with 5 μM MG-132 and 1 μg/ml cytochalasin D. Bacteria (90 μl) were added to HeLa cells to produce a multiplicity of infection of 50:1 (1 × 108 bacteria:2 × 106 HeLa cells).
The infection was allowed to proceed for 150 min at 37°C and 5% CO2. Nonadherent cells were removed with three 15-ml washes of cold PBS, and following the aspiration of the last wash, papain was added to digest extracellular proteins (150 μl from a 0.25-mg/ml papain stock in PBS with 2 mM dithiothreitol). After 30 s, 140 μl of the papain solution was removed, leaving a thin layer of papain coating the HeLa cell monolayer. This digestion proceeded for 20 min at 26°C and was stopped by the addition of E-64 (90 μl of a 10-μg/ml solution). The cells were incubated in E-64 for 5 min before the addition of 400 μl lysis buffer (1% Triton X-100, 10% glycerol, 150 mM NaCl, 10 mM Tris, pH 8.0), which remained on the cells for 5 min. Cell lysates were collected by scraping and then clarified by centrifugation (10 min, 16,000 × g, 4°C). The clarified lysate was passed through a 0.22-μm-cutoff filter to remove intact cells and debris. The total protein concentration of the lysate was quantified by a Lowry assay (DC protein detection kit; Bio-Rad). Lysate volumes were adjusted with lysis buffer to make protein concentrations equivalent among samples. Typical concentrations were 1 to 2 mg/ml, determined by comparison to a BSA standard. SDS-PAGE sample buffer (4×) was added, and samples were boiled for 10 min and centrifuged (1 min, 16,000 × g, 26°C). Samples were analyzed by Western blotting.
Transfection.
HeLa cells were cultured as described above and plated at a density of 1.5 × 105 cells per 9.6-cm2 well 18 h prior to transfection. Thirty minutes prior to transfection, adherent cells were washed twice with PBS (37°C) and incubated in 1.9 ml unsupplemented DMEM. Two micrograms of the expression vector pcDNA3.1+ expressing YopE, YopE-3Ala, or YopE-5Ala was diluted to 95 μl with unsupplemented DMEM, and 5 μl Fugene HD (Roche) was added. The resulting 100-μl solution was added to each well, and cells were incubated for 5 h at 37°C and 5% CO2. The medium was then removed by aspiration and replaced with DMEM supplemented with 10% FBS, glutamine, streptomycin, and penicillin. Cells were incubated for an additional 22 h (37°C, 5% CO2), and the culture medium was removed by aspiration. Cells were lysed by the addition of 200 μl lysis buffer, which remained on the cells for 5 min. Cell lysates were collected by scraping, and SDS-PAGE sample buffer (4×) was added to the samples prior to analysis by SDS-PAGE and Western blotting.
RESULTS
We targeted residues in the YopE Cb region for alanine substitution, choosing residues that are solvent exposed when the Cb region is bound to SycE (5). Based on previously noted similarities between the Cb regions of YopE and the Salmonella effector SptP (5), we focused on YopE Val23, Glu25, and Ser32. These residues were replaced with alanine, and the triple-alanine substitution mutant (V23A/E25A/S32A) was designated YopE-3Ala. The side chains of these residues do not contact SycE (5), and thus their substitution was not expected to affect binding to SycE. The surface patch defined by these three residues is neighbored by Ser27 and Arg29 (Fig. 1D), which make polar contacts with SycE but also have solvent-exposed portions. Because polar interactions usually contribute more to specificity than to affinity, we replaced Ser27 and Arg29 with alanine along with Val23, Glu25, and Ser32 to increase the chances of disrupting a potential receptor-binding site. The quintuple-alanine substitution mutant (V23A/E25A/S27A/R29A/S32A) was designated YopE-5A.
Affinity for SycE.
The affinities of the YopE-3Ala and YopE-5Ala alanine substitution mutants for SycE were determined and compared to that of wild-type YopE. To carry this out, complexes containing SycE dimers and either a wild-type or mutant version of a YopE Cb fragment (i.e., Cb-3Ala or Cb-5Ala) were coexpressed in E. coli and purified. The Cb fragment (residues 1 to 80) was used in lieu of intact YopE to simplify the analysis. The Cb region is the only portion of YopE that interacts with SycE, and this interaction is independent of other portions of YopE (5, 37). The version of SycE used in this experiment, called SycEΔ122, was originally designed for structure determination and lacks the last eight residues. These last eight residues are flexible and do not contact YopE (5).
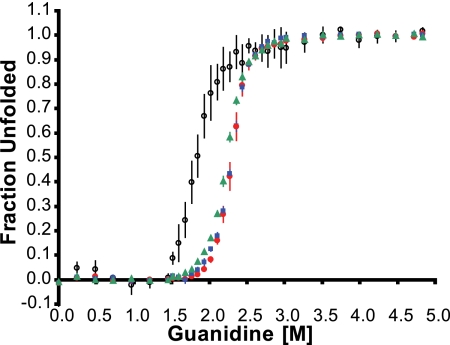
The purified complexes were subjected to reversible unfolding with guanidine, and the progress of unfolding was monitored by circular dichroism analysis at 222 nm. As shown previously (5), the midpoint concentration for unfolding of free SycE was lower than that for SycE-YopE(Cb) (Fig. 2). This difference reflects the binding energy derived from the association between SycE and YopE and is thus a measure of the binding affinity between SycE and YopE. A mutation that results in a decrease in the SycE-YopE affinity would be manifested as a decrease in the guanidine concentration required for unfolding (compared to the guanidine concentration required for unfolding of wild-type SycE-YopE). As predicted from structural grounds, the triple-alanine substitution in YopE had no effect on SycE-YopE affinity. SycEΔ122-YopE(Cb-3Ala) had a midpoint concentration for unfolding of 2.30 ± 0.01 M guanidine (mean ± standard error of the mean), which is identical to the 2.30 ± 0.02 M guanidine midpoint for wild-type SycEΔ122-YopE(Cb) (Fig. 2). For comparison, SycEΔ122-YopE(Cb-5Ala) had a slightly lower midpoint concentration for unfolding of 2.23 ± 0.01 M guanidine, indicative of a small loss in affinity for SycE (Fig. 2). These results indicate that the combined alanine substitutions of YopE Val23, Glu25, and Ser32 had no influence on the affinity of the Cb region for SycE and that the additional alanine substitutions of Ser27 and Arg29 had only a slight effect.
