Abstract
Although the genome of Haloferax volcanii contains genes (flgA1-flgA2) that encode flagellins and others that encode proteins involved in flagellar assembly, previous reports have concluded that H. volcanii is nonmotile. Contrary to these reports, we have now identified conditions under which H. volcanii is motile. Moreover, we have determined that an H. volcanii deletion mutant lacking flagellin genes is not motile. However, unlike flagella characterized in other prokaryotes, including other archaea, the H. volcanii flagella do not appear to play a significant role in surface adhesion. While flagella often play similar functional roles in bacteria and archaea, the processes involved in the biosynthesis of archaeal flagella do not resemble those involved in assembling bacterial flagella but, instead, are similar to those involved in producing bacterial type IV pili. Consistent with this observation, we have determined that, in addition to disrupting preflagellin processing, deleting pibD, which encodes the preflagellin peptidase, prevents the maturation of other H. volcanii type IV pilin-like proteins. Moreover, in addition to abolishing swimming motility, and unlike the flgA1-flgA2 deletion, deleting pibD eliminates the ability of H. volcanii to adhere to a glass surface, indicating that a nonflagellar type IV pilus-like structure plays a critical role in H. volcanii surface adhesion.
To escape toxic conditions or to acquire new sources of nutrients, prokaryotes often depend on some form of motility. Swimming motility, a common means by which many bacteria move from one place to another, usually depends on flagellar rotation to propel cells through liquid medium (24, 26, 34). These motility structures are also critical for the effective attachment of bacteria to surfaces.
As in bacteria, rotating flagella are responsible for swimming motility in archaea, and recent studies suggest that archaea, like bacteria, also require flagella for efficient surface attachment (37, 58). However, in contrast to bacterial flagellar subunits, which are translocated via a specialized type III secretion apparatus, archaeal flagellin secretion and flagellum assembly resemble the processes used to translocate and assemble the subunits of bacterial type IV pili (34, 38, 54).
Type IV pili are typically composed of major pilins, the primary structural components of the pilus, and several minor pilin-like proteins that play important roles in pilus assembly or function (15, 17, 46). Pilin precursor proteins are transported across the cytoplasmic membrane via the Sec translocation pathway (7, 20). Most Sec substrates contain either a class I or a class II signal peptide that is cleaved at a recognition site that lies subsequent to the hydrophobic portion of the signal peptide (18, 43). However, the precursors of type IV pilins contain class III signal peptides, which are processed at recognition sites that precede the hydrophobic domain by a prepilin-specific peptidase (SPase III) (38, 43, 45). Similarly, archaeal flagellin precursors contain a class III signal peptide that is processed by a prepilin-specific peptidase homolog (FlaK/PibD) (3, 8, 10, 11). Moreover, flagellar assembly involves homologs of components involved in the biosynthesis of bacterial type IV pili, including FlaI, an ATPase homologous to PilB, and FlaJ, a multispanning membrane protein that may provide a platform for flagellar assembly, similar to the proposed role for PilC in pilus assembly (38, 44, 53, 54). These genes, as well as a number of others that encode proteins often required for either flagellar assembly or function (flaCDEFG and flaH), are frequently coregulated with the flg genes (11, 26, 44, 54).
Interestingly, most sequenced archaeal genomes also contain diverse sets of genes that encode type IV pilin-like proteins with little or no homology to archaeal flagellins (3, 39, 52). While often coregulated with pilB and pilC homologs, these genes are never found in clusters containing the motility-specific flaCDEFG and flaH homologs; however, the proteins they encode do contain class III signal peptides (52). Several of these proteins have been shown to be processed by an SPase III (4, 52). Moreover, in Sulfolobus solfataricus and Methanococcus maripaludis, some of these archaeal type IV pilin-like proteins were confirmed to form surface filaments that are distinct from the flagella (21, 22, 56). These findings strongly suggest that the genes encode subunits of pilus-like surface structures that are involved in functions other than swimming motility.
In bacteria, type IV pili are multifunctional filamentous protein complexes that, in addition to facilitating twitching motility, mediate adherence to abiotic surfaces and make close intercellular associations possible (15, 17, 46). For instance, mating between Escherichia coli in liquid medium has been shown to require type IV pili (often referred to as thin sex pili), which bring cells into close proximity (29, 30, 57). Recent work has shown that the S. solfataricus pilus, Ups, is required not only for efficient adhesion to surfaces of these crenarchaeal cells but also for UV-induced aggregation (21, 22, 58). Frols et al. postulate that autoaggregation is required for DNA exchange under these highly mutagenic conditions (22). Halobacterium salinarum has also been shown to form Ca2+-induced aggregates (27, 28). Furthermore, conjugation has been observed in H. volcanii, which likely requires that cells be held in close proximity for a sustained period to allow time for the cells to construct the cytoplasmic bridges that facilitate DNA transfer between them (35).
To determine the roles played by haloarchaeal flagella and other putative type IV pilus-like structures in swimming and surface motility, surface adhesion, autoaggregation, and conjugation, we constructed and characterized two mutant strains of H. volcanii, one lacking the genes that encode the flagellins and the other lacking pibD. Our analyses indicate that although this archaeon was previously thought to be nonmotile (14, 36), wild-type (wt) H. volcanii can swim in a flagellum-dependent manner. Consistent with the involvement of PibD in processing flagellins, the peptidase mutant is nonmotile. Unlike nonhalophilic archaea, however, the flagellum mutant can adhere to glass as effectively as the wild type. Conversely, the ΔpibD strain fails to adhere to glass surfaces, strongly suggesting that in H. volcanii surface adhesion involves nonflagellar, type IV pilus-like structures.
MATERIALS AND METHODS
Reagents.
All enzymes used for standard molecular biology procedures were purchased from New England Biolabs, except for iProof High-Fidelity DNA Polymerase, which was purchased from Bio-Rad. An ECL Plus Western blotting system, horseradish peroxidase-linked sheep anti-mouse antibody, and donkey anti-rabbit antibody were purchased from Amersham Biosciences. Polyvinylidene difluoride membrane was purchased from Millipore. DNA and plasmid purification kits and anti-His antibodies were purchased from Qiagen. NuPAGE gels, buffers, and reagents were purchased from Invitrogen. Difco agar and Bacto yeast extract were purchased from Becton, Dickinson, and Company. Peptone was purchased from Oxoid. 5-Fluoroorotic acid (5-FOA) was purchased from Toronto Research Biochemicals. All other chemicals and reagents were purchased from either Fisher or Sigma.
Strains and growth conditions.