FIG. 2.
Affinity of YopE Cb region for SycE. The dependence of the fraction of unfolded protein on the guanidine concentration was determined by the circular dichroism signal at 222 nm. Red circles, wild-type SycEΔ122-YopE(Cb); blue squares, SycEΔ122-YopE(Cb-3Ala); green triangles, SycEΔ122-YopE(Cb-5Ala); empty circles, SycE-His. Data were averaged from at least three independent experiments, and bars represent standard errors of the means.
Expression and secretion by Y. pseudotuberculosis.
We next asked whether the alanine substitutions affected expression and secretion of YopE by Y. pseudotuberculosis. Wild-type YopE, YopE-3Ala, and YopE-5Ala were expressed under the inducible control of the tac promoter from a low-copy-number plasmid (9) in Y. pseudotuberculosis 126 ΔyopE. In this ΔyopE strain, the entire coding sequence for yopE was replaced by that of aminoglycoside 3′-phosphotransferase (Kanr), and the loss of YopE expression was verified by Western blotting. Strain 126, strain 126 ΔyopE, and the 126 ΔyopE strains expressing plasmid-encoded wild-type YopE, YopE-3Ala, and YopE-5Ala all had equivalent growth rates (data not shown).
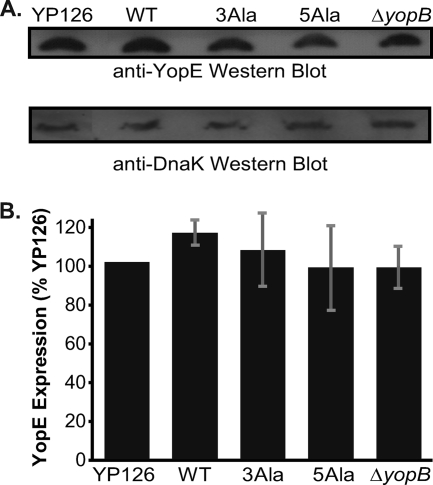
The steady-state expression level of plasmid-encoded YopE was compared to that of YopE expressed from its native promoter on the pIB plasmid of Y. pseudotuberculosis 126 (34). These experiments were carried out under nonsecreting conditions through the inclusion of Ca2+ in the growth medium at 37°C. A Western blot showed that plasmid-encoded, wild-type YopE was produced at a level equivalent to that of natively encoded YopE (Fig. 3A). This steady-state expression level was also equivalent to those of YopE-3Ala and YopE-5Ala. The number of cells analyzed for these experiments was normalized, as verified by detecting DnaK on the same blot. Quantification of Western blots from multiple independent experiments showed that the steady-state levels of YopE expression were statistically indistinguishable among these strains (Fig. 3B) (P value = 0.85 by analysis of variance [ANOVA]). These results are consistent with the observation that the YopE-3Ala and YopE-5Ala substitution mutants had the same or nearly the same affinity for SycE as wild-type YopE. Loss of binding to SycE has previously been demonstrated to lower the level of YopE due to degradation (12, 13, 17, 48).
FIG. 3.
Expression of YopE in Y. pseudotuberculosis. (A) (Top) Western blot of steady-state expression level of YopE in Y. pseudotuberculosis grown at 37°C under nonsecreting conditions. YopE was detected with anti-YopE polyclonal antibodies. (Bottom) The number of cells analyzed was normalized, and as a loading control, DnaK was detected on the same blot with a monoclonal anti-DnaK antibody. YP126 is wild-type YopE expressed from its native locus on the pIB plasmid by Y. pseudotuberculosis 126 (YP126). WT, 3Ala, and 5Ala are wild-type YopE, YopE-3Ala, and YopE-5Ala, respectively, expressed from a low-copy-number plasmid by Y. pseudotuberculosis 126 ΔyopE. The last sample, ΔyopB, is wild-type YopE expressed from its native locus on the pIB plasmid by Y. pseudotuberculosis 126 ΔyopB. (B) Quantification of YopE expression levels under nonsecreting conditions, assessed by Western blotting. Mean values from four independent experiments carried out in triplicate are depicted. These values were normalized to the expression level of YP126 and are expressed as percentages of the YP126 value. Gray bars represent standard errors of the means.
Type III secretion of alanine substitution mutants.
We next asked whether the alanine substitutions had an effect on the secretion of YopE. The process of type III secretion into the extrabacterial milieu in the absence of host cells has similarities to translocation, but the requirements for the two processes differ. For example, while SycE is required for translocation of YopE, it is not required for secretion (12, 13). A SycE-deficient strain contains ∼2-fold lower steady-state levels of YopE than those in a wild-type strain, but the SycE-deficient strain secretes YopE at the same steady-state efficiency as the wild-type strain (12). In contrast, the SycE-deficient strain is entirely incapable of translocating YopE into HeLa cells (13).
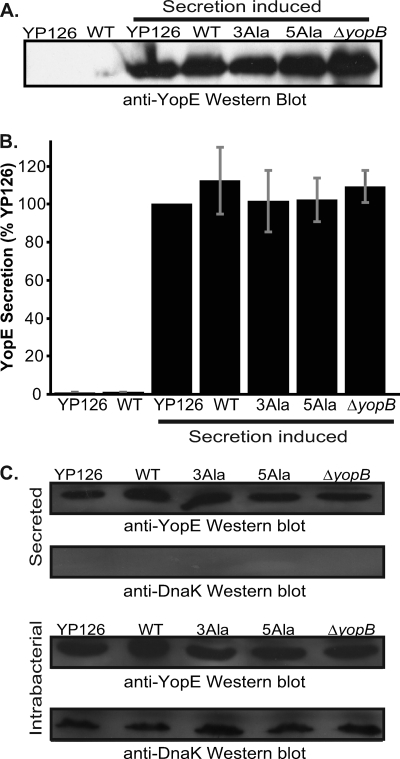
Type III secretion was induced by growing Y. pseudotuberculosis in low-calcium medium at 37°C, and secretion of YopE was monitored by Western blotting of culture supernatants. YopE secretion occurred only when calcium was absent from the medium, consistent with the secretion being type III (Fig. 4A). The levels of secreted wild-type YopE were similar regardless of whether YopE was expressed from its native locus on the pIB plasmid or from the low-copy-number plasmid, as determined from quantification of Western blots (Fig. 4B). Most importantly, this experiment showed that the levels of secreted YopE-3Ala and YopE-5Ala were indistinguishable from that of wild-type YopE (Fig. 4A and B).
FIG. 4.
Secretion of YopE. (A) Western blot of wild-type YopE secreted by Y. pseudotuberculosis 126 (YP126); wild-type YopE (WT), YopE-3Ala (3Ala), and YopE-5Ala (5Ala) secreted by Y. pseudotuberculosis 126 ΔyopE; and wild-type YopE secreted by Y. pseudotuberculosis 126 ΔyopB. Type III secretion was inhibited for the samples in the two left lanes by growth at 37°C in 2.5 mM CaCl2, while type III secretion was induced in the remaining samples by the addition of 10 mM EGTA at 37°C. Supernatants from Y. pseudotuberculosis cultures were separated by SDS-PAGE and probed with anti-YopE polyclonal antibodies. (B) Quantification of YopE secretion, assessed by Western blotting. Mean values from three independent experiments carried out in triplicate are depicted. These values were normalized to the secretion level of YP126 under T3S-inducing conditions and are expressed as percentages of the YP126 value. Gray bars report standard errors of the means. (C) Western blots of supernatants (top two blots) and cell pellets (bottom two blots) from Y. pseudotuberculosis strains induced to secrete due to a shift to a low Ca2+ concentration and growth at 37°C. Bacterial cultures were centrifuged to separate supernatant from pellet fractions. The first panel in each set shows YopE, detected by anti-YopE polyclonal antibodies, and the bottom panel in each set shows DnaK, detected on the same membrane by an anti-DnaK monoclonal antibody.
We verified that equivalent numbers of bacterial cells were examined in these experiments, as shown by immunodetection of DnaK from the intrabacterial pool (Fig. 4C). As expected from the equivalent steady-state expression levels of YopE, the levels of intrabacterial YopE were similar among these strains under secreting conditions (Fig. 4C). Furthermore, DnaK was present only in the intrabacterial fraction and was not detected in the secreted fraction, verifying proper separation of secreted from intrabacterial pools.
In summary, these experiments demonstrated that the five residues in the Cb region targeted for mutagenesis are not consequential for secretion.
Translocation of alanine substitution mutants.
Having established that the alanine substitutions in YopE had no or little effect on the affinity for SycE and that the expression and secretion of YopE were not altered, we turned to examining translocation. Y. pseudotuberculosis strains expressing either wild-type or alanine-substituted YopE were incubated with HeLa cells at a multiplicity of infection of 50:1. Prior to and during infection, HeLa cells were treated with the proteasome inhibitor MG-132 to prevent ubiquitin-mediated YopE degradation (38). HeLa cells were also treated with the actin polymerization inhibitor cytochalasin D to prevent invasion of HeLa cells and to ensure that actin disruption, a downstream effect of YopE action, was equivalent in all infections (32, 39).