Unless noted otherwise, all H. volcanii strains (Table 1) were grown at 45°C in complex medium (CX) or defined liquid medium (CDM or CA medium) or on solid medium containing agar (19, 40). The CX (complex medium) contains the following (per liter): 125 g of NaCl, 45 g of MgCl2·6H2O, 10 g of MgSO4·7H2O, 10 g of KCl, 1.34 g of CaCl2·2H2O, 3 g of yeast extract, and 5 g of tryptone (19). For high- and low-salt CX preparations, total salt concentrations (proportional amounts of each salt) were 23% and 14%, respectively, compared to the 19% total salt concentration of the CX described above. CA medium contains the following (per liter): 5 g of Casamino acids, 2.4 ml of 1 M KOH, 600 ml of 30% salt water solution (described below), 6 ml of 0.5 M CaCl2, 900 μl of thiamine and biotin solution (9.6 ml of thiamine [1 mg/ml], 1.2 ml of biotin [1 mg/ml] in 100 ml). Salt water solution (30%) contains the following (per liter): 240 g of NaCl, 30 g of MgCl2·6H2O, 35 g of MgSO4·7H2O, 7 g of KCl, 5 ml of 1 M CaCl2·2H2O, 2 ml of 1 M Tris-HCl buffer (pH 7.5) (40). CDM contains the following (per liter): 125 g of NaCl2, 50 g of MgCl2·6H2O, 5 g of K2SO4, 0.26 g of CaCl2, 5 ml of 1 M NH4Cl, 22.5 ml of Na succinate (10g/100 ml of double-distilled H2O [ddH2O]), 2.5 ml of 10% glycerol, 2 ml of 0.5 M K2HPO4 (8.7g/100 ml of ddH2O), 1 ml of trace elements (36 mg of MnCl2·4H2O, 44 mg of ZnSO4·7H2O, 230 mg of FeSO4·7H2O, and 5 mg of CuSO4·5H2O per 100 ml of ddH2O), 0.8 ml of 1 mg/ml thiamine, 0.1 ml of 1 mg/ml biotin (19). Fifteen grams of agar was added for regular plates, and 3 g of agar was added for motility plates. To ensure equal concentration of agar in all plates, the agar was completely dissolved prior to autoclaving, and the autoclaved medium was stirred before the plates were poured. H. volcanii strain H53 (6) was grown in CDM and CA medium supplemented with tryptophan and uracil (50 μg/ml final concentration). H. volcanii strain H98 (6) was grown in CX supplemented with thymidine (40 μg/ml) or in CDM and CA medium supplemented with thymidine, hypoxanthine (40 μg/ml), and uracil (50 μg/ml, unless noted differently). 5-FOA, for the selection of ΔflgA1 ΔflgA2 and ΔpibD deletion mutants (see below), was added at a final concentration of 150 μg/ml. H. volcanii strains transformed with pRVI-ptna (31) and its derivatives were grown in medium supplemented with novobiocin (final concentration, 2 μg/ml). To induce protein expression from the pRV10-ptna plasmid, tryptophan (final concentration, 100 μg/ml) was added to the medium. E. coli strains (Table 1) were grown at 37°C in NZCYM medium (Fisher; 22 g NZCYM per 1 liter of ddH2O), supplemented with ampicillin (200 μg/ml) when necessary (12).
TABLE 1.
Strains and plasmids
| Plasmid or strain | Relevant properties or description | Reference or source |
|---|---|---|
| Plasmids | ||
| pTA131 | Ampr; pBluescript II with BamHI-XbaI fragment from pGB70 containing pfdx-pyrE2 | 6 |
| pRV1-ptna | Ampr Novr ptna | 31 |
| pMT1 | pTA131 containing chromosomal flgA1-flgA2 region | This study |
| pMT2 | pTA131 containing chromosomal pibD region | This study |
| pMT3 | pRV1-ptna containing flgA1-flgA2-His | This study |
| pMT4 | pRV1-ptna containing pibD-His | This study |
| pMT5 | pRV1-ptna containing Hvo_A0632-His; type IV pilin-like gene | This study |
| pMT6 | pRV1-ptna containing Hvo_2451-His; type IV pilin-like gene | This study |
| Strains | ||
| E. coli | ||
| DH5α | F− 80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | Invitrogen |
| DL739 | MC4100 recA dam-13::Tn9 | 13 |
| H. volcanii | ||
| H98 | ΔpyrE2 ΔhdrB | 6 |
| H53 | ΔpyrE2 Δtrp | 6 |
| MT1 | H98 ΔflgA1 ΔflgA2 | This study |
| MT2 | H53 ΔflgA1 ΔflgA2 | This study |
| MT3 | H98 ΔpibD | This study |
| MT4 | H53 ΔpibD | This study |
| MT5 | H98 containing pMT3 | This study |
| MT6 | H98 containing pMT4 | This study |
| MT7 | MT3 containing pMT3 | This study |
| MT8 | MT3 containing pMT4 | This study |
| MT9 | H98 containing pMT5 | This study |
| MT10 | H98 containing pMT6 | This study |
| MT11 | MT3 containing pMT5 | This study |
| MT12 | MT3 containing pMT6 | This study |
| aglB strain | H53 ΔaglB | 1 |
Assembly of knockout constructs.
Plasmid constructs for use in homologous recombination were generated as previously described (5). In brief, approximately 700 nucleotides flanking each end of the gene of interest were PCR amplified (primers are listed in Table 2). The amplified flanking regions were sequentially cloned into the haloarchaeal suicide vector pTA131. First, the upstream flanking region was cloned into pTA131 digested with restriction enzymes EcoRI and HindIII. The construct was then digested with BamHI and XbaI, and the downstream flanking region was inserted. The final constructs, pMT1 and pMT2, contain upstream and downstream region inserts that are identical to the corresponding chromosomal regions of flgA1-flgA2 and pibD, respectively.
TABLE 2.
Primers used for PCR amplification
| Primer name | Sequences (5′-3′)a | Target sequence |
|---|---|---|
| FlgA1/2KO up for HindIII | GCCGTAAAAGCTTGCTCGTCCTCGACTC | 705 bp upstream of flgA1 start codon, extension toward gene |
| FlgA1/2KO up rev EcoRI | TTACTTGGAATTCGTAACCCGGCACCGA | 1 bp upstream of flgA1 start codon, extension away from gene |
| FlgA1/2KOd for BamHI | ACGTCGGGATCCAGATTTCGTGGGTTT | 1 bp downstream of flgA2 stop codon, extension away from gene |
| FlgA1/2KOd rev XbaI | GCGACGTCTAGAACGTTCGTCCTCGTC | 705 bp downstream of flgA2 stop codon extension toward gene |
| PibDKOup for HindIII | TTATTAAGCTTCTCTCGCGCCGGGATC | 695 bp upstream of pibD start codon, extension toward gene |
| PibDKOup rev EcoRI | GCAGTGAATTCTACCCGGAGACTGGGG | 1 bp upstream of pibD start codon, extension away from gene |
| PibDKOd for BamHI | ATTATGGATCCGTCGCTGCGGAGTCG | 1 bp downstream of pibD stop codon, extension away from gene |
| PibDKOd for XbaI | GGATAGTCTAGAGTCGTCCGGCGGGTC | 705 bp downstream of pibD stop codon extension toward gene |
| FlgA1/2for | CGCCCGTGCGAGAGGTGATT | 71 bp downstream of flgA1 start codon, extension toward gene |
| FlgA1/2rev | CGAGGCGGACCGTCAGGAAA | 262 bp upstream of flgA2 stop codon, extension toward gene |
| PibDfor | TTCGCCTTCGGACTGCTCGGT | 283 upstream of pibD start codon, extension toward gene |
| PibDrev | CGCGACGAACTCTGGGAGAAG | 255 downstream of pibD stop codon, extension toward gene |
| FlgA1for NdeI | ATTAATCATATGTTCGAAAACATCAACGAA | 1 bp downstream of flgA1 start codon, extension toward gene |
| FlgA2 6-His EcoRI rev | TATATTGAATTCTCAGTGATGGTGATGGTGAT GCGGGCCGCCCAGTTCGACGGGTT | 1 bp upstream of flgA2 stop codon, extension toward gene |
| PibDfor NdeI | GCTATCCATATGTTCGGTATCGGCACG | 1 bp downstream of pibD start codon, extension toward gene |
| PibD 6-His EcoRI rev | TATATTGAATTCTCAGTGATGGTGATGGTGAT GCGGGCCGCCGACGATACCGAGCGCG | 1 bp upstream of pibD stop codon, extension toward gene |
| Hvo_A0632 forNdeI | CCCGCGCATATGGATATCAAGAAATTCCTTAG | 1 bp downstream of Hvo_A0632 start codon, extension toward gene |
| Hvo_A0632 6-His EcoRI rev | TATATTGAATTCTCAGTGATGGTGATGGTGAT GCGGGCCGCCGTGCGTGTACGTTTG | 1 bp upstream of Hvo_A0632 stop |
| Hvo_2451 for NdeI | AATTACCATATGCAACTCAAACATCTCTTCAC | 1 bp downstream of Hvo_2451 start codon, extension toward gene |
| Hvo_2451 6-His EcoRI rev | GTAATTGAATTCTCAGTGATGGTGATGGTGA TGCGGGCCGCCTCCGTTGTAGGTCCA | 1 bp upstream of Hvo_2451 stop |
Restriction endonuclease sites are underlined.
Generating chromosomal deletions.