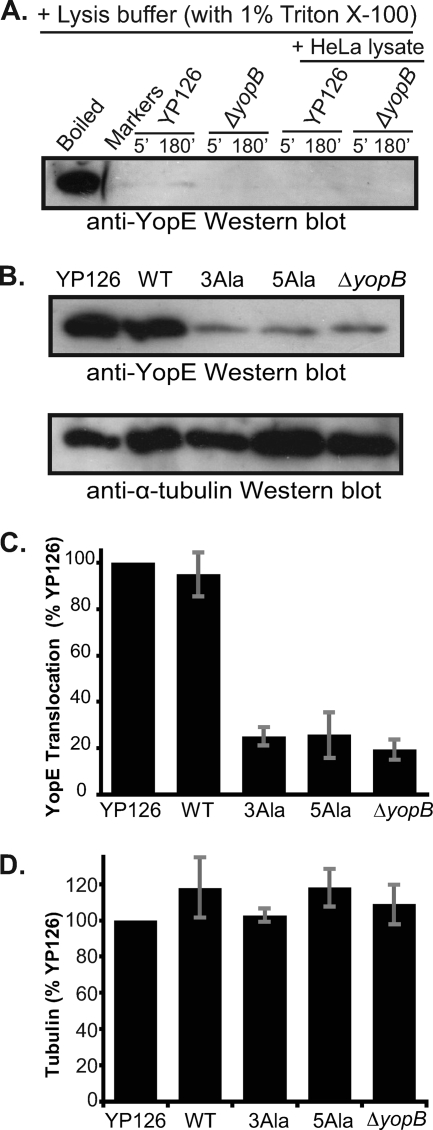
After infection for 150 min, HeLa cells were selectively lysed with Triton X-100, leaving bacterial cells intact. We verified previously published evidence that Triton X-100 does not lyse Y. pseudotuberculosis (32). We incubated Y. pseudotuberculosis in Triton X-100 and examined the soluble (i.e., supernatant) fraction resulting from this incubation by Western blotting (Fig. 5A). We also included HeLa cell lysates in some of these incubations, in case components in the HeLa cell lysate promoted lysis of Y. pseudotuberculosis. In neither case did we detect lysis of Y. pseudotuberculosis, as assayed by the release of YopE (Fig. 5A). We further verified that Triton X-100 did not lyse Y. pseudotuberculosis by looking for the presence of SycE, which is not translocated (17), in the supernatant and pellet fractions by Western blotting. SycE was detected only in the pellet (i.e., intrabacterial) fraction, not in the supernatant (i.e., translocated) fraction, of a Triton X-100 lysate of infected HeLa cells (data not shown). As expected, YopE was detected in both fractions, confirming the existence of both intrabacterial and translocated pools of YopE (data not shown).
FIG. 5.
Translocation of YopE. (A) Western blot of YopE in the supernatant fraction of Y. pseudotuberculosis 126 or Y. pseudotuberculosis 126 ΔyopB incubated for 5 or 180 min in HeLa cell lysis buffer (containing 1% Triton X-100) in the absence or presence of a HeLa cell lysate. As a positive control for YopE, YP126 was incubated in PBS and lysed by boiling in SDS-PAGE sample buffer (boiled). (B) (Top) Western blot of wild-type YopE translocated into HeLa cells by Y. pseudotuberculosis 126 (YP126); wild-type YopE (WT), YopE-3Ala (3Ala), and YopE-5Ala (5Ala) translocated by Y. pseudotuberculosis 126 ΔyopE; and wild-type YopE translocated by Y. pseudotuberculosis 126 ΔyopB. Translocated fractions were separated by SDS-PAGE and probed with anti-YopE polyclonal antibodies. (Bottom) The blot at the top was stripped and reprobed with anti-α-tubulin monoclonal antibodies. (C) Quantification of YopE translocation, assessed by Western blotting. Mean values from three independent experiments carried out in triplicate are depicted. These values were normalized to the translocation level of YP126 and are expressed as percentages of the YP126 value. Gray bars report standard errors of the means. (D) Quantification of α-tubulin in translocation samples, assessed by Western blotting. Mean values from three independent experiments carried out in triplicate are depicted. These values were normalized to the level of tubulin in the YP126 sample and are expressed as percentages of the YP126 value. Gray bars report standard errors of the means.
Having verified the experimental format, we then quantified the amount of translocated YopE. To establish the level of background contamination in HeLa cell lysate supernatants by untranslocated YopE, the same experiment was carried out with Y. pseudotuberculosis 126 ΔyopB (34). YopB is required for translocation (21, 33) and most likely contributes to forming a pore in the host cell membrane through which effectors are transported. YopB also promotes translocation via a signaling mechanism in mammalian cells (32). Consistent with prior results, deletion of yopB had no effect on the steady-state expression level of YopE or the type III secretion of YopE (Fig. 3) (21, 33).
Strikingly, a major difference in translocation level between wild-type and alanine-substituted YopE mutants was seen. Compared to wild-type YopE, encoded either natively or on a low-copy-number plasmid, YopE-3Ala and YopE-5Ala were translocated at levels that matched the background contamination level set by the ΔyopB strain (Fig. 5B). The differences between the alanine substitution mutants and wild-type YopE were statistically significant (Fig. 5C) (P < 0.01 by ANOVA). These differences do not reflect loading errors, as equivalent amounts of total protein were loaded in each lane and equal loading was further verified by quantification of HeLa cell α-tubulin in these Western blots (Fig. 5B and D).
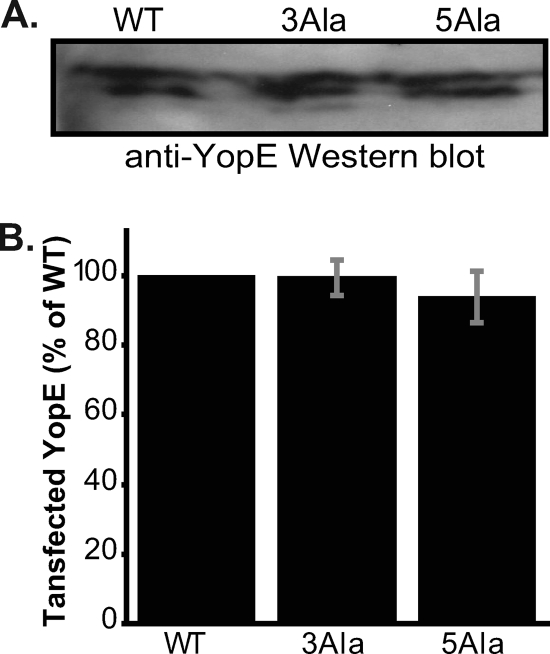
Although the alanine substitutions did not affect stability in Y. pseudotuberculosis, it remained possible that the differences observed between levels of translocated wild-type and alanine-substituted YopE proteins were not due to translocation but instead to differential stability in HeLa cells. To evaluate this possibility, we transfected HeLa cells with a mammalian expression vector encoding YopE, YopE-3Ala, or YopE-5Ala. We found that the steady-state levels of wild-type and alanine-substituted YopE proteins were similar in HeLa cells (Fig. 6A and B), indicating that the alanine substitutions did not alter the stability of YopE in HeLa cells.
FIG. 6.
Transfection of YopE. (A) Western blot of HeLa cells expressing transfected wild-type YopE (WT), YopE-3Ala (3Ala), and YopE-5Ala (5Ala). (B) Quantification of YopE expressed in HeLa cells. Mean values from three independent experiments are reported. These values were normalized to the expression level of WT YopE and are expressed as percentages of the WT level. Gray bars report standard errors of the means.
We concluded from these results that the surface patch defined by Val23, Glu25, and Ser32 has a significant role in translocation. Mutation of these residues to alanine reduced the translocation level of YopE to background values but did not alter other features of YopE.
DISCUSSION
Despite a wealth of structural and biochemical data on T3S chaperones and their interactions with effectors, the mechanism by which T3S chaperones promote translocation of effectors remains elusive. A growing body of evidence supports the notion that the specific structure of the effector Cb region bound to the chaperone acts as a three-dimensional translocation signal. The surface of the highly extended Cb region, in combination with the adjoining chaperone surface, is envisaged to constitute a binding site for a T3S component required for translocation, i.e., a receptor. The identity of this putative receptor is currently unknown. Although E. coli and Salmonella T3S ATPases have been shown to bind chaperone-effector complexes, these events have not been shown to be dependent on the chaperone (2, 19). Indeed, the E. coli effector Tir binds the T3S ATPase EscN in the absence of a chaperone (19). In addition, SycE-YopE complexes have not been detected to bind the Yersinia T3S ATPase YscN (37). Thus, the T3S ATPase is unlikely to be the chaperone-dependent receptor that explains the necessity for the chaperone in translocation.
In this study, we tested a prediction of the translocation signal model. We reasoned that if binding of SycE-YopE to a receptor was required for translocation, then disruption of the potential receptor-binding site should disrupt translocation of YopE but should leave other properties of YopE unaltered. We targeted residues that are solvent exposed in the chaperone-bound conformation of the YopE Cb region and constructed two alanine substitution mutants, namely, YopE-3Ala (V23A/E25A/S32A) and YopE-5Ala (V23A/E25A/S27A/R29A/S32A). We found that the triple-alanine substitution in YopE had no effect on the affinity for SycE and that the quintuple-alanine substitution had only a small effect. Furthermore, these substitutions did not alter the steady-state expression level in Y. pseudotuberculosis, the efficiency of secretion by Y. pseudotuberculosis, or stability in HeLa cells. They did, however, have a large effect on translocation. The level of translocation for these substitution mutants was equivalent to that found for Y. pseudotuberculosis 126 ΔyopB, a strain that secretes but does not translocate effectors.
We chose a biochemical assay of translocation in lieu of mammalian cell reporter fusion assays (3, 11, 15, 24, 25, 40, 45). This was due to concerns regarding potential artifacts of fusion partners. An example of such an artifact comes from studies of the fusion of dihydrofolate reductase (DHFR) to YopE. While biophysical evidence indicates that SycE does not promote unfolding of YopE, but rather the opposite, in bringing about structuring of YopE (37), the opposite seems to be true for YopE-DHFR fusions (15). In addition, the biochemical assay provides a direct means for evaluating levels of translocated YopE.
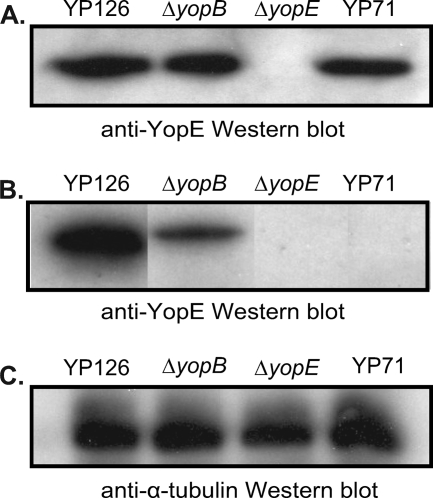
One potential issue with the biochemical assay is possible contamination of fractions of translocated YopE with extrabacterial but nontranslocated YopE (e.g., from lysis of bacterial cells). The biochemical assay depends on the selective lysis of mammalian cells by Triton X-100 (32). The selectivity of this detergent was confirmed in our experiments. Following published precedents, we used a ΔyopB strain to control for other sources of YopE contamination in the translocated fraction. While the level of translocated YopE (i.e., YopE found in the supernatant of the Triton X-100 lysate) was always much higher for the wild-type strain than for the ΔyopB strain, a small amount of YopE was consistently found in the translocated fraction of infections carried out with the ΔyopB strain, as has been seen previously (1). Further experiments showed that the small amount of YopE observed for the ΔyopB strain was T3S dependent. This was surmised from the fact that no YopE was detected in the translocated fraction from an infection with Y. pseudotuberculosis 71 (7), a strain containing a random transposon insertion that renders the T3S system inactive (Fig. 7). It is possible that in the absence of YopB, a small amount of YopE may leak into the extracellular medium due to the lack of a tight junction between the T3S needle and the host cell membrane. Notably, this small amount of YopE was resistant to papain, which was added to remove extracellular YopE prior to lysis of HeLa cells. The papain resistance of this fraction of YopE contrasts with the papain sensitivity of soluble YopE and may be due to insertion of some small amount of YopE into the host cell membrane. Membrane insertion of YopE has been observed previously (26).