Chromosomal deletions were generated using the homologous recombination (pop-in-pop-out) method as previously described (5). The pMT1 and pMT2 constructs were isolated from E. coli DH5α and transformed into E. coli DL739 (13). Using the standard polyethylene glycol method, nonmethylated plasmid DNA isolated from E. coli DL739 was used to transform H. volcanii strains H98 and H53 (6). A single homologous recombination event between one of the flanking regions cloned into the plasmid and the chromosome (pop in) was selected for by growth on CA agar lacking uracil that was supplemented with either thymidine and hypoxanthine or tryptophan, depending on the strain. pMT1 and pMT2 contain pyrE2, a gene that catalyzes the biosynthesis of uracil, which allows positive selection for recombination events (Table 1). PCR was used to confirm that recombination occurred at the proper location on the chromosome. Recombinants were grown for 48 h in liquid CA medium supplemented with thymidine, hypoxanthine, and uracil (H98) or with tryptophan and uracil (H53) to allow a second recombination event, which results in excision of the plasmid from the chromosome (pop out). Plasmid excision results in either reversion to the parental locus or replacement of the chromosomal gene with the insert. After 48 h, H98 liquid cultures were transferred to CA agar plates supplemented with thymidine, hypoxanthine, uracil (10 μg/ml), and 5-FOA while H53 strains were transferred to CA agar plates supplemented with tryptophan, uracil (10 μg/ml), and 5-FOA. These growth conditions permit the growth only of cells in which the plasmid has been excised from the chromosome. Colonies derived from these cultures were screened by PCR to confirm the chromosomal replacement event (primers used are listed in Table 2). The correct identities of the PCR products were confirmed by sequencing using the same primers. The ΔflgA1 ΔflgA2 deletion mutants generated in strains H98 and H53 were designated MT1 and MT2, respectively, and the pibD deletion mutants generated in strains H98 and H53 were designated MT3 and MT4, respectively (Table 1).
Construction of expression vectors, expression and extraction of proteins, and Western blotting.
To investigate protein expression, we created gene fusions that encode proteins containing a C-terminal His tag; the inducible tryptophan promoter (tna) drives expression of these gene fusions (31). Genes were amplified using the primers listed in Table 2 and inserted into pRV1-ptna digested with NdeI and EcoRI. pMT3, pMT4, pMT5, and pMT6 (pRV1-ptna containing C-terminally His-tagged FlgA2, PibD, Hvo_A0632, and Hvo_02451, respectively) were isolated from E. coli DH5α and transformed into E. coli DL739 (Table 1). Using the standard polyethylene glycol method, nonmethylated plasmid DNA isolated from E. coli DL739 was used to transform H. volcanii strains (19). Tryptophan was added to the CX to a final concentration of 100 μg/ml to induce gene expression from these plasmids.
All protein samples were stored in NuPAGE lithium dodecyl sulfate sample buffer, supplemented with 50 mM dithiothreitol. Samples were run on Bis-Tris NuPAGE gels (Invitrogen) under denaturing conditions using morpholinepropanesulfonic acid (MOPS). Proteins were transferred from the gel to a polyvinylidene difluoride membrane using a Bio-Rad Transblot-SD semidry transfer cell at 15 V for 30 min.
Western blots of whole-cell lysates from H. volcanii strains expressing His-tagged constructs were probed with anti-His antibody at a dilution of 1:1,000, followed by a secondary anti-mouse antibody at a dilution of 1:10,000. Cell lysates isolated from wild-type or ΔpibD H. volcanii were also probed with anti-H. volcanii S-layer (cell wall) glycoprotein (anti-CWG) antibody at a dilution of 1:1,000 followed by a secondary anti-rabbit antibody at a dilution of 1:10,000 (23). Antibody-labeled protein bands were identified using an Amersham ECL Plus Western blotting detection system.
Surface adhesion assay.
Surface adhesion was assayed using a modified air-liquid interface (ALI) assay protocol (42) as follows: 3 ml of culture in CX at an optical density at 600 nm (OD600) of ∼0.3, was incubated in each well of a 12-well plate. Glass coverslips (22 by 22 mm; 0.19 to 0.25 mm thick) were inserted into each well at an angle of 90°. Lids were placed over the wells; the plates were then placed on wet paper towels and wrapped in polyvinyl paper to limit evaporation. The wrapped plates were incubated at 45°C with or without shaking. After various time intervals, coverslips were removed from the wells with forceps, submerged for 3 min in 2% acetic acid, and then allowed to air dry. When the coverslips were completely dry, they were stained in 0.1% crystal violet for 10 min. The coverslips were then washed three times with distilled water. Stained coverslips were air dried and then examined using light microscopy.
Aggregation assays.
To identify conditions under which H. volcanii can aggregate, 3 ml of H. volcanii cultures grown in CX or CA medium to an OD600 of ∼0.3 were incubated at 45°C in each well of a 12-well plate. To test the effect of calcium on cell aggregation, various concentrations of CaCl2 were added to the cultures. To test the effect of magnesium, MgCl2 was added to the cultures. CA medium was also incubated with CaCl2 to control for nonspecific precipitation of calcium. Twelve-well plates were examined for cell aggregates at various time intervals.
Motility assays.
The motility assay was performed in 0.3% motility agar plates of CX, CA medium, or CDM. Agar media were supplemented with tryptophan, thymidine, hypoxanthine, uracil, and novobiocin, depending on the H. volcanii strain being assayed. A toothpick was used to stab inoculate the agar.
Conjugation assay.
Conjugation was assayed using a modified version of a protocol previously described by Mevarech and Werczberger (35). In brief, equal volumes (1 to 5 ml) from cultures of two different auxotrophic H. volcanii strains (H53 and H98) at OD600s of 0.5 to 0.9 were mixed and filtered through Millipore Swinnex 25 filter units with Millipore 0.45-μm-pore-size type HA 25-mm filters using a 5-ml syringe. The filter unit was then disassembled, and filter discs were placed cell side up on CX supplemented with thymidine and incubated at 45°C overnight. After incubation, the filters were removed from the medium, placed in 2-ml Eppendorf tubes containing 1 ml of 18% salt water (19), and shaken on a rotator for 1 h. An undiluted sample and a sample that was diluted 1:100 were plated on CA agar medium supplemented with uracil. Twenty microliters was also plated on CX agar supplemented with thymidine for viable cell counts.
A liquid conjugation assay was performed as follows: 1 ml from each H53 and H98 culture at equal cell density was mixed; 3 ml of CX supplemented with thymidine was added to the mix, and the mixed cultures were grown overnight at 45°C in a shaking incubator. After the incubation, 2 ml from the mixed cultures was centrifuged, and pellets were resuspended in 100 μl of CA medium, which was then plated on CA agar supplemented with uracil. Conjugation assays using H98 and H53 containing the flgA1-flgA2 or pibD deletions were carried out the same way. Conjugation frequencies were expressed as the number of transconjugants per CFU of the donor. The reported results are the mean values from three separate experiments.
RESULTS
H. volcanii exhibits swimming motility.
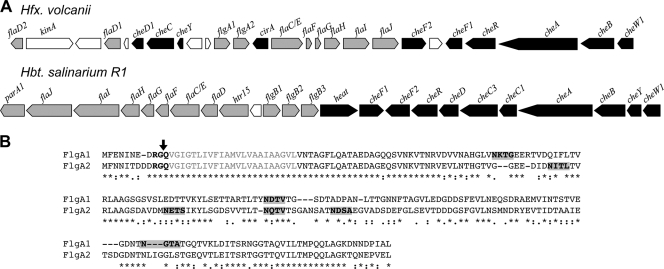
Although the current description of H. volcanii notes that this haloarchaeon is nonmotile (14), in silico analyses of the H. volcanii genome have identified homologs of genes that encode flagellins in other archaea, as well as genes that encode proteins involved in archaeal flagellum biosynthesis (25, 47, 49) (Fig. 1A). Moreover, with the exception of the gene encoding the SPase III, these genes are clustered with genes that encode proteins involved in chemotaxis, reminiscent of the organization of a similar fla-che cluster of genes in Halobacterium salinarum sp. R1 (48) (Fig. 1A). Similar to other reported archaeal flagellins, the H. volcanii FlgA1 and FlgA2 flagellin homologs contain a predicted class III Sec signal peptide and several predicted N-glycosylation sites (33, 38, 54) (Fig. 1B).