FIG. 7.
Expression and translocation of YopE by strain 71. (A) Western blot of YopE expressed by Y. pseudotuberculosis 126 (YP126), Y. pseudotuberculosis 126 ΔyopB, Y. pseudotuberculosis 126 ΔyopE, and Y. pseudotuberculosis 71 (YP71). (B) Western blot of YopE translocated by the same strains as those in panel A. Translocated fractions were separated by SDS-PAGE and probed with anti-YopE polyclonal antibodies. Samples are from the same blot, but lanes were rearranged for the sake of consistency. (C) Western blot of samples from panel B probed with anti-α-tubulin monoclonal antibodies.
Most striking in our findings were the low translocation levels for YopE-3Ala and YopE-5Ala compared to that of wild-type YopE. Having found no differences between these alanine substitution mutants and wild-type YopE in processes that precede translocation (i.e., expression and SycE binding), we asked whether there was a difference in the behaviors of the proteins following translocation, namely, in the stability of these proteins in HeLa cells. Y. enterocolitica YopE is degraded by the ubiquitin-proteasome pathway, with Lys75 being the site of ubiquitination (38). While Y. pseudotuberculosis YopE has Gln instead of Lys at position 75, we nevertheless utilized the proteasome inhibitor MG-132 during the infection process. Furthermore, we transfected and expressed wild-type YopE and the alanine substitution mutants in HeLa cells and saw no differences in the levels of these proteins.
These results altogether indicate that the site defined by Val23, Glu25, and Ser32 is crucial for translocation. The location of this site at the N terminus of the Cb region is consistent with recent results showing that the C-terminal portion of the YopE Cb region, i.e., residues 54 to 75, is not required for translocation (23). Instead, this C-terminal portion of the Cb region has been identified to promote localization of YopE to host cell membranes (26). Thus, the chaperone may serve two purposes: it may template the formation of a specific structure in the N-terminal portion of the Cb region that is required for translocation, and it may also maintain the solubility in the bacterial cytosol of a C-terminal portion of the Cb region that is lipophilic (26, 27).
We chose residues 23, 25, 27, 29, and 32 due to the structural and chemical similarities between chaperone-bound YopE and Salmonella SptP Cb regions. These were the first two effector Cb regions to be visualized in their chaperone-bound conformations (5, 46). Succeeding work on other chaperone-bound Cb regions has shown a looser correspondence of residues at these positions, suggesting the existence of interactions that differ in detail among effectors. While we favor the possibility that these residues constitute part of a receptor-binding site in the chaperone-effector complex, other possibilities cannot be excluded. Among these is the possibility that these residues are important in steps following dissociation of the chaperone, for example, in transiting from the T3S needle to the host cell membrane-inserted translocon. In either case, our work identifies a crucial set of functional residues in the Cb region and provides guidance for the direction of future mechanistic investigation.
Acknowledgments
This work was supported by NIH grant T32 GM007240 (L.R.), by the university-wide AIDS Research Program (L.R.), and by NIH grant R01 AI061452 (P.G.).
Footnotes
Published ahead of print on 9 April 2010.
REFERENCES
- 1.Aili, M., E. L. Isaksson, B. Hallberg, H. Wolf-Watz, and R. Rosqvist. 2006. Functional analysis of the YopE GTPase-activating protein (GAP) activity of Yersinia pseudotuberculosis. Cell. Microbiol. 8:1020-1033. [DOI] [PubMed] [Google Scholar]
- 2.Akeda, Y., and J. E. Galan. 2005. Chaperone release and unfolding of substrates in type III secretion. Nature 437:911-915. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, D. M., and O. Schneewind. 1997. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science 278:1140-1143. [DOI] [PubMed] [Google Scholar]
- 4.Birtalan, S., and P. Ghosh. 2001. Structure of the Yersinia type III secretory system chaperone SycE. Nat. Struct. Biol. 8:974-978. [DOI] [PubMed] [Google Scholar]
- 5.Birtalan, S. C., R. M. Phillips, and P. Ghosh. 2002. Three-dimensional secretion signals in chaperone-effector complexes of bacterial pathogens. Mol. Cell 9:971-980. [DOI] [PubMed] [Google Scholar]
- 6.Bolin, I., L. Norlander, and H. Wolf-Watz. 1982. Temperature-inducible outer membrane protein of Yersinia pseudotuberculosis and Yersinia enterocolitica is associated with the virulence plasmid. Infect. Immun. 37:506-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolin, I., and H. Wolf-Watz. 1984. Molecular cloning of the temperature-inducible outer membrane protein 1 of Yersinia pseudotuberculosis. Infect. Immun. 43:72-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyd, A. P., I. Lambermont, and G. R. Cornelis. 2000. Competition between the Yops of Yersinia enterocolitica for delivery into eukaryotic cells: role of the SycE chaperone binding domain of YopE. J. Bacteriol. 182:4811-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brosius, J., and A. Holy. 1984. Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc. Natl. Acad. Sci. U. S. A. 81:6929-6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buttner, C. R., G. R. Cornelis, D. W. Heinz, and H. H. Niemann. 2005. Crystal structure of Yersinia enterocolitica type III secretion chaperone SycT. Protein Sci. 14:1993-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charpentier, X., and E. Oswald. 2004. Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 beta-lactamase as a new fluorescence-based reporter. J. Bacteriol. 186:5486-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng, L. W., D. M. Anderson, and O. Schneewind. 1997. Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Mol. Microbiol. 24:757-765. [DOI] [PubMed] [Google Scholar]
- 13.Cheng, L. W., and O. Schneewind. 1999. Yersinia enterocolitica type III secretion. On the role of SycE in targeting YopE into HeLa cells. J. Biol. Chem. 274:22102-22108. [DOI] [PubMed] [Google Scholar]
- 14.Cornelis, G. R. 2006. The type III secretion injectisome. Nat. Rev. Microbiol. 4:811-825. [DOI] [PubMed] [Google Scholar]
- 15.Feldman, M. F., S. Muller, E. Wuest, and G. R. Cornelis. 2002. SycE allows secretion of YopE-DHFR hybrids by the Yersinia enterocolitica type III Ysc system. Mol. Microbiol. 46:1183-1197. [DOI] [PubMed] [Google Scholar]
- 16.Ferreiro, D. U., C. F. Cervantes, S. M. Truhlar, S. S. Cho, P. G. Wolynes, and E. A. Komives. 2007. Stabilizing IkappaBalpha by “consensus” design. J. Mol. Biol. 365:1201-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frithz-Lindsten, E., R. Rosqvist, L. Johansson, and A. Forsberg. 1995. The chaperone-like protein YerA of Yersinia pseudotuberculosis stabilizes YopE in the cytoplasm but is dispensible for targeting to the secretion loci. Mol. Microbiol. 16:635-647. [DOI] [PubMed] [Google Scholar]
- 18.Galan, J. E., and H. Wolf-Watz. 2006. Protein delivery into eukaryotic cells by type III secretion machines. Nature 444:567-573. [DOI] [PubMed] [Google Scholar]
- 19.Gauthier, A., and B. B. Finlay. 2003. Translocated intimin receptor and its chaperone interact with ATPase of the type III secretion apparatus of enteropathogenic Escherichia coli. J. Bacteriol. 185:6747-6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh, P. 2004. Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68:771-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hakansson, S., K. Schesser, C. Persson, E. E. Galyov, R. Rosqvist, F. Homble, and H. Wolf-Watz. 1996. The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell plasma membrane and displays a contact-dependent membrane disrupting activity. EMBO J. 15:5812-5823. [PMC free article] [PubMed] [Google Scholar]
- 22.Hu, X., M. S. Lee, and A. Wallqvist. 2009. Interaction of the disordered Yersinia effector protein YopE with its cognate chaperone SycE. Biochemistry 48:11158-11160. [DOI] [PubMed] [Google Scholar]
- 23.Isaksson, E. L., M. Aili, A. Fahlgren, S. E. Carlsson, R. Rosqvist, and H. Wolf-Watz. 2009. The membrane localization domain is required for intracellular localization and autoregulation of YopE in Yersinia pseudotuberculosis. Infect. Immun. 77:4740-4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobi, C. A., A. Roggenkamp, A. Rakin, R. Zumbihl, L. Leitritz, and J. Heesemann. 1998. In vitro and in vivo expression studies of yopE from Yersinia enterocolitica using the gfp reporter gene. Mol. Microbiol. 30:865-882. [DOI] [PubMed] [Google Scholar]
- 25.Jaumouille, V., O. Francetic, P. J. Sansonetti, and G. Tran Van Nhieu. 2008. Cytoplasmic targeting of IpaC to the bacterial pole directs polar type III secretion in Shigella. EMBO J. 27:447-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krall, R., Y. Zhang, and J. T. Barbieri. 2004. Intracellular membrane localization of Pseudomonas ExoS and Yersinia YopE in mammalian cells. J. Biol. Chem. 279:2747-2753. [DOI] [PubMed] [Google Scholar]