FIG. 1.
H. volcanii flgA1 and flgA2 genomic loci and amino acid sequences of FlgA1 and FlgA2. (A) H. volcanii motility and chemotaxis gene cluster compared to that of Halobacterium salinarum R1 as described by Schlesner et al. and the HaloLex Web server (25, 47, 48). The arrows represent the open reading frames (ORFs) encoding H. salinarum R1 and H. volcanii homologs of motility (gray) and chemotaxis (black) genes. The orientation of the arrows corresponds to the orientation of the ORFs relative to other genes within the putative operons. Each H. volcanii gene is annotated based on significant BLASTP alignment scores with experimentally verified homologous proteins. White arrows represent ORFs of unknown function. H. volcanii genes correspond to Hvo_1200 (flaD2) to Hvo_1225 (cheW1). (B) Amino acid alignment of the flagellins FlgA1 and FlgA2, as determined using ClustalW2 (32). Asterisks indicate identical residues, colons indicate conserved substitutions, and periods indicate semiconserved substitutions. The PibD-processing motif (bold) was predicted by FlaFind (52); the arrow shows the predicted PibD cleavage site. Signal peptide hydrophobic stretches (gray) were predicted by TMHMM (50). Putative glycosylation sites of mature FlgA1 and FlgA2 (bold and shaded gray) were predicted using the NetNGlyc, version 1.0, server (http://www.cbs.dtu.dk/services/NetNGlyc/).
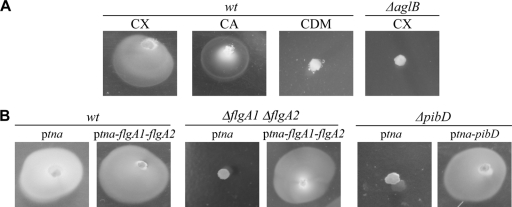
Here, we show that H. volcanii produces a growth halo after stab inoculation in a complex 0.3% agar motility medium (Fig. 2A). Consistent with microarray data, which indicate that genes in this fla cluster are expressed at low levels in a defined medium (CDM) relative to the expression levels observed in the CX (C. Daniels, personal communication), H. volcanii is nonmotile in the CDM (Fig. 2A). However, we have identified a distinct defined medium (CA medium) in which H. volcanii is motile (see Materials and Methods for medium composition) (Table 3 and Fig. 2A). The growth halos observed on CA motility plates are smaller than those on CX plates, likely reflecting the longer doubling time for H. volcanii grown in the CA medium (Table 3 and Fig. 2A).
FIG. 2.
H. volcanii swims in a flagellum-dependent manner. (A) H. volcanii H98 (wt) was stab inoculated into complex medium (CX) or defined medium (CDM or CA medium), while the ΔaglB mutant was stab inoculated into only CX (0.3% agar). Cells were then incubated for three days. (B) H. volcanii H98 (wt) transformed with pRV1-ptna (ptna) or pMT1 (ptna-flgA1-flgA2), MT1 (ΔflgA1 ΔflgA2) transformed with pRV1-ptna or pMT3 (ptna-flgA1-flgA2), or MT3 (ΔpibD) transformed with pRV1-ptna or pMT4 (ptna-pibD) was stab inoculated into complex (CX-tryptophan) medium containing novobiocin (0.3% agar). Cells were incubated at 45°C for 5 days. Note that growth of H. volcanii is significantly slower in medium containing novobiocin. Strains lacking flagellins or PibD did not produce halos, even after 14 days of incubation (data not shown).
TABLE 3.
Phenotypic characterization of H. volcanii ΔflgA1 ΔflgA2 and ΔpibD deletion strains
| Straina | Value for the characteristic on the indicated medium |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Minimal doubling time (h)b,c |
Competition assay (H53/H98) |
Conjugation frequency |
Ca2+-dependent aggregationb |
Motility (halo diam [cm] after 72 h)b,d |
Adhesionb |
|||||||
| CX | CA | CX | CA | CA + filter | Liquid CA | CX | CA | CX | CA | CX | CA | |
| WT | 3.5 | 4.5 | 50/50 | 50/50 | 10−4 | 10−8 | − | + | 0.9 | 0.7 | + | + |
| ΔflgA1 ΔflgA2 strain | 3.5 | 4.5 | 50/50 | 50/50 | 10−4 | 10−8 | − | + | − | − | + | + |
| ΔpibD strain | 3.5 | 4.5 | 50/50 | 50/50 | 10−4 | 10−8 | − | + | − | − | − | − |
Both the H53 and H98 backgrounds were used for all strains.
No differences were observed whether H98 or H53 background was used.
Standard deviation, 0.5 h.
Standard deviation, 0.22 and 0.29 cm in CX and CA motility media, respectively.
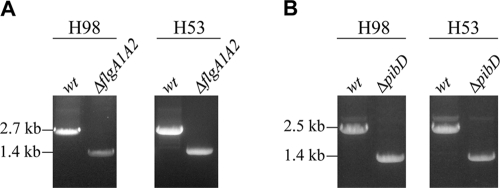
To confirm that H. volcanii swimming motility requires functional flagella, we used homologous recombination to construct a strain in which flgA1 and flgA2 are deleted. The deletion was verified using PCR. In PCRs that included primers designed to amplify adjacent upstream and downstream regions, including the flgA1-flgA2 genes, the PCR products generated for the deletion strain were smaller than those obtained for the parent strain, as expected (Fig. 3A).
FIG. 3.
Disruption of the chromosomal flgA1-flgA2 and pibD loci in H. volcanii strains H98 and H53. PCR amplification was performed using primers against the flanking regions. (A) Amplification of approximately 700 nucleotides upstream and 700 bp downstream of flgA1-flgA2, using template DNA isolated from the wild-type (H98 or H53) or the ΔflgA1 ΔflgA2 H. volcanii strains (MT1 or MT2, respectively). (B) Amplification of approximately 700 nucleotides upstream and 700 bp downstream of pibD, using template DNA isolated from the wild-type (H98 and H53) or the ΔpibD H. volcanii strains (MT3 o MT4, respectively). As expected, the amplicons are approximately 1,300 and 1,100 nucleotides smaller for the ΔflgA1 ΔflgA2 and ΔpibD deletion strains, respectively, than the amplicons obtained using DNA isolated from the parental strains.
We determined that the H. volcanii ΔflgA1 ΔflgA2 strain is not motile in CX-0.3% agar medium (Fig. 2B). To confirm that this phenotype is due to the absence of flagellins, the deletion strain was complemented with a plasmid that expresses FlgA1/FlgA2-His under the control of an inducible trp promoter (pMT3) (31) (Table 1). The ability of this complemented strain to swim is restored when cells are grown in the presence of tryptophan, which induces flagellin expression (Fig. 2B and 4A). These results reaffirm that H. volcanii swimming motility requires functional flagella.
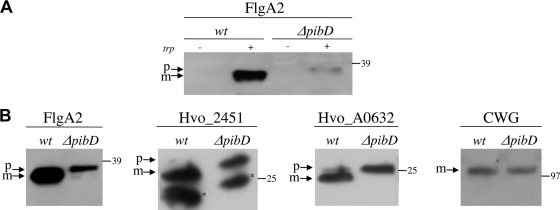
FIG. 4.
H. volcanii PibD processes class III signal peptide-containing preproteins, including flagellins and other pilin-like proteins. (A) H. volcanii H98 (wt) and MT3 (ΔpibD) strains expressing FlgA1/FlgA2-His were grown to mid-log phase in CA medium in which the promoter was turned on and off by the addition of tryptophan. (B) H. volcanii H98 (wt) and MT3 (ΔpibD) strains expressing FlgA1/FlgA2-His, Hvo_A0632-His, or Hvo_02451-His in a tryptophan-inducible manner were grown to mid-log phase in CX plus tryptophan. Equivalent amounts of whole-cell lysates were subjected to SDS-PAGE and visualized by immunoblotting using an anti-His antibody. Cell extracts of H98 (wt) and ΔpibD strains expressing C-terminally His-tagged Hvo_A0632 were also tested for S-layer (cell-wall) glycoprotein (CWG) migration by immunoblotting using an anti-CWG antibody (23). Predicted positions of precursor (p) and mature (m) proteins are indicated. The lower bands in the Hvo_2451 lanes, indicated by an asterisk, are likely degradation products of Hvo_2451 precursor and mature proteins. The migration of molecular mass standards (expressed in kilodaltons) is indicated on the right.
N-glycosylation has been shown to be critical for stable expression of flagella (16, 33, 38, 54, 55). Consistent with the identification of predicted N-glycosylation sites in the FlgA1 and FlgA2 flagellins, we determined that the lack of the H. volcanii aglB gene, which encodes a known oligosaccharide transferase (1, 2), renders this strain nonmotile in CX-0.3% agar (Fig. 2A), demonstrating that the identification of conditions under which H. volcanii is motile will also be valuable for the characterization of pathways that indirectly affect motility.
ΔflgA1 ΔflgA2 H. volcanii exhibits no conjugation, aggregation, or surface adhesion defects.
Bearing in mind the similarities between archaeal flagella and bacterial type IV pili, as well as the recent report suggesting that Pyrococcus furiosus (37) and S. solfataricus (22, 58) flagella are involved in biofilm formation and autoaggregation, we attempted to determine whether our flagellin deletion mutants have phenotypes similar to those of bacterial type IV pili and archaeal flagellum deletion mutants. We first determined that the H. volcanii flagellin mutants have no obvious growth defects. Under standard laboratory growth conditions, we found that the flagellin deletion mutant grows as well as the wild type in either complex or defined medium (Table 3). The ΔflgA1 ΔflgA2 deletion strain also grows as well as the wild-type in CX containing a high (23%) or low (14%) salt concentration (5.5- and 6-h minimal doubling times, respectively). Moreover, similar growth rates were observed in CX at high (48°C) and low (30°C) temperatures (4- and 8-h minimal doubling times, respectively).
Since bacterial type IV pili are involved in conjugation (29, 30, 57), we tested whether the H. volcanii conjugation also requires the type IV pilus-like flagella by comparing the rate of conjugation between the H53 (Δtrp ΔpyrE2) and H98 (ΔhdrB ΔpyrE2) parental strains to the conjugation rates for the ΔflgA1 ΔflgA2 deletion mutants of the corresponding strains, allowing us to identify both the dominant and recessive effects of the ΔflgA1 ΔflgA2 mutation on conjugation. Upon confirming that the flagellin deletion mutants grew as well as the wild type (Table 3), we used the conjugation assay described by Mevarech and Werczberger to investigate the effect of the flagellin gene deletions on conjugation (35). We observed that the rates of conjugation between the modified ΔflgA1 ΔflgA2 strains are similar to the rates between the parental strains when the strains are cocultured on a filter placed on a complex medium (CX) plate and subsequently cocultured in minimal medium (CA medium) lacking tryptophan and thymidine (conjugation frequency, 10−4) (Table 3). This result suggests that flagella are not involved in H. volcanii conjugation when cells are grown in close proximity. As the thin pilus of E. coli is required only for conjugation in liquid (29), we developed a liquid conjunction assay, which to the best of our knowledge is the first reported use of this type of assay in an archaeon. Although the frequency was significantly lower (10−8), we determined that conjugation does occur between cocultured H. volcanii auxotrophic strains. Interestingly, the conjugation frequency for the flagellin deletion mutants is similar to the frequency determined for the auxotrophic parent stains (Table 3).
Kawakami et al. have reported that H. volcanii can aggregate in a Ca2+-dependent manner, albeit with low efficiency (28). We determined that H. volcanii Ca2+-induced aggregation is highly efficient when cells are grown in CA medium but not in CX (Fig. 5). This aggregation was not observed when the divalent cation Mg2+ is added to the cultures or when CA medium was incubated with CaCl2 (Fig. 5). To determine whether this Ca2+-induced H. volcanii aggregation is flagellum-dependent, we compared aggregation of the H98 and the ΔflgA1 ΔflgA2 strains using the modified aggregation protocol and determined that both strains aggregate in a Ca2+-dependent manner within a few minutes (Table 3).
FIG. 5.
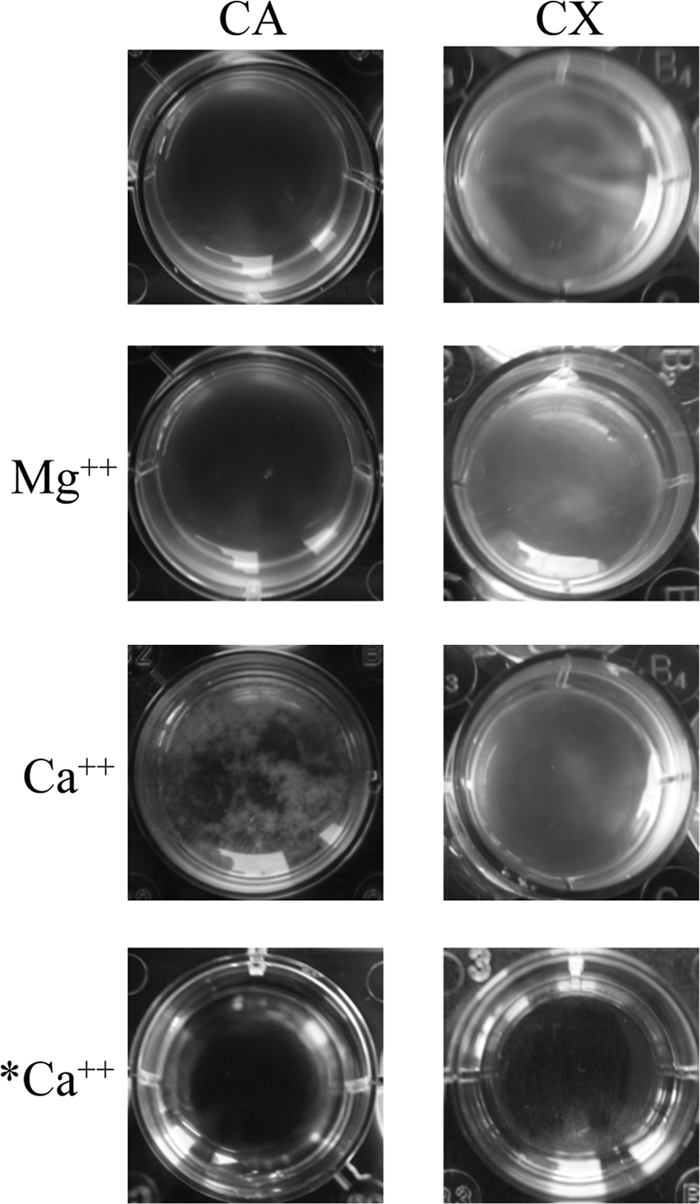
H. volcanii Ca2+-induced aggregation in a defined medium. Adding 10 mM Ca2+ induced aggregation of H. volcanii (H98) in mid-log phase cultures. While the majority of cells aggregated within 20 min after Ca2+ addition in defined medium (CA), aggregation was not apparent when Mg2+ was added to the cultures or in complex medium (CX). No aggregation was observed when Ca2+ was added to medium lacking H. volcanii (*Ca2+).