- 27.Letzelter, M., I. Sorg, L. J. Mota, S. Meyer, J. Stalder, M. Feldman, M. Kuhn, I. Callebaut, and G. R. Cornelis. 2006. The discovery of SycO highlights a new function for type III secretion effector chaperones. EMBO J. 25:3223-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lilic, M., M. Vujanac, and C. E. Stebbins. 2006. A common structural motif in the binding of virulence factors to bacterial secretion chaperones. Mol. Cell 21:653-664. [DOI] [PubMed] [Google Scholar]
- 29.Lloyd, S. A., M. Sjostrom, S. Andersson, and H. Wolf-Watz. 2002. Molecular characterization of type III secretion signals via analysis of synthetic N-terminal amino acid sequences. Mol. Microbiol. 43:51-59. [DOI] [PubMed] [Google Scholar]
- 30.Locher, M., B. Lehnert, K. Krauss, J. Heesemann, M. Groll, and G. Wilharm. 2005. Crystal structure of the Yersinia enterocolitica type III secretion chaperone SycT. J. Biol. Chem. 280:31149-31155. [DOI] [PubMed] [Google Scholar]
- 31.Luo, Y., M. G. Bertero, E. A. Frey, R. A. Pfuetzner, M. R. Wenk, L. Creagh, S. L. Marcus, D. Lim, F. Sicheri, C. Kay, C. Haynes, B. B. Finlay, and N. C. Strynadka. 2001. Structural and biochemical characterization of the type III secretion chaperones CesT and SigE. Nat. Struct. Biol. 8:1031-1036. [DOI] [PubMed] [Google Scholar]
- 32.Mejia, E., J. B. Bliska, and G. I. Viboud. 2008. Yersinia controls type III effector delivery into host cells by modulating Rho activity. PLoS Pathog. 4:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nordfelth, R., and H. Wolf-Watz. 2001. YopB of Yersinia enterocolitica is essential for YopE translocation. Infect. Immun. 69:3516-3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmer, L. E., S. Hobbie, J. E. Galan, and J. B. Bliska. 1998. YopJ of Yersinia pseudotuberculosis is required for the inhibition of macrophage TNF-alpha production and downregulation of the MAP kinases p38 and JNK. Mol. Microbiol. 27:953-965. [DOI] [PubMed] [Google Scholar]
- 35.Phan, J., J. E. Tropea, and D. S. Waugh. 2004. Structure of the Yersinia pestis type III secretion chaperone SycH in complex with a stable fragment of YscM2. Acta Crystallogr. D 60:1591-1599. [DOI] [PubMed] [Google Scholar]
- 36.Quenee, L. E., and O. Schneewind. 2007. Ubiquitin-Yop hybrids as probes for post-translational transport by the Yersinia type III secretion pathway. Mol. Microbiol. 65:386-400. [DOI] [PubMed] [Google Scholar]
- 37.Rodgers, L., A. Gamez, R. Riek, and P. Ghosh. 2008. The type III secretion chaperone SycE promotes a localized disorder-to-order transition in the natively unfolded effector YopE. J. Biol. Chem. 283:20857-20863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruckdeschel, K., G. Pfaffinger, K. Trulzsch, G. Zenner, K. Richter, J. Heesemann, and M. Aepfelbacher. 2006. The proteasome pathway destabilizes Yersinia outer protein E and represses its antihost cell activities. J. Immunol. 176:6093-6102. [DOI] [PubMed] [Google Scholar]
- 39.Runco, L. M., S. Myrczek, J. B. Bliska, and D. G. Thanassi. 2008. Biogenesis of the fraction 1 capsule and analysis of the ultrastructure of Yersinia pestis. J. Bacteriol. 190:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russmann, H., T. Kubori, J. Sauer, and J. E. Galan. 2002. Molecular and functional analysis of the type III secretion signal of the Salmonella enterica InvJ protein. Mol. Microbiol. 46:769-779. [DOI] [PubMed] [Google Scholar]
- 41.Schesser, K., E. Frithz-Lindsten, and H. Wolf-Watz. 1996. Delineation and mutational analysis of the Yersinia pseudotuberculosis YopE domains which mediate translocation across bacterial and eukaryotic cellular membranes. J. Bacteriol. 178:7227-7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schubot, F. D., M. W. Jackson, K. J. Penrose, S. Cherry, J. E. Tropea, G. V. Plano, and D. S. Waugh. 2005. Three-dimensional structure of a macromolecular assembly that regulates type III secretion in Yersinia pestis. J. Mol. Biol. 346:1147-1161. [DOI] [PubMed] [Google Scholar]
- 43.Singer, A. U., D. Desveaux, L. Betts, J. H. Chang, Z. Nimchuk, S. R. Grant, J. L. Dangl, and J. Sondek. 2004. Crystal structures of the type III effector protein AvrPphF and its chaperone reveal residues required for plant pathogenesis. Structure 12:1669-1681. [DOI] [PubMed] [Google Scholar]
- 44.Sory, M. P., A. Boland, I. Lambermont, and G. R. Cornelis. 1995. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc. Natl. Acad. Sci. U. S. A. 92:11998-12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sory, M. P., and G. R. Cornelis. 1994. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol. Microbiol. 14:583-594. [DOI] [PubMed] [Google Scholar]
- 46.Stebbins, C. E., and J. E. Galan. 2001. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature 414:77-81. [DOI] [PubMed] [Google Scholar]
- 47.van Eerde, A., C. Hamiaux, J. Perez, C. Parsot, and B. W. Dijkstra. 2004. Structure of Spa15, a type III secretion chaperone from Shigella flexneri with broad specificity. EMBO Rep. 5:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wattiau, P., and G. R. Cornelis. 1993. SycE, a chaperone-like protein of Yersinia enterocolitica involved in Ohe secretion of YopE. Mol. Microbiol. 8:123-131. [DOI] [PubMed] [Google Scholar]
- 49.Woestyn, S., M. P. Sory, A. Boland, O. Lequenne, and G. R. Cornelis. 1996. The cytosolic SycE and SycH chaperones of Yersinia protect the region of YopE and YopH involved in translocation across eukaryotic cell membranes. Mol. Microbiol. 20:1261-1271. [DOI] [PubMed] [Google Scholar]
- 50.Wulff-Strobel, C. R., A. W. Williams, and S. C. Straley. 2002. LcrQ and SycH function together at the Ysc type III secretion system in Yersinia pestis to impose a hierarchy of secretion. Mol. Microbiol. 43:411-423. [DOI] [PubMed] [Google Scholar]