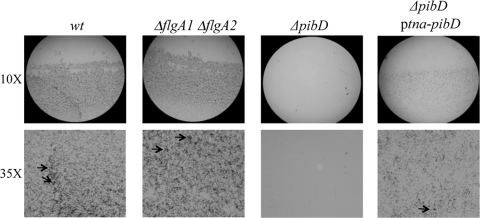
Finally, to determine whether H. volcanii surface adhesion involves flagella, we used an air-liquid interface (ALI) assay modified from O'Toole et al. (42) (see Materials and Methods) to compare the ability of the parent and mutant strains to adhere to glass coverslips. These experiments were performed in cultures grown with shaking and cultures grown without shaking as S. solfataricus surface adhesion was flagellum dependent only in shaking cultures (58; also V. Albers, personal communication). Crystal violet staining of H. volcanii adhering to the air-liquid interface indicated that both the H. volcanii wild-type and the ΔflgA1 ΔflgA2 strains adhere efficiently to glass at the air-liquid interface whether the cells are shaken or not (Fig. 6). Our results, for what we believe is the first description of an assay demonstrating haloarchaeal surface adhesion, suggest that surface structures other than flagella are involved in H. volcanii conjugation, surface adhesion, and Ca2+-induced aggregation.
FIG. 6.
H. volcanii adheres to glass in a FlgA1/FlgA2-independent, PibD-dependent manner. Adhesion to glass coverslips was tested using a modified ALI assay (42). Light micrographs of the coverslips taken at ×10 and ×35 magnifications are shown. Coverslips were placed in individual wells of 12-well plates, each containing 3 ml of a mid-log phase H. volcanii liquid culture (H98, wt, MT1, ΔflgA1 ΔflgA2; MT3, ΔpibD; and MT7, ΔpibD ptna-pibD). After overnight incubation, cells were fixed with 2% acetic acid, stained with 0.1% crystal violet, and observed by light microscopy. Attachment of H98 and MT1 to coverslips was observed within 10 min of incubation (data not shown). Multicellular structures reminiscent of microcolonies (a subset indicated by arrows) were observed.
The H. volcanii PibD homolog is required for processing a flagellin precursor, as well as precursors of other pilin-like proteins.
In M. maripaludis, FlaK, an SPase III, specifically processes preflagellins, while EppA, a distinct SPase III, processes the precursors of its pilins, all of which belong to a specific subgroup of type IV pilins (9, 52). Conversely, the S. solfataricus SPase III, PibD, has broader substrate specificity, processing both preflagellins and prepilins (4, 9). Like the genomes of the Sulfolobales, haloarchaeal genomes sequenced to date encode only a single SPase III and contain a number of predicted type IV pilins (52). To confirm that the H. volcanii SPase III cleaves preflagellins and to determine whether this proposed PibD homolog also cleaves the precursors of type IV pilin-like proteins, H. volcanii strain H98 containing a pibD deletion was constructed (MT3). The pibD deletion in this strain was confirmed in accordance with the methods used to confirm the ΔflgA1 ΔflgA2 deletions (see above) (Fig. 3B). We then transformed H98 (wt) and MT3 with a pRV1-ptna plasmid that expresses FlgA1/FlgA2-His when cells are grown in the presence of tryptophan (Table 1, pMT3). Western blot analyses showed that protein fractions isolated from the transformed H98 strain cultured under inducing conditions contained a band migrating faster than the band observed in fractions isolated from the corresponding ΔpibD transformant (Fig. 4A). MT3 transformants grown in CX also displayed a second band that migrated faster than the principal band, possibly representing a degradation product. In fact, in both CA medium and CX, the His-tagged FlgA2 observed in MT3 was significantly less abundant than that observed in H98 (Fig. 4A and B). These results are consistent with the idea that the H. volcanii prepilin peptidase processes its flagellins. Moreover, preflagellins appear not to be incorporated into surface filaments, resulting in a minor subset of unprocessed membrane-associated FlgA2.
Using FlaFind, a program that identifies genes that are likely to encode archaeal type IV pilin-like proteins, in silico analyses of the H. volcanii genome have identified genes that encode proteins resembling type IV pilins (25, 47, 48). To determine whether PibD could process nonflagellin subunits, we overexpressed two of the H. volcanii FlaFind positives (Hvo_A0632 or Hvo_02451) fused to a C-terminal His tag in H. volcanii H98 as well as MT3. Consistent with the facts that these putative pilin-like proteins contain predicted class III signal peptides at their N termini and that PibD has a broad substrate specificity, the putative pilins expressed in H98 were processed while those expressed in MT3 appeared to be unprocessed (Fig. 4B). Conversely, the H. volcanii S-layer glycoprotein, which has been shown to have a class I Sec signal peptide (51), migrated at the same position in an SDS-polyacrylamide gel, regardless of whether it was from an H98 or MT3 cell extract. This result suggests that, as expected, the lack of the prepilin peptidase does not have an indirect effect on class I signal peptide cleavage of the glycoprotein precursor (Fig. 4B).
PibD is required for swimming motility and surface adhesion.
Similar to the ΔflgA1 ΔflgA2 strains (MT1 and MT2), H. volcanii ΔpibD strains (MT3 and MT4) grow as well as the parental strains (H98 and H53, respectively) in complex and defined media (Table 3). These deletion strains are also competitively equivalent to the wild-type strain under both standard and modified growth conditions (Table 3). As with the ΔflgA1 ΔflgA2 strain, Ca2+-induced aggregation as well as conjugation of the H98 ΔpibD and H53 ΔpibD (MT3 and MT4) strains seems unaffected by the pibD deletion (Table 3), and consistent with a lack of flagella, the ΔpibD strains are not motile in CX-0.3% agar medium (Fig. 2B). However, unlike the ΔflgA1 ΔflgA2 strain, the adherence of the ΔpibD strain to the glass coverslip is severely defective as only a negligible number of cells are observed at the air-liquid interface (Fig. 6). Since the MT8 strain containing a plasmid that expresses PibD under the control of the inducible trp promoter is motile and can adhere to glass when grown in the presence of tryptophan, the phenotypes of the ΔpibD strain are thus strongly correlated to the lack of PibD (Fig. 2B and 6). Hence, while surface structures other than type IV pili appear to be involved in aggregation and conjugation, this strongly suggests that type IV pilus-like structures are involved in H. volcanii surface adhesion.
DISCUSSION
Microorganisms have developed many strategies to escape stress conditions. Motility allows the cells to move toward nutrients and away from toxins; aggregation on surfaces (biofilms) or in liquid medium protects cells from harmful extracellular conditions and promotes DNA exchange. Neither swimming motility nor surface adhesion had been reported for H. volcanii prior to this study. In addition to determining medium conditions that support these cellular processes in this widely used archaeal model organism, we have also identified a number of genes that encode proteins which play critical roles in these processes.
While the conditions used previously to scrutinize H. volcanii motility produced a negative result, we have now defined two distinct media in which this haloarchaeon is motile. We have also shown that an in-frame deletion of the flgA1 and flgA2 flagellin genes renders cells nonmotile and that complementing this deletion mutant with the same genes in trans from a plasmid rescues the motility phenotype. Similarly, consistent with the fact that we have demonstrated that the H. volcanii prepilin peptidase homolog processes the FlgA2 flagellin, a deletion mutant lacking this peptidase is also nonmotile, clearly demonstrating that H. volcanii motility is dependent on its flagella.
Bacterial and archaeal flagella are also involved in biofilm formation, perhaps providing the motive force necessary to overcome the repulsive forces that exist between a bacterium and a typical abiotic surface (41). Modifying the ALI assay to conform to the high-salt concentrations required by H. volcanii, we have shown that H. volcanii cells bind to a glass coverslip at the air-liquid interface and that under low-oxygen conditions adhesion is inhibited. It is intriguing that although the H. volcanii mutant is unable to swim, the mutant bacteria adhere to a glass surface just as effectively as the wild type. Perhaps haloarchaea, distinct from nonhalophilic prokaryotes, do not require flagella to adhere to surfaces because the high salt concentration in the medium masks the repulsive forces between the charged surfaces.
Interestingly, pibD deletion mutants do not bind to a glass surface, and complementing this strain in trans with pibD expression from a plasmid restores its ability to bind to glass. This result clearly demonstrates that the observed adhesion of H. volcanii is not due to nonspecific surface attachment. Moreover, by showing that a pibD deletion mutant does not bind to glass coverslips and that PibD is responsible for processing H. volcanii pilin-like proteins (Hvo_2451 and Hvo_A0632), we have demonstrated that the observed flagellum-independent surface adhesion is very likely to involve type IV pilus-like structures. Interestingly, while our results for the pibD deletion mutant are consistent with results produced by studies of prepilin peptidase homologs in bacteria as well as with results obtained for S. solfataricus (21, 26-28, 37), to date, we have not observed adherence to plastic surfaces, nor have we seen UV-induced autoaggregation. Our previous comprehensive in silico analysis of the H. volcanii genome identified five operons, each of which contains at least two genes encoding type IV pilin-like proteins (25). Under the conditions that we have tested for surface adhesion, microarray data gathered by Charles Daniels' group indicates that two of these operons are either not expressed or are expressed at very low levels (C. Daniels, personal communication). Finding the conditions under which these operons are expressed will be crucial to a complete understanding of the roles type IV pilus-like structures play in H. volcanii.
While we have not yet identified conditions under which UV light induces autoaggregation, we have observed Ca2+-dependent aggregation. Interestingly, effective H. volcanii Ca2+-induced aggregation requires neither flagella nor the prepilin peptidase. Moreover, the conjugation frequencies between two distinct auxotrophic H. volcanii pibD deletion strains, whether determined on filter plates or in liquid medium, are comparable to the frequencies observed for the wild type. Consistent with these results, we have observed protein complexes on the cell surfaces of H. volcanii ΔflgA1 ΔflgA2 and ΔpibD strains (data not shown) using electron microscopy. In fact, we have not yet discerned any differences between the surfaces of these deletion strains and the wild type. Clearly, the knockout and complementation studies carried out reveal that motility is flagellum dependent. Perhaps the structures encoded by the deleted genes are sheered off during the experimental procedures prior to electron microscopy observations, or, alternatively, the missing structures may be very similar in appearance to some of the structures that remain. The presence of a diverse set of type IV pilus-like structures and other distinct types of filaments on the surface of H. volcanii reflects the critical roles that cell surface structures play in cellular processes in all domains of life. Comparing and contrasting the roles and relationships of these various structures, both within and between domains, will have a significant impact on our understanding of cell biology and all that this entails, from pathogenesis to survival in extreme environments.
Acknowledgments
Our work was supported by a National Science Foundation grant (reference MCB02-39215).
We thank Charles Daniels for providing us with the microarray data, Jerry Eichler for providing us with the H. volcanii aglB mutant strain as well as for helpful hints to identify predicted glycosylation sites, and Friedhelm Pfeiffer for helpful information concerning the annotation of the fla-che operon.
Footnotes
Published ahead of print on 2 April 2010.
REFERENCES
- 1.Abu-Qarn, M., and J. Eichler. 2006. Protein N-glycosylation in Archaea: defining Haloferax volcanii genes involved in S-layer glycoprotein glycosylation. Mol. Microbiol. 61:511-525. [DOI] [PubMed] [Google Scholar]
- 2.Abu-Qarn, M., S. Yurist-Doutsch, A. Giordano, A. Trauner, H. R. Morris, P. Hitchen, O. Medalia, A. Dell, and J. Eichler. 2007. Haloferax volcanii AglB and AglD are involved in N-glycosylation of the S-layer glycoprotein and proper assembly of the surface layer. J. Mol. Biol. 374:1224-1236. [DOI] [PubMed] [Google Scholar]
- 3.Albers, S. V., and M. Pohlschroder. 2009. Diversity of archaeal type IV pilin-like structures. Extremophiles 13:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Albers, S. V., Z. Szabo, and A. J. Driessen. 2003. Archaeal homolog of bacterial type IV prepilin signal peptidases with broad substrate specificity. J. Bacteriol. 185:3918-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allers, T., and H. P. Ngo. 2003. Genetic analysis of homologous recombination in Archaea: Haloferax volcanii as a model organism. Biochem. Soc. Trans. 31:706-710. [DOI] [PubMed] [Google Scholar]
- 6.Allers, T., H. P. Ngo, M. Mevarech, and R. G. Lloyd. 2004. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 70:943-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arts, J., R. van Boxtel, A. Filloux, J. Tommassen, and M. Koster. 2007. Export of the pseudopilin XcpT of the Pseudomonas aeruginosa type II secretion system via the signal recognition particle-Sec pathway. J. Bacteriol. 189:2069-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bardy, S. L., J. Eichler, and K. F. Jarrell. 2003. Archaeal signal peptides—a comparative survey at the genome level. Protein Sci. 12:1833-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bardy, S. L., and K. F. Jarrell. 2002. FlaK of the archaeon Methanococcus maripaludis possesses preflagellin peptidase activity. FEMS Microbiol. Lett. 208:53-59. [DOI] [PubMed] [Google Scholar]
- 10.Bardy, S. L., T. Mori, K. Komoriya, S. Aizawa, and K. F. Jarrell. 2002. Identification and localization of flagellins FlaA and FlaB3 within flagella of Methanococcus voltae. J. Bacteriol. 184:5223-5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bardy, S. L., S. Y. Ng, and K. F. Jarrell. 2004. Recent advances in the structure and assembly of the archaeal flagellum. J. Mol. Microbiol. Biotechnol. 7:41-51. [DOI] [PubMed] [Google Scholar]
- 12.Blattner, F. R., B. G. Williams, A. E. Blechl, K. Denniston-Thompson, H. E. Faber, L. Furlong, D. J. Grunwald, D. O. Kiefer, D. D. Moore, J. W. Schumm, E. L. Sheldon, and O. Smithies. 1977. Charon phages: safer derivatives of bacteriophage lambda for DNA cloning. Science 196:161-169. [DOI] [PubMed] [Google Scholar]
- 13.Blyn, L. B., B. A. Braaten, and D. A. Low. 1990. Regulation of pap pilin phase variation by a mechanism involving differential dam methylation states. EMBO J. 9:4045-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boone, D. R., R. W. Castenholz, and G. M. Garrity (ed.). 2001. Bergey's manual of systematic bacteriology. The Archaea and the deeply branching and phototrophic bacteria, 2nd ed., vol. 1. Springer Verlag, Berlin, Germany.
- 15.Burrows, L. L. 2005. Weapons of mass retraction. Mol. Microbiol. 57:878-888. [DOI] [PubMed] [Google Scholar]
- 16.Chaban, B., S. Voisin, J. Kelly, S. M. Logan, and K. F. Jarrell. 2006. Identification of genes involved in the biosynthesis and attachment of Methanococcus voltae N-linked glycans: insight into N-linked glycosylation pathways in Archaea. Mol. Microbiol. 61:259-268. [DOI] [PubMed] [Google Scholar]
- 17.Craig, L., and J. Li. 2008. Type IV pili: paradoxes in form and function. Curr. Opin. Struct. Biol. 18:267-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Driessen, A. J., and N. Nouwen. 2008. Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 77:643-667. [DOI] [PubMed] [Google Scholar]
- 19.Dyall-Smith, M. March 2008, posting date. Halohandbook: protocols for haloarchaeal genetics. University of Melbourne, Victoria, Australia. http://haloarchaea.com/resources/halohandbook/Halohandbook_2008v7.pdf.
- 20.Francetic, O., N. Buddelmeijer, S. Lewenza, C. A. Kumamoto, and A. P. Pugsley. 2007. Signal recognition particle-dependent inner membrane targeting of the PulG pseudopilin component of a type II secretion system. J. Bacteriol. 189:1783-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frols, S., M. Ajon, M. Wagner, D. Teichmann, B. Zolghadr, M. Folea, E. J. Boekema, A. J. Driessen, C. Schleper, and S. V. Albers. 2008. UV-inducible cellular aggregation of the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by pili formation. Mol. Microbiol. 70:938-952. [DOI] [PubMed] [Google Scholar]
- 22.Frols, S., P. M. Gordon, M. A. Panlilio, I. G. Duggin, S. D. Bell, C. W. Sensen, and C. Schleper. 2007. Response of the hyperthermophilic archaeon Sulfolobus solfataricus to UV damage. J. Bacteriol. 189:8708-8718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hand, N. J., R. Klein, A. Laskewitz, and M. Pohlschroder. 2006. Archaeal and bacterial SecD and SecF homologs exhibit striking structural and functional conservation. J. Bacteriol. 188:1251-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harshey, R. M. 2003. Bacterial motility on a surface: many ways to a common goal. Annu. Rev. Microbiol. 57:249-273. [DOI] [PubMed] [Google Scholar]
- 25.Hartman, L. A., C. Norais, J. Badger, S. Delmas, S. Haldenby, R. Madupu, J. Robinson, H. Khouri, Q. Ren, T. Lowe, J. Maupin-Furlow, M. Pohlschroder, C. Daniels, F. Pfeiffer, T. Allers, and J. A. Eisen. 2010. The complete genome sequence of Haloferax volcanii DS2, a model archaeon. PLoS One 5(3):e9605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarrell, K. F., and M. J. McBride. 2008. The surprisingly diverse ways that prokaryotes move. Nat. Rev. Microbiol. 6:466-476. [DOI] [PubMed] [Google Scholar]
- 27.Kawakami, Y., N. Hayashi, M. Ema, and M. Nakayama. 2007. Effects of divalent cations on Halobacterium salinarum cell aggregation. J. Biosci. Bioeng. 104:42-46. [DOI] [PubMed] [Google Scholar]
- 28.Kawakami, Y., T. Ito, M. Kamekura, and M. Nakayama. 2005. Ca2+-dependent cell aggregation of halophilic archaeon, Halobacterium salinarum. J. Biosci. Bioeng. 100:681-684. [DOI] [PubMed] [Google Scholar]
- 29.Kim, S. R., and T. Komano. 1997. The plasmid R64 thin pilus identified as a type IV pilus. J. Bacteriol. 179:3594-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komano, T., S. R. Kim, and T. Yoshida. 1995. Mating variation by DNA inversions of shufflon in plasmid R64. Adv. Biophys. 31:181-193. [DOI] [PubMed] [Google Scholar]
- 31.Large, A., C. Stamme, C. Lange, Z. Duan, T. Allers, J. Soppa, and P. A. Lund. 2007. Characterization of a tightly controlled promoter of the halophilic archaeon Haloferax volcanii and its use in the analysis of the essential cct1 gene. Mol. Microbiol. 66:1092-1106. [DOI] [PubMed] [Google Scholar]
- 32.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 33.Logan, S. M. 2006. Flagellar glycosylation—a new component of the motility repertoire? Microbiology 152:1249-1262. [DOI] [PubMed] [Google Scholar]
- 34.Macnab, R. M. 2004. Type III flagellar protein export and flagellar assembly. Biochim. Biophys. Acta 1694:207-217. [DOI] [PubMed] [Google Scholar]
- 35.Mevarech, M., and R. Werczberger. 1985. Genetic transfer in Halobacterium volcanii. J. Bacteriol. 162:461-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mullakhanbhai, M. F., and H. Larsen. 1975. Halobacterium volcanii spec. nov., a Dead Sea halobacterium with a moderate salt requirement. Arch. Microbiol. 104:207-214. [DOI] [PubMed] [Google Scholar]
- 37.Nather, D. J., R. Rachel, G. Wanner, and R. Wirth. 2006. Flagella of Pyrococcus furiosus: multifunctional organelles, made for swimming, adhesion to various surfaces, and cell-cell contacts. J. Bacteriol. 188:6915-6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng, S. Y., B. Chaban, and K. F. Jarrell. 2006. Archaeal flagella, bacterial flagella and type IV pili: a comparison of genes and posttranslational modifications. J. Mol. Microbiol. Biotechnol. 11:167-191. [DOI] [PubMed] [Google Scholar]
- 39.Ng, S. Y., B. Zolghadr, A. J. Driessen, S. V. Albers, and K. F. Jarrell. 2008. Cell surface structures of Archaea. J. Bacteriol. 190:6039-6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nieuwlandt, D. T., and C. J. Daniels. 1990. An expression vector for the archaebacterium Haloferax volcanii. J. Bacteriol. 172:7104-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 42.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 43.Paetzel, M., A. Karla, N. C. J. Strynadka, and R. E. Dalbey. 2002. Signal peptidases. Chem. Rev. 102:4549-4580. [DOI] [PubMed] [Google Scholar]
- 44.Patenge, N., A. Berendes, H. Engelhardt, S. C. Schuster, and D. Oesterhelt. 2001. The fla gene cluster is involved in the biogenesis of flagella in Halobacterium salinarum. Mol. Microbiol. 41:653-663. [DOI] [PubMed] [Google Scholar]
- 45.Peabody, C. R., Y. J. Chung, M. R. Yen, D. Vidal-Ingigliardi, A. P. Pugsley, and M. H. Saier, Jr. 2003. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149:3051-3072. [DOI] [PubMed] [Google Scholar]
- 46.Pelicic, V. 2008. Type IV pili: e pluribus unum? Mol. Microbiol. 68:827-837. [DOI] [PubMed] [Google Scholar]
- 47.Pfeiffer, F., A. Broicher, T. Gillich, K. Klee, J. Mejia, M. Rampp, and D. Oesterhelt. 2008. Genome information management and integrated data analysis with HaloLex. Arch. Microbiol. 190:281-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schlesner, M., A. Miller, S. Streif, W. F. Staudinger, J. Muller, B. Scheffer, F. Siedler, and D. Oesterhelt. 2009. Identification of Archaea-specific chemotaxis proteins which interact with the flagellar apparatus. BMC Microbiol. 9:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneider, K. L., K. S. Pollard, R. Baertsch, A. Pohl, and T. M. Lowe. 2006. The UCSC Archaeal Genome Browser. Nucleic Acids Res. 34:D407-D410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sonnhammer, E. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:175-182. [PubMed] [Google Scholar]
- 51.Sumper, M., E. Berg, R. Mengele, and I. Strobel. 1990. Primary structure and glycosylation of the S-layer protein of Haloferax volcanii. J. Bacteriol. 172:7111-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szabo, Z., A. O. Stahl, S. V. Albers, J. C. Kissinger, A. J. Driessen, and M. Pohlschroder. 2007. Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases. J. Bacteriol. 189:772-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas, N. A., S. Mueller, A. Klein, and K. F. Jarrell. 2002. Mutants in flaI and flaJ of the archaeon Methanococcus voltae are deficient in flagellum assembly. Mol. Microbiol. 46:879-887. [DOI] [PubMed] [Google Scholar]
- 54.Trachtenberg, S., and S. Cohen-Krausz. 2006. The archaeabacterial flagellar filament: a bacterial propeller with a pilus-like structure. J. Mol. Microbiol. Biotechnol. 11:208-220. [DOI] [PubMed] [Google Scholar]
- 55.VanDyke, D. J., J. Wu, S. Y. Ng, M. Kanbe, B. Chaban, S. Aizawa, and K. F. Jarrell. 2008. Identification of a putative acetyltransferase gene, MMP0350, which affects proper assembly of both flagella and pili in the archaeon Methanococcus maripaludis. J. Bacteriol. 190:5300-5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, Y. A., X. Yu, S. Y. Ng, K. F. Jarrell, and E. H. Egelman. 2008. The structure of an archaeal pilus. J. Mol. Biol. 381:456-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshida, T., N. Furuya, M. Ishikura, T. Isobe, K. Haino-Fukushima, T. Ogawa, and T. Komano. 1998. Purification and characterization of thin pili of IncI1 plasmids ColIb-P9 and R64: formation of PilV-specific cell aggregates by type IV pili. J. Bacteriol. 180:2842-2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zolghadr, B., A. Klingl, A. Koerdt, A. J. Driessen, R. Rachel, and S. V. Albers. 2010. Appendage-mediated surface adherence of Sulfolobus solfataricus. J. Bacteriol. 192:104-110. [DOI] [PMC free article] [PubMed] [Google Scholar